Colloquium
Genetic complexity of pathogen perception by plants: The example of Rcr3, a tomato gene required specifically by Cf-2
Mark S. Dixon*, Catherine Golstein, Colwyn M. Thomas, Erik A. van der Biezen†, and Jonathan D. G. Jones‡
The Sainsbury Laboratory, John Innes Centre, Norwich Research Park, Colney Lane, Norwich NR4 7UH, United Kingdom
Genetic analysis of plant–pathogen interactions has demonstrated that resistance to infection is often determined by the interaction of dominant plant resistance (R) genes and dominant pathogen-encoded avirulence (Avr) genes. It was postulated that R genes encode receptors for Avr determinants. A large number of R genes and their cognate Avr genes have now been analyzed at the molecular level. R gene loci are extremely polymorphic, particularly in sequences encoding amino acids of the leucine-rich repeat motif. A major challenge is to determine how Avr perception by R proteins triggers the plant defense response. Mutational analysis has identified several genes required for the function of specific R proteins. Here we report the identification of Rcr3, a tomato gene required specifically for Cf-2-mediated resistance. We propose that Avr products interact with host proteins to promote disease, and that R proteins “guard” these host components and initiate Avr-dependent plant defense responses.
M any plant pathogens are highly adapted biotrophic parasites that require living hosts to complete their life cycle. Flor, in his pioneering work in the 1940s, studied the interaction between flax and flax rust. He showed that recessive virulence genes enable mutant strains to overcome specific disease resistance (R) genes, and that avirulence (Avr) genes are dominant (1). This “gene-for-gene” interaction was subsequently demonstrated for many other plant–pathogen interactions. Plant R proteins are postulated to provide a surveillance system that can detect Avr determinants from diverse viral, prokaryotic, and eukaryotic pathogens. Here we review recent studies that reveal the genetic complexity of pathogen perception by plants, and we report the identification and preliminary characterization of Rcr3, a gene specifically required for tomato Cf-2 function.
Virulence and Avirulence
It would be surprising if pathogens were to carry Avr genes that had no function other than to enable recognition by plants that carry the matching R genes. During infection, pathogens make an array of virulence factors. If plant R genes can evolve to recognize one of these molecules to trigger a defense response, any such virulence factor could become an avirulence determinant. Avr genes of the bacterial plant pathogens Xanthomonas campestris and Pseudomonas syringae encode hydrophilic proteins that are delivered inside the plant cell by a specialized type III secretion mechanism (2). In the absence of recognition by corresponding R genes, some of these Avr genes have been shown to confer enhanced virulence (3, 4). Viral pathogens also present potential ligands intracellularly, and for some R genes, the corresponding viral Avr protein has been defined as the replicase domain (5) or the coat protein (6).
We study resistance to two biotrophic pathogens, the tomato leaf mold fungus Cladosporium fulvum and the oomycete Peronospora parasitica, which causes downy mildew on cruciferous plants, including Arabidopsis. Studies on C. fulvum, which colonizes the intercellular spaces of infected leaves, led to the cloning of Avr4 and Avr9 that confer avirulence on tomato plants carrying the corresponding Cf-4 and Cf-9 R genes (see below). Avr4 and Avr9 encode small secreted peptides that trigger Cf gene-dependent resistance (7, 8). No evidence to support a role for Avr4 or Avr9 as virulence determinants has been demonstrated. In contrast, ECP2 is a secreted peptide that has a virulence function in all C. fulvum strains analyzed (9) and is recognized as an avirulence factor in certain tomato lines (10). P. parasitica, like many other parasites, forms hyphae that produce feeding structures inside the plant cell wall, termed haustoria, that provide intimate association with the plant plasma membrane (11). Avr effectors may be delivered through haustoria inside the plant cell. This is plausible because their cognate R proteins are predicted to be intracellular (see next section).
R Proteins
Tomato Cf-2, Cf-4, Cf-5, and Cf-9 genes confer recognition of different C. fulvum Avr genes. They encode an extracellular leucine-rich repeat (eLRR) domain, a transmembrane domain, and a short cytoplasmic domain with no sequence similarity to known signaling domains (Fig. 1A) (12). Cf-9 has recently been shown to be a plasma membrane-localized glycoprotein (13). Xa21 is a rice gene that confers resistance to bacterial blight caused by X. campestris pv. oryzae and also encodes eLRRs and a transmembrane domain, but in addition has a cytoplasmic Ser/Thr protein kinase domain (Fig. 1B) (14). The presence of eLRRs in these R proteins is consistent with a presumed role in the recognition of extracellular ligands. Interestingly, Pto, a tomato gene that confers resistance to P. syringae pv. tomato strains expressing the AvrPto gene, also encodes a Ser/Thr protein kinase (Fig. 1C) (15). Pto lacks a signal peptide (but carries a putative myristoylation site), and its interaction with AvrPto in yeast suggests a cytoplasmic recognition capacity (16, 17).
This paper was presented at the National Academy of Sciences colloquium “Virulence and Defense in Host–Pathogen Interactions: Common Features Between Plants and Animals, ” held December 9–11, 1999, at the Arnold and Mabel Beckman Center in Irvine, CA.
Abbreviations: AFLP, amplified restriction fragment polymorphism; (e)LRR, (extracellular) leucine-rich repeat; NB, nucleotide-binding; ARC, Apaf-1, R proteins, and CED4 homology; R, resistance; TIR, Toll and interleukin-1 receptors homology; TLR, Toll-like receptor; LZ, leucine zipper; CARD, caspase recruitment domain; GUS, β-glucuronidase.
|
* |
Present address: School of Biological Sciences, University of Southampton, Southampton SO16 5YA, U.K. |
|
† |
Present address: Aventis CropScience, B-9000 Gent, Belgium. |
|
‡ |
To whom reprint requests should be addressed. E-mail: jonathan.jones@bbsrc.ac.uk. |
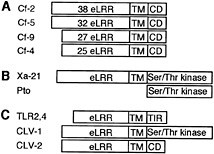
Fig. 1. Tomato Cf proteins and structurally related proteins. (A) Cf proteins contain different numbers of N-terminal eLRRs, a transmembrane (TM) domain, and a short cytoplasmic domain (CD). (B) Rice Xa-21 and tomato Pto bacterial R proteins. (C) eLRR-TM domain-containing proteins with different cytoplasmic domains (CD). See text for descriptions.
Both the Cf class and Xa21 resemble the Drosophila Toll receptor and its human homologs, the Toll-like receptors (TLRs) (18). TLR2 and TLR4 (Fig. 1C) function in innate immunity and activate microbial defense pathways to trigger inflammatory responses after recognition of conserved molecular structures of distinct microbial pathogen classes (19). TLRs have therefore been designated “pattern recognition receptors” (20) and serve an analogous role to plant R proteins in detecting pathogen molecules and activating defense pathways. Interestingly, TLR-mediated inflammatory responses resemble plant defense responses, including production of antimicrobial active oxygen species and nitric oxide and ultimately cell death (21, 22). The significance of these homologies for plant R protein mechanisms has yet to be established, but it is tempting to suggest that, whereas vertebrates use the TLRs to recognize conserved structures of different pathogen classes, plants have evolved these to detect strain-specific pathogen Avr products.
Arabidopsis RPP1 and RPP5 genes confer resistance to P. parasitica and belong to the largest class of R genes that encode nucleotide-binding leucine-rich repeat (NB-LRR) proteins (Fig. 2A). Genome sequencing has shown that approximately 200 NB-LRR-encoding genes are present in Arabidopsis (23). NBLRR proteins can be divided into two subclasses on the basis of their N-terminal domain. The leucine zipper (LZ)-NB-LRR is a broad class of NB-LRR proteins (with perhaps two subclasses) that contains an N-terminal putative heptad LZ or coiled-coil domain (23). Members of this class have been identified for resistance to bacteria, viruses, fungi, oomycetes, and even nematodes and aphids. Examples include Arabidopsis RPS2, RPM1, and RPS5 for resistance to P. syringae (24), RPP8 for resistance to P. parasitica (25), and maize Rp1 for resistance to Puccinia sorghi (26).
Members of the TIR-NB-LRR class carry N-terminal homology with the cytoplasmic domain of the Toll and interkeukin-1 receptors (Fig. 2B) involved in vertebrate innate immunity (27, 28). Besides RPP1 and RPP5, this class also includes tobacco N for resistance to tobacco mosaic virus, flax L6 for resistance to flax rust, and Arabidopsis RPS4 for resistance to P. syringae (24, 29). After activation, the TIR domain of human TLR2 and TLR4 binds, through homophilic interactions, to the TIR domain of the MyD88 adaptor protein (Fig. 2B) (30). MyD88 also carries a death domain (DD) that then binds to the DD of the Ser/Thr protein kinase IRAK (the human ortholog of Drosophila Pelle, and interestingly, also homologous to tomato Pto; Fig. 1B), leading to the translocation of the transcription factor NF-κB and induction of the inflammatory response (27). These homologies may suggest a role for the TIR domain of the plant R proteins in homophilic TIR–TIR interactions with TIR domaincontaining plant adaptor proteins, but these have yet to be reported in plants.
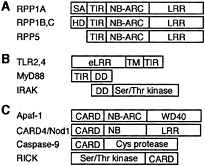
Fig. 2. Arabidopsis RPP1, RPP5, and structurally related proteins. (A) RPP1 and RPP5 have an N-terminal TIR domain, a nucleotide-binding Apaf-1, R proteins and CED4 homology (NB-ARC) domain, and a LRR domain. The RPP1 family differ by their N-terminal domains, which are absent in RPP5: RPP1A has a putative signal anchor (SA) domain, RPP1B,C have hydrophobic domains (HD). (B) Human proteins that function in the NF-κB pathway. eLRR, extracellular LRR domain; TM, transmembrane domain. (C) Human proteins with CARDs that function in NF-κB and/or apoptosis pathways. See text for descriptions.
An intriguing homology has been noted between plant NB-LRR proteins and the apoptotic adaptor CED4 from Caenorhabditis elegans (31). This region of homology also extends to its human ortholog Apaf-1 (Fig. 2C) (32) and has been designated the NB-ARC domain (Fig. 2C) (33) or Ap-ATPase domain (34). The NB-ARC domain is an ancient motif, because it is also present in proteins of Gram-positive bacteria (34). Another protein that resembles plant NB-LRR proteins is human caspase recruitment domain (CARD)4/Nod1 (35, 36), which contains LRRs and a NB site (the homology to the rest of the NB-ARC domain is less convincing). Instead of TIR or LZ domains at their N termini, Apaf-1 and CARD4/Nod1 carry CARDs (Fig. 2C). Induction of apoptosis by both Apaf-1 and CARD4/Nod1 occurs through homophilic CARD–CARD interactions with caspase-9, a CARD-containing cysteine protease (36, 37). CARD4/Nod1, in addition, plays a role in activation of the NF-κB pathway through homophilic CARD–CARD interactions with the Ser/Thr kinase RICK (Fig. 2C) (35, 36). Caspases and CARDs have not yet been reported in plants; however, the use of peptide inhibitors specific to caspases in animal cells suggests plants may possess caspase activity (38). Although the significance of these homologies is not clear, they provide interesting paradigms for NB-LRR protein mechanisms. For example, the WD40 repeat domain of Apaf-1 negatively regulates its NBARC domain, which is alleviated after binding to cytochrome c, then allowing ATP-dependent oligomerization and caspase-9 activation (37). By analogy, pathogen Avr signals perceived by the LRR domain of plant NB-LRR proteins might alleviate negative regulation of their NB-ARC domain such that ATP hydrolysis and oligomerization can be initiated, possibly leading to homo- or heterodimerization (through
homophilic interactions) of the N-terminal TIR or LZ domains.
R Gene Evolution
Plants can activate both localized and systemic defense mechanisms in response to pathogen infection. This activation is often a consequence of R protein perception of a pathogenencoded Avr determinant in cells targeted by the pathogen (24). This contrasts with the situation in mammals where circulating defender cells migrate to the sites of pathogen ingress. Furthermore, in mammals, the elaboration of recognition capacity and the consequent resistance to an array of potential pathogens occurs somatically through the adaptive immune system and germinally in genes encoding the MHC (39). In plants, R gene sequence evolution takes place exclusively through the germline. Population geneticists have proposed that in natural populations, where plants and their pathogens coevolve, disease is kept in check by “balancing polymorphisms” at R gene loci (40). In this model, no single R gene allele is present at high frequency because, as a result of selection pressure, it may be overcome by a novel pathogen variant (40). For example, R gene monocultures in crops eventually succumb to pathogen infection, and breeding durable disease resistance remains a major challenge. Overdominance, or heterozygote advantage, has been proposed to explain the polymorphism at the human MHC locus. However, overdominance cannot explain polymorphism at R gene loci in inbreeding species. We and others have proposed that frequency-dependent selection may account for the maintenance of sequence polymorphism at the Arabidopsis RPP5, RPM1, and other R gene loci (41, 42).
Recent analyses have revealed the molecular organization of a number of R gene loci and provided insight to the molecular mechanisms that generate sequence diversity. We have analyzed two Cf loci in tomato and two RPP loci in Arabidopsis. The Cf-0, Cf-9, and Cf-4 haplotypes of tomato contain distinct complements of tandemly duplicated Cf-9 homologs (Hcr9s; refs.43 and 44). Several Hcr9s have been shown to confer resistance to C. fulvum through recognition of distinct Avr determinants (43, 45). A similar comparison has revealed the polymorphic nature of Cf-2 homologs (Hcr2s) at the tomato Cf-2/Cf-5 locus (46). The sequence polymorphism at Cf loci is because of interspecific variation (12). However, in Arabidopsis the RPP1 and RPP5 loci (41, 47), which are also comprised of linked multigene families, show pronounced intraspecific polymorphism when different landraces are compared by DNA gel blot analysis (Fig. 3).
DNA sequence analysis of tomato Hcr9s and Hcr2s (43, 46), the Arabidopsis RPP1, RPP5, and RPP8 families (25, 41, 47, 48) and NB-LRR homologs at the lettuce Dm3 locus (49) have recently been reported. Like RPP5 and RPP1 homologs, Hcr2s contain variable numbers of LRRs, suggesting this is an important mechanism to generate novel recognition specificities (12, 46). Hcr9s, however, contain similar numbers of LRRs, and most sequence variation occurs in nucleotides that encode the putative solvent-exposed amino acids of a conserved β-strand/β-turn structural motif of LRR proteins. These amino acids are thought to be critical for the recognition specificity of ligand binding in LRR proteins (50). Sequence comparisons of NB-LRR multigene families such as RPP1, RPP5, and Dm3 have also shown high ratios of nonsynonymous to synonymous substitutions in sequences encoding the corresponding amino acids of their LRRs. These sequences within the LRRs have clearly undergone diversifying selection, whereas sequences encoding putative signaling domains have undergone purifying selection (23, 41, 47).
As in studies on the evolution of the mammalian MHC locus (39), the relative contribution of interallelic recombination, gene conversion, unequal crossing over, and the accumulation of point mutations in generating R gene diversity is controversial. Classical genetic experiments suggested that interallelic sequence exchange at the flax L locus (51) or chromosomal mispairing and unequal crossing over between homologs at the maize Rp1 locus (52) could generate R gene variation. This was subsequently verified by molecular analyses of the L and Rp1 loci (26, 51). At some R gene loci, a “birth-and-death” model of R gene evolution has been proposed (53), based on a model for evolution of the mammalian MHC locus (54). In this model, expansion or contraction of members of a multigene family is caused by unequal crossing over, and evolution of alleles proceeds by sequence exchange between gene orthologs, random mutation and selection.
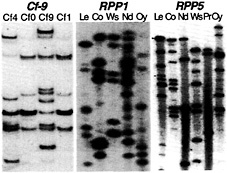
Fig. 3. Polymorphic R gene families revealed by DNA gel blot analysis. Comparisons of tomato lines with introgressed chromosome segments containing different Cf genes show interspecific polymorphisms (44). Comparisons of different Arabidopsis landraces show intraspecific polymorphisms at the RPP1 (47) and RPP5 loci (48).
A comparison of the Cf-0, Cf-4 and Cf-9 haplotypes in tomato showed that the copy number of Hcr9s at this locus can vary. In the Cf-4 and Cf-9 haplotypes (each containing five Hcr9s), evidence for extensive intergenic sequence exchange between Hcr9 paralogs was shown that generated recombinant Hcr9s. This sequence exchange could have contributed to the creation of novel recognition specificities. Also, the order of putative gene orthologs differed in the two haplotypes (43) and is inconsistent with the proposal that negligible sequence exchange occurs between paralogs. Hcr9s have also accumulated multiple point mutations that generate sequence diversity in the solvent-exposed residues of their LRRs (see above), and this appears to be another important source of sequence variation. The comparison of Col-0 and La-er haplotypes of the Arabidopsis RPP5 locus also revealed extensive sequence exchange between RPP5 homologs (41). With the exception of homologs at the ends of the cluster, orthologous genes were difficult to discern. Furthermore, many “mutated” homologs were identified. It is possible that these sequences provide a reservoir for intergenic sequence exchange and the generation of sequence diversity.
It is likely that the rate and molecular mechanism for generating R gene novelty will depend on a number of factors. Further insight into these mechanisms will come from comparative analysis of natural R gene variants such as the numerous recombinant Rp1 alleles in maize (26, 52). These analyses should reveal the range of molecular mechanisms that generate R gene diversity in different plants, at different R gene loci within plants,
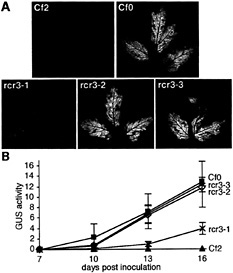
Fig. 4. Infection phenotypes of rcr3 mutants. Seedlings were immersed in a solution containing 3.5 × 105 ml−1 spores of the appropriate C. fulvum race. Inoculated plants were kept for 3 to 6 days in a phytochamber at saturated humidity and subsequently at 70% relative humidity, 24°C, during the 16-hr day-light, 18°C overnight. (A) Comparison of infection phenotype of the rcr3 mutants and the Cf0-sensitive and Cf2-resistant control lines. Plants were inoculated with C. fulvum race 5 15 days after sowing, and leaves were photographed 17 days after inoculation. Fungal colonization is visible on the abaxial surface of the leaves as white mycelium. (B) Comparison of fungal biomass accumulation in rcr3–1 (X), rcr3–2 (◊), rcr3–3 (●), Cf0 (▪), and Cf2 (▴) lines by using a fluorometric GUS assay (56) over a time course after inoculation. Twelve-day-old plants were inoculated with C. fulvum race 4 GUS (57). No GUS activity (4-methyl umbelliferone mg−1 protein min−1) was detected on Cf2 controls in contrast to Cf0 controls. Three cotyledons were harvested and analyzed separately for each sample at each time point (the mean value for each set of triplicate samples is shown and error bars indicate the standard deviation).
and the molecular basis for R protein-mediated pathogen perception.
Results and Discussion
Mutational Analysis Identifies Rcr3, a Gene Required for Cf-2 Function. Mutational analysis is a powerful tool to dissect signaling pathways and has been used to identify genes required for R gene function. Mutational analysis of genes required for Cf-9 function has revealed two loci required for full C. fulvum resistance, Rcr1 and Rcr2 (55). Here, we report the results of mutagenesis of the tomato Cf2 line. Twenty-five M2 seeds from 3,200 diepoxybutane- or ethyl methanesulfonate-treated Cf2 M1 plants were screened for their response to C. fulvum race 4 β-glucuronidase (GUS) infection, and four M2 families that segregated for resistance or sensitivity to infection were identified (Table 1). PCR analysis of disease-sensitive plants confirmed these plants contained Cf-2. To determine the genetic basis of disease sensitivity in these mutants, all four were crossed to the Cf0 near-isogenic line that lacks known resistance genes to C. fulvum. In crosses to Cf0, mutants at Cf-2 should give susceptible F1 progeny as neither parent would have contained a functional gene for resistance to C. fulvum infection. Crosses between the four mutant lines and Cf0 gave resistant F1 progeny, confirming the mutations were not in Cf-2. Progeny from intercrosses between all four mutants were sensitive to C. fulvum race 4 GUS infection, demonstrating that these mutations are allelic. This gene has been designated Rcr3 (required for C. fulvum resistance 3; Table 1).
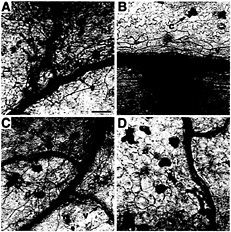
Fig. 5. Comparison of fungal development and plant cell death response during the C. fulvum infection of rcr3 mutants and control lines. At least three cotyledons and two leaves were sampled every 3 days from each genotype during the experiment described in Fig. 4A and were stained with lactophenol-trypan blue (58). Photomicrographs of cotyledons were taken 7 days after inoculation at the same magnification. Scale bar = 100 µm. (A) Fungal growth in the Cf0 line: C. fulvum hyphae (h) tend to localize close to plant veins (v) where they start swelling, (B) Characteristic early resistance response in the Cf2 line: patches of dead mesophyll cells (p) develop at the vicinity of the plant veins, whereas the fungus is undetectable. (C) Infection phenotype of rcr3–3, and rcr3–2 (not shown): once the fungus has penetrated the leaf apoplast, the fungus develops as in Cf0. However, these fungal foci appear to occur at a lower frequency on rcr3–3 and rcr3–2 than on Cf0. (D) rcr3–1 intermediate phenotype: although rcr3–1 allows sparse fungal growth in some areas; the fungus remains undetectable on the major part of the cotyledon. Discrete dead cells (d) were scattered around the upper mesophyll of the mutants but not in wild type, irrespective of C. fulvum infection (noninoculated material not shown). This cell death phenotype was poorly reproducible. Calcium oxalate crystals (c) in some mesophyll cells are common in tomato leaves. These phenotypes were observed in three independent infections with C. fulvum race 5 and in an infection with C. fulvum race 4 GUS, where over 300 infection sites were analyzed in cotyledon and leaf samples.
Strong and Weak Mutant Alleles of Rcr3. Inoculation with C. fulvum race 5 revealed that rcr3–2 (the line carrying the rcr3–2 mutation) and rcr3–3 allowed as much fungal growth as Cf0, whereas rcr3–1 appeared significantly less susceptible (Fig. 4). From 2 weeks after inoculation, C. fulvum growth on rcr3–1 was intermediate to Cf0 and Cf2 (Fig. 4B). Fungal development was barely detectable on rcr3–1 at 1 week after inoculation, whereas hyphae had already invaded the mesophyll layers and were proliferating around the vascular tissue of rcr3–2, rcr3–3, and Cf0 cotyledons (Fig. 5). Microscopic inspection of 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acidstained tissue confirmed that the fungus was able to sporulate on all three mutants (not shown).
The rcr3–1, rcr3–2, and rcr3–3 mutants were each back crossed
to Cf 2 and selfed to produce F2 seed. Approximately one-quarter of the F2 progeny from crosses between Cf2 and rcr3–2 or rcr3–3 appeared to be fully susceptible to C. fulvum race 4 GUS infection. One-quarter of the F2 progeny from the Cf2 × rcr3–1 cross showed an intermediate level of disease sensitivity as observed in the original M2 seedlings, suggesting this phenotype is caused by allelic variation at rcr3 and not by another unlinked mutation (data not shown). Therefore, rcr3–1 is a weak, and rcr3–2 and rcr3–3 are strong, suppressors of Cf-2 function.
Rcr3 Maps to Chromosome 2. To generate a mapping population where rcr3 and molecular markers were segregating, rcr3–1 was crossed to Lycopersicon pennellii (59). F1 progeny from this cross were resistant to C. fulvum race 4 GUS infection, demonstrating that the L. pennellii Rcr3 allele can complement the rcr3 mutation. A single F1 plant was back crossed to rcr3–1 to give back cross (BC1) individuals that segregated 1:1 for resistance or susceptibility to C. fulvum race 4 GUS infection. DNA gel blot analysis was used to screen resistant BC1 plants for Cf-2 homozygotes, and one was used as male parent in a back cross to rcr3–1. In the resulting BC2 population, all plants were homozygous for Cf-2 and, as expected, segregated 1:1 for resistance or susceptibility to C. fulvum race 4 GUS infection. The chromosomal location of Rcr3 was determined by amplified restriction fragment polymorphism (AFLP) analysis (60) of bulked segregant pools. Fifty-eight BC2 individuals were inoculated with C. fulvum race 4 GUS and screened for resistance or susceptibility to infection. DNA was extracted from pools of 24 resistant and 34 susceptible plants and subjected to AFLP analysis. Several AFLP markers linked to the L. pennellii Rcr3 allele were identified (Fig. 6A). AFLP analysis of L. esculentum × L. pennellii introgression lines (61) showed these markers were located on tomato chromosome 2 (Fig. 6B).
Rcr3 Is Required Specifically for Cf-2 Function. To determine whether Rcr3 is required for the function of other Cf genes, rcr3 mutants were crossed to Cf5 and Cf9 near isogenic lines. Rcr3 will segregate independently of Cf-5 and Cf-9, because they map to different chromosomes (12). In a cross between Cf9 and rcr3 lines, F2 progeny should segregate 3 resistant to 1 susceptible when inoculated with C. fulvum race 2,4 if Rcr3 is not required for Cf-9 function (C. fulvum race 2,4 lacks Avr2 and Avr4 and is virulent on tomato lines carrying either Cf-2 or Cf-4, so in a cross between Cf9 and rcr3, Cf-2 confers no effective resistance to C. fulvum race 2,4). If Rcr3 is required for Cf-9 function, the progeny should segregate 9 resistant to 7 susceptible when inoculated with C. fulvum race 2,4. Progeny from this cross segregated 3 resistant to 1 susceptible, demonstrating Rcr3 is not required for Cf-9-mediated resistance (Table 2). Further confirmation of this result was obtained from analysis of back-cross progeny. As predicted, the progeny segregated 1:1 for resistance and susceptibility to C. fulvum race 2,4 infection (Table 2), confirming that Rcr3 is not required for Cf-9 function. DNA gel blot analysis confirmed that none of the susceptible progeny contained Cf-9 (data not shown).
Table 1. C. fulvum-sensitive Cf2 mutants
|
Mutant allele |
Mutagen |
Frequency¶ |
|
rcr3-1* |
DEB‡ |
5/23 |
|
rcr3-2* |
EMS§ |
2/22 |
|
rcr3-3† |
EMS |
5/23 |
|
rcr3-4† |
EMS |
4/23 |
|
* Identified with fluorometric GUS assay. † Identified by visual inspection. ‡ Diepoxybutane. § Ethyl methanesulfonate. ¶ Number of susceptible individuals in total M2 family. |
||
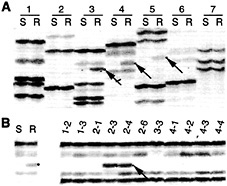
Fig. 6. Rcr3 maps to chromosome 2. AFLP analysis was carried out as previously described (59). (A) Identification of AFLP markers linked to Rcr3 (arrows). Susceptible (S) and resistant (R) pools of the BC2 mapping populations were subjected to AFLP analysis. Pairs of lanes (1 to 7) represent analysis with different AFLP primer combinations. (B) Mapping of a linked AFLP marker by using L. pennellii introgression lines (1–2 to 4–4) with primer combination 4. Susceptible and resistant pools of the BC2 mapping populations were analyzed as controls. The arrows indicate the AFLP fragment linked to L. pennellii Rcr3 present in introgression lines 2–3 and 2–4 that localizes that marker to chromosome 2.
A similar analysis was used to determine whether Rcr3 is required for Cf-5-mediated resistance. Cf-5 and Cf-2 are allelic, and their products are more than 90% identical (46). F2 progeny from a cross between rcr3–2 and Cf5 were screened by inoculation with C. fulvum race 2,4 and segregated at a ratio of 3 resistant to 1 susceptible (Table 2), demonstrating Rcr3 is not required for Cf-5-mediated resistance. PCR analysis confirmed that all C. fulvum race 2,4 susceptible plants lacked Cf-5 (data not shown). To confirm the authenticity of the cross, the same F2 plants were reinoculated with C. fulvum race 5 (a race that lacks Avr5 and is virulent on lines carrying only Cf-5, so only Cf-2 will confer resistance to infection in this test). The segregation ratio of resistant to sensitive progeny was close to a 9:7 ratio predicted on the basis of independent segregation of Cf-2 and Rcr3 (Table 2).
Cf Protein Function. One approach to understanding Cf protein function is to analyze the biochemical and physiological re-
Table 2. Effect of rcr3 mutations on Cf-5 and Cf-9 functions
|
Segregation |
Goodness of fit (χ2)¶ |
||||
|
Cf line |
Cross |
R:S |
1:1 |
3:1 |
9:7 |
|
Cf9* |
F2‡ |
41:17 |
– |
0.57 |
4.9 |
|
Cf9* |
BC1‡ |
16:10 |
1.38 |
18.5 |
– |
|
Cf5† |
F2¶ |
91:32 |
– |
0.068 |
15.7 |
|
Cf5† |
F2§ |
61:52 |
– |
25 |
0.24 |
|
S, susceptible; R, resistant. * Crosses with rcr3-1. † Crosses with rcr3-2. ‡ Screened with C. fulvum race 2,4. § Screened with C. fulvum race 5. ¶ Significance level at P = 0.05 for a χ2 with one degree of freedom is 3.84. |
|||||
sponses that are activated on Avr-dependent elicitation. R proteins appear to activate similar cellular responses that are effective against a broad range of plant pathogens. For example, in transgenic tobacco, Cf-9 can confer resistance to Potato Virus X expressing Avr9 (62). We have defined five main Avr9-dependent responses that ensue on elicitation in tobacco Cf-9 plants and cell cultures. These include stimulation of ion fluxes (63), production of active oxygen species (64), activation of two mitogen-activated protein kinases (65), and a calciumdependent protein kinase (66). We have also defined a large set of rapidly induced genes (67) and are currently trying to identify plasma membrane proteins that interact with epitope-tagged Cf-9 in tobacco (13).
Another approach is to dissect the signaling pathway of Cf-mediated resistance by using mutational analysis. Previously we identified two genes (Rcr1 and Rcr2) that are required for full Cf-9 function (55). In the present study, mutations in Rcr3, a gene required for Cf-2 function, were identified. No Cf-2 mutants were recovered because the Cf2 line carries two functional copies of the gene (68). Four mutations in Rcr3 were identified, and no mutations were identified in other genes, indicating that the genetic screen for suppressors of Cf-2 function may be saturated. The lack of additional mutants in the Cf-2-mediated signaling pathway may reflect functional redundancy or lethality. Genetic analysis demonstrated that Rcr3 is not required for Cf-9 function, therefore Rcr3 is not a component of a conserved Cf signal transduction pathway. We suggested previously that Cf-2 and Cf-5 may interact with a common partner to activate an Avr-dependent plant defense response (12, 46). Because Rcr3 is not required for Cf-5 function, Rcr3 cannot be a Cf-2/Cf-5 common interacting partner.
In Arabidopsis, several genes appear to be required for the function of either of the two classes of NB-LRR genes. EDS1 is required for the function of the TIR-NB-LRR genes RPP5, RPP1, and RPS4, whereas NDR1 and Pbs2 are required for the function of the LZ-NB-LRR genes RPS2, RPS5 and RPM1 (29, 69, 70). These studies appeared to identify at least two parallel signaling pathways for R genes from the two different NB-LRR classes. Pbs3 is required for the function of RPS2, RPS5 and RPM1 (all LZ-NB-LRR class) and also RPS4 (TIR-NB-LRR class), thus establishing a possible convergence point for these pathways (70). Another gene, Pbs1, has been identified that is specifically required for RPS5 (70). Rcr3 and Pbs1 are therefore unique because they appear to suppress the function of single R genes. These analyses have revealed additional genetic complexity in the pathogen perception mechanism of plants.
There are several possible roles for Rcr3 in Cf-2/Avr2-mediated resistance to C. fulvum. Rcr3 could be specifically required for the expression, stability, or modification of Cf-2. Rcr3 may also play a role in the perception of Avr2 or in its proteolytic processing/modification to generate a mature ligand that can be recognized by Cf-2. It is known that maturation of Avr9 and Avr4 requires the activity of both fungal and plant proteases (71, 72). Proteolytic processing of ligand molecules is common in other systems, e.g., processing of the Spätzle ligand of the Drosophila Toll receptor (73). However, if Rcr3 functions solely as an extracellular protease or modifying enzyme, infiltrating cotyledons of rcr3 mutants with intercellular fluids isolated from a compatible tomato –C. fulvum race 5 interaction should restore the characteristic chlorotic response observed in Cf2 plants (74). No complementation of the mutant phenotype was observed, suggesting Rcr3 does not function as an extracellular modifying enzyme (data not shown).
It is possible that Cf-2 and Cf-5 utilize independent signaling pathways to activate the plant defense response, and that Rcr3 is a component of the Cf-2-dependent signaling pathway, downstream of Avr2 perception. An analogous situation may occur in barley, where at least two distinct classes of Mla alleles can be identified on the basis of their requirement for the Rar1 gene (75). Alternatively, Rcr3 may interact directly with Cf-2 to function as a signaling partner. A paradigm for Cf protein function was suggested from studies on three genes (CLV1, CLV2, and CLV3) that condition the Arabidopsis “clavata” mutant phenotype (76, 77 and 78). CLV1 is an Xa21-like extracellular receptor kinase, and CLV2 is an extracellular membrane anchored Cf-like protein (Fig. 1C). CLV1 and CLV2 are proposed to form a heterodimer that may be activated on binding of a peptide ligand (CLV3) resulting in phosphorylation of the kinase domain of CLV1 and association with downstream factors such as a kinase-associated protein phosphatase, KAPP, and the small GTPase, Rop (77, 79). Likewise, the truncated rice Xa21D protein for resistance to X. campestris pv. oryzae may signal through a full-length Xa21 homolog (80). A similar model has been postulated for the sporophytic self-incompatibility mechanism in Brassica species to explain the requirement of SLG, a secreted S-locus glycoprotein, and SRK, an S-locus receptor kinase that contains an extracellular SLG domain.
We with others have investigated whether Cf-9 and Avr9 directly interact with each other. These experiments have failed to reveal a direct interaction between Avr9 and Cf-9, suggesting that other factors are required for Avr9 perception (J.D.G.J., P. J. G. M. De Wit & T. Nürnberger, unpublished work). These factors and other proteins required for Cf-function are conserved in solanaceous plants because Cf-9 confers responsiveness to Avr9 in tobacco and potato (81) and Cf-2 to Avr2 in tobacco (M.S.D., unpublished work). Furthermore, studies using labeled Avr9 have identified a high-affinity binding site on plasma membranes of tomato and tobacco that is present irrespective of their Cf-9 genotype (82). Similarly in soybean, a 34-kDa protein has been identified that binds syringolide elicitors, avirulence factors recognized by Rpg4, irrespective of the Rpg4 genotype (83). By analogy, Avr2 may not interact directly or exclusively with Cf-2 but may interact with Rcr3. Another example where accessory proteins are required for full function of recognition proteins is provided by the mammalian CD14 surface protein. CD14 contributes to the perception of microbial products and acts in concert with specific TLRs that discriminate between pathogens and initiate transmembrane signaling (84).
Pathogen-encoded (a)virulence factors could interact with host proteins to modify their functions to access nutrients or to suppress defense mechanisms. For example, the human pathogen Yersinia uses a type III secretion system, in common with many plant pathogenic bacteria (2), to deliver YopJ into host cells to suppress host defense mechanisms (85). R proteins may have evolved to specifically recognize the physical association of pathogen-encoded virulence factors with their plant cellular targets to subsequently activate defense mechanisms, i.e., R proteins may “guard” host proteins and initiate an Avr-dependent defense response (86, 87). The physical interaction of Pto with AvrPto provides the only published evidence for an Avr product binding to its corresponding R protein (16, 17). We have proposed that Prf, a NB-LRR protein required for Pto function (88), might best be thought of as an R protein that recognizes the AvrPto/Pto pathogenicity complex to activate a resistance response (86). A similar model has been proposed to account for yeast two-hybrid interactions between RPP5 and an Arabidopsis RelA/SpoT homolog (87). Accordingly, Rcr3 may be the pathogenicity target of AVR2, and Cf-2 could specifically “guard” Rcr3. The tendency for a lower disease sensitivity observed 10 days after C. fulvum infection in rcr3–2 and rcr3–3 mutants compared with the Cf0 susceptible control (Fig. 4B) could be caused by
absence of the Rcr3 pathogenicity target. The characterization of a Cf0/rcr3 mutant will reveal whether Rcr3 is necessary for full pathogenicity of C. fulvum.
The Guard hypothesis predicts that for each R protein there is both a corresponding pathogen Avr product and a host target. Such a model would explain the dual recognition capacity of some NB-LRR proteins such as RPM1 and Mi-1 if they “guard” the same host component targeted by unrelated Avr products. Evolutionary mechanisms sustaining R gene diversity are essential for the plant to be able to detect distinct pathogen (a)virulence products that target R protein-“guarded ” host components.
All members of the Jones lab are thanked for useful discussion. We thank Sara Perkins, Margaret Shailer, and Justine Campling for their excellent horticultural service. This work was supported in part by the United Kingdom Biotechnology and Biological Sciences Research Council (M.S.D.) and by a Gatsby Ph.D. studentship (C.G.). The Sainsbury Laboratory is supported by the Gatsby Charitable Foundation.
1. Flor, H. H. ( 1971) Annu. Rev. Phytopathol. 9, 275–296.
2. Galan, E. J. & Collmer, A. ( 1999) Science 284, 1322–1328.
3. Kearney, B. & Staskawicz, B. J. ( 1990) Nature ( London) 346, 385–386.
4. Ritter, C. & Dangl, J. L. ( 1995) Mol. Plant-Microbe Interact. 8, 444–453.
5. Erickson, F. L., Holzberg, S., Calderon-Urrea, A., Handley, V., Axtell, M., Corr, C. & Baker, B. ( 1999) Plant J. 18, 67–75.
6. Bendahmane, A., Kohm, B. A., Dedi, C. & Baulcombe, D. C. ( 1995) Plant J. 8, 933–941.
7. Van den Ackerveken, G. F. J. M., Van Kan, J. A. L. & De Wit, P. J. G. M. ( 1992) Plant J. 2, 359–366.
8. Joosten, M. H. A. J., Cozijnsen, T. J. & De Wit, P. J. G. M. ( 1994) Nature ( London) 367, 384–386.
9. Lauge, R., Joosten, M. H. A. J., Van den Ackerveken, G. F. J. M., Van den Broek, H. W. J. & De Wit, P. J. G. M. ( 1997) Mol. Plant-Microbe Interact. 10, 725–734.
10. Lauge, R., Joosten, M. H. A. J., Haanstra, J. P. W., Goodwin, P. H., Lindhout, P. & De Wit, P. J. G. M. ( 1998) Proc. Natl Acad. Sci. USA 95, 9014–9018.
11. Koch, E. S. & Slusarenko, A. ( 1990) Plant Cell 2, 437–445.
12. Thomas, C. M., Dixon, M. S., Parniske, M., Golstein, C. & Jones, J. D. G. ( 1998) Philos. Trans. R. Soc. London 353, 1413–1424.
13. Piedras, P., Rivas, S., Droge, S., Hillmer, S. & Jones, J. D. G. ( 2000) Plant J., 21, 529–536.
14. Song, W.-Y., Wang, G.-L., Chen, L.-L., Kirn, H.-S., Pi, L.-Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.-X., Zhu, L.-H., et al. ( 1995) Science 270, 1804–1806.
15. Bogdanove, A. J. & Martin, G. B. ( 2000) Proc. Natl Acad. Sci. USA 97, 8836–8840.
16. Scofield, S. R., Tobias, C. M., Rathjen, J. P., Chang, J. H., Lavelle, D. T., Michelmore, R. W. & Staskawicz, B. J. ( 1996) Science 274, 2063–2065.
17. Tang, X., Frederick, R. D., Zhou, J., Halterman, D. A., Jia, Y. & Martin, G. B. ( 1996) Science 274, 2060–2063.
18. Rock, F. L., Hardiman, G., Timans, J. C., Kastelein, R. A. & Bazan, J. F. ( 1998) Proc. Natl. Acad. Sci. USA 95, 588–593.
19. Janeway, C. A. & Medzhitov, R. ( 1999) Curr. Biol. 9, R879–R882.
20. Medzhitov, R. & Janeway, C. A. ( 1997) Curr. Opin. Immunol. 9, 4–9.
21. Brightbill, H. D., Libraty, D. H., Krutzik, S. R., Yang, R. B., Belisle, J. T., Bleharski, J. R., Maitland, M., Norgard, M. V., Plevy, S. E., Smale, S. T., et al. ( 1999) Science 285, 732–736.
22. Aliprantis, A. O., Yang, R. B., Mark, M. R., Suggett, S., Devaux, B., Radolf, J. D., Klimpel, G. R., Godowski, P. & Zychlinsky, A. ( 1999) Science 285, 736–739.
23. Meyers, B. C., Dickerman, A. W., Michelmore, R. W., Sivaramakrishnan, S., Sobral, B. W. & Young, N. D. ( 1999) Plant J. 20, 317–332.
24. Hammond-Kosack, K. E. & Jones, J. D. G. ( 1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607.
25. McDowell, J. M., Dhandaydham, M., Long, T. A., Aarts, M. G. M., Goff, S., Holub, E. B. & Dangl, J. L. ( 1998) Plant Cell 10, 1861–1874.
26. Collins, N., Drake, J., Ayliffe, M., Sun, Q, Ellis, J., Hulbert, S. & Pryor, T. ( 1999) Plant Cell 11, 1365–1376.
27. O'Neill, L. A. J. & Greene, C. ( 1998) J. Leukocyte Biol. 63, 650–657.
28. Hoffmann, J. A., Kafatos, F. C., Janeway, C. A. & Ezekowitz, R. A. B. ( 1999) Science 284, 1313–1318.
29. Gassmann, W., Hinsch, M. E. & Staskawicz, B. J. ( 1999) Plant J. 20, 265-277.
30. Medzhitov, R., Preston-Hurlburt, P., Kopp, E., Stadlen, A., Chen, C. Q., Ghosh, S. & Janeway, C. A. ( 1998) Mol. Cell 2, 253–258.
31. Chinnaiyan, A. M., Chaudhary, D., O'Rourke, K., Koonin, E. V. & Dixit, V. M. ( 1997) Nature ( London) 388, 728–729.
32. Zou, H., Henzel, W. J., Liu, X. S., Lutschg, A. & Wang, X. D. ( 1997) Cell 90, 405–413.
33. Van der Biezen, E. A. & Jones, J. D. G. ( 1998) Curr. Biol. 8, 226–227.
34. Aravind, L., Dixit, V. M. & Koonin, E. V. ( 1999) Trends Biochem. Sci. 24, 47–53.
35. Bertin, J., Nir, W. J., Fischer, C. M., Tayber, O. V, Errada, P. R., Grant, J. R., Keilty, J. J., Gosselin, M. L., Robison, K. E., Wong, G. H. W., et al. ( 1999) J. Biol. Chem. 274, 12955–12958.
36. Inohara, N., Koseki, T., Del Peso, L., Hu, Y. M., Yee, C., Chen, S., Carrio, R., Merino, J., Liu, D., Ni, J. & Nunez, G. ( 1999) J. Biol. Chem. 274, 14560–14567.
37. Hu, Y. M., Benedict, M. A., Ding, L. Y. & Nunez, G. ( 1999) EMBO J. 18, 3586–3595.
38. Del Pozo, O. & Lam, E. ( 1998) Curr. Biol. 8, 1129–1132.
39. Hughes, A. L. & Yeager, M. ( 1998) Annu. Rev. Genet. 32, 415–435.
40. Leonard, K. J. ( 1997) in The Gene-for-Gene Relationship in Plant-Parasite Interactions, eds. Crute, I. R., Holub, E. B. & Burdon, J. J. (CAB, Wallingford, U. K.), pp. 211–230.
41. Noel, L., Moores, T. L., Van der Biezen, E. A., Parniske, M., Daniels, M. J., Parker, J. E. & Jones, J. D. G. ( 1999) Plant Cell 11, 2099–2012.
42. Stahl, E. A., Dwyer, G., Mauricio, R., Kreitman, M. & Bergelson, J. ( 1999) Nature ( London) 400, 667–671.
43. Parniske, M., Hammond-Kosack, K. E., Golstein, C, Thomas, C. M., Jones, D. A., Harrison, K., Wulff, B. B. H. & Jones, J. D. G. ( 1997) Cell 91, 821–832.
44. Thomas, C. M., Jones, D. A., Parniske, M., Harrison, K., Balint-Kurti, P. J., Hatzixanthis, K. & Jones, J. D. G. ( 1997) Plant Cell 9, 2209–2224.
45. Takken, F. L. W., Thomas, C. M., Joosten, M., Golstein, C., Westerink, N., Hille, J., Nijkamp, H. J. J., De Wit, P. J. G. M. & Jones, J. D. G. ( 1999) Plant J. 20, 279–288.
46. Dixon, M. S., Hatzixanthis, K., Jones, D. A., Harrison, K. & Jones, J. D. G. ( 1998) Plant Cell 10, 1915–1925.
47. Botella, M. A., Parker, J. E., Frost, L. N., BittnerEddy, P. D., Beynon, J. L., Daniels, M. J., Holub, E. B. & Jones, J. D. G. ( 1998) Plant Cell 10, 1847–1860.
48. Parker, J. E., Coleman, M. J., Szabo, V., Frost, L. N., Schmidt, R., Van der Biezen, E. A., Moores, T., Dean, C., Daniels, M. J. & Jones, J. D. G. ( 1997) Plant Cell 9, 879–894.
49. Meyers, B. C., Shen, K. A., Rohani, P., Gaut, B. S. & Michelmore, R. W. ( 1998) Plant Cell 10, 1833–1846.
50. Kobe, B. & Deisenhofer, J. ( 1994) Trends Biochem. Sci. 19, 415–421.
51. Ellis, J., Lawrence, G., Ayliffe, M., Anderson, P., Collins, N., Finnegan, J., Frost, D., Luck, J. & Pryor, T. ( 1997) Annu. Rev. Phytopathol. 35, 271–291.
52. Hulbert, S. H. ( 1997) Annu. Rev. Phytopathol. 35, 293–310.
53. Michelmore, R. W. & Meyers, B. C. ( 1998) Genome Res. 8, 1113–1130.
54. Nei, M., Gu, X. & Sitnikova, T. ( 1997) Proc. Natl. Acad. Sci. USA 94, 7799–7806.
55. Hammond-Kosack, K. E., Jones, D. A. & Jones, J. D. G. ( 1994) Plant Cell 6, 361–374.
56. Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. ( 1987) EMBO J. 6, 3901–3907.
57. Oliver, R. P., Farman, M. L., Jones, J. D. G. & Hammond-Kosack, K. E. ( 1993) Mol. Plant-Microbe Interact. 6, 521–525.
58. Keogh, R. C., Deverall, B. J. & Mcleod, S. ( 1980) Trans. Brit. Mycol. Soc. 74, 329–333.
59. Thomas, C. M., Vos, P., Zabeau, M., Jones, D. A., Norcott, K. A., Chadwick, B. P. & Jones, J. D. G. ( 1995) Plant J. 8, 785–794.
60. Vos, P., Hogers, R., Bleeker, M., Reijans, M., Van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M. & Zabeau, M. ( 1995) Nucleic Acids Res. 23, 4407–4414.
61. Eshed, Y. & Zamir, D. ( 1994) Euphytica 79, 175–179.
62. Thomas, C. M., Tang, S., Hammond-Kosack, K. E. & Jones, J. D. G. ( 2000) Mol. Plant-Microbe Interact. 13, 465–469.
63. Blatt, M. R., Grabov, A., Brearley, J., Hammond-Kosack, K. E. & Jones, J. D. G. ( 1999) Plant J. 19, 453–462.
64. Piedras, P., Hammond-Kosack, K. E., Harrison, K. & Jones, J. D. G. ( 1998) Mol. Plant-Microbe Interact. 11, 1155–1166.
65. Romeis, T., Piedras, P., Zhang, S. Q., Klessig, D. F., Hirt, H. & Jones, J. D. G. ( 1999) Plant Cell 11, 273–287.
66. Romeis, T., Piedras, P. & Jones, J. D. G. ( 2000) Plant Cell 12, 803–815.
67. Durrant, W., Rowland, O., Piedras, P. & Jones, J. D. G. ( 2000) Plant Cell 12, 963–977.
68. Dixon, M. S., Jones, D. A., Keddie, J. S., Thomas, C. M., Harrison, K. & Jones, J. D. G. ( 1996) Cell 84, 451–459.
69. Aarts, N., Metz, M., Holub, E., Staskawicz, B. J., Daniels, M. J. & Parker, J. E. ( 1998) Proc. Natl. Acad. Sci. USA 95, 10306–10311.
70. Warren, R. F., Merritt, P. M., Holub, E. & Innes, R. W. ( 1999) Genetics 152, 401–412.
71. Van den Ackerveken, G. F. J. M., Vossen, J. P. M. J. & De Wit, P. J. G. M. ( 1993) Plant Physiol. 103, 91–96.
72. Joosten, M. H. A. J., Vogelsang, R., Cozijnsen, T. J., Verberne, M. C. & De Wit, P. J. G. M. ( 1997) Plant Cell 9, 367–379.
73. Van Eeden, F. & St Johnston, D. ( 1999) Curr. Opin. Genet. Dev. 9, 396–404.
74. Hammond-Kosack, K. E. & Jones, J. D. G. ( 1994) Mol. Plant-Microbe Interact. 7, 58–70.
75. Jorgensen, J. H. ( 1996) Genome 39, 492–498.
76. Clark, S. E., Williams, R. W. & Meyerowitz, E. M. ( 1997) Cell 89, 575–585.
77. Jeong, S., Trotochaud, A. E. & Clark, S. E. ( 1999) Plant Cell 11, 1925–1933.
78. Fletcher, L. C., Brand, U., Running, M. P., Simon, R. & Meyerowitz, E. M. ( 1999) Science 283, 1911–1914.
79. Trotochaud, A. E., Hao, T., Wu, G., Yang, Z. B. & Clark, S. E. ( 1999) Plant Cell 11, 393–405.
80. Wang, G. L., Ruan, D. L., Song, W. Y., Sideris, S., Chen, L. L., Pi, L. Y., Zhang, S. P., Zhang, Z., Fauquet, C, Gaut, B. S., et al. ( 1998) Plant Cell 10, 765–779.
81. Hammond-Kosack, K. E., Tang, S. J., Harrison, K. & Jones, J. D. G. ( 1998) Plant Cell 10, 1251–1266.
82. Kooman-Gersmann, M., Honee, G., Bonnema, G. & De Wit, P. J. G. M. ( 1996) Plant Cell 8, 929–938.
83. Ji, C, Boyd, C., Slaymaker, D., Okinaka, Y., Takeuchi, Y., Midland, S. L., Sims, J. J., Herman, E. & Keen, N. ( 1998) Proc. Natl. Acad. Sci. USA 95, 3306–3311.
84. Ulevitch, R. J. ( 1999) Nature ( London) 401, 755–756.
85. Orth, K., Palmer, L. E., Bao, Z. Q., Stewart, S., Rudolph, A. E., Bliska, J. B. & Dixon, J. E. ( 1999) Science 285, 1920–1923.
86. Van der Biezen, E. A. & Jones, J. D. G. ( 1998) Trends Biochem. Sci. 23, 454–456.
87. Van der Biezen, E. A., Sun, J., Coleman, M. J., Bibb, M. J. & Jones, J. D. G. ( 2000) Proc. Natl. Acad. Sci. USA 97, 3747–3752.
88. Salmeron, J. M., Oldroyd, G. E. D., Rommens, C. M. T., Scofield, S. R., Kim, H.-S., Lavelle, D. T., Dahlbeck, D. & Staskawicz, B. J. ( 1996) Cell 86, 123–133.








