Spatial and temporal control of RNA stability
Arash Bashirullah*†, Ramona L.Cooperstock*‡, and Howard D.Lipshitz*‡§
*Program in Developmental Biology, Research Institute, The Hospital for Sick Children, and ‡Department of Molecular and Medical Genetics, University of Toronto, 555 University Avenue, Toronto, ON M5G 1×8, Canada
Maternally encoded RNAs and proteins program the early development of all animals. A subset of the maternal transcripts is eliminated from the embryo before the midblastula transition. In certain cases, transcripts are protected from degradation in a subregion of the embryonic cytoplasm, thus resulting in transcript localization. Maternal factors are sufficient for both the degradation and protection components of transcript localization. Cisacting elements in the RNAs convert transcripts progressively (i) from inherently stable to unstable and (ii) from uniformly degraded to locally protected. Similar mechanisms are likely to act later in development to restrict certain classes of transcripts to particular cell types within somatic cell lineages. Functions of transcript degradation and protection are discussed.
The early development of all animals is programmed by maternally synthesized RNAs and proteins that are loaded into the developing oocyte by the mother (reviewed in ref. 1). The volumes of mature oocytes from mammals, amphibians, insects, and sea urchins range over several orders of magnitude, as do their total mass of RNA. However, studies several decades ago showed that the complexity of maternal RNA populations in the oocytes of different species—a measure of the number of different classes of transcripts present—varies at most a few fold. For example, the RNA complexity in the mature Drosophila oocyte is 1.2×107 nt (2), whereas that in sea urchin or Xenopus oocytes is ˜4×107 nt (3, 4). In Drosophila, the measured RNA complexity represents ˜5,000 different classes of transcripts of average length 2.5 kb. With the completion of the Drosophila genome sequence (5–8), we now know that this complexity represents the products of more than a third of all of the genes in the fly.
At the midblastula transition (MBT), control of development passes from maternally encoded molecules to proteins synthesized from zygotically transcribed mRNAs. The maternal transcripts that are present in the early embryo can be subdivided into two classes according to whether they are destroyed before the MBT or are stable through this transition (reviewed in ref. 1). It is widely presumed—although there is little evidence that addresses this presumption—that degradation of certain maternal mRNAs is necessary so that the zygotically synthesized transcripts and proteins can take control of development at the MBT. To date, quantitative analyses of individual transcripts have been very limited. In Drosophila, for example, the ribosomal protein-encoding mRNA, rpA1, is stable through the MBT whereas nanos, string, Pgc, and Hsp83 transcripts are unstable (9).
Transcript stability is regulated in space as well as in time. In the early embryo of Drosophila, spatial control of transcript stability functions as a novel RNA localization mechanism (9, 10). For example, string transcripts are degraded throughout the embryo before the MBT. In contrast nanos, Hsp83, and Pgc transcripts are degraded everywhere except in the posterior polar plasm and the pole cells (9, 11, 12).
Here the focus is on the spatial and temporal control of RNA stability in Drosophila. The default state of maternal transcripts in the early embryo is stability; specific cis-acting sequences tag certain classes of transcripts for degradation. Further, if a transcript is targeted for degradation, then the default is generalized degradation throughout the embryo. Localization of a subset of the unstable classes of transcripts is achieved through cis-acting elements that allow protection of these RNAs from degradation in particular cytoplasmic domains. The genetic requirements for transcript degradation and protection are discussed. Preliminary evidence is presented that transcript localization to mother versus daughter cells in the neuronal cell lineage also is accomplished through a degradation-protection mechanism. The mechanisms that regulate transcript stability are likely to be evolutionarily conserved in all metazoa. Possible functions of temporal and spatial control of transcript stability are considered.
Temporal Control of Maternal RNA Stability in Drosophila
Two transcript degradation pathways function together to eliminate maternal transcripts from the early Drosophila embryo (9). One of these pathways begins to function at or shortly after egg activation, independent of fertilization (Fig. 1) (9). This “maternal” pathway is active in unfertilized eggs and thus must be exclusively maternally encoded because there is no “zygotic” transcription in this situation. For example, degradation of maternal Hsp83, string, nanos, and Pgc transcripts occurs in activated unfertilized eggs (in contrast to rpA1 transcripts, which are stable; Fig. 1) (9). A second transcript degradation pathway, which has been termed the zygotic pathway, requires fertilization and becomes active 2 h after this event (9). At the present time there is no definitive evidence that addresses whether the zygotic pathway in fact requires zygotic transcription either to produce the degradation machinery or to activate it. Reinterpretation of drug inhibition experiments carried out before discovery of the two pathways suggests that zygotic transcription is indeed required; for example, inhibition of zygotic transcription using a-amanitin before 1 h after fertilization partially stabilizes transcripts such as string (9, 13). The time course of degradation is thus biphasic: the degradation rate is significantly slower before 2 h after fertilization (when only the maternal pathway is active; during this period the half-life of transcripts such as Hsp83 is ˜75 min) than after 2 h (when both the maternal and zygotic pathways function; the half-life of Hsp83 transcripts is reduced to ˜25 min) (9).
Stability is the default state of transcripts in the early embryo of Drosophila. Unstable transcripts contain cis-acting sequences that target them for degradation. For example, it has been possible to define small elements (˜100 nt) in the 3' untrans
|
† |
Present address: Department of Human Genetics, University of Utah, 15 North 2030 East, Salt Lake City, UT 84112–5331. |
|
§ |
To whom reprint requests should be addressed. E-mail: lipshitz@sickkids.on.ca. |
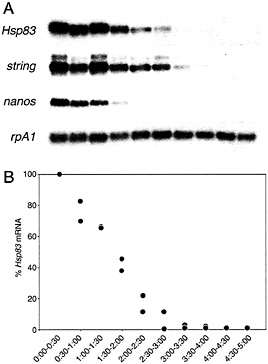
Fig. 1. Time course of maternal transcript degradation in activated, unfertilized eggs. (A) The same Northern blot probed for rpA1 (stable) and string, Hsp83, or nanos (unstable) transcripts. (B) Quantitative analysis of the time course of Hsp83 transcript degradation. The points represent the ratio of Hsp83 transcripts to stable rpA1 transcripts relative to the initial (0-0.5 h) concentration. It can be seen that more than 95% of the Hsp83 transcripts have disappeared by 3.0 to 3.5 h after egg activation. Data from two independent experiments are presented. Half-hour time windows are shown. See ref. 9 for details.
lated region (UTR) of Hsp83 and nanos that, when deleted, result in substantial stabilization of corresponding transgenic transcripts in both unfertilized and fertilized eggs (Fig. 2) (9, 14-16). It was comparison of the stability of the cis element-deleted transcripts in unfertilized versus fertilized eggs that led to the discovery of the zygotic degradation pathway (9): transcripts deleted for an element required for maternal degradation are fully stabilized in unfertilized eggs (Fig. 2 A) but are destabilized starting 2 h after fertilization in developing embryos (Fig. 2 B). Thus, there must be additional cis-acting elements that are still present in these transcripts and that mediate zygotic degradation.
Spatial Control of Maternal RNA Stability in Drosophila Although unstable maternal transcripts such as string and Hsp70 are eliminated throughout the egg or early embryo (Fig. 3 A and B), unstable transcripts such as Pgc, Hsp83, and nanos are eliminated from the bulk cytoplasm of the egg or embryo but remain stable at the posterior (Fig. 3 C-F) (9). Uniform instability is the default state for the unstable classes of transcripts: if the Hsp83 3′ UTR is replaced with a 3′ UTR from a uniformly degraded transcript (e.g., Hsp70) or if a cis-acting "protection" element is deleted (see below), then the resulting transgenic Hsp83 transcripts are degraded throughout the embryo (see Fig. 5
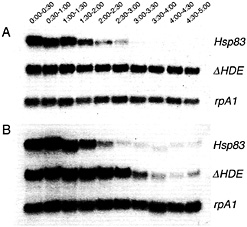
Fig. 2. Removal of a maternal Hsp83 degradation element stabilizes transgenic transcripts in unfertilized (A) but not fertilized (B) eggs. Northern blots are shown that were simultaneously probed for (i) endogenous Hsp83 transcripts; (ii) transgenic reporter transcripts carrying the Hsp83 5′ UTR, the first 111 codons of the Hsp83 ORF, an Escherichia coli ß-galactosidase RNA tag, and the Hsp83 3' UTR deleted for a 97-nt element referred to as the Hsp83 degradation element (HDE) (for a detailed description of this transgene see ref. 9); (iii) endogenous rpA1 transcripts. On both blots it can be seen that endogenous Hsp83 transcripts are unstable and endogenous rpA1 transcripts are stable. However, although transgenic ∆HDE transcripts are stable in unfertilized eggs (1A), they are degraded commencing 2 h after fertilization in developing embryos (B). Half-hour time windows after egg activation or fertilization are shown. See ref. 9 for details.
A and B) (9). This experiment proves that the endogenous Hsp83, nanos, and Pgc transcripts that remain at the posterior of the egg and early embryo are protected from degradation in that region of the cytoplasm (i.e., the degradation machinery is
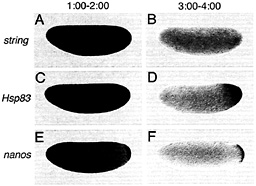
Fig. 3. Certain classes of maternal transcripts are degraded throughout the cytoplasm of activated, unfertilized eggs whereas others are protected from degradation in the posterior polar plasm, string transcripts are initially present throughout the egg (A) and are subsequently degraded (B). In contrast, whereas Hsp83 (C) and nanos (E) transcripts are initially present in both the posterior polar plasm and the presumptive somatic region (C and E), degradation is limited to the somatic region whereas transcripts are protected from degradation in the posterior polar plasm (D and F). (A, C, and E) One to 2 h after egg activation; (B, D, and F) 3-4 h after egg activation. Whole-mount RNA in situ hybridizations are shown, with anterior to the left and dorsal toward the top of the page. See ref. 9 for details.
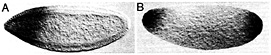
Fig. 4. The posterior polar plasm is necessary and sufficient for transcript protection. (A) Posterior protection of maternal Hsp83 transcripts fails in an embryo from a cappuccino mutant female because posterior polar plasm is not assembled. The anterior expression of Hsp83 is zygotic and serves as an internal control for the in situ hybridization, (B) Protection of Hsp83 transcripts occurs at both poles of an unfertilized egg derived from a female carrying an osk-bcd 3' UTR transgene. Posterior polar plasm is ectopically assembled at the anterior pole of such embryos and is sufficient for transcript protection. (A) Stage 5 embryo, about 2.5 h after fertilization; (B) unfertilized egg ˜3–4 h after egg activation. Whole-mount RNA in situ hybridizations are shown, with anterior to the left and dorsal toward the top of the page. See ref. 18 for details.
present in the posterior but the transcripts are masked from the machinery).
Cis-acting protection elements can be mapped within the 3' UTRs of these localized transcripts; deletion of such an element converts an RNA from one localized by degradation/protection into one that is eliminated from the entire embryo (9). These mapping experiments also demonstrate that degradation elements and protection elements map to distinct regions within the 3' UTR.
The fact that transcript protection occurs in activated, unfertilized eggs (9), together with the absence of zygotic transcription in the pole plasm and pole cells of the early embryo (17), indicates that transcript protection is carried out exclusively by maternally encoded molecules.
Genetic Control of Maternal RNA Stability in Drosophila
Given the fact that maternally encoded molecules are sufficient for both degradation and protection of transcripts in the early embryo, genetic screens initially focused on identification of maternal effect degradation or protection mutants (9). To date, several degradation mutants have been identified, each of which is defective in aspects of egg activation per se. These mutants confirm the importance of egg activation as a prerequisite for transcript degradation (i.e., if the egg is not activated normally, transcript degradation is not triggered). However, they also emphasize the need for larger scale maternal effect screens as well as screens that do not focus exclusively on maternal effect mutants, if mutations in the degradation machinery itself are to be obtained.
Transcript protection at the posterior requires assembly of the posterior polar plasm and its constituent polar granules, which are a crucial component of this specialized cytoplasmic domain (9, 18). Any mutant that eliminates the polar granules results in failure of transcript protection at the posterior (Fig. 4A) (18). Similarly, ectopic assembly of polar plasm and polar granules at the anterior of the egg or early embryo results in ectopic protection of transcripts at the anterior (Fig. 4B) (18). Whether RNA-binding proteins that are known to reside in, and be required for assembly of, the posterior polar plasm (e.g., Staufen, Vasa) interact directly with protection elements in the localized RNAs is not yet known. It is, however, clear that certain RNA-binding proteins that reside in the polar plasm but are not required for polar granule assembly (e.g., Nanos) also are not required for protection, because protection occurs normally in mutants that eliminate these proteins (18).
Control of RNA Stability in a Somatic Cell Lineage
Evidence is beginning to accumulate that transcript localization mechanisms are conserved among different cell types. For example, it has been shown that, in Drosophila, the Staufen
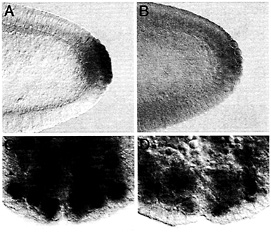
Fig. 5. Maternally synthesized Hsp83 transcripts are protected from degradation in the pole cells (A and B) whereas zygotically synthesized Hsp83 transcripts are protected from degradation in neuroblasts (NB) but not their daughter cells, the ganglion mother cells (GMC) (C and D). (A and C) Endogenous Hsp83 transcripts accumulate in the pole cells but are degraded in the somatic cells of a stage 5 embryo (A, maternal transcripts) and in the NBs but not their daughter cells, the GMCs, of a stage 10 embryo (C, zygotic transcripts; the arrowhead points to a GMC). (D) Transgenic transcripts carrying a degradation element but not a protection element are degraded in both the somatic cells and the pole cells of a stage 5 embryo (B, maternal transcripts). Such transcripts are expressed in the NBs of a stage 10 embryo but are degraded in both the NBs and GMCs. The transgenic transcripts comprise the Hsp83 5' UTR, the first 111 codons of the Hsp83 ORF, an E.coli ß-galactosidase RNA tag, and the Hsp70 3' UTR (for a detailed description of this transgene see ref. 9). The Hsp70 3' UTR lacks a protection element but carries a degradation element. Whole-mount RNA in situ hybridizations are shown.
RNA-binding protein functions in transcript localization in the egg as well as in neuroblasts (reviewed in ref. 10). Of interest here is the possibility that the degradation-protection mechanisms that operate to localize certain maternally encoded transcripts to the germ plasm and germ cells of the early embryo also act at other stages and in other cell types. It was therefore particularly tantalizing to find that zygotically synthesized Hsp83 transcripts accumulate at high levels in the embryonic neuroblasts, stem cells that divide asymmetrically to give rise to the central nervous system (Fig. 5). Strikingly, Hsp83 transcripts are absent from the neuroblasts’ small daughter cells, the ganglion mother cells (Fig. 5 C). This finding suggested that zygotic Hsp83 transcripts might be protected from degradation in the neuroblasts but not in the ganglion mother cells.
To address this possibility, zygotically synthesized transgenic transcripts with a 3' UTR carrying a degradation element but not a protection element were examined in the neuroblast lineage. Transcripts lacking the protection element are expressed in neuroblasts but rapidly disappear, suggesting that removal of this element results in transcript degradation in both neuroblasts and ganglion mother cells rather than only in the ganglion mother cells (Fig. 5 D). This finding suggests that the same degradation-protection machinery acts on maternal transcripts in the early embryo as well as on zygotically synthesized transcripts later in development. In each case, the result is to restrict transcripts to one cell type: in the early embryo, to primordial germ cells but not somatic cells; in the neuronal lineage, to stem cells (neuroblasts) but not their daughter cells (ganglion mother cells).
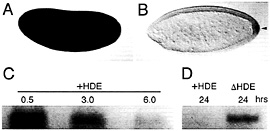
Fig. 6. Transcript degradation and protection are evolutionary conserved processes. Localization of maternal Hsp83 transcripts to the pole cells of D. virilis occurs by degradation and protection as in D. melanogaster. (A) Maternal Hsp83 transcripts are initially uniformly distributed throughout a syncytial stage D. virilis embryo. (B) Subsequently, these transcripts are degraded in the somatic region but are protected from degradation in the pole cells that bud from the posterior (arrowhead). Note that Bicoid-dependent zygotic expression of Hsp83 that occurs in the anterior of D. melanogaster embryos (see Fig. 4 A and ref. 18) does not occur in D. virilis. (C) In vitro-transcribed D. melanogaster Hsp83 3′ UTR transcripts that carry the HDE(+HDE) are highly unstable when injected into X. laevis stage 6 oocytes (the time points are hours after injection). (D) In contrast, Hsp83 3′ UTR transcripts lacking the HDE (∆HDE) are stable for at least 24 h after injection. (A and B) Whole-mount RNA in situ hybridizations are shown, with anterior to the left and dorsal toward the top of the page. (C and D) Blots are shown of digoxigenin-labeled transcripts recovered the specified number of hr after injection into Xenopus oocytes. See ref. 9 for details.
Evolutionary Conservation of Degradation-Protection Mechanisms
To determine whether transcript localization by degradationprotection is conserved between distant Drosophila species, Hsp83 transcripts were examined in early embryos of Drosophila virilis, which has diverged ≈60 million years from D. melanogaster (19). Both components of the localization mechanism—degradation and protection—are conserved (Fig. 6 A and B).
When in vitro-synthesized transcripts comprising the D. melanogaster Hsp83 3′ UTR (i.e., carrying the degradation element) are injected into Xenopus laevis stage 6 oocytes or early embryos, the transcripts are unstable (Fig. 6 C and D) (9). Strikingly, deletion of the degradation element increases the half-life of injected transcripts approximately an order of magnitude (9). Thus, Drosophila cis-acting sequences can be recognized by the Xenopus trans-acting machinery, which suggests that the fundamental maternal transcript degradation machinery is conserved throughout the metazoa.
Functions of Transcript Degradation and Protection
As discussed above, maternal transcript degradation in the early embryo has been presumed to be necessary for the passage of developmental control to the zygotic genome. However, there have been few experiments that address whether this is in fact the case. Perhaps the best data derives from dosage analyses in which the concentration of maternal string transcripts was either halved or doubled (13) (string encodes the Drosophila Cdc25 cell cycle regulator). The transition from uniform maternally regulated to spatially patterned zygotically regulated cell divisions was delayed (double dose) or induced prematurely (half dose). Now that the pathways of transcript instability in the early Drosophila embryo have been defined, it should be possible to delete instability elements from string transcripts to ask whether stabilization of maternal string mRNA results in delay of the cell cycle transition at the MBT.
In contrast to the poorly defined role of maternal RNA degradation in the early embryo, the functions of posterior protection are better understood. For example, it is known that posterior protection of nanos and Pgc transcripts is crucial for the biological roles of these RNAs in germ-cell differentiation; elimination of posterior localization of these transcripts results in abnormal differentiation of the germ cells (12, 20, 21). The role of posterior protection of Hsp83 transcripts has not yet been defined.
Conclusions
The analyses summarized above have shown that transcript stability is exquisitely regulated both in time and in space in the early Drosophila embryo as well as at other developmental stages and in other cell types. Transcript localization by degradation-protection represents a distinct mechanism from that involving directed cytoplasmic transport via cytoskeletal motors, although these two mechanisms are not mutually exclusive (reviewed in refs. 10 and 22). Further analyses of degradation and protection using the combination of genetic and biochemical methods available in Drosophila are expected to lead to general insights into the mechanisms and functions of these processes during animal development.
R.L.C. has been supported in part by a Medical Research Council of Canada Graduate Scholarship and a University of Toronto Open Scholarship. Our research on RNA localization mechanisms is funded by an operating grant to H.D.L. from the Canadian Institutes of Health Research (formerly the Medical Research Council of Canada).
1. Davidson, E.H. (1986) Gene Activity in Early Development (Academic, Orlando, FL).
2. Hough-Evans, B.R., Jacobs-Lorena, M., Cummings, M.R., Britten, R.J. & Davidson, E.H. (1980) Genetics 95, 81–94.
3. Davidson, E.H. & Hough, B.R. (1971) J. Mol. Biol. 56, 491–506.
4. Hough-Evans, B.R., Wold, B.J., Ernst, S.G., Britten, R.J. & Davidson, E.H. (1977) Dev. Biol. 60, 258–277.
5. Rubin, G.M., Hong, L., Brokstein, P., Evans-Holm, M., Frise, E., Stapleton, M. & Harvey, D.A. (2000) Science 287, 2222–2224.
6. Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. (2000) Science 287, 2185–2195.
7. Myers, E.W., Sutton, G.G., Delcher, A.L., Dew, I.M., Fasulo, D.P., Flanigan, M.J., Kravitz, S.A., Mobarry, C.M., Reinert, K.H., Remington, K.A., et al. (2000) Science 287, 2196–2204.
8. Rubin, G.M., Yandell, M.D., Wortman, J.R., Gabor Miklos, G.L., Nelson, C.R. , Hariharan, I.K., Fortini, M.E., Li, P.W., Apweiler, R., Fleischmann, W., et al. (2000) Science 287, 2204–2215.
9. Bashirullah, A., Halsell, S.R., Cooperstock, R.L., Kloc, M., Karaiskakis, A., Fisher, W.W., Fu, W., Hamilton, J.K., Etkin, L.D. & Lipshitz, H.D. (1999) EMBO J. 18, 2610–2620.
10. Lipshitz, H.D. & Smibert, C.A. (2000) Curr. Opin. Genet. Dev. 10, 476–488.
11. Bergsten, S.E. & Gavis, E.R. (1999) Development (Cambridge, U.K.) 126, 659–669.
12. Nakamura, A., Amikura, R., Mukai, M., Kobayashi, S. & Lasko, P.F. (1996) Science 274, 2075–2079.
13. Edgar, B.A. & Datar, S.A. (1996) Genes Dev. 10, 1966–1977.
14. Gavis, E.R., Lunsford, L., Bergsten, S.E. & Lehmann, R. (1996) Development (Cambridge, U.K.) 122, 2791–2800.
15. Dahanukar, A. & Wharton, R.P. (1996) Genes Dev. 10, 2610–2620.
16. Smibert, C.A., Wilson, J.E., Kerr, K. & Macdonald, P.M. (1996) Genes Dev. 10, 2600–2609.
17. Van Doren, M., Williamson, A.L. & Lehmann, R. (1998) Curr. Biol. 8, 243–246.
18. Ding, D, Parkhurst, S.M., Halsell, S.R. & Lipshitz, H.D. (1993) Mol. Cell. Biol. 13, 3773–3781.
19. Beverley, S.M. & Wilson, A.C. (1984) J. Mol. Evol 21, 1–13.
20. Forbes, A. & Lehmann, R. (1998) Development (Cambridge, U.K.) 125, 679–690.
21. Kobayashi, S., Yamada, M., Asaoka, M. & Kitamura, T. (1996) Nature (London) 380, 708–711.
22. Bashirullah, A., Cooperstock, R.L. & Lipshitz, H.D. (1998) Annu. Rev. Biochem. 67, 335–394.




