Vasopressin mRNA localization in nerve cells: Characterization of cis-acting elements and trans-acting factors
Evita Mohr*†, Nilima Prakash‡, Kerstin Vieluf*, Carola Fuhrmann*, Friedrich Buck*, and Dietmar Richter*
*Universität Hamburg, Institut für Zellbiochemie und klinische Neurobiologie, Martinistrasse 52, 20246 Hamburg, Germany; and ‡Department of Molecular Genetics, The Weizmann Institute of Science, 76100 Rehovot, Israel
mRNA localization is a complex pathway. Besides mRNA sorting per se, this process includes aspects of regulated translation. It requires protein factors that interact with defined sequences (or sequence motifs) of the transcript, and the protein/RNA complexes are finally guided along the cytoskeleton to their ultimate destinations. The mRNA encoding the vasopressin (VP) precursor protein is localized to the nerve cell processes in vivo and in primary cultured nerve cells. Sorting of VP transcripts to dendrites is mediated by the last 395 nucleotides of the mRNA, the dendritic localizer sequence, and it depends on intact microtubules. In vitro interaction studies with cytosolic extracts demonstrated specific binding of a protein, enriched in nerve cell tissues, to the radiolabeled dendritic localizer sequence probe. Biochemical purification revealed that this protein is the multifunctional poly(A)-binding protein (PABP). It is well known for its ability to bind with high affinity to poly(A) tails of mRNAs, prerequisite for mRNA stabilization and stimulation of translational initiation, respectively. With lower affinities, PABP can also associate with non-poly(A) sequences. The physiological consequences of these PABP/RNA interactions are far from clear but may include functions such as translational silencing. Presumably, the translational state of mRNAs subject to dendritic sorting is influenced by external stimuli. PABP thus could be a component required to regulate local synthesis of the VP precursor and possibly of other proteins.
Neurons of the central and peripheral nervous system have the capacity to deliver distinct mRNA species to locations outside their cell bodies. In the majority of cases, defined transcripts are sorted to the dendrites. The functional significance of this transport process appears obvious: dendrites are principally able to synthesize proteins on site because they are equipped with ribosomes and many of the components required for translation (1–4). Indeed, local translation in dendrites may play an important role in establishing at least certain forms of synaptic plasticity (5). By dendritic mRNA targeting, synthesis of components detrimental for nerve cell functions may be modulated in a spatial and possibly in a temporal manner.
In mammalian nerve cells, unequivocal evidence for local translation in the axonal compartment, with the exception of the unmyelinated initial segment, has not been obtained (3, 6). Yet specific sorting of distinct mRNA species in vivo to axons of nerve cells, such as primary sensory neurons projecting to the olfactory bulb and hypothalamic magnocellular neurons, has been described (reviewed in ref. 3). Their functional role remains elusive. In rare cases, the same transcript species is targeted to axons and dendrites. An example to be discussed here is the mRNA encoding the vasopressin (VP) precursor protein.
Work on RNA sorting in nerve cells has remained descriptive for many years. In the past, the “neuronal RNA localization community” had to recognize the pioneering role of other scientific disciplines that defined the molecular entities of the RNA localization machinery in non-neuronal cells, particularly in developing systems such as Drosophila oocytes and early embryos, but also in terminally differentiated cell types in species ranging from yeast to human. From these studies, it became clear that subcellular mRNA transport is highly complex and includes several components: (i) sequences within the RNA molecule, referred to as cis-acting elements, that may adopt secondary, tertiary, or even quaternary structures; (ii) a whole array of proteins, trans-acting factors, which mediate RNA sorting by binding either directly or indirectly to the mRNA to be transported; (iii) mechanisms of translational silencing and derepression; and (iv) components of the cytoskeleton as railway tracks and anchor sites of the ribonucleoproteins (reviewed in refs. 7–11). Conceivably, mRNA sorting in nerve cells should operate similarly. Indeed, cis-acting signals mediating dendritic transport of the mRNAs encoding the microtubule-associated protein 2 (MAP2; ref. 12), the VP precursor (13), and the a-subunit of Ca2+/calmodulin-dependent protein kinase II (14) and of the noncoding brain cytosolic 1 RNA (15) have been deciphered. In addition, trans-acting factors have been characterized that interact in vitro with the dendritic localizer elements of MAP2- (16, 17) and VP (18) mRNAs, even though their functional role in mRNA trafficking remains to be elucidated.
Here, current knowledge of the molecular determinants of VP mRNA sorting in vivo and in primary cultured sympathetic nerve cells microinjected with eukaryotic expression vector constructs will be summarized.
Materials and Methods
Preparation of Protein Extracts. All steps were performed at 4°C or on ice. Cytosolic extracts were prepared by homogenizing (Dounce homogenizer) 30 g of rat brain tissue in 150 ml of homogenization buffer A containing 1 mM K-acetate, 1.5 mM Mg-acetate, 2 mM DTT, 10 mM Hepes, pH 7.8, and protease inhibitor (Complete, Boehringer Mannheim). The homogenate was centrifuged for 10 min at 16,500×g, and the supernatant fraction was saved. The sediment was resuspended in 100 ml of buffer A and centrifuged as above. The supernatant fractions were combined and centrifuged above a cushion of 30% sucrose in buffer A for 3.5 h at 90,000×g. The supernatant fraction
|
† |
To whom reprint requests should be addressed. E-mail: emohr@uke.uni-hamburg.de. |
Table 1. Biochemical purification and peptide sequence analyses reveal that VP-RBP is the rat PABP
|
Protein purification |
Peptide sequence |
Percent identity to mouse PABP1 |
|
Heparin column: 0.2 M NaCl Affinity purification: 0.1% SDS |
P-I: EFSPFGTITSAK |
100%, amino acid, 313–324 |
|
Heparin column: 0.5 M NaCl Affinity purification: 1 M NaCl |
P-II: GYGFVHFETQEAAER |
100%, amino acid, 139–153 |
|
Heparin column: 0.5 M NaCl Affinity purification: 0.1% SDS |
P-III: NFGEDMDDERL |
100%, amino acid, 197–207 |
(S-90) was carefully removed. Solid ammonium sulfate was added to a final concentration of 45% saturation at 0°C. If necessary, the pH was adjusted to 7.8 with 1 M Tris-base. Precipitated proteins were sedimented for 30 min at 3,500×g. The supernatant fractions were discarded. Proteins were dissolved in 18 ml of 10 mM Hepes, pH 7.8/10 mM NaCl/2 mM DTT/1 mM EDTA/2% glycerol (vol/vol)/0.5 mM PMSF and desalted by using the same buffer and a 5-ml HiTrap desalting column (Amersham Pharmacia Biotech), according to the manufacturer’s instructions. Typically, the desalted eluent had a concentration of ˜7 mg/ml of protein, as determined with the Protein Assay Reagent (Bio-Rad) and BSA as a standard. Twenty-five milligrams of protein was subjected to heparin column chromatography (5 ml of HiTrap heparin column, Amersham Pharmacia Biotech). Proteins were eluted with 10.5 ml each (7 fractions, 1.5 ml each) of 10 mM Hepes, pH 7.8/2 mM DTT/1 mM EDTA/2% (vol/vol) glycerol/0.5 mM PMSF containing 0.1, 0.2, 0.3, 0.4, and 0.5 M NaCl, respectively. Protein fractions were snap-frozen in liquid nitrogen and stored at –80°C. Each fraction (1.5 µl) was tested for VP mRNA-binding activity by performing U V-crosslinking assays, as described (18). Fractions containing binding activity were further purified by affinity chromatography.
Affinity Chromatography. Preparation of biotinylated VP mRNA. Full-size VP mRNA was prepared by using 5 µg of linearized template DNA and the RiboMAX T7-system (Promega) in a 100-µl assay. Biotin-16-UTP (Boehringer Mannheim) was included at a final concentration of 0.3 mM. In vitro transcripts were purified by using the RNeasy midi kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions.
Coupling of biotinylated RNA to streptavidin-coated paramagnetic particles. Streptavidin-coated paramagnetic particles [0.6 ml (1 mg/ml); Promega] was washed three times with 1 ml each of PBS (10 mM Na-phosphate, pH 7.4/150 mM NaCl). Three hundred picomol of biotinylated VP RNA in 1 ml of PBS was incubated with the particles for 30 min at room temperature on a rotating wheel. A coupling efficiency of 20–30% was routinely achieved. After removal of the RNA solution, the beads were washed twice with 1 ml of PBS and twice with 1 ml of binding buffer [10 mM Tris-HCl, pH 7.8/2 mM DTT/1.5 mM EDTA/10 mM KCl/4% (vol/vol) glycerol/6.7 µg/µl yeast tRNA] and Complete protease inhibitor.
Affinity chromatography. Fractions obtained during heparin column chromatography containing binding activity were desalted as described by using binding buffer. For analytical affinity purification, 1.5–2.0 ml of desalted protein fractions (˜1 mg of protein) was incubated with biotinylated VP RNA coupled to 0.6 ml of streptavidin-coated paramagnetic particles for 20 min at room temperature after addition of heparin (final concentration 2.5 mg/ml) on a rotating wheel. Protein solution (unbound proteins) was removed and saved for later analysis. The particles were washed (2 min each) once with 1 ml of binding buffer containing heparin (2.5 mg/ml), twice with 1 ml each of binding buffer, and once with 200 µl of binding buffer. Bound protein was eluted with 100 µl of 0.1% SDS for 10 min at room temperature and stored at—20°C. In some cases, proteins were eluted first with the same volume of 1 M NaCl/10 mM Tris-HCl, pH 7.8/1.5 mM EDTA/2 mM DTT and Complete protease inhibitor for 30 min at room temperature, concentrated 10-fold by using Vivaspin columns (Sartorius), snap-frozen in liquid nitrogen, and stored at –80°C. For preparative purposes, affinity purification assays were scaled up 10-fold.
Peptide Sequencing. Proteins purified by affinity chromatography were separated by SDS/PAGE and stained for several hours with colloidal Coomassie blue (Roti-Blue, Roth, Karlsruhe, Germany) according to the protocol recommended by the manufacturer. The protein bands were cut out, washed in water (3×2 h), and treated with acetonitrile for 30 min. The shrunken gel pieces were rehydrated by addition of 1 µg of endoproteinase LysC (Roche Molecular Biochemicals) in 100 µl of digestion buffer (50 mM Tris-HCl, pH 8.5/1 mM EDTA) and incubated overnight at 37°C. The reaction was stopped by adding 1 µl of trifluoroacetic acid (TFA), and the supernatant fraction was collected. The gel pieces were sequentially incubated for 1 h with 100 µl each of reaction buffer, TFA/acetonitrile (50:50, vol/vol), and acetonitrile. All solutions were combined, and the proteolytic fragments were separated by narrowbore HPLC (130A, Applied Biosystems) on a C4 reverse-phase column (Vydac C4, 300 A pore size, 5 mm particle size, 2.1×250 mm). Peptides were eluted with a linear gradient (0–100% B in 50 min; solvent A: water/0.1% TFA, solvent B:70% acetonitrile/0.09% TFA) at a flow rate of 200 ml/min. Peptide-containing fractions detected at 210 nm were collected into siliconized tubes and frozen immediately.
Peptide sequences (Table 1) were determined by standard Edman degradation on an automatic sequencer (476A, Applied Biosystems).
Cloning of Rat Poly(A)-Binding Protein (PABP). A fragment of the rat PABP cDNA was amplified by the PCR after reverse transcription of rat brain RNA. Two fully degenerate primers were designed: forward primer, 5'-TT[TC]GT[GATC]CA[TC]TT-[TC]GA[GA]AC[GATC]CA[GA]GA[GA]GC-3', deduced from the amino acid sequence FVHFETQEA of peptide P-II, and reverse primer, 5'-[GAT]AT[GATC]GT[GATC]-CC[GA]AA[GATC]GG[GATC]GA[GA]AA[TC]TC-3', deduced from the amino acid sequence EFSPFGTI of peptide P-I.
Table 2. Comparison of axonal and dendritic VP mRNAs
|
Axonal VP transcripts |
|
|
• |
have shorter poly(A) tails than transcripts located in the perikarya |
|
• |
are transported to axons after translation |
|
• |
are located in varicosities devoid of peptide hormones |
|
• |
are not associated with ribosomes and are therefore not translated. |
|
Dendritic VP transcripts |
|
|
• |
are identical in size to transcripts located in the perikarya |
|
• |
are transported to dendrites before translation |
|
• |
are located in parts of dendrites that contain ribosomes and small cisterns of rough endoplasmic reticulum |
|
• |
are most likely translated on-site. |
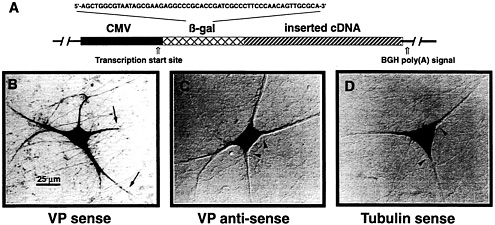
Fig. 1. VP mRNA is sorted to the dendrites of primary cultured SCG neurons. (A) Schematic view of eukaryotic expression vectors microinjected into the cell nuclei of in vitro cultured SCG neurons. The expression of any inserted cDNA is driven by the cytomegalovirus (CMV) promoter. A short sequence of the bacterial ß-galactosidase (ß-gal) gene was included such that it forms part of the 5′ untranslated region of the resulting transcripts. This sequence permits discrimination of mRNAs that are endogenously expressed in SCG neurons from those derived by transcription of the expression vector by performing in situ hybridizations with ß-gal anti-sense oligodeoxyribonucleotides. Addition of a poly(A) tail is mediated by the bovine GH (BGH) gene poly(A) addition sequence. (B-D) In situ hybridization analyses of cells microinjected with three different expression vector constructs. (B) Injection of a construct containing the rat VP cDNA inserted in sense orientation leads to transport of the mRNA into proximal and distal parts of the dendrites (arrows). Labeling of the axon has not been observed. (C) A cell is shown that expresses VP anti-sense transcripts. In this case, sorting out of the cell somata does not occur (arrowheads). (D) The vector-derived mRNA encoding α-tubulin is confined to the cell somata (arrowheads). Microinjected cells have been processed for in situ hybridization ≈18 h after injections. For experimental details, see ref. 13.
The amplification product was cloned into the SmaI site of pBluescript SK(+) (Stratagene). The sequence was determined by fluorescence-based dideoxy sequencing, by using the PRISM377 DNA Sequencer and PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems), according to the manufacturer’s protocol. A 453-bp insert with a very high degree of identity to the mouse PABP cDNA sequence [European Molecular Biology Laboratory (EMBL)/ GenBank accession no. X65553; nucleotides 758–1210] was isolated. A rat brain λ gt10 cDNA library was screened with the 32P-labeled rat PABP cDNA fragment by standard procedures (20). cDNA inserts from positive phage clones were excised with EcoRI and cloned into the EcoRI site of pBluescript SK (+). These constructs were analyzed by DNA sequencing. One clone consisting of 2,190 bp was obtained that contained the entire coding region for the rat PABP (EMBL/GenBank accession no. AJ298278). Analysis of cDNA was performed with the software package LASERGENE DNASTAR (Madison, WI). Nucleotide and amino acid sequences were compared with sequences in the EMBL and Swiss-Prot data libraries by using the program BLAST from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Sitemap/index.html#BLAST).
Other Experimental Procedures. All other experimental procedures, such as preparation of primary cultured neurons, nuclear microinjections of eukaryotic expression vector constructs, preparation of riboprobes, in situ hybridizations, and UV-crosslinking assays, have been described in detail (13, 18).
Results and Discussion
In Vivo, VP mRNA Is Sorted to Axons and Dendrites. The genes encoding the VP and structurally closely related oxytocin (OT) precursors are expressed in different populations of hypothalamic magnocellular neurons. The cells are peculiar because the peptide hormones are not only secreted from the nerve terminals in the posterior pituitary into the systemic circulation. Substantial amounts of VP and OT are also released from the dendrites into the brain. Thus, VP and OT have dual functions: first, they act as peptide hormones on diverse peripheral organs, and second, they have a role as neurotransmitters and/or neuromodulators in the central nervous system (21). Both VP and OT mRNAs are sorted to the axonal domain and to dendrites. Unlike other nerve cells, do magnocellular neurons possibly lack specific mRNA sorting mechanisms, for instance as a consequence of their secretory activity? Because the axonal and dendritic transcripts exhibit different characteristics (Table 2), this does not appear to be the case (3). (i) Axonal VP and OT transcripts have snorter poly(A) tracts than their counterparts in the cell bodies. (ii) Although the poly(A) tail of both mRNA species located in the cell somata significantly increases in length in response to osmotic challenge, this is not the case for the RNAs residing in the axon. (iii) In situ hybridizations combined with immunocytochemical analyses revealed that the peptide hormones, and their mRNAs are not colocalized within the axon. Hence, mRNA targeting to the axon is unlikely to result from sticking unspecifically to the neurosecretory granules, (iv) Transcripts are transported to the axon subsequent to translation, (v) There is no evidence for local translation of both mRNAs within the axonal compartment because they are not associated with ribosomes. The dendritic VP and OT mRNAs exhibit distinct characteristics: (i) A variant poly(A) tail length was not evident. The length of VP and OT mRNAs was of the same size in dendrites and cell somata. (ii) The mRNAs are sorted to dendrites before translation. (iii) Immunohistochemical studies at the ultrastructural level have confirmed synthesis of the VP and OT precursors in dendrites, in small cisterns of rough endoplasmic reticulum (ER). This observation is in line with the detection of VP mRNA by electron microscopic in situ hybridization in dendritic
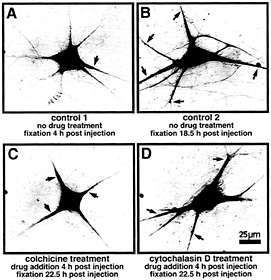
Fig. 2. Localization of VPRNA to the dendrites of microinjected SCG neurons is mediated by microtubules. Primary cultured SCG neurons have been injected with a eukaryotic expression vector containing the complete rat VP cDNA. The subcellular distribution of VP transcripts was detected by in situ hybridization by using a digoxigenin-labeled in vitro synthesized VP antisense RNA probe (for details, see ref. 13). Because the microinjection represents a stressful condition, the experiments were designed to allow for a period of recovery (4 h) before drugs were added. (A) Control 1 shows a nontreated cell fixed 4 h after injection. The VP RNA is largely confined to the cell body. Minor amounts are detectable in basal parts of the dendrites close to the cell body (arrow). (B) Control 2 shows a nontreated cell fixed 18.5 h after injection. The VP RNA has been transported to distal parts of the dendrites (arrows). (C) This neuron has been subjected to cokhicine treatment (0.2 µg/ml) for 18.5 h. The drug was added after a recovery time of 4 h after injection. VP RNA remains largely located in the cell body and in basal and very proximal parts of the dendrites (arrows). (D) Example of a neuron subjected to cytochalasin D treatment (0.15 µg/ml) for 18.5 h. The drug was added after a recovery time of 4 h after injection. Cytochalasin D does not inhibit transport of VP RNA to distal dendritic segments (arrows).
segments containing rough ER (for review, see ref. 3). A currently unresolved problem is the apparent lack of Golgi-like structures in dendrites of nerve cells. Even though Golgi marker proteins of the cis-, intermediate-, and trans-Golgi compartments are detectable immunocytochemically, these molecules are usually located in only one of the major dendrites, and they are frequently restricted to parts proximal to the cell body (22).
Characterization of Dendritic Localizer Sequences Within VP mRNA.
To define cis-acting sequences mediating VP mRNA sorting to nerve cell processes, eukaryotic expression vector constructs (Fig. 1 A) have been designed and introduced by nuclear microinjections into primary cultured neurons isolated from embryonic rat superior cervical ganglia (SCG) (13). When injections were done with a construct containing the cDNA in sense orientation, VP transcripts were detectable in the cell somata as well as in dendrites (Fig. 1 B). In contrast, VP antisense RNA remained confined to the cell bodies (Fig. 1 C), indicating that either a sequence motif or a secondary structure of the VP mRNA harbors information essential for mRNA transport. Vector-expressed a-tubulin transcripts (Fig. 1 D), like endogenous mRNA, are also entirely located in the perikarya (13). Further analyses revealed that the last 395 nucleotides, termed
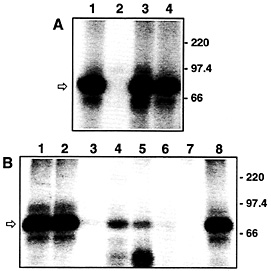
Fig. 3. VP-RBP binds specifically to the DLS of the VP mRNA and is enriched in brain tissues. (A) Autoradiogram of UV-crosslinking analyses performed with 7.5 µg of rat brain cytosolic protein extract and 5 fmol of the radiolabeled DLS riboprobe [lacking a poly(A) tail]. Unlabeled competitor RNAs were added at a 100-fold molar excess. Lane 1, no competitor; lane 2, full-size VP RNA (GenBank accession no. M25646); lane 3, dendritic targeting element of rat MAP2 mRNA (GenBank accession no. X51842, nucleotide residues 5383–5552); lane 4, full-size rat a-tubulin RNA (GenBank accession no. V01227). The positions of molecular size marker proteins is indicated on the Right. The arrow denotes the 85-kDa VP-RBP/RNA complex. All competitor RNAs represent the sense strands; none of the transcripts possess poly(A) tails, (B) Autoradiogram of UV-crosslinking analyses performed with 7.5 µg each of various rat tissue/cell line cytosolic protein extracts and 5 fmol of the radiolabeled DLS riboprobe [lacking a poly(A) tail]. Proteins were prepared from: lane 1, total brain; lane 2, hypothalamus; lane 3, heart; lane 4, lung; lane 5, spleen; lane 6, liver; lane 7, Rat I cells; lane 8, pheochromocytoma 12 cells. The positions of molecular size marker proteins are indicated on the Right. The arrow denotes the 85-kDa VP-RBP/RNA complex.
dendritic localizer sequence (DLS), containing part of the coding region and the complete 3'-untranslated region (UTR), were able to confer dendritic targeting to a normally nonlocalized reporter transcript comparable to that achieved by the VP mRNA alone. Partial DLS segments spanning either its proximal or its distal half or the 3'-UTR alone each confer only a moderate degree of dendritic localization to very proximal parts of the dendrites. Hence, the DLS contains several weak localizer elements, and these have to act in concert to mediate an efficient transport of the VP mRNA to the dendritic domain (3, 13).
In vivo but not in SCG neurons, VP mRNA is sorted to dendrites as well as to axons (13). Thus, SCG neurons apparently lack the machinery essential for sorting of distinct mRNA species to the axonal domain. Alternatively, low levels of mRNA might prevent their detection in axons.
Dendritic Transport of VP mRNA Is Mediated by Microtubules. With rare exceptions, the cytoskeleton is indispensable when mRNAs are localized to distinct subcellular destinations. Microfilaments or microtubules are needed for mRNA sorting in non-neuronal cells in yeast, Xenopus oocytes, Drosophila oocytes and early embryos, and in mammalian cells (23, 24). Present knowledge about the type of cytoskeletal element as part of the mRNA transport machinery in nerve cells is still preliminary. Yet its
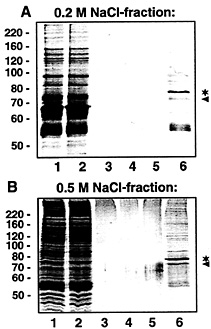
Fig. 4. Purification of VP-RBP by affinity chromatography. VP-RBP-containing fractions obtained after heparin column chromatography (eluted with 0.2 and 0.5 M NaCl, respectively) were individually subjected to affinity purification by using biotinylated full-size VP RNA [but lacking a poly(A) tail] immobilized on streptavidin-coated paramagnetic particles. Protein fractions obtained during affinity purification were separated by SDS gel electrophoresis and stained with Coomassie blue. (A) Affinity purification with proteins eluted with 0.2 M NaCl from the heparin column. (B) Affinity purification with proteins eluted with 0.5 M NaCl from the heparin column. Lane 1, 5 µg of protein eluted before affinity purification; lane 2, 5 µg of protein after incubation with VP RNA immobilized on streptavidin-coated paramagnetic particles (unbound proteins); lanes 3–5, 20 µl each of the wash fractions; lane 6, protein (20 µl) bound to VP RNA and eluted with 0.1% SDS. The asterisk denotes the major protein with an apparent molecular mass of ˜78 kDa. The protein marked by the arrowhead probably represents BSA (for details, see Fig. 6 legend). The positions of marker proteins (in kilodaltons) are indicated on the Left.
identification is important because it provides valuable information concerning the motor proteins that link the ribonucleoprotein unit to microfilaments or microtubules. Earlier studies (25) suggest the involvement of microtubules in sorting newly synthesized poly(A)-RNA to dendrites. Transport of a defined newly synthesized mRNA species in the presence of cytoskeleton-disrupting drugs has not been investigated. We have analyzed dendritic targeting of VP mRNA in primary cultured SCG neurons subjected to depolymerization of either microtubules or microfilaments by colchicine and cytochalasin D treatment, respectively. As shown in Fig. 2, VP mRNA is directed toward the dendritic compartment along microtubules. Because microtubules in dendrites exhibit a mixed polarity (26), plus-end-and/or minus-end-directed motor proteins such as kinesins and dynein are likely partners of the translocation complex. Microfilaments do not appear to be required for the transport process per se. However, the data do not rule out the possibility that the actin-based cytoskeleton may be necessary for mRNA anchoring within the neurites. Recently, evolutionarily conserved RNA-binding proteins have been identified that may play a role in
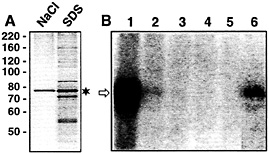
Fig. 5. The 78-kDa protein purified by affinity chromatography and eluted with 1 M NaCl retains binding activity to radiolabeled VP RNA. (A) Protein was first eluted with 1 M NaCl (left lane). Afterward, protein that remained bound to the VP RNA was eluted with 0.1% SDS (right lane). One-fiftieth each of the eluted protein was applied to the gel. The Coomassie-stained SDS gel is shown. The asterisk denotes the major protein with an apparent molecular mass of ˜78 kDa. The positions of marker proteins (in kilodaltons) are indicated on the Left, (B) Autoradiogram of UV-crosslinking analyses performed with fractions obtained during affinity purification and 5 fmol of the radiolabeled full-size VP RNA [lacking a poly(A) tail]. Lane 1, protein enriched by the heparin column chromatography before affinity purification; lane 2, protein after incubation with VP RNA immobilized on streptavidin-coated paramagnetic particles (unbound proteins); lanes 3–5, wash fractions; lane 6, protein bound to VP RNA, eluted with 1 M NaCl. The arrow denotes the 85-kDa protein/RNA complex.
transporting ß-actin transcripts in fibroblasts (27) and vegetal pole 1 (Vg1) mRNA in Xenopus oocytes (28, 29), respectively. Although ß-actin mRNA is transported along microfilaments, microtubules are prerequisite for Vg1 mRNA sorting to the vegetal hemisphere. On the basis of their high degree of similarity, it is conceivable that, by the interaction of such proteins (either directly or via additional adapter proteins) with different motor proteins, the ribonucleoprotein cargo could be transferred from microtubules to microfilaments and vice versa.
Characterization of Trans-acting Factors. Proteins (trans-acting factors) play an active role in sorting mRNA molecules to defined cytoplasmic locations. By using UV-crosslinking analyses, we have identified a protein enriched in brain tissue, termed VP-RNA-binding protein (VP-RBP), which interacts in a specific manner with the DLS of the VP mRNA (Fig. 3 A and B) but not with the 5'-end of the VP mRNA, which lacks a role in dendritic mRNA localization (18). Moreover, the protein fails to bind to a variety of other transcripts, including a-tubulin mRNA and the dendritic targeting element of the MAP2 transcript (Fig. 3 A). These findings are complemented by recently published data that two trans-acting factors, MARTA1 and MARTA2, interact in vitro with the dendritic targeting element of MAP2 mRNA but not with the rat VP mRNA or with other transcripts known to be sorted to dendrites (16). Hence, the molecular determinants required for sorting of different mRNAs to dendrites appear to be surprisingly specific for a given transcript species. Presumably, this finding reflects the existence of different pathways that govern the correct temporal and spatial distribution of defined mRNAs that have been observed in nerve cells (1, 3).
Purification of VP-RBP. Biochemical purification, including precipitation with 45% (NH4)2SO4 from cytosolic brain extracts and subsequent heparin column chromatography, was used to identify the molecular nature of VP-RBP. Binding activity, as revealed by formation of the 85-kDa protein/RNA complex, was detected in fractions eluted with 0.2 M and 0.5 M NaCl, respectively (data not shown). These fractions were separately
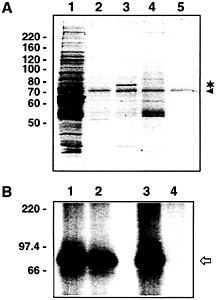
Fig. 6. The 78-kDa protein binds specifically to VP RNA. (A) Coomassiestained SDS gel of affinity chromatography assays performed with proteins obtained during heparin column chromatography (0.2 M NaCl-eluent) and different biotinylated RNAs immobilized on streptavidin-coated paramagnetic particles. Lane 1, proteins before affinity purification; lane 2, proteins eluted from paramagnetic particles coupled with biotinylated rat SSTR3 RNA (GenBank accession no. X63574, partial sequence corresponding to nucleotides 3010–3855); lane 3, proteins eluted from paramagnetic particles coupled with biotinylated rat VP RNA [lacking a poly(A) tail]; lane 4, proteins eluted from paramagnetic particles without RNA coupling; lane 5, coupling of biotinylated rat VP RNA but without addition of protein. The 78-kDa protein is denoted by the asterisk. The protein marked by the arrowhead probably represents BSA. The paramagnetic particles are stored in a buffer containing BSA. Residual amounts of this protein are obviously eluted from the particles with 0.1% SDS. (B) Autoradiogram of UV-crosslinking analyses performed with affinity-purified proteins and radiolabeled VP RNA [lacking a poly(A) tail]. Proteins obtained during heparin column chromatography (0.2 M NaCl-eluent) were affinity-purified with streptavidin-coated paramagnetic particles coupled with SSTR3 RNA (lanes 1 and 2) or full-size VP RNA (lanes 3 and 4). Lanes 1 and 3, protein before affinity purification; lanes 2 and 4, proteins after affinity purification (proteins not binding to the immobilized RNAs). The arrow denotes the 85-kDa VP-RBP/VP mRNA complex. The positions of molecular size marker proteins (in kilodaltons) are indicated on the Left.
subjected to affinity chromatography with biotinylated VP transcripts immobilized on streptavidin-coated paramagnetic particles. Proteins bound to VP RNA were eluted with 0.1% SDS and separated by denaturing PAGE. In both cases, that is when the 0.2 M and 0.5 M heparin column eluents were used as starting material, a major protein with an apparent molecular weight of about 78 kDa was eluted (Fig. 4 A and B, lane 6*). UV-crosslinking analyses performed with individual fractions obtained during affinity purifications revealed almost complete binding of the 78-kDa protein to the immobilized VP RNA (data not shown).
In repeated experiments, elution of the 78-kDa protein by buffers containing high concentrations of salt turned out to be variable and often was rather ineffective. Most of the protein came off the matrix by incubation with detergent (Fig. 5 A). However, as demonstrated by UV-crosslinking analysis, the salt-eluted protein retained RNA-binding activity forming a
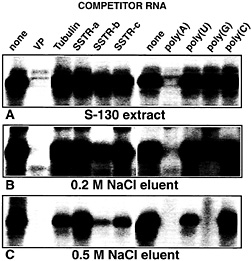
Fig. 7. PABP present in cytosolic brain extracts and in partially purified chromatographic fractions exhibits different binding properties to VP mRNA. Autoradiogram of UV-crosslinking competition analyses performed with 5 fmol of radiolabeled full-size VP RNA [lacking a poly(A) tail] and (A) rat brain cytosolic proteins (S-130) or protein partially purified by precipitation with 45% (NH4)2SO4followed by heparin column chromatography and eluted with 0.2 M NaCl (B) or 0.5 M NaCl (C). Unlabeled competitor RNAs were added at 100-fold molar excess, and ribohomopolymers poly(A), poly(U), poly(G), and poly(C) were added at a concentration of 100 ng/assay. The following competitor RNAs were used: VP, complete rat vasopressin RNA; Tubulin, complete rat a-tubulin RNA; SSTRa-c, partial sequences derived from the rat SSTR3 mRNA (SSTR-a: nucleotide residues 451–1650; SSTR-b: nucleotide residues 1660–3010; SSTR-c: nucleotide residues 3010–3855). All competitor RNAs represent the sense strands; none of the transcripts possess poly(A) tails.
complex with radiolabeled VP mRNA identical in size to that seen with protein extracts before affinity purification (Fig. 5 B). Thus, the 78-kDa protein purified from rat cytosolic brain extracts most likely represents VP-RBP that, in UV-crosslinking analyses, gives rise to the protein/RNA complex with an apparent molecular weight of ˜85 kDa (ref. 18; see also Fig. 3).
Further control affinity purifications demonstrate specificity of VP-RBP interaction with VP mRNA (Fig. 6 A, lane 3*). (i) The protein fails to associate with an RNA fragment corresponding to part of the mRNA encoding the G-protein-coupled somatostatin receptor 3 (SSTR3, lane 2). As shown earlier, this RNA does not inhibit formation of the VP-RBP/VP RNA complex in competition UV-crosslinking assays (18). (ii) It shows no unspecific interaction with paramagnetic particles in the absence of RNA-coupling (lane 4). (iii) The protein is no constituent of particles coupled with VP mRNA and processed identically in the absence of protein (lane 5). Subsequent UV-crosslinking analyses (Fig. 6 B) give further confirmation: when affinity purification was done with coupling of SSTR3 RNA, binding of VP-RBP to VP mRNA is readily detectable in the protein pool before and after incubation with SSTR3 RNA-coated particles (lanes 1 and 2), whereas residual binding activity is largely depleted in the unbound protein pool when VP RNA was coupled to the matrix (lanes 3 and 4).
VP-RBP Is the Rat PABP. Large-scale affinity purification by using liganded VP RNA and subsequent separation of eluted protein by SDS gel electrophoresis was used to isolate the 78-kDa protein in quantities sufficient for peptide sequencing. The
following preparations were processed for protein identification: (i) Proteins eluted with 0.2 M NaCl from a heparin column, affinity purified and eluted with 0.1% SDS; (ii) proteins eluted with 0.5 M NaCl from a heparin column, affinity purified and eluted with 1 M NaCl; and (iii) proteins eluted with 0.5 M NaCl from a heparin column, affinity purified and eluted with 0.1% SDS. Proteins were digested separately with endoproteinase Lys-C. The resulting peptides were resolved by HPLC. The elution profiles were very similar, indicating that the proteins are either identical or at least highly related (data not shown). One peptide from each chromatographic separation (but exhibiting different retention times) was subjected to sequence analysis, revealing that the 78-kDa protein is the rat PABP. All peptides are 100% identical to parts of mouse PABP1 (Table 1). Cloning and cDNA sequencing showed that the rat PABP consists of 636-aa residues with a high degree of identity when compared with mouse (99.7%; ref. 19) and human (99.5%; ref. 30) PABP. Hence, VP-RBP will be referred to as PABP.
We do not know why PABP elutes from the heparin column with 0.2 and 0.5 M NaCl, but several explanations are possible. First, covalent modifications, for example phosphorylated forms, of PABP with reduced affinity for the negatively charged ligand could exist. Second, PABP may be part of larger complexes displaying distinct binding characteristics to heparin. Currently, we cannot discriminate between these possibilities. However, UV-crosslinking competition analyses performed with VP mRNA indeed reveal different binding behaviors of PABP in cytosolic extracts and in partially purified protein pools (Fig. 7). PABP present in rat brain cytosolic extracts exhibits the highest degree of binding specificity. Complex formation is inhibited by a molar excess of unlabeled VP mRNA but not by a-tubulin transcripts and segments spanning various parts of the SSTR3 mRNA. With ribohomopolymer competitors, poly(A) competed as would be expected, whereas poly(U), poly(G), and poly(C) were inefficient (Fig. 7 A). When the same series of experiments was done with proteins eluted with 0.2 M NaCl from a heparin column, additional, albeit minor, competition was observed with one of the SSTR3 RNA segments (SSTR-b probe) and with poly(G) (Fig. 7 B). The lowest degree of specificity with respect to interaction with VP mRNA is observed for PABP present in the protein pool eluted with 0.5 M NaCl from the column (Fig. 1 C). These data clearly demonstrate that binding specificity of PABP to VP mRNA is determined by additional parameters, for instance a covalent modification or by other proteins that could alter PABP’s sequence selection by protein/protein interactions. Apparently, this “specificity factor,” whatever its nature, is brain specific. Evidence stems from UV-crosslinking analyses shown in Fig. 3 B. Even though PABP is known to be extremely abundant (31), most peripheral tissues and non-neuronal cell lines harbor much lower amounts of the protein in a form that is able to interact with VP mRNA compared with brain tissue.
Possible Role of PABP in VP mRNA Metabolism. PABP harbors four highly conserved RNA recognition motifs (RRM; 80–100 aa in length) at the N-terminal part of the protein and a more divergent C-terminal auxiliary domain (32). It binds with high affinity to the poly (A) tail of mRNAs, thereby enhancing translation via interaction with initiation factors bound at the 5' end (33). Furthermore, it stabilizes mRNAs in a translation-dependent manner (34). Binding studies with individual RRMs or combinations thereof revealed several interesting features: single RRMs are unable to bind to poly (A). RRMs 1 and 2 have a high affinity for poly(A) identical to that of the full-size protein, whereas RRMs 3 and 4 have a much lower affinity for poly(A). In fact, binding of RRMs 3 and 4 to poly(U)- and poly(G)-sequences is much better than to poly(A) (35, 36). In yeast, PABP is essential for cell viability. Yet, whereas deletion of RRMs 1 and 2 alone still supported growth, removal of RRM 4 joined with C-terminal amino acid residues did not, suggesting critical functions of this sequence (35). Taken together, RRMs 1–4 are functionally diverse, and features other than high-affinity binding to poly(A) sequences are essential for cell viability. PABP is an extremely abundant protein. HeLa cells, for instance, have a roughly 3-fold excess of protein over binding sites on poly(A) mRNAs (31). Because PABP interacts with sequences other than poly(A) in vitro (31, 36), it probably has additional functions in mRNA metabolism. For instance, PABP is able to control translation of its own mRNA, and it does so by specific association with sequences of the 5'-UTR (37, 38).
Given the heterogeneous roles of PABP, it is conceivable that it may be involved in regulating the translational state of the VP (and possibly of other) mRNA. Accumulating evidence suggests that dendritically localized mRNAs are not translated until external stimuli trigger the activation of protein synthesis (5, 39). Translational silencing by PABP could, for instance, be accomplished by its binding to the DLS of the VP mRNA. A direct or indirect interaction of this (or these) molecule(s) with those that are bound to the poly(A) tract could inhibit translational stimulation, because it interferes with the interaction of poly(A) tail-bound PABP with translational initiation factors at the 5'-end of the mRNA. A similar model involving PABP as part of a larger and preexisting protein complex has been proposed as a mechanism that regulates translation-dependent turnover of the c-fos mRNA (40).
Several questions to be addressed in future experiments remain open, (i) Which RRMs of PABP participate in its interaction with DLS of the VP mRNA? (ii) What type of molecule (or modification) determines its binding specificity to the DLS? (iii) Does PABP also play a role in the metabolism of other dendritically localized mRNAs, and what exactly is that role?
We thank Susanne Franke for expert technical assistance. This work is supported by the Deutsche Forschungsgemeinschaft and the Volkswagenstiftung (to D.R. and E.M.). Part of this work forms the Ph.D. thesis of Carola Fuhrmann.
1. Steward, O. (1997) Neuron 18, 9–12.
2. Kuhl, D. & Skehel, P. (1998) Curr. Opin. Neurobiol. 8, 600–606.
3. Mohr, E. (1999) Prog. Neurobiol. 57, 507–525.
4. Tiedge, H., Bloom, F.E. & Richter, D. (1999) Science 283, 186–187.
5. Schuman, E.M. (1999) Neuron 23, 645–648.
6. Alvarez, J., Giuditta, A. & Koenig, E. (2000) Prog. Neurobiol. 62, 1–62.
7. Bashirullah, A., Cooperstock, R.L. & Lipshitz H.D. (1998) Annu. Rev. Biochem. 67, 335–394.
8. Barbarese, E., Brumwell, C., Kwon, S., Cui, H. & Carson, J.H. (1999) J. Neurocytol. 28, 263–270.
9. Gonzalez, I., Buonomo, S.B.C., Nasmyth, K. & von Ahsen, U. (1999) Curr. Biol. 9, 337–340.
10. King, M.L., Zhou, Y. & Bubunenko, M. (1999) BioEssays 21, 546–557.
11. Lipshitz, H.D. & Smibert, C.A. (2000) Curr. Opin. Genet. Dev. 10, 476–488.
12. Blichenberg, A., Schwanke, B., Rehbein, M., Garner, C.C., Richter, D. & Kindler, S. (1999) J. Neurosci. 19, 8818–8829.
13. Prakash, N., Fehr, S., Mohr, E. & Richter, D. (1997) Eur. J. Neurosci. 9, 523–532.
14. Mori, Y., Imaizumi, K., Katayama, T., Yoneda, T. & Tohyama, M. (2000) Nat. Neurosci. 3, 1079–1084.
15. Muslimov, I.A., Santi, E., Hamel, P., Perini, S., Higgins, D. & Tiedge, H. (1997) J. Neurosci. 17, 4722–4733.
16. Rehbein, M., Kindler, S., Horke, S. & Richter, D. (2000) Mol. Brain Res. 79, 192–201.
17. Monshausen, M., Putz, U., Rehbein, M., Schweizer, M., DesGroseillers, L., Kuhl, D., Richter, D. & Kindler, S. (2001) J. Neurochem. 76, 155–165.
18. Mohr, E., Fuhrmann, C. & Richter, D. (2001) Eur. J. Neurosci., 13, 1107–1112.
19. Wang, M., Cutler, M., Karimpour, I. & Kleene, K.C. (1992) Nucleic Acids Res. 20, 3519.
20. Sambrook, J., Fritsch, E.F. & Maniatis, T. (1989) in Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
21. Morris, J.F., Pow, D.V., Sokol, H.W. & Ward, A. (1993) in Vasopressin, eds. Gross, P., Richter D. & Robertson, G.L. (John Libbey Eurotext, Paris), pp. 171–182.
22. Torre, E.R. & Steward, O. (1996) J. Neurosci. 16, 5967–5978.
23. Nasmyth, K. & Jansen, R.-P. (1997) Curr. Opin. Cell Biol. 9, 396–400.
24. Arn, E.A. & Macdonald, P.M. (1998) Cell 95, 151–154.
25. Bassell, G.J., Singer, R.H. & Kosik, K.S. (1994) Neuron 12, 571–582.
26. Baas, P.W., Black, M.M. & Banker, G.A. (1989) J. Cell Biol. 109, 3085–3094.
27. Ross, A.F., Oleynikov, J., Kislauskis, E.H., Taneja, K.L. & Singer, R.H. (1997) Mol. Cell. Biol. 17, 2158–2165.
28. Deshler, J.O., Highett, M.I., Abramson, T. & Schnapp, B.J. (1998) Curr. Biol. 8, 489–496.
29. Havin, L., Git, A., Elisha, Z., Oberman, F., Yaniv, K., Pressman-Schwartz, S., Standart, N. & Yisraeli, J.K. (1998) Genes Dev. 12, 1593–1598.
30. Grange, T., de Sa, C.M., Oddos, J. & Pictet, R. (1987) Nucleic Acids Res. 15, 4771–4787.
31. Görlach, M., Burd, C.G. & Dreyfuss, G. (1994) Exp. Cell Res. 211, 400–407.
32. Burd, C.G. & Dreyfuss, G. (1994) Science 265, 615–621.
33. Preiss, T., Muckenthaler, M. & Hentze, M.W. (1998) RNA 4, 1321–1331.
34. Coller, J.M., Gray, N.K. & Wickens, M.P. (1998) Genes Dev. 12, 3226–3235.
35. Burd, C.G., Matunis, E.L. & Dreyfuss, G. (1991) Mol. Cell Biol. 11, 3419–3424.
36. Kühn, U. & Pieler, T. (1996) J. Mol. Biol. 256, 20–30.
37. de Melo Neto, O.P., Standart, N. & de Sa, C.M. (1995) Nucleic Acids Res. 23, 2198–2205.
38. Bag, J. & Wu, J. (1996) Eur. J. Biochem. 237, 143–152.
39. Marin, P., Nastiuk, K.L., Daniel, N., Girault, J.-A., Czernik, A.J., Glowinski, J., Nairn, A.C. & Prémont, J. (1997) J. Neurosci. 17, 3445–3454.
40. Grosset, C., Cheu, C.A., Xu, N., Sonenberg, N., Jaquemin-Sablou, H. & Shyu, A. (2000) Cell 103, 29–40.








