Colloquium
AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato
Adam J. Bogdanove and Gregory B. Martin*
Boyce Thompson Institute For Plant Research and Department of Plant Pathology, Cornell University, Ithaca, NY 14853
The plant-intracellular interaction of the avirulence protein AvrPto of Pseudomonas syringae pathovar tomato, the agent of bacterial speck disease, and the corresponding tomato resistance protein Pto triggers responses leading to disease resistance. Pto, a serine/threonine protein kinase, also interacts with a putative downstream kinase, Pto-interactor 1, as well as with members of a family of transcription factors Pto-interactors 4, 5, and 6. These proteins are likely involved, respectively, in a phosphorylation cascade resulting in hypersensitive cell death, and in defense gene activation. The mechanism by which the interaction of AvrPto and Pto initiates defense response signaling is not known. To pursue the hypothesis that tertiary interactions are involved, we modified the yeast two-hybrid protein interaction trap and conducted a search for tomato proteins that interact with Pto only in the presence of AvrPto. Five classes of AvrPto-dependent Pto interactors were isolated, and their interaction specificity confirmed. Also, to shed light on a recently demonstrated virulence activity of AvrPto, we conducted a standard two-hybrid screen for tomato proteins in addition to Pto that interact with AvrPto: i.e., potential virulence targets or modifiers of AvrPto. By constructing an N-terminal rather than a C-terminal fusion of AvrPto to the LexA DNA binding domain, we were able to overcome autoactivation by AvrPto and identify four classes of specific AvrPto-interacting proteins.
I n many plant pathosystems, an important form of disease resistance known as gene-for-gene resistance depends on the pathogen delivering a specific “avirulence signal,” generated by the expression of an avirulence (avr) gene, and on the plant perceiving and responding to that signal, an ability conferred by a corresponding resistance (R) gene (1, 2). We study gene-for-gene resistance in bacterial speck of tomato caused by Pseudomonas syringae pathovar tomato. In this system, disease resistance is triggered by strains that express the avr gene avrPto (3, 4) and is mediated in tomato by the corresponding R gene Pto (5). The resistance response includes expression of defense-related genes (6), a rapid generation of reactive oxygen species known as the oxidative burst (7), and rapid, localized cell death termed the hypersensitive response (8). It has been demonstrated indirectly (9, 10) that the pathogen delivers the product of the avrPto gene, a small hydrophilic protein, into the plant cell, where it interacts directly with the product of the Pto R gene, a serine/threonine protein kinase (11). The interaction is highly specific and sensitive to single amino acid changes in the Pto kinase activation domain (12). Subsequent events ultimately thwart further advance of the pathogen.
Another required component of the AvrPto-Pto resistance response pathway, the Prf gene, was identified in a mutant screen (13). Prf encodes a protein with a leucine zipper, a nucleotide binding site, and leucine-rich repeats, motifs common to R gene products in other systems (14). The role of Prf remains unclear, although based on studies with tomato plants that overexpress Prf, it might act downstream of Pto (15).
Other putative effectors of Pto-mediated disease resistance have been identified by their specific interaction with Pto in the yeast two-hybrid system. These Pto-interacting (Pti) proteins include Pti1, a serine/threonine protein kinase that is specifically phosphorylated in vitro by Pto and is involved in the hypersensitive response (16), and Pti4, Pti5, and Pti6, putative transcription factors that are similar to the tobacco ethylene-responsive element binding proteins and may be involved in activation of defense-related genes (17). Recent results indicate that Pti4 also is phosphorylated specifically by Pto, and that this phosphorylation enhances Pti4 binding to a defined cis element present in the promoter of many pathogenesis-related genes associated with plant defense (18).
The interaction between AvrPto and Pto might expose certain domains of Pto, which are not available in the resting state, to autophosphorylation or to phosphorylation by another kinase, thereby activating the resistance protein. AvrPto might stabilize Pto, raising steady-state levels of the kinase. AvrPto might act as a bridge or increase the affinity of Pto for its substrates, or for an accessory protein [e.g., Prf (19)] that directs or enhances Pto kinase activity toward substrates involved in resistance to Pseudomonas.
But genes such as avrPto are not likely to have been maintained through evolution because of their function in triggering plant defense. Indeed, in the absence of a corresponding R gene, some avr genes have been shown to promote disease (see ref.2 for a review), and recently, a virulence-enhancing effect has been observed for AvrPto in Pseudomonas strains inoculated to tomato plants lacking Pto (X. Tang, personal communication)
To elucidate early events in AvrPto/Pto-mediated activation of defense responses, we modified the yeast two-hybrid system and conducted a search for tomato proteins that interact with Pto only when AvrPto is also expressed in the yeast cell. Also, to shed light on the virulence function of AvrPto, we constructed a non-autoactivating AvrPto-LexA fusion and conducted a search for tomato proteins in addition to Pto that interact with AvrPto: i.e., potential virulence targets or other proteins.
Isolation and Characterization of Proteins That Interact with Pto in an AvrPto-Dependent Fashion
The LexA-based yeast two-hybrid system (20) was modified such that AvrPto fused only to a nuclear localization signal sequence would be expressed in yeast under the control of the same inducible promoter as the prey. First the 1.3-kb KpnI/BamHI fragment of prey plasmid pJG4-5 containing the prey expression
This paper was presented at the National Academy of Sciences colloquium “Virulence and Defense in Host–Pathogen Interactions: Common Features Between Plants and Animals, ” held December 9–11, 1999, at the Arnold and Mabel Beckman Center in Irvine, CA.
Abbreviations: Adi, AvrPto-dependent Pto-interacting; Api, AvrPto-interacting; Pti, Pto-interacting; avr, avirulence.
|
* |
To whom reprint requests should be addressed at: Boyce Thompson Institute, Tower Road, Ithaca, NY 14853. E-mail: gbm7@cornell.edu. |
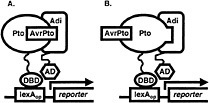
Fig. 1. Yeast three-hybrid hunt for AvrPto-dependent Pto-interacting (Adi) proteins. Coexpression of AvrPto fused to a nuclear localization signal and of Pto fused to the DNA-binding domain (DBD) of the LexA transcriptional activator (bait fusion) made it possible to screen a library of tomato cDNA clones fused to the activation domain (AD) of LexA (prey fusion) for proteins that interact with an AvrPto-Pto complex (A) or with Pto that had assumed an AvrPto-dependent conformation (B). Association of the bait and prey reconstitutes the LexA transcriptional activator and drives expression of reporter genes (lacZ and leu2) under the control of the LexA operator (LexAop). Pto interaction with candidate Adi clones is later tested for AvrPto-dependence.
cassette (21) was subcloned into pBluescript II SK(−) (Stratagene), yielding plasmid pBOG5. avrPto was amplified from pPTE6 (4) by the PCR by using primers avrPtoFP2 (5′ GGGTATACAGCTGGGAAATATATGTGTCGG 3′), which contains a PvuII site (underlined) and a Met to Leu change at the initiator codon (italicized), and avrPtoRP2 (5′ ACGCACTCGAGAACCTCTGCACTCACC 3′), which contains an XhoI site (underlined). The portion of pBOG5 encoding the activation domain and the HA epitope tag was excised by cutting with EcoRV and XhoI, and was replaced with the avrPto PCR product cut with PvuII and XhoI. The ligation destroyed the EcoRV and PvuII sites and resulted in an in-frame fusion of avrPto to the nuclear localization signal sequence of the prey construct. Nucleotide sequence of the fusion junction and the avrPto gene was confirmed. The construct was excised by digestion with PvuII and SacI, which cut in the vector, and was cloned into the reporter plasmid pSH18-34 (21) that had been cut with SmaI and SacI, releasing a 2.8-kb fragment containing the 5′ end of the lacZ reporter gene. This plasmid was designated as pBOG7. Finally, the reporter gene was restored by reinsertion of the 2.8-kb SmaI/SacI fragment between the unique NotI and SacI sites of pBOG7 to yield pBOG8. The tomato cDNA library previously used to isolate Pti proteins was then screened by using Pto as a bait as described (16, 21) but substituting pBOG8 for pSH18-34. In this way, we were able to search for clones encoding proteins that interacted with an AvrPto-Pto complex or with Pto that had assumed an AvrPto-dependent conformation (Fig. 1). Sixty million primary transformants were obtained, and after plating to obtain a 3× representation, 3,780 leucine prototrophs were obtained. Of these, 84 showed strong activation of the lacZ reporter gene, and 155 weak. Partial DNA sequence was determined for each of these and compared against the GenBank database by using the BLASTX algorithm (22). Clones showing similarity to abundant housekeeping transcripts such as RUBISCO and ubiquitin, as well as clones representing 2 isopropylmalate dehydrogenase (functionally equivalent to the leu2 gene) were eliminated. Remaining clones were grouped into classes (26 total) according to identity based on BLASTX results or DNA sequence identity. For a representative clone of each class, interaction with Pto was retested for AvrPto-dependence and was tested for specificity, including possible interaction with AvrPto alone (using the non-autoactivating AvrPto bait construct described herein) as shown in Fig. 2A. Five classes of AvrPto-dependent Pto-interacting (Adi) proteins were identified (Table 1). Some of the Adi proteins showed weak interaction with Pto alone based on activation of the leu2 gene (Fig. 2A; Table 2), which is a more sensitive indicator of interaction than the lacZ gene in our experience. Nevertheless, the interaction in each case was greatly enhanced by AvrPto.
Adi1. Adi1 is tomato catalase 1 (23), and represents the largest class in the screen (17 isolates). Protein expressed from a full-length cDNA was active (Fig. 3). Truncated cDNAs encoding the C terminus of the highly similar tomato catalase 2 (GenBank accession no. AF112368) were also isolated that showed a weak AvrPto-dependent interaction with Pto, suggesting that the interacting domain of Adi1 may reside in the C terminus. An interaction of Adi1 with Pto in the plant resulting in inactivation or turnover of this H2O2-scavenger might contribute to the oxidative burst. Adi1 is a member of the class II catalase group that includes the salicylic acid-binding catalase of

Fig. 2. Test for AvrPto-dependence and specificity of Adi protein interactions with Pto, and for specificity of Api protein interactions with AvrPto. Individual yeast transformants expressing each of the indicated three-or two-hybrid protein combinations were streaked to minimal medium agar plates containing 40 µg/ml X-gal to assay expression of the lacZ reporter gene, indicated by a developing blue color, and to minimal medium lacking leucine to assay expression of the leu2 reporter gene, indicated by growth. The Drosophila proteins Bicoid and Dorsal were used as arbitrary bait and prey fusions (kindly provided by R. Brent, Massachusetts General Hospital) for testing the specificity of the interactions observed. (A) Adi clones were tested in yeast containing the indicated constructs. Shown are results for Adi1. Results for all of the Adi proteins are summarized in Table 1. The interaction of Pto and AvrPto is shown for reference. (B) Lack of autoactivation and confirmed interaction with Pto by the AvrPto-LexA fusion used in the hunt for AvrPto interactors. Pto prey fusion was constructed previously by X. Tang in our laboratory. ( C) Api clones were tested in yeast containing the indicated constructs. Shown are results for Api1. Results for all of the Adi proteins are summarized in Table 2.
Table 1. Identity, number of clones isolated, and interaction specificity of Adi proteins
|
Relative lacZ activation by interaction with various bait proteins in yeast with and without AvrPto |
||||||
|
Adi |
Identity |
Isolates |
Pto with AvrPto |
Pto |
Bicoid with AvrPto |
AvrPto |
|
1 |
Tomato catalase 1* |
17 |
+++ |
+ |
− |
− |
|
2 |
Pti1† homolog |
2 |
+++ |
+ |
− |
− |
|
3 |
Similar to Ser/Thr protein kinases |
1 |
+++ |
− |
− |
− |
|
4 |
Unknown (“Mackerel”)‡ |
4 |
+++++ |
+ |
− |
− |
|
5 |
Full-length Pti2§ |
6 |
++++ |
− |
− |
− |
|
* Ref.23. † Ref.16. ‡ Adi4 has no similarity to sequences in published databases and was given this arbitrary designation. § Ref.27. |
||||||
tobacco (24). The putative role of SA binding to members of this class of catalase is not understood, but the interaction of Adi1 with Pto could mediate an effect on defense signaling involving this compound.
Adi2. Adi2 is a putative serine/threonine protein kinase, 46% identical to Pti1. Of particular note among the conserved residues are those in the ATP-binding site, as well as a threonine in the predicted activation domain that corresponds to the major site for Pto phosophorylation of Pti1(25, Fig. 4). Based on the high degree of amino acid sequence identity surrounding this latter site, Adi2 seems a likely substrate of Pto. It will be of interest to determine whether AvrPto enables or enhances Pto phosphorylation of Adi2. There are many examples of direct, ligand-dependent activation of tyrosine kinases and instances of induced serine/threonine protein kinase activation (26), but to our knowledge, phosphorylation of a substrate that depends on ligand interaction with a serine/threonine protein kinase would be novel.
Adi3. Adi3 is another putative serine/threonine protein kinase with no other informative similarities. We view this protein as a possible candidate for a kinase involved in activation of Pto.
Adi4. Adi4, tentatively designated as “Mackerel,” is a predicted 25-kDa, largely hydrophilic protein with no similarity to know proteins. The interactor that showed the greatest activation of reporter genes for interaction, it represents a total of four isolated clones in its class.
Adi5. Adi5 is a full-length cDNA of Pti2 (27). Pti2 is a truncated proteasome alpha subunit previously isolated that interacts strongly with Pto (J. Zhou and G.B.M., unpublished work). Curiously, the interaction of Pto with the truncated protein does not require AvrPto. Our results suggest the possibility that an AvrPto/Pto complex may play a role in regulation of the proteasome via interaction with the alpha subunit. Phosphorylation of this subunit has been postulated to affect localization of the proteasome (28). Other mechanisms, including stimulation of interaction of the proteasome with other cellular proteins (29), are also conceivable and could play a role in the events that lead to disease resistance.
Table 2. Identity, number of clones isolated, and interaction specificity of Api proteins
|
Relative lacZ activation by interaction with different bait proteins in yeast |
|||||
|
Api |
Identity |
Isolates |
AvrPto |
Bicoid |
Pto |
|
1 |
PvSRP* homolog |
53 |
+++ |
− |
− |
|
2 |
Similar to small GTP-binding protein Rab8† |
3 |
+++ |
− |
− |
|
3 |
Similar to small GTP-binding proteins |
4 |
++ |
+ |
− |
|
4 |
Similar to myristyl-CoA protein N-myristyltransferases |
1 |
++ |
+ |
− |
|
* P. vulgaris stress-related protein, GenBank accession no. U54704. † Ref.31. |
|||||
Isolation and Characterization of Proteins That Interact with AvrPto
A standard AvrPto bait construct (C-terminal fusion of AvrPto to the LexA DNA-binding domain) made previously in our laboratory was found to activate both the leu2 and the lacZ reporter genes in the absence of prey (R. Frederick and G.B.M., unpublished work). To overcome this problem of autoactivation, we constructed an N-terminal fusion of AvrPto to the LexA DNA-binding domain by using the vector pNLexA (21), kindly provided by E. Golemis (Fox Chase Cancer Center). AvrPto was amplified from pPTE6 (4) by PCR using oligonucleotide primers AJB5 and AJB6. AJB5 (5′ CAGTGAATTCCGAACCATGGGAAATATATGTGTCGG 3′) contains a minimal Kozak consensus sequence (double underlined) (30) just upstream of the initiator codon (italics) of AvrPto for efficient translation in a

Fig. 3. Alignments of the predicted ATP-binding sites (A) and the predicted activation domains of Adi2 and Pti1 (B). Residues that are highly conserved in similar protein kinases and that constitute the two motifs are underlined (34). T233, the major site of Pti1 autophosphorylation and phosphorylation by Pto (25), is marked with a caret.
eukaryotic cell, and an EcoRI site (underlined). AJB6 (5′ GGCTTCTCGAGGTTGCCAGTTACGGTACG 3′) introduces a mutation to change the stop codon of avrPto to a Pro codon (italics), and contains an XhoI site (underlined) for in-frame fusion to the LexA DNA-Binding domain in pNLexA. The PCR product was cloned in pBluescript II SK(−) by using these restriction sites, and its DNA sequence was confirmed. It was then transferred to pNLexA, using the same restriction sites, to create pBOG10. No autoactivation by the new AvrPto bait construct in yeast was detected, and, as hoped, it showed strong interaction with a Pto prey fusion (Fig. 2B). A standard two-hybrid screen (21) of the tomato cDNA library was conducted by using pBOG10. Thirty million primary transformants, 1,480 leucine prototrophs, and ultimately 58 clones showing strong lacZ activation and 94 clones showing weak lacZ activation were obtained. Clones were selected and grouped as for the three-hybrid screen above. Representative clones were retested for specific interaction with AvrPto (Fig. 2C). Four specific AvrPto-interacting (Api) proteins were identified.
Api1. Api1 is homologous with GenBank no. U54704, annotated in the accession as a stress-related protein of Phaseolus vulgaris regulated by heavy metal-, wound-, and virus-induced stress. Api1 was strongly induced similarly in near isogenic lines of tomato with and without the Pto locus inoculated with a P. syringae strain expressing AvrPto (not shown). Overwhelmingly the most frequent isolate, greater than 50 Api1 clones were obtained in the screen. As a likely component of general defense responses, Api1 stands out as a potential virulence target of AvrPto.

Fig. 4. Complementation of catalase activity in yeast by expression of Adi1. Catalase-deficient Saccharomyces cerevisiae strain 578 (A. Hartig, University of Vienna) was transformed with the expression vector pIB61 (J. Pardo, Yale University School of Medicine) or with pIB61 containing the Adi1 cDNA. Transformed cells were suspended in a solution containing 1% Triton X-100 and 3% H2O2. Catalase activity is indicated by bubbling attributable to the conversion of H2O2 to H2O and O2.
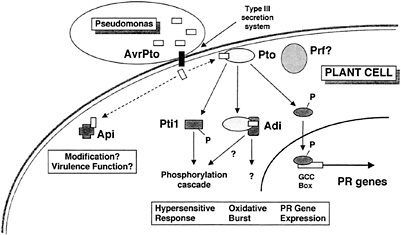
Fig. 5. A model for disease resistance mediated by the interaction of AvrPto and Pto, and for a role of AvrPto in disease.
Api2. Api2 is a Ras-related small, putative GTP-binding protein most similar to Rab8. Rab8 is involved in vesicular protein trafficking from the Golgi apparatus to the plasma membrane (31). Isolation of Api2 suggests the possibility that the AvrPto contributes to disease by interfering with plant protein trafficking, possibly blocking the extracellular release of plant antimicrobial peptides.
Api3. Api3 is another small, putative GTP-binding protein highly similar to Api2.
Api4. Api4 was isolated once in the screen, as a partial cDNA. Its interaction with AvrPto was relatively weak. The predicted amino acid sequence is roughly 50% identical to N-myristyltransferases isolated from yeast and mammals, and it also aligns with a putative N-myristyltransferase of Arabidopsis thaliana (GenBank accession no. AL049711). This interactor suggests the possibility, as has been proposed for other Avr proteins, that AvrPto is modified in planta after delivery by the pathogen. Both AvrPto and Pto have a consensus myristylation motif (32) at their N termini. Mutant forms
of AvrPto lacking this motif lack avirulence function (X. Tang, personal communication). Mutant forms of Pto lacking this motif are still functional when overexpressed, but the motif might be required when Pto is present only at endogenous levels (33). Interestingly, Pto appeared to interact weakly with Api4 (Table 2).
Discussion
We have not established whether any of the Adi and Api proteins actually are involved in speck disease resistance or play a role in the manifestation of AvrPto virulence function. Further characterization of the Adi and Api proteins, including in vitro binding and phosphorylation assays, in planta transient expression assays, localization, and other studies, will be required to confirm the functional significance of the interactions we have observed in yeast. Based on the stringency of the screens, the specificity of the interactions, and the potential functions of the proteins isolated in our screens, however, we view the Adi and Api proteins as strong candidates for plant components involved in the responses of tomato to AvrPto. A model for disease resistance mediated by the interaction of AvrPto and Pto and for a role of AvrPto in disease is shown in Fig. 5, and discussed below. We have incorporated the Adi and Api proteins in the model, but their placement should be viewed as tentative.
Upon delivery into the tomato cell via the type III secretion system of Pseudomonas, AvrPto binds to Pto. This event likely activates Pto, stimulating its phosphorylation of substrates such as the transcription factors Pti4/5/6 and the serine/threonine protein kinase Pti1. Phosphorylation of Pti4/5/6 may target these factors to the nucleus or activate them otherwise, leading to the expression of pathogenesis-related proteins involved in plant defense via interaction with the GCC box promoter element (18). Phosphorylation of Pti1 may constitute an early event in a phosphorylation cascade leading to the oxidative burst and hypersensitive response (16). The interaction of AvrPto and Pto may also recruit other putative components of the defense response pathway, the Adi proteins. The interactions with these proteins may be involved in the generation of the oxidative burst (Adi1?), the hypersensitive response (Adi2?), or other processes leading to plant defense. AvrPto may also interact with Api proteins other than Pto in the tomato cell. These interactions might lead to modification and consequent membrane localization of AvrPto (Api4?) or to the disruption of plant stress responses (Api1?) or other metabolic processes such as protein transport (Api2?). In the absence of Pto, some or all of these could constitute a virulence effect of AvrPto. The role of the Prf protein, although required for disease resistance, is currently unknown.
We did not isolate a Prf cDNA in either of our screens, but we do not discount the possibility that Prf interacts with Pto, AvrPto, or an AvrPto/Pto complex. Low transcript abundance or the large transcript size of Prf could have resulted in poor representation in our oligo(dT)-primed cDNA library. Even if present, the large size of the protein could have precluded the entry of the prey fusion into the nucleus necessary for reporter gene activation. Elucidation of the role of Prf awaits a more directed set of experiments.
Ultimately, we hope, further characterization of the Adi and Api proteins will yield a yet more detailed understanding of the molecular mechanisms involved in bacterial speck disease and disease resistance, and contribute toward the development of means to control this disease, and others, through genetic manipulation.
We thank Elizabeth Suryaatmaja and Philip Pusey for assistance conducting the yeast screens and Yuko Nakajima for specificity screening of Adi5 and helpful discussion. This research was supported by U.S. Department of Agriculture grant NRI-9735303-5299 (A.J.B.) and a David and Lucile Packard Foundation Fellowship (G.B.M.).
1. Keen, N. T. ( 1990) Annu. Rev. Genet. 24, 447–463.
2. Dangl, J. L. ( 1994) in Curr. Top. Microbiol. Immunol. 192, 99–118.
3. Ronald, P. C., Salmeron, J. M., Carland, F. M. & Staskawicz, B. J. ( 1992) J. Bacteriol. 174, 1604–1611.
4. Salmeron, J. M. & Staskawicz, B. J. ( 1993) Mol. Gen. Genet. 239, 6–16.
5. Martin, G. B., Brommonschenkel, S. H., Chunwongse, J., Frary, A., Ganal, M. W., Spivey, R., Wu, T., Earle, E. D. & Tanksley, S. D. ( 1993) Science 262, 1432–1436.
6. Jia, Y. & Martin, G. B. ( 1999) Plant Mol. Biol. 40, 455–65.
7. Chandra, S., Martin, G. B. & Low, P. S. ( 1996) Plant Biol. 93, 13393–13397.
8. Goodman, R. N. & Novacky, A. J. ( 1994) in The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon (Am. Phytopathol. Soc., St. Paul, MN), pp. 117–173.
9. Scofield, S. R., Tobias, C. M., Rathjen, J. P., Chang, J. H., Lavelle, D. T., Michelmore, R. W. & Staskawicz, B. J. ( 1996) Science 274, 2063–2065.
10. Tang, X., Frederick, R. D., Zhou, J., Halterman, D. A., Jia, Y. & Martin, G. B. ( 1996) Science 274, 2060–2063.
11. Loh, Y.-T. & Martin, G. B. ( 1995) Plant Physiol. 108, 1735–1739.
12. Frederick, R. D., Thilmony, R. L., Sessa, G. & Martin, G. B. ( 1998) Mol. Cell 2, 241–5.
13. Salmeron, J. M., Oldroyd, G. E. D., Rommens, C. M. T., Scofield, S. R., Kim, H.-S., Lavelle, D. T., Dahlbeck, D. & Staskawicz, B. J. ( 1996) Cell 86, 123–133.
14. Staskawicz, B. J., Ausubel, F. M., Baker, B. J., Ellis, J. G. & Jones, J. D. G. ( 1995) Science 268, 661–667.
15. Oldroyd, G. E. D. & Staskawicz, B. J. ( 1998) Proc. Natl Acad. Sci. USA 95, 10300–10305.
16. Zhou, J., Loh, Y.-T., Bressan, R. A. & Martin, G. B. ( 1995) Cell 83, 925–935.
17. Zhou, J., Tang, X. & Martin, G. B. ( 1997) EMBO J. 16, 3207–3218.
18. Gu, Y.-Q., Yang, C., Thara, V. K., Zhou, J. & Martin, G. B. ( 2000) Plant Cell 12, 771–785.
19. van der Biezen, E. A. & Jones, J. D. G. ( 1998) Trends Biochem. Sci. 23, 454–6.
20. Gyuris, J., Golemis, E., Chertkov, H. & Brent, R. ( 1993) Cell 75, 791–803.
21. Golemis, E., Gyuris, J. & Brent, R. ( 1996) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Green & Wiley, New York), pp. 13.14.1– 13.14.17.
22. Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. ( 1990) J. Mol. Biol. 215, 403–410.
23. Drory, A. & Woodson, W. R. ( 1992) Plant Physiol. 100, 1605–1606.
24. Chen, Z., Silva, H. & Klessig, D. F. ( 1993) Science 262, 1883–1885.
25. Sessa, G., D'Ascenzo, M. & Martin, G. B. ( 2000) Eur. J. Biochem. 267, 1–9.
26. Mufson, R. A. ( 1997) FASEB J. 11, 37–44.
27. Zhou, J., Tang, X., Frederick, R. & Martin, G. ( 1998) J. Plant Sci. Res. 111, 353–356.
28. Umeda, M., Manabe, Y. & Uchimiya, H. ( 1997) FEBS Lett. 403, 313–317.
29. Castano, J. G., Mahillo, E., Arizti, P. & Arribas, J. ( 1996) Biochemistry 35, 3782–3789.
30. Kozak, M. ( 1989) J. Cell Biol. 229, 241.
31. Peranen, J., Auvinen, P., Virta, H., Wepf, R. & Simons, K. ( 1996) J. Cell Biol. 135, 153–167.
32. Towler, D. A., Gordon, J. I., Adams, S. P. & Glaser, L. ( 1988) Am. Rev. Biochem. 57, 69–99.
33. Loh, Y. T., Zhou, J. & Martin, G. B. ( 1998) Mol. Plant–Microbe Interact. 11, 572–6.
34. Hanks, S. K., Quinn, A. M. & Hunter, T. ( 1988) Science 241, 42–52.





