The physiological significance of ß-actin mRNA localization in determining cell polarity and directional motility
Elena A.Shestakova, Robert H.Singer*, and John Condeelis
Department of Anatomy and Structural Biology, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461
ß-actin mRNA is localized near the leading edge in several cell types, where actin polymerization is actively promoting forward protrusion. The localization of the ß-actin mRNA near the leading edge is facilitated by a short sequence in the 3' untranslated region, the “zip code.” Localization of the mRNA at this region is important physiologically. Treatment of chicken embryo fibroblasts with antisense oligonucleotides complementary to the localization sequence (zip code) in the 3' untranslated region leads to delocalization of ß-actin mRNA, alteration of cell phenotype, and a decrease in cell motility. To determine the components of this process responsible for the change in cell behavior after ß-actin mRNA delocalization, the Dynamic Image Analysis System was used to quantify movement of cells in the presence of sense and antisense oligonucleotides to the zip code. It was found that net path length and average speed of antisense-treated cells were significantly lower than in sense-treated cells. Total path length and the velocity of protrusion of antisense-treated cells were not affected compared with those of control cells. These results suggest that a decrease in persistence of direction of movement and not in velocity results from treatment of cells with zip code-directed antisense oligonucleotides. To test this, direct analysis of directionality was performed on antisense-treated cells and showed a decrease in directionality (net path/total path) and persistence of movement. Less directional movement of antisense-treated cells correlated with a unpolarized and discontinuous distribution of free barbed ends of actin filaments and of ß-actin protein. These results indicate that delocalization of ß-actin mRNA results in delocalization of nucleation sites and ß-actin protein from the leading edge followed by loss of cell polarity and directional movement.
Dynamic Image Analysis System | directionality | antisense oligonucleotides
Beta-actin mRNA is localized to the leading lamella in chicken embryo fibroblasts (CEFs) and several other cell types, just proximal to the lamellipodia (1–4). Localization of ß-actin mRNA depends on an intact actin cytoskeleton in CEFs (5). The nucleotide sequence that determines the localization of ß-actin mRNA was found in the 3' untranslated region (UTR) and is composed of 54 nt 3' of the stop codon (the “zip code,” ref. 6). A protein of 68 kDa (zip code binding protein 1, ZBP1) binds the zip code in ß-actin mRNA (7). Binding of ZBP1 to the zip code correlated with localization of ß-actin mRNA; a mutated zip code unable to localize was unable to bind ZBP1. Delocalization of ß-actin mRNA with antisense oligonucleotides complementary to the zip code (zip code antisense) suppresses cell polarity (6) and motility (2). Likewise, inhibition of protein synthesis also slowed cell motility (2). These results suggested that there was some aspect of cell motility that was enhanced by the synthesis of ß-actin near the leading edge. In this work, we elucidate the significance of this localization of ß-actin mRNA and show that it plays a role in determining the polarity of nucleation sites for actin polymerization.
There could be several reasons for suppression of cell motility upon delocalization of ß-actin mRNA. Cell motility requires actin polymerization in the leading edge (8–10). Cells with delocalized ß-actin mRNA may not polymerize actin filaments at the same rate if ß-actin is not synthesized at sites of polymerization. As a result the cells would have a lower velocity of protrusion, which is driven by actin polymerization. Alternatively, the rate of actin polymerization may be unaffected by actin synthesis. Instead, the site of actin synthesis may affect the location of nucleation of actin polymerization that would define the direction of protrusion and, therefore, polarity of movement.
To test which of these hypotheses was likely to be correct, several parameters of movement were measured in cells treated with zip code antisense oligonucleotides to delocalize ß-actin mRNA. The measurements were correlated with the sites of actin polymerization to determine how delocalization of ß-actin mRNA affected the location of protrusive activity. Our results indicate that the delocalization of the mRNA does not substantially change the rate of protrusion, but rather it significantly alters the sites where this protrusion occurs.
Materials and Methods
Cell Culture. Primary CEFs were prepared as described (11), cultured 72–96 h in alpha-modified MEM (GIBCO) containing 10% FBS and antibiotics (penicillin, streptomycin). For further experiments, cells were replated on 0.5% gelatin-coated 12-mm gridded coverslips (Eppendorf). Cells were used for motility analysis after plating on coverslips and oligonucleotide treatment for 12 h. For in situ hybridization, cells were fixed at 37°C in 4% paraformaldehyde in PBS (1 mM KH2PO4, 10 mM N2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.0), washed in PBS, and dehydrated in 70% ethanol at 4°C overnight.
Oligodeoxynucleotide (ODN) Treatment of Cells. Phosphorothioate-modified ODNs comprising the antisense and sense sequence to 18 nt of the zip code (6) were synthesized on an Applied Biosystems 394 DNA/RNA Synthesizer and purified by electophoresis through polyacrylamide gel, eluted, lyophilized, resuspended in water, and additionally purified by gel filtration on Sephadex G50. Purified ODNs were lyophilized and resus
|
* |
To whom reprint requests should be addressed. E-mail: rhsinger@aecom.yu.edu. |
pended in diethyl-pyrocarbonate-treated water. ODNs (8 µM) were added to a cell medium three times at 4-h intervals (6).
Rhodamine-Actin-Based Detection of Barbed Ends of Actin Filaments. Stock rhodamine-labeled actin was thawed and diluted with 1 mM Hepes (pH 7.5), 0.2 mM MgCl2, and 0.2 mM ATP, sonicated, and clarified in Beckman centifuge at 95 krpm, for 20 min. Cells were permeabilized with 20 mM Hepes (pH 7.5), 138 mM KCl, 4 mM MgCl2, 9 mM EGTA, 0.25 mg/ml saponin, 1 mM ATP, 1% BSA containing 0.45 µm rhodamine-actin that was added to the lysis buffer just before application to cells. One to three minutes after incubation, cells were fixed for 5 min with 3.7% formaldehyde in PBS, incubated with 0.1 M glycine in PBS for 10 min, and washed with PBS. Cells were stained with 1 µM fluorescein phalloidin in buffer for 40 min in humidified chamber, washed, and mounted on 0.1 M N-propylgallate in 50% glycerol in PBS, pH 7.0.
Immunofluorescence. Cells were plated on coverslips, fixed in 3.7% formaldehyde, permeabilized with 0.5% Triton in PBS, and incubated with primary antibodies to ß-actin (a gift of Ira Herman, Tufts Medical School, Boston) and secondary fluorescein-labeled antibodies to rabbit IgG for 1 h and mounted as described (12).
In Situ Hybridization. Chicken ß-actin-specific 3' UTR probes (five probes of 50 nt each, with five amino linkers per probe spaced ˜10 nt apart) were synthesized on an Applied Biosystems 394 DNA/RNA Synthesizer. Chicken ß-actin probes were labeled with CY3. To detect ß-actin mRNAs, coverslips were rehydrated in PBS, permealized with 0.5% Triton in PBS for 10 min, and then hybridized for 3 h at 37°C with 5 ng of the mixture of five oligonucleotides. Each oligonucleotide can hybridize independently with ß-actin mRNA so as to increase the signal to noise when all five have hybridized to a single molecule (25 fluorochromes total; ref. 13). Coverslips were washed twice with 50% formamide in 2×SSC (300 mM NaCl, 30 mM sodium citrate, pH 7.0), then in 2×SSC, 1×SSC, and mounted.
High-Resolution Microscopy. An Olympus BX60 microscope was used with a×60 planapo objective numerical aperture 1.4. Digital images were captured by using a Photometries camera and CELLSCAN software.
Computer-Assisted Analysis of Cell Behavior. Cells were recorded with an Olympus microscope equipped with a charge-coupled device camera through a×10 objective with a 1-min time interval between image frames over 60 min. Images were processed with DIAS (Dynamic Image Analysis System) software (14). Cell motility data were displayed as an overlay of cell perimeters, i.e., as a stack of every fifth video frame (cell perimeter plot) and as a centroid plot showing the location of the geometrical center of the cell as a function of time.
Results
Antisense Treatment of Cells. It was shown previously that cisacting elements in the 3' UTR of chicken ß-actin mRNA were responsible for the localization of this mRNA. The 54 nt 3' of the stop codon were most potent in localizing ß-actin mRNA. This region is called the zip code and can be divided into A, B, and C regions (6). In this study, an antisense ODN, complementary to the 3' 18 nt of the zip code was used (C–). For a control, the sense strand (C+) was used.
Effects of Antisense ODNs on Cell Motility. It was reported previously that the velocity of cell locomotion is reduced by treatment of cells with antisense-oligos directed against the zip code of ß-actin mRNA (2). To confirm and extend this observation we
Table 1. Percent of cells with ß-actin mRNA, ß-actin protein, and nucleation (barbed ends) localized to the leading edge as a function of zip code antisense (C–) or sense (C+) oligonucleotide treatments
|
|
C– % localization |
C+ % localization |
||
|
|
Leading edge |
Diffuse |
Leading edge |
Diffuse |
|
ß-actin mRNA (n) |
33 (186) |
67 (375) |
58 (392) |
42 (288) |
|
Barbed ends (n) |
30 (130) |
70 (303) |
70 (210) |
30 (90) |
|
ß-actin protein (n) |
32 (70) |
68 (149) |
68 (128) |
32 (60) |
repeated these experiments. CEFs were treated with zip code antisense (C–) or sense (C+) ODNs. The distribution of ß-actin mRNA was determined in each population by using fluorescence in situ hybridization. Cells treated with antisense showed decreased localization of ß-actin mRNA to the leading edge, whereas cells treated with sense ODNs (control) localized the mRNA to the leading edge to an extent that was statistically indistinguishable from untreated cells (Table 1, ß-actin mRNA).
The average path length migrated by antisense-treated and control cells was measured as a change in the nuclear position during a 60-min observation period. In Table 2, the average path lengths for 100 antisense-treated and 64 control cells are presented. The antisense-treated cells migrated shorter distances, and this difference was statistically significant. This result is similar to that reported previously by Kislauskis et al. (2). However, the underlying mechanism for this observation has not been investigated. Therefore, we subjected the cells to a more rigorous analysis of their motility to ascertain which of the components of cell motility was most affected. To do this we correlated various aspects of cell motility with ß-actin mRNA localization. Treated and control cells were monitored by using an inverted microscope supplemented with a heating chamber. Time-lapse movies were obtained over 60 min with 1-min intervals between frames. Fig. 1 demonstrates the difference in behavior between these two cell populations. In the presence of the zip code antisense, cells did not translocate appreciably, whereas in the presence of sense ODNs, the cells continued to migrate. The movies obtained in this way were analyzed by using the Dynamic Image Analysis System (Materials and Methods). Several cell motility parameters were determined: net path length, average speed, average instantaneous speed (protrusion velocity), directionality, and persistence (Table 3). Delocalization of ß-actin mRNA in CEFs correlated with a significant decrease in net path length and average speed. Total path length and average protrusive velocity were not statistically different from control cells (Table 3). These results are explained by a decrease in the directionality and persistence of movement
Table 2. Average path length migrated by CEFs (measured as a change in the nuclear position) during 60 min as a function of zip code antisense (C–) or sense (C+) oligonucleotide treatment
|
|
C– (n=100) |
C+ (n=64) |
|
Average net path length, µm |
12.67 |
16.32 |
|
SE |
0.87 |
1.56 |
|
t test |
4.39% |
|
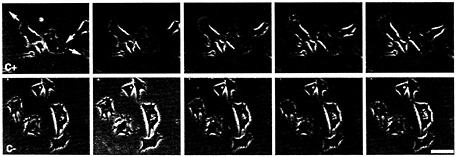
Fig. 1. Movement of CEFs in the presence of (Upper) zip code sense (C–) and (Lower) antisense oligonucleotides(C+). Pictures depict frames 5 min apart from the video analysis of two fields of cells. Arrows indicate direction of movement of each cell over the subsequent frames. Note that cells move in the presence of sense, but not antisense, oligonucleotides.
without a decrease in rate of locomotion. Consistent with this possibility is the comparison of centroid plots of antisense-treated and control cells. An example of one of these analyses is represented in Fig. 2, which shows that cells with delocalized ß-actin mRNA exhibit random directionality of motility and have less persistence in the direction of motility whereas control cells move with fixed polarity and linear directionality and are more persistent in the direction of motility (Table 3). This finding indicates that mRNA localization is not necessary for the ability of the cells to move, but rather for their ability to maintain this movement in one direction.
The Distribution of Free Barbed Ends Is Randomized in Antisense-Treated CEFs. The above results indicate that the decrease in apparent cell velocity observed by Kislauskis et al. (2) was due to a decrease in net path traveled over the time of observation. This resulted from a decrease in persistence rather than a decrease in speed of locomotion. This observation is consistent with a conclusion that the rate of protrusion of the leading edge was unaffected by delocalization of ß-actin mRNA. In contrast, it is the persistence of the direction of protrusion that is affected by delocalization of ß-actin mRNA. We speculated that the mechanism behind the loss of polarity of protrusion in antisense-treated cells involved loss of polarized nucleation of actin polymerization. To test this hypothesis, permeabilized CEFs were incubated with a concentration of rhodamine-actin close to the critical concentration of barbed end addition to label the barbed ends of actin filaments. Labeling of barbed ends demonstrates sites of nucleation of actin polymerization (9, 12). The sites of rhodamine-actin incorporation were not polarized in antisense-treated CEFs but rather were around the entire periphery (Fig. 3) whereas in sense-treated CEFs the sites of rhodamine-actin incorporation were polarized and distributed ontinuously along the leading edge of the lamellipod (Fig. 3; Table 1, barbed ends).
The peripheral, nonpolarized distribution of the nucleation sites would predict that ß-actin would be distributed likewise peripherally, but not in a polarized distribution. To test whether the change in the distribution of barbed ends resulted in changes in ß-actin distribution, we used isoform-specific antibodies to determine the ß-actin protein location in sense- and antisense-treated cells. Fig. 4 demonstrates the common observation between these two populations; treatment with the zip code antisense results in a peripheral, nonlocalized distribution of ß-actin whereas the sense-treated cells show the characteristic concentration of ß-actin at the leading and retracting poles of the fibroblast. Therefore, in these two populations of cells the distribution of ß-actin protein, which normally is localized to the leading edge, was unaffected by sense treatment but became diffusely distributed in antisense-treated cells (Fig. 4; Table 1, ß-actin protein). This finding indicates that the distribution of barbed ends and the synthesis of ß-actin are likely to be related functionally. This functionality would derive directly from the distribution of the site of synthesis of the ß-actin.
Discussion
This study was undertaken to elucidate the observation whereby delocalization of ß-actin mRNA can affect cell motility and polarity in fibroblasts. Delocalization of ß-actin mRNA with antisense oligonucleotides has been observed to reduce the motility of CEFs (2) and smooth muscle cells (15).
Table 3. Delocalization of ß-actin mRNA with zip code antisense oligonucleotides (C–) but not sense (C+) oligonucleotides causes loss of polarized cell movement but no significant change in protrusion rates
|
|
Net path length, µm |
Average speed, µm/min |
Total path length, µm |
Protrusion velocity |
Directionality, net/total |
Persistence, µm/min×deg |
|
C– (n=20) |
12.72 |
0.22 |
84 |
0.79 |
0.18 |
0.12 |
|
SE |
0.33 |
0.0056 |
2.3 |
0.017 |
0.0063 |
0.003 |
|
C+ (n=20) |
20.92 |
0.35 |
85.64 |
0.88 |
0.26 |
0.16 |
|
SE |
0.53 |
0.009 |
1.19 |
0.011 |
0.0064 |
0.002 |
|
t test (%) |
1 |
1 |
89 |
33 |
6 |
4 |
|
|
Explanation of categories (see ref. 14 for details): net path length, the distance traveled between the beginning and ending points of the analysis (60 min); average speed, the average distance traveled divided by the observation time (60 min); total path length, the entire path summed from all movements at each interval (1 min); protrusion velocity, the movement of protrusions during each interval (1 min) and averaged; directionality and persistence are measurements as to how consistently the cells stay on a course (1-min intervals) determined either by length of the net path relative to the total path or number of turns the cell makes in degrees per min, respectively. |
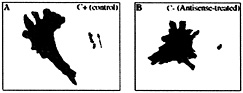
Fig. 2. ß-actin mRNA localization in the presence of zip code sense oligonucleotides (C+) or antisense oligonucleotides (C–). Perimeter and centroid (dots) plots are from ODN-treated cells over the time frames of analysis (1 min). These results are representative of the analysis of populations of cells depicted in Fig. 1. (A) Sense-treated cell. (B) Antisense-treated cell. Note that the antisense oligonucleotides cause loss of polarized cell movement defined as a linear centroid track (arrow in A).
Our results show that antisense but not control (sense) oligos caused a delocalization not only of ß-actin mRNA, but also of ß-actin protein and barbed ends from the leading edge of fibroblasts and resulted in a random distribution of all three. This was reversible upon removal of the antisense oligonucleotides. By investigating sites of actin filament nucleation, we showed that they were delocalized as a result of disrupting the targeting of ß-actin mRNA. This result reveals a possible mechanism for establishing cell polarity: ß-actin protein, and/or proteins with related zip codes, define the location of nucleation of actin
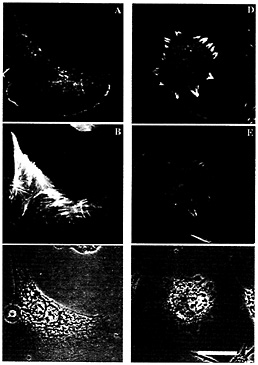
Fig. 3. Sites of rhodamine actin incorporation in zip code sense-treated (C+) and antisense treated (C–) cells. (A–C) Sense treatment. (D–F) Antisense treatment. (A and D) Rhodamine actin incorporation showing the location of free barbed ends on actin filaments. (B and E) FITC-phalloidin-labeling of all actin filaments. (C and F) Phase-contrast image. (Bar, 10µm.) Note that rhodamine actin incorporation sites are unpolarized in antisense-treated cells.
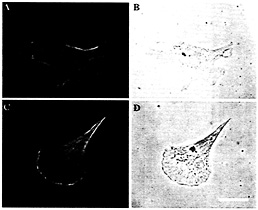
Fig. 4. Localization of ß-actin protein in zip code antisense (A and B) compared with sense-treated cells (C and D). (A and C) Staining with anti-ß-actin antibodies. (B and D) Nomarski optics. (Bar, 10 µm.) Note that the ß-actin staining is not as prominently localized to the leading edge in antisense-treated cells.
polymerization and consequently, cell polarity and directional motility.
The molecular mechanism by which polarity of cell crawling is affected by ß-actin mRNA localization could depend on several interdependent events: (i) Localized synthesis of ß-actin from localized mRNA drives protrusion of the lamellipod. (ii) ß-actin isoform specific protein interactions are responsible for the protrusion. (iii) Localization to the leading edge of the mRNAs for other proteins in addition to ß-actin, e.g., the nucleating complex containing Arp 3 mRNA. A discussion of the evidence for each of these is detailed below.
Localized Synthesis of ß-Actin from Localized mRNA. Based on the estimated 2,500 ß-actin mRNA molecules per cell, at the established translational rate of 1.5 actins per sec per mRNA molecule, the cell would synthesize 3,900 actin molecules per sec or 2.34×105 per min (2). In moving cells, the polymerization zone uses a minimum of 3.6×106 actin molecules per min (9). Therefore, it is unlikely that a 6.5% contribution of newly synthesized ß-actin will significantly contribute to the rate of actin polymerization at the leading edge. However, if all of the ß-actin is synthesized in a restricted volume, a consequence of localizing the ß-actin mRNA to the leading edge, the local rate of synthesis of ß-actin might significantly impact the actin polymerization events in this restricted volume and, therefore, establish a preferred location for actin polymerization.
The above model would be further supported if newly synthesized actin would have a faster rate of polymerization, or a higher affinity for a nucleation complex than “older” actin, which may be posttranslationally modified. For instance, interaction of a chaperone with the ß-actin nascent chain (16) could promote assembly of a nucleation complex near the site of synthesis.
ß-Actin Isoform-Specific Protein Interactions. The ß isoform of actin may be preferentially stored as the monomer used for polymerization at the leading edge. In this hypothesis, local accumulation of a nonfilamentous form of actin that could be released suddenly upon stimulation of motility would determine the location of actin polymerization. Potential storage particles containing nonfilamentous actin have been identified by comparing the localization patterns of vitamin D-binding protein,
which binds to G-actin with 5 nM kd, and phalloidin, which binds to actin (17). These stores of nonfilamentous actin are found at the leading edge and are located adjacent to sites of actin polymerization and in the region of the cell where the ß-actin mRNA is also present. Possibly these sites could result from islands of ß-actin synthesis.
ß-actin is found at the leading edge of crawling cells. ß-actin does not substitute for muscle actin in either the formation of stress fibers (17) or myofibrils in cardiomyocytes (18). In addition, it seems to interact more tightly with certain actin binding proteins that may function at the leading edge of crawling cells. Ezrin (19), profilin (20), thymosin ß 4 (21), and L-plastin (22) bind more strongly to ß-actin than a-actin. A capping protein, ß-cap 73, may cap the barbed end in an isoform-specific manner (23). There is growing evidence that the Arp2/3 complex is required for nucleation of actin filaments at the leading edge (12, 24–27). If the Arp2/3 complex is the dominant nucleation activity at the leading edge, a possible preference for the ß-actin isoform by the Arp2/3 complex would require local synthesis of ß-actin to supply the preferred monomer for polymerization. Therefore, the localization of ß-actin synthesis at the leading edge may be functionally important for polarity and motility.
Localization to the Leading Edge of Motility-Related mRNAs. The localization of ß-actin mRNA may be representative of the localization of a family of mRNAs with related 3' UTR zip codes, many of which function synergistically at the leading edge. Proteins coded for by these mRNAs therefore might have related functions. We have analyzed the 3' UTRs of mRNAs, which code for proteins believed to have actin binding functions at the leading edge, for the presence of the zip code consensus sequence. This sequence GACUX7–38ACACC is found in ß-actin mRNAs known to target to the leading edge from all vertebrates. Besides ß-actin mRNAs, mRNA for Arp3 and myosin IIB heavy chain contain the consensus sequence and are predicted to be recognized by the localization mechanism that targets ß-actin mRNA to the leading edge. It is known that the ACACCC consensus sequence, when mutated in ß-actin mRNA, results in a failure to localize the mRNA to the leading edge of cells (2, 7), even if the ß-actin coding sequence remains intact and is used as the reporter mRNA. Preliminary results indicate that Arp3 mRNA, like ß-actin mRNA, also localizes to the leading edge (G.Liu, W.Grant, D.Persky, V.L.Lathaur, R.H.S., and J.C., unpublished work). Serum-dependent localization of ß-actin mRNA suggests that signaling mechanisms are involved in the localization of motility-related mRNAs, thereby coordinating their temporal and spatial distribution and expression (28). Furthermore, it is possible that localized synthesis of, for instance, Arp3 could determine the localization of Arp2/3 complex in the leading edge of the cells even if mRNAs coding other components of Arp2/3 complex were more diffusely distributed. Arp2/3 complex and ß-actin, both localized in the leading edge, could determine the nucleation sites for actin polymerization. Newly formed actin filaments could interact with ß-actin isoform-specific binding proteins, thereby stabilizing the cell polarity and consequent directional motility (29).
The leading edge of the cell is a complex composite of asymmetrically distributed proteins many of which function in concert to produce the motility response. It is likely that other proteins like ß-actin also are synthesized asymmetrically and therefore would provide not only a differential concentration of these proteins but also an increased likelihood of interactions among relevant proteins in a cellular region where function depends on these interactions. We presume therefore that a panoply of mRNAs comprising a significant complexity of sequences is localized to the lamella to effect the complex events required by motility. It is our expectation that these sequences will contain a common motif and/or structure in the 3' UTR characterizing them as mRNAs for motility-related proteins. It is likely that further investigations will reveal the consensus sequences (see below).
The localization of ß-actin mRNA is not restricted to fibroblasts, but seems to be a feature of other localized cells. Neurons localize ß-actin mRNA to the growth cone of developing neurites (30, 31). The presence of the mRNA results in the specific translation of ß-actin protein in the growth cone. Like fibroblasts, the delocalization of the mRNA results in growth cone retraction and nondirectionality of growth cone guidance (37). In addition to the neuronal growth cones, embryonic neural crest cells might localize ß-actin mRNA to the front of the cell, in the direction of their migration. Disruption of the Xenopus homolog of ZBP1 appears to inhibit their migration and result in severe embryological defects in forebrain development (J.Yisraeli, personal communication). Furthermore, if the zip code for ß-actin mRNA is transferred to another protein, not normally at the leading edge, in this case vimentin, a distorted morphology results wherein the cell structure at the leading edge is branched and attenuated (32). These results argue that synthesis of the correct protein in the correct place (near the leading edge) is an important requirement for cell structure and polarity.
In addition to ß-actin mRNA localization in fibroblasts (1), the field of RNA localization has been advanced by the discovery of a number of systems where mislocalization of the RNA can lead to a significantly altered phenotype or lethality (33–36). In many of these cases mRNA localization is required for normal development and differentiation because the localized mRNA codes for nuclear factors and the resultant cell divisions segregate the mRNAs for these morphogenic determinants. However, the nature of the localization we describe here is important for a different reason: it determines the spatial orientation, morphology, and behavior of these somatic cells. In this second aspect of RNA localization, the complex of proteins involved in cell migration, cellular reaction to the environment and development of cell polarity are organized within the cytoplasm by virtue of the spatial segregation of their cognate mRNAs, and are not in the short term related to transcription of genes. In this way, components of the mechanism controlling cell behavior and structure can rapidly reassemble within the cell. In this model, the proteins involved in forming these multipolypeptide complexes (the nucleation complex, for instance) would be compartmentalized in response to environmental cues and subsequent signal transduction events and then synthesized in proximity to each other where they would interact preferentially because of their higher local concentrations. Possibly these higher concentrations of proteins could autoregulate their own synthesis. In this way, we propose that the localization of ß-actin mRNA represents one mechanism for the spatially compartmentalized assembly of cellular complexes.
We thank Michael Cammer in the Einstein Analytical Imaging Facility and Jeff Wyckoff and Shailesh M.Shenoy for technical help with light microscopy and the Dynamic Image Analysis System, Wayne Grant for technical help with experiments, and Steve Braut for synthesis of oligonucleotides. This work was supported by National Institutes of Health grants to R.H.S. and J.C.
1. Lawrence, J.B. & Singer, R.H. (1986) Cell 45, 407–415.
2. Kislauskis, E.H., Zhu, X. & Singer, R.H. (1997) J. Cell Biol. 136, 1263–1270.
3. Hill, M.A., Schedlich, L. & Gunning, P. (1994) J. Cell Biol. 126, 1221–1230.
4. Hoock, T.C, Newcomb, P.M. & Herman, I.M. (1991) J. Cell Biol. 112, 653–664.
5. Sundell, C. & Singer, R.H. (1991) Science 253, 1275–1277.
6. Kislauskis, E.H., Zhu, X. & Singer, R.H. (1994) J. Cell Biol. 127, 441–451.
7. Ross, A.F., Oleynikov, Y., Kislauskis, E.H., Taneja, K.L. & Singer, R.H. (1997) Mol. Cell. Biol. 17, 2158–2165.
8. Bailly, M., Macaluso, F., Cammer, M., Segall, J. & Condeelis, J. (1998) J. Cell Biol. 145, 331–345.
9. Chan, A.Y., Raft, S., Bailly, M., Wyckoff, J.B., Segall, J.E. & Condeelis, J.S. (1998) J. Cell Sci. 111, 199–211.
10. Segall, J.E., Tyerech, S., Boselli, L., Masseling, S., Helft, J., Chan, A., Jones, J. & Condeelis, J. (1996) Clin. Exp. Metastasis 14, 61–72.
11. Kislauskis, E.H., Li, Z., Singer, R.H. & Taneja, K.L. (1993) J. Cell Biol. 123, 165–172.
12. Bailly, M., Yan, L., Whitesides, G., Condeelis, J.S. & Segall, J.E. (1999) Exp. Cell Res. 241, 285–299.
13. Femino, A.N., Fay, F.S., Fogarty, K. & Singer, R.H. (1998) Science 280, 585–590.
14. Soll, D.R. (1995) Int. Rev. Cytol. 163, 43–104.
15. Schedlich, L., Hill, M. & Lockett, T. (1997) Biol. Cell 89, 113–122.
16. Hansen, W.J., Cowan, N.J. & Welch, W.J. (1999) J. Cell Biol. 145, 265–277.
17. Cao, L., Fishkind, D.J. & Wang, Y. (1993) J. Cell Biol. 123, 173–181.
18. von Arx, P., Bantle, S., Soldati, T. & Perriard, J.C. (1995) J. Cell Biol. 131, 1759–1773.
19. Shuster, C. & Herman, I. (1995) J. Cell Biol. 128, 837–848.
20. Segura, M. & Lindberg, U. (1984) J. Biol. Chem. 259, 3949–3954.
21. Weber, A., Nachmias, V.T., Pennise, C.R., Pring, M. & Safer, D. (1992) Biochemistry 31, 6179–6185.
22. Namba, Y., Ito, M., Zu Y., Shigesada, K. & Maruyama, K. (1992) J. Biochem. 112, 503–507.
23. Shuster, C.B., Lin, A.Y., Nayak, R. & Herman, I.M. (1996) Cell Motil. Cytoskeleton 35, 175–187.
24. Machesky, L.M., Reeves, E., Wientjes, F., Mattheyse, F.J., Grogon, A., Totty, N.F., Burlingame, A.L., Hsuan, J.J. & Segal, A.W. (1997) Biochem. J. 328, 105–112.
25. Mullins, R.D., Heuser, J.A. & Pollard, T.D. (1998) Proc. Natl. Acad. Sci. USA 95, 6181–6186.
26. Welch, M.D., Rosenblatt, J., Skoble, J., Portnoy, D.A. & Mitchison, T.J. (1998) Science 281, 105–108.
27. Blanchoin, L., Pollard, T.D. & Mullins, R.D. (2000) Curr. Biol. 10, 1273–1282.
28. Latham, V.M., Jr., Kislauskis, E.H., Singer, R.H. & Ross, A.F. (1994) J. Cell Biol. 126, 1211–1219.
29. Liu, G., Edmonds, B. & Condeelis, J. (1996) Trends Cell Biol. 6, 168–171.
30. Zhang, H.L., Singer, R.H. & Bassell, G.J. (1999) J. Cell Biol. 147, 59–70.
31. Bassell, G. & Singer, R.H. (2001) in Results and Problems in Cell Differentiation, ed. Richter, D. (Springer, Berlin), Vol. 34, pp. 41–56.
32. Morris, E.J., Evason, K., Wiand, C., L’Ecuyer, T.J. & Fulton, A.B. (2000) J. Cell Biol. 113, 2433–2443.
33. St. Johnston, D. (1995) Cell 81, 161–170.
34. Long, R.M., Singer, R.H., Meng, X., Gonzalez, I., Nasmyth, K. & Jansen, R.-P. (1997) Science 277, 383–387.
35. Hazelrigg, T. (1998) Cell 95, 451–460.
36. Carson, J.H., Cui, H., Krueger, W., Schlepchenko, B., Brunwell, C. & Barbarese, E. (2001) in Results and Problems in Cell Differentiation, ed. Richter, D. (Springer, Berlin), Vol. 34, pp. 69–81.
37. Zhang, H.L., Eom, T., Oleynikov, Y., Shenoy, S.M., Liebelt, D.A., Singer, R. H. & Bassell, G.J. (2001) Neuron, in press.






