4
Addressing the Threats: Conclusions and Recommendations
Ten years after the 1992 Institute of Medicine report Emerging Infections: Microbial Threats to Health in the United States was issued, it has become even more apparent that infectious diseases continue to have a dramatic impact on the United States and the world. The response to microbial threats—from detection to prevention and control—requires a multidisciplinary effort involving all sectors of the public health, clinical medicine, and veterinary medicine communities. The committee’s recommendations, which emerged from focused deliberations and the application of the criteria of urgency, priority, and amenability to immediate action, are presented in this chapter. Given that infectious diseases are a significant threat to the health of the world’s population, several of the committee’s recommendations could be justified solely on the basis of humanitarian need; all are justified as being in the best interest of the United States to protect the health of its own citizens.
ENHANCING GLOBAL RESPONSE CAPACITY
The emergence of infectious diseases reflects complex social, economic, political, environmental, ecological, and microbiological factors that are globally linked. A number of forces operating in developing countries in particular, including urbanization, deforestation, changes in land use and climate, population growth, poverty, malnutrition, political instability, and even terrorism, have created the conditions for several infectious diseases to become new or recurrent threats. To devise and implement effective preven-
tion and control strategies, therefore, the factors influencing the emergence of infectious disease must be recognized and addressed at a global level.
Disease burdens—such as those incurred as a result of HIV, tuberculosis, and malaria—can contribute to the destabilization of nations, damaging their social and political infrastructures (National Intelligence Council, 2000; Denver Summit of the Eight, 1997). The past decade has seen the HIV epidemic besieged but entrenched in the United States, and spread globally with a catastrophic social and economic impact on many developing countries. Affecting adults in their productive years disproportionately, HIV has led to a grievous decrease in per capita gross domestic product (GDP) across Africa, resulting in a vicious spiral of decreased investment in public health and worsening of the epidemic. The resurgence of tuberculosis is devastating many countries, particularly Russia and other former Soviet republics, where tuberculosis rates have increased an astounding 70 percent in less than a decade. Antimicrobial resistance has become a major barrier to treatment of tuberculosis and malaria worldwide, threatens the effectiveness of antiretroviral therapy in persons with AIDS, and has made treatment of common bacterial infections more difficult in the United States and elsewhere. Infectious diseases are appearing abruptly in new locations and claiming hundreds of lives; a case in point is West Nile encephalitis, which spread to most parts of the United States within 3 years following its sudden appearance in the Northeast. Certain risks to health, such as contamination of food products, have resulted in enormous economic consequences, along with implications for human disease. Infectious diseases have even been used to intentionally terrorize populations, further dramatizing the need for a comprehensive assessment of and response to microbial threats.
Amelioration of major health risks and problems in any country, therefore, is a global good that may indirectly benefit the United States. Moreover, in an era of heightened concern regarding international networks of terrorism and nations with weapons of mass destruction, leadership in addressing the infectious disease problems of other countries can build trust and goodwill toward the United States. Repeatedly, U.S. efforts to monitor and address infectious disease threats in other countries have been welcomed and have increased understanding and improved relationships between countries. The need for an adequate global response to infectious disease threats, therefore, derives from the United States’ humanitarian, economic, and national security interests.
According to a recent analysis by the National Intelligence Council (2000), newly emerging infectious diseases, including the intentional use of a biological agent, will pose an increasing global health threat and will complicate U.S. and global security over the next 20 years. As outlined in that report, the future impact of infectious diseases will be heavily influ-
enced by three sets of variables: (1) the relationship between increasing antimicrobial resistance and the success of research to develop new antibiotics and vaccines; (2) the trajectory of developing and transitional economies, especially concerning the basic quality of life of the poorest groups among the population; and (3) the degree of success of global and national efforts to create public health infrastructure with effective systems of surveillance and response. The interplay among these variables will determine the overall outlook regarding the impact of infectious diseases.
In this context, it is clear that the response to emerging infectious diseases at a global level requires an investment in the capacity of developing countries to address these diseases as they arise. Such investments should take the form of financial and technical assistance, operational research, enhanced surveillance, and efforts to share both knowledge and best public health practices across national boundaries. For example, the World Health Organization (WHO) has developed a program for ensuring global health security by strengthening country capacity in microbiology and epidemiology to improve national preparedness (see Box 4-1). Financial and technical assistance to international agencies, governments, and nongovernmental organizations has already proven to be an effective means of addressing global disease threats. The Centers for Disease Control and Prevention (CDC) continues to support reference laboratories and provide technical assistance for disease outbreaks. Likewise, the National Institutes of Health (NIH) has expanded the number of international research and treatment centers. Financial and technical support has also come from private foundations and other U.S. agencies and organizations, and has been particularly effective in supporting efforts to combat HIV, tuberculosis, malaria, and polio.
The United States should seek to enhance the global capacity for response to infectious disease threats, focusing in particular on threats in the developing world. Efforts to improve the global capacity to address microbial threats should be coordinated with key international agencies such as the World Health Organization (WHO) and based in the appropriate U.S. federal agencies (e.g., the Centers for Disease Control and Prevention [CDC], the Department of Defense [DOD], the National Institutes of Health [NIH], the Agency for International Development [USAID], the Department of Agriculture [USDA]), with active communication and coordination among these agencies and in collaboration with private organizations and foundations. Investments should take the form of financial and technical assistance, operational research, enhanced surveillance, and efforts to share both knowledge and best public health practices across national boundaries.
|
BOX 4-1 The World Health Organization Office in Lyon Epidemics and emerging infections continue to threaten human health worldwide, and many developing countries lack the capacity and expertise necessary to address these threats effectively. The World Health Organization (WHO), head-quartered in Geneva, Switzerland, is working to ensure global health security. In 2001, WHO’s Department of Communicable Diseases, Surveillance, and Response opened an office in Lyon, France. To strengthen country capacity in microbiology and epidemiology, this new office provides a training program focused on enhancing the capacity of national public health laboratories, supporting field epidemiology training programs, and improving the capacity to detect and respond to disease outbreaks. The overall objective of the program is to strengthen diagnostic and surveillance capabilities at all levels. This goal can be achieved through an increase in reference diagnostic capabilities for communicable diseases; the development of appropriate core public health administrative practices; the development of rapid, sustainable national and international laboratory communications networks; the development of rapid, efficient, and safe means for shipment of diagnostic materials and laboratory specimens; and the establishment of appropriate quality control principles and practices. The 2-year training program is designed for senior laboratory staff. Throughout the course of the program, participants receive training in essential laboratory diagnostic practices and techniques, biosafety, data collection and management, statistical analysis, basic disease epidemiology, and personnel management and administration. Following an initial 8-week session in Lyon, the trainees return to their home organizations. Over the course of the next 2 years, they are followed up in their home countries and return to Lyon for two shorter visits. Upon completion of the program, participants should be able to contribute effectively to the rapid detection of epidemic and emerging diseases in their countries. Each year the program enrolls 15 participants for two sessions. It is estimated that after 5 years, the program will have trained 150 specialists from 45 countries. The first training cohort consisted of participants from 7 African countries who were selected for their senior roles in the management of their country’s national public health reference laboratory. The first training session consisted of three modules: laboratory, surveillance, and information technology; laboratory response; and laboratory management. The second group of trainees was selected from Middle Eastern and North African countries and began training in 2002. SOURCE: World Health Organization, 2001h. |
Improving the global capacity to respond to microbial threats will require sustained efforts over time. Given the imminent nature of many infectious disease threats, however, it is critical that immediate action be taken toward achieving this capacity. Mobilization of young graduates in the health sciences has proven to be a successful strategy for meeting the
goals identified by government agencies responsible for improving health domestically. For example, the National Health Service Corps, administered by the Health Resources and Services Administration (HRSA), has created a mechanism for dedicated health professionals to work in underserved communities where they are most needed nationwide. A similar mechanism could be used to established a Global Health Services Corps, offering loan forgiveness in exchange for service in areas of global public health need. Such a program could provide the stimulus for an immediate U.S. workforce to serve as a means of increasing global response capacity by assisting developing countries in creating the infrastructure, knowledge, and skills necessary to sustain long-term independent success. In addition to building developing-country capacity to respond, the program could enable U.S. public health agencies (e.g., CDC, NIH) to maintain expertise in epidemiology and laboratory issues related to diseases no longer endemic in the United States through training of U.S. scientists within countries where these diseases remain endemic. The same is true for diseases that are potential bioterrorist agents, particularly since the cadre of U.S. experts in rare diseases has declined (see the later discussion of educating and training the microbial threats workforce).
Expansion of programs in infectious disease research and training for health professionals from other countries is also needed. Notable successes in this area include the NIH Fogarty International Center for Advanced Study in the Health Sciences (FIC) that sponsors U.S. schools of medicine and public health in providing training for foreign scientists from developing countries through its AIDS International Training and Research Program (NIH, 1999). Since the center’s inception, more than 2,000 scientists from more than 100 countries and territories have received training. In addition, over 46,000 students and health professionals have been provided short-term training through courses conducted in 65 countries. FIC also supplies funding for competitive supplemental awards under the Tuberculosis International Training and Research Program, a collaborative program with the National Institute of Allergy and Infectious Diseases (NIAID), CDC, and USAID. An aim of this funding is to foster global health research efforts and public health capacity to better respond to the threat posed by tuberculosis and multidrug-resistant tuberculosis. In yet another collaborative program, FIC and NIAID provide awards to U.S. universities under the International Training and Research Program in Emerging Infectious Diseases, which expands NIH research training efforts in the study of microbial threats. The long-term objective is to train teams of scientists in regions of the world that offer unique opportunities to understand the fundamental biology, epidemiology, and control of emerging microbial diseases.
IMPROVING GLOBAL INFECTIOUS DISEASE SURVEILLANCE
The need to strengthen global infectious disease surveillance is vital. As noted earlier, in addition to the United States’ humanitarian objective of aiding countries in crisis, it is critical to U.S. national security that quality population-based data on disease burden and trends in the developing world be obtained through global surveillance (Hyder and Morrow, 2000). Yet disease burden estimates and projections are often based on only fragmentary data (Murray and Lopez, 1997). The reality in many developing societies is that deaths and births are not recorded, and a formal system of medical care is unavailable to most of the population (Cooper et al., 1998). Health care infrastructures that lack simple diagnostic tests for diseases such as tuberculosis or that have insufficient resources to perform diagnostic tests add to the lack of knowledge of disease burden. Developing countries in which high proportions of the population experience morbidity and/ or mortality from infectious diseases may be the least likely to be encompassed by official statistics because of this lack of resources. Basic health indices, such as death rates or causes of death, are unknown in such contexts. Health ministries may generate health reports, but the data are generally unreliable. Such numbers have been used as the basis for broad policy recommendations; if the numbers are incorrect, however, the resulting policies can be damaging.
In addition to monitoring disease burden, surveillance efforts should be expanded and diversified to include the capacity to recognize previously unknown illnesses or unusual outbreaks of disease that may have global significance. With today’s rapid and often mass global movements of people, animals, and goods, the transnational spread of infectious diseases can occur quickly and easily. Global surveillance, especially for newly recognized infectious diseases, is therefore crucial in responding to and containing microbial threats before isolated outbreaks develop into regional or worldwide epidemics.
U.S. agencies have been working with WHO and other partners to achieve the goal of a comprehensive global surveillance system, and efforts to date are aptly described as creating a “network of networks” (see Figure 4-1). In Europe, countries have made significant progress through the development of networks such as those for travel-related Legionnaires’ disease, enteric organisms (Enter-net), and drug resistance. The United States has also supported efforts to establish regional networks. An example is DOD’s support for laboratory-based surveillance in the 21 countries of the Caribbean Epidemiology Center, in collaboration with the Pan American Health Organization and CDC. Likewise, CDC and others have worked in many areas to assist regional surveillance networks. Examples include the Amazon and Southern Cone networks, which encompass eight laboratories
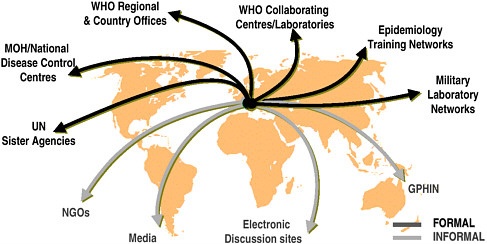
FIGURE 4-1 Global surveillance of communicable diseases: a network of networks.
SOURCE: WHO, Communicable Diseases.
in six countries of South America (CDC, 2002r), and the MeKong Delta Surveillance Network, which includes five countries of Asia, as well as the province of Yunan in China.
In 1996, DOD was mandated to use its long-standing and well-respected overseas research laboratories in Egypt, Indonesia, Kenya, Peru, and Thailand to establish the Global Emerging Infections Surveillance (GEIS) program. GEIS is a critical and unique resource for the United States in the context of global infectious disease surveillance; it is the only U.S. entity with broad-based laboratory capacity in overseas settings. GEIS has already demonstrated its excellent potential to detect the emergence of disease in those and surrounding countries (IOM, 2001e). CDC has assigned several epidemiologists to GEIS to provide increased epidemiologic capacity at these overseas sites. CDC plans to establish multiple international programs to address emerging infections, the first of which was established in Thailand in 2001 (CDC, 2002r). As more DOD overseas laboratories and CDC Emerging Infections Programs are established, increased collaboration between the two agencies will be beneficial, and serious consideration must be given to which geographic sites will fill the most critical gaps in surveillance worldwide.
Also important for global surveillance are novel training programs initiated by NIAID that provide the opportunity for field training in Asia, Africa, and South America, along with laboratory-based training in the United States, with incentives to return trainees to their home countries. Such programs require expansion in particular in the “hot zones” of Africa and Asia that are recognized as epicenters for the emergence of such agents
as Ebola, HIV, Nipah, and influenza. NIAID initiatives on pandemic preparedness for influenza in Asia, which promote zoonotic surveillance and preparation of the necessary reagents, are prototypes for the programs necessary for global surveillance.
The WHO Global Influenza Surveillance Program, now 50 years old, was responsible for the early identification of the H5N1 influenza A virus, as well as the H9N2 virus that occurred later—viruses that had previously been detected only in birds. The reagents necessary for identification of these viruses were developed by NIAID and were made available to the WHO program. Because WHO must issue recommendations for the composition of influenza vaccines twice a year—once for the Northern Hemisphere in February and once for the Southern Hemisphere in September— data must be gathered throughout the year. The infrastructure in place allows the identification of new variants, whether they are new epidemic variants or new variants with pandemic potential. The infrastructure rests on a number of national influenza centers that serve as the key laboratories for the isolation and identification of influenza viruses, using a kit of reagents produced by CDC and distributed globally. The laboratories also collect epidemiological information for transmittal to WHO headquarters in Geneva. International collaborating centers, including CDC, conduct comparative analyses of influenza viruses from around the world. Collaboration with industry is essential because the strains that are identified as vaccine candidates are provided free of charge to the pharmaceutical industry for vaccine production.
Globally, advances in information technology have also allowed novel uses of the Internet in disease surveillance. The Program for Monitoring Infectious Diseases (Pro-Med) uses electronic communications to provide up-to-date news on disease outbreaks and is open to all users. A team of experts in human, animal, and plant diseases screens, reviews, and investigates reports before posting notices. The system was designed to promote communication among the international infectious disease community, and to provide for the exchange of information about outbreaks and other matters of interest regarding emerging infectious diseases (International Society for Infectious Diseases, 2001). PacNet, an Asian network of health professionals on 20 Pacific Islands, is another such network, established to allow the exchange of information among health professionals regarding epidemics in that region. An even more innovative system, established by Health Canada in collaboration with WHO, is the Global Public Health Intelligence Network (GPHIN), an Internet-based application that continuously scans global electronic media (news wires, websites) for information on global public health risks, including infectious disease outbreaks (WHO, 1998b) (see Box 4-2). In line with the growth of electronic media, approximately 65 percent of the world’s first news about infectious disease events
|
BOX 4-2 Global Outbreak Alert and Response Network The Global Outbreak Alert and Response Network enables WHO to monitor disease outbreaks continuously. This network was formally launched in 2000 and links over 72 existing networks around the world, some of which are able to diagnose and detect unusual agents and handle dangerous pathogens. The four critical tasks of the network are epidemic intelligence and detection, verification of rumors and reports, immediate alert, and rapid response. The Global Outbreak Alert and Response Network gathers global disease intelligence using a number of sources, such as ministries of health, WHO country offices and collaborating centers, laboratories, academic institutes, and nongovernment organizations. The Global Public Health Intelligence Network (GPHIN), an electronic system that constantly performs surveillance of worldwide communications for disease events, is one of the most important informal sources from which the network gathers data. GPHIN was developed for WHO through a collaboration with Health Canada in 1996. The intelligence gathered is converted by the WHO Outbreak Alert and Response team, which then determines whether a reported disease event constitutes cause for international concern. The team meets each morning to review reports and rumors, assess their epidemiological significance, and determine actions needed. The team creates a detailed report that is distributed electronically each day to specific WHO staff around the world. From 1998 to 2001, WHO verified 578 outbreaks in 132 countries. The network electronically connects WHO member countries, disease experts, institutions, agencies, and laboratories to keep them constantly informed of outbreak events, rumored and confirmed. The network also provides real-time alerts through an outbreak verification list, offering detailed information on current outbreaks that is regularly updated and maintained. In addition, WHO posts information on outbreaks on its Disease Outbreak News website. Rapid response is a critical task of the Global Outbreak Alert and Response Network. Once an outbreak has been verified, the Outbreak Alert and Response team determines whether an international response is needed to contain it. When an international response is necessary, partners in the global health network are called upon to provide specific support, from investigations and patient management to logistics, including the provision of necessary staff and supplies. WHO and the Nuclear Threat Initiative recently partnered to create an Emergency Outbreak Response Fund to ensure that the rapid response teams can be at a designated site within 24 hours of a detected outbreak. Since 2000, WHO and the network have launched effective international responses to outbreaks in Afghanistan, Cote d’Ivoire, Egypt, Ethiopia, and other countries. SOURCE: World Health Organization, 2003b. |
during the past 4 years has come not from official country notifications, but from informal sources, including press reports and the Internet (Heymann, 2001). Recent efforts to increase capacity for translation to the six official United Nations languages will further enhance the GPHIN system.
As described earlier, surveillance of and response to emerging infectious disease threats in other parts of the world can directly benefit the United States as well as the country in which an occurrence is detected. For example, the investigation of hantavirus in Korea in the 1970s and the development of a diagnostic test were useful in the identification of and response to the epidemic of hantavirus infection in the southwestern United States in 1993. Similarly, the investigation of the H5N1 influenza virus in Hong Kong in 1997 alerted the United States and the world to the threat posed by influenza viruses in avian species as sources of pandemic influenza viruses in humans, and highlighted the urgency of influenza pandemic planning globally. The rapid measures taken to control H5N1 influenza in Hong Kong exemplify increasing global cooperation in disease surveillance. WHO, together with experts from the United States, Europe, and the Pacific region, provided information to the Hong Kong authorities on the virological and epidemiological properties of the H5N1 threat, and as a consequence, the local authorities decided to slaughter all poultry in Hong Kong. This decision resulted in a dramatic cessation of human cases of H5N1, providing a direct benefit to Hong Kong, China, and the global community. Similar steps to stamp out the epidemic of Nipah viruses among livestock and humans in Malaysia provide yet another example of the importance of global disease surveillance and the benefits to global health. Likewise, liaisons between the U.S. and European sentinel surveillance networks have led to the identification and removal of products being marketed in numerous countries, including the United States, that were contaminated with bacterial pathogens.
Several national and international groups, including the National Science and Technology Council (1995) and the Denver Summit of the Eight (1997), have echoed the 1992 IOM recommendation to establish a global disease and outbreak surveillance system. Significant efforts have been made to enhance global surveillance, but the system remains skeletal and is inadequate to monitor disease incidence and prevalence in most parts of the world.
The United States should take a leadership role in promoting the implementation of a comprehensive system of surveillance for global infectious diseases that builds on the current global capacity of infectious disease monitoring. This effort, of necessity, will be multinational and will require regional and global coordination, advice, and resources from participating nations. A comprehensive
system is needed to accurately assess the burden of infectious diseases in developing countries, detect the emergence of new microbial threats, and direct prevention and control efforts. To this end, CDC should enhance its regional infectious disease surveillance; DOD should expand and increase in number its Global Emerging Infections Surveillance (GEIS) overseas program sites; and NIH should increase its global surveillance research. In addition, CDC, DOD, and NIH should increase efforts to develop and arrange for the distribution of laboratory diagnostic reagents needed for global surveillance, transferring technology to other nations where feasible to ensure self-sufficiency and sustainable surveillance capacity. The overseas disease surveillance activities of the relevant U.S. agencies (e.g., CDC, DOD, NIH, USAID, USDA) should be coordinated by a single federal agency, such as CDC. Sustainable progress and ultimate success in these efforts will require health agencies to broaden partnerships to include nonhealth agencies and institutions, such as the World Bank.
REBUILDING DOMESTIC PUBLIC HEALTH CAPACITY
The U.S. capacity to respond to microbial threats to health is contingent on a public health infrastructure that has suffered years of neglect. Upgrading current public health capacities will require considerably increased investments across differing levels of government. Most important, this support will have to be sustained over time. Such an investment will have lasting and measurable benefits for all humankind. With recent increased funding for bioterrorism preparedness, the United States has an opportunity to develop programs and policies that will both protect against acts of bioterrorism and improve the U.S. public health response to all microbial threats. However, it is alarming that some of these funds have been diverted from multipurpose infrastructure building to single-agent preparedness.
The threat of bioterrorism is intimately related to that of naturally occurring infectious diseases. The response to bioterrorism is much like the response to any microbial threat to health, and the necessary resources for building the public health infrastructure are, in essence, the same as those needed to respond to bioterrorism. It would be counterproductive to develop an ancillary system for bioterrorist threats. Rather, such efforts must be integrated with those addressing the continuum of infectious disease concerns and potential disasters to which public health agencies are already charged to respond. While preparedness for bioterrorist-inflicted outbreaks will require certain specialized program elements and policies (related, e.g., to law enforcement, evidence collection), the human health aspects of this
new challenge mirror many of the requirements for preventing and responding to a range of naturally occurring infectious disease threats. Wherever possible, therefore, effective strategies should build on existing systems that are used routinely and can be useful for both purposes. In short, the objectives of the funding that has been allocated for bioterrorism will be met only if the public health infrastructure is enhanced first and foremost. Otherwise, preparedness programs will be inadequate, and critical opportunities to protect both human populations and agriculture (food animals and plants) from a range of disease threats, both naturally occurring and maliciously caused, may be missed.
Strong and well-functioning local, state, and federal public health agencies working together represent the backbone of effective response to a major outbreak of infectious disease, including a bioterrorist attack. How quickly public health agencies can recognize and respond to an emerging threat dramatically influences the ability to reduce casualties, control contagion, and minimize panic and disruption. Unfortunately, an overall shortage of qualified public health workers makes it difficult to meet this demand. Following the events of 2001, public health agencies were asked to develop new programs and add new staff despite the lack of available candidates. An estimated 3,200 to 4,000 new positions were requested in the bioterrorism cooperative agreements submitted to CDC. In addition, an estimated 13,000 to 15,000 persons are needed to provide 24-hour emergency coverage at the local level (Center for Infectious Disease Research and Policy, 2002). Yet a wide range of administrative barriers prevent public health agencies from obtaining qualified staff. These include non-competitive pay scales, cumbersome hiring procedures, lack of system flexibility, and inadequate incentives for retaining qualified personnel. Local health departments range in coverage from small areas served by part-time staff with little or no formal public health training to large urban health districts with inadequate resources to support the continuing education and training of their workforce. Some of the smaller local health departments could be consolidated and strengthened to ensure needed professional expertise and coverage on a more regional basis. To strengthen the public health infrastructure for infectious disease detection and response, it will be necessary to train, equip, and expand the workforce to provide both on-the-ground epidemiologic expertise and laboratory capability.
Communication, including computer connectivity, must also be strengthened to efficiently collect, analyze, and share information among public health and other officials at the local, state, and federal levels. Enabling public health agencies to obtain fast and secure Internet access is key in facilitating linkages between health departments and health care providers. For example, the Health Alert Network (HAN) is being developed by
CDC as a nationwide, integrated, secure, electronic communications system that will provide high-speed Internet connections, enabling public health officials to engage in distance learning and share laboratory findings, health advisories, and other information relevant to disease outbreaks. The network’s primary goal is to improve the information technology infrastructure of local and state health departments. The Health Alert Network is designed to be the nation’s rapid online system for health communication and will serve as the electronic platform for the National Electronic Disease Surveillance System (NEDSS) (discussed later), Epi-X (see Box 4-3), and
|
BOX 4-3 Epidemic Information Exchange The Epidemic Information Exchange (Epi-X) is a secure, web-based communications network for public health officials. Developed by CDC in 2001, it enables health officials to rapidly report and discuss public health information on disease outbreaks and other health events as they are identified and investigated. Since its launch, Epi-X has provided health officials throughout the United States with up-to-the-minute information, reports, alerts, and discussions about terrorist events, disease outbreaks, and other events of public health significance. Public health officials and other designated users can use Epi-X to post reports, notify colleagues, and receive feedback on ongoing epidemiological investigations, as well as research current and past outbreaks. Epi-X will strengthen bioterrorism preparedness efforts by supporting information sharing about disease outbreaks and other health events over a secure communications system. The network includes a Forum area in which state epidemiologists can post information on surveillance and response activities for approximately 500 public health officials in the United States, including those in the U.S. military. As of 2002, Epi-X had posted over 1000 reports of disease outbreaks, other public health activities, and requests for epidemiologic assistance from CDC. Over 1,000 public health officials at the federal, state, and local levels had used Epi-X to communicate with colleagues and experts across a secure, encrypted web-based network; track information for outbreak investigations and response; conduct online conferences to discuss such topics as West Nile virus and anthrax investigations; alert health officials by pager, phone, and e-mail to urgent events; request CDC assistance in investigations; and communicate with bioterrorism preparedness programs. Plans to expand Epi-X are under way and include increasing its user base and expanding secure communications for public health and safety officials, as well as expanding the network to provide information on international outbreaks that might affect public health in the United States. SOURCE: Center for Disease Control and Prevention, 2002s. |
other applications. HAN can assist health agencies in assessing their technology needs, acquiring equipment to help meet these needs, establishing Internet connection and e-mail capabilities, and developing training programs. The network was activated on September 11, 2001, and within 4 hours of the terrorist attacks in New York and Washington, D.C., was transmitting health messages to 250 top health officials in the United States. The network has continued to transmit health alerts, advisories, and updates, and has been expanded to reach an estimated 1 million recipients, including public health officials, physicians, nurses, laboratory staff, and other health professionals.
To rebuild the public health workforce needed to respond to microbial threats, health profession students (especially those in the medical, nursing, veterinary, and laboratory sciences) must be educated in public health as a science and as a career. Even for students within schools of public health, education has traditionally focused on academic research training, not public health practice. A 1988 IOM report notes that “many observers feel that some [public health] schools have become somewhat isolated from public health practice and therefore no longer place a sufficiently high value on the training of professionals to work in health agencies” (IOM, 1988:15). A more recent IOM report states that in 1998, only 56 of 125 medical schools required courses on such topics as public health, epidemiology, or biostatistics (IOM, 2002e). The report recommends that all medical students receive basic public health training. It also concludes that all nurses should have at least an introductory grasp of their role in public health, and that all undergraduates should have access to education in public health. Educational strategies in which applied epidemiology programs provide exposure to state and local health departments may help increase awareness of the role of public health in population-based infectious disease control and prevention, and provide for exposure to public health as a potential career choice (see the later discussion on educating and training the microbial threats workforce).
Managing and controlling epidemic diseases requires deep engagement and coordination on the part of both the public health and the medical communities. Recent experiences with both anthrax (intentionally caused) and West Nile virus (naturally occurring) reinforced the importance of links between educated, alert medical providers and a responsive public health system. Rapid recognition of an event requires that health care providers be trained to recognize unusual symptoms of disease that may reflect an emerging health problem, whatever the source. The experience with anthrax and West Nile virus demonstrated the potential difficulties involved in distinguishing naturally occurring from intentionally caused disease outbreaks early on. In fact, in some instances, the source of an outbreak and whether the infectious agent was intentionally introduced may never be known.
A strengthened relationship between public health and clinical medicine is also vitally important to the development of plans for a surge of patients in the nation’s health care system, whose facilities routinely operate at or near capacity. The need to have such plans in place is just as important for preparedness for a severe flu season as for preparedness for a bioterrorist attack. To control most infectious disease epidemics, public health agencies must be closely linked with those who can deliver medical care to persons in need and provide prophylactic treatment or vaccines that may be required for disease control.
Looking to the future, the nation’s public health system will continue to be challenged to combat both routine and unexpected outbreaks of disease. In fact, we may anticipate discoveries of an increasing array of previously unknown infectious disease threats, including newly bioengineered microbial agents for which we may have no effective control or treatment strategies. A successful response to these new threats will require that the nation make a renewed and much-needed commitment to public health and address the threat of bioterrorism in the broader context of infectious disease. We must recognize and act on the understanding that public health is an essential aspect of public safety and a critical pillar in our national security framework. Our programs and policies must reflect this recognition; adequate public health and infectious disease expertise must be present at the table when critical decisions are made; and public health professionals must be part of our national security team.
U.S. federal, state, and local governments should direct the appropriate resources to rebuild and sustain the public health capacity necessary to respond to microbial threats to health, both naturally occurring and intentional. The public health capacity in the United States must be sufficient to respond quickly to emerging microbial threats and monitor infectious disease trends. Prevention and control measures in response to microbial threats must be expanded at the local, state, and national levels and be executed by an adequately trained and competent workforce. Examples of such measures include surveillance (medical, veterinary, and entomological); laboratory facilities and capacity; epidemiological, statistical, and communication skills; and systems to ensure the rapid utility and sharing of information.
IMPROVING DOMESTIC SURVEILLANCE THROUGH BETTER DISEASE REPORTING
Surveillance is the foundation for infectious disease prevention and control. Surveillance provides information crucial to monitoring the health of the public, identifying public health problems and priorities, taking pub-
lic health actions to prevent further illness, and evaluating the effectiveness of these actions. Surveillance of infectious diseases is dependent largely on timely and accurate diagnosis by health care providers and prompt reporting of disease to relevant public health authorities. Open lines of communication and good working relationships between health care providers and public health authorities are essential to a robust system of surveillance and effective implementation of disease investigation and response activities.
No single surveillance system captures all the information required to monitor the health of the public. Such a capability is impossible given the existence of multiple data sources, differing information requirements, multiple distinct users, and different partners with which CDC collaborates to obtain data for specific programs areas (CDC, 2000f). To better manage and enhance the large number of current surveillance systems and allow the public health community to respond more quickly to public health threats, CDC has developed the National Electronic Disease Surveillance System (NEDSS). NEDSS is an initiative designed to promote the use of data and information system standards to advance the development of efficient, integrated, and interoperable surveillance systems at the federal, state, and local levels (CDC, 2002t). The vision of NEDSS is to have integrated surveillance systems that can transfer appropriate public health, laboratory, and clinical data efficiently and securely over the Internet. Gathering and analyzing information quickly and accurately will help improve the nation’s ability to identify and track emerging infectious diseases and potential bioterrorism attacks, as well as to investigate outbreaks and monitor disease trends.
The long-term vision for NEDSS is that of complementary electronic information systems that automatically gather health data from a variety of sources on a real-time basis; facilitate monitoring of the health of communities; assist in the ongoing analysis of trends and detection of emerging public health problems; and provide information for setting public health policy. CDC is focusing on the development, testing, and implementation of standards to serve as the framework that will support more complete and comprehensive integration of systems in the future. While the various systems developed by CDC and state and local health departments will remain distinct from one another, the use of standards will ensure that surveillance data can readily be shared, that users familiar with one system can easily use another, and that software can be shared across programs.
Largely in response to recommendations in Emerging Infections: Microbial Threats to Health in the United States (IOM, 1992), CDC initiated several new surveillance programs, including the Emerging Infections Program for population-based surveillance and research (see Box 4-4). In addition, several sentinel surveillance systems were established for various infectious diseases or conditions in emergency departments, in travelers’ clinics,
and through a network of infectious disease clinicians (see Table 4-1). These systems have been extremely useful in improving surveillance, particularly for invasive bacterial diseases, including foodborne illnesses, and in several other specific areas of infectious disease control.
Notification of public health officials of the occurrence of an unusual illness has been, and will continue to be, vital to the detection of emerging microbial threats. Health care providers are an essential component of surveillance programs. Astute clinicians are the first line of defense for the identification of most emerging microbial threats. Health care providers are critical in recognizing unusual presentations of illness or clusters of unusual illnesses, and report their observations to local or state health officials. Reports are likely to be generated because of close clustering, unusual morbidity and mortality, novel clinical features, or the availability of medical expertise. Recent diseases identified as initial clusters of unusual illness include Legionnaires’ disease, Lyme disease, hantavirus pulmonary syndrome, and West Nile encephalitis in North America. At the same time, other medical personnel, such as infection control professionals, could play an enhanced role in detecting outbreaks and increases in emergency department visits or hospital admissions for diagnoses that may be of public health importance; some played this role effectively during the anthrax events of 2001. Infection control professionals are well situated to detect unusual disease clusters throughout a hospital, including the emergency department and the intensive care unit; they have a close collaboration with both the infectious disease specialists and the microbiologists within a health system. This potentially critical link with local and state public health agencies must be supported within the health care environment. Unfortunately, the ranks of these professionals are thinning.
CDC monitors disease burden in the United States through the National Notifiable Diseases Surveillance System (NNDSS), implemented in 1961. The list of nationally notifiable diseases is maintained and revised as needed by the Council of State and Territorial Epidemiologists (CSTE) in collaboration with CDC (see Box 4-5). Regulatory authority for disease surveillance in the United States is provided through state legislation; health officials in every state report voluntarily to CDC. All states generally report the internationally quarantinable diseases (yellow fever, cholera, and plague) in compliance with WHO’s International Health Regulations (CDC, 2002t). Most states include within their disease reporting requirements a provision for the reporting of any unusual presentation of illness or death (in an individual or cluster of individuals), especially those for which a cause cannot be identified. In addition, states can elect to add other diseases to their list that may be relevant for their geographic area. Some states include conditions other than infectious diseases. Current data on nationally re-
|
BOX 4-4 Emerging Infections Program In 1994, CDC developed a new proposal to improve and strengthen infectious disease surveillance. This initiative, outlined and expanded in CDC’s 1998 publication Preventing Emerging Infectious Diseases: A Strategy for the 21st Century, has been implemented in collaboration with many public health partners (CDC, 1998c). CDC implemented the Emerging Infections Program (EIP) as a result of this strategic initiative. The EIP is a collaboration among CDC, state health departments, and other public health partners for the purpose of conducting population-based surveillance and research on infectious diseases. The EIP network comprises nine EIP sites: California (San Francisco Bay area), Colorado, Connecticut, Georgia, Maryland, Minnesota, New York, Oregon, and Tennessee. These sites conduct population-based surveillance and research that go beyond the routine functions of local health departments to address important issues in infectious diseases and public health. The primary objective of the EIP is to act as a national resource for the surveillance, prevention, and control of emerging infectious diseases (CDC, 2002u). The EIP network is able to achieve this objective by addressing important issues in infectious diseases, participating in emergency responses to outbreaks, providing public health agencies with new information, recognizing the importance of training in all EIP activities, and making prevention of infectious diseases a priority (Schuchat et al., 2001). The EIP sites have performed investigations of meningococcal and streptococcal disease, and have also established surveillance for unexplained deaths and severe illness in an attempt to identify diseases and infectious agents, known and unknown, that can lead to severe illness or death (CDC, 1998c). Two projects are conducted through the entire EIP network: Active Bacterial Core surveillance (ABCs) and Foodborne Diseases Active Surveillance Network (FoodNet). ABCs is a population-based surveillance system that conducts active surveillance for |
portable diseases are available in CDC’s Morbidity and Mortality Weekly Report.
Efforts to educate physicians and other health care professionals are critical to improving national surveillance through disease reporting. As noted earlier, practicing health care providers have detected and reported many recent emerging threats, including inhalational anthrax in Florida. In addition, it is essential that strong links be established between animal care providers (e.g., veterinarians, wildlife officials) and public health officials to enhance reporting of animal infections of relevance to human health; the emergence of West Nile virus has clearly demonstrated this need.
Many health care providers do not fully understand their role in infectious disease surveillance, including their role as a source of data (IOM, 2000). Health care providers receive little formal education in infectious disease surveillance: few medical or other health science schools include the
|
invasive disease caused by Streptococcus pneumoniae, group A streptococcus, group B streptococcus, Neisseria meningitidis, and Haemophilus influenzae. A population of 17 to 30 million is actively surveyed for the presence of these bacterial pathogens which were the cause of 10,000 deaths in the United States in 1998 (Schuchat et al., 2001). In 1999, the most recent complete year of surveillance, 7,632 cases of invasive disease due to the five pathogens were reported (7,067 isolates collected). Other ABCs accomplishments include a study of the risk of invasive group A streptococcal (GAS) infections among household contacts of index patients, analysis of a population-based case-control study of other risk factors for invasive GAS, and the development of a procedures manual and database for use in a post-licensure efficacy study of a pneumococcal conjugate vaccine. FoodNet is a collaboration among the CDC EIP sites, the U.S. Food and Drug Administration, and the U.S. Department of Agriculture, created in 1996 to conduct population-based, active surveillance for foodborne infections. The primary objectives of FoodNet are to (1) determine the epidemiology of bacterial, parasitic, and viral food-borne diseases; (2) determine the prevalence of foodborne diseases in the United States; and (3) investigate the link between certain foods and the proportion of food-borne disease caused by their ingestion (Yang, 1998). FoodNet conducts surveillance for E. coli O157:H7, Campylobacter, Listeria, Salmonella, Shigella, Yersinia, Vibrio, Cryptosporidium, and Cyclospora. The EIP network has scored several accomplishments. From 1993 to 1998, ABCs detected a decline in the incidence of group B streptococcal disease in newborns in the monitored population. The results of this surveillance provided the basis for guidelines for the prevention of mother-to-child transmission of group B streptococcus through the use of intrapartum antibiotics (Schrag et al., 2000). FoodNet has been successful in monitoring, tracking trends, and defining risk factors for causes of foodborne illnesses, and in estimating the burden of foodborne illnesses in the United States (CDC, 2002u). |
importance of and requirements for reporting diseases of public health significance to public health authorities in their curricula; residency programs seldom address the need for provider participation in public health surveillance; and little, if any, continuing medical education exists on the topic, nor is it widely integrated into board certification exams.
CDC should take the necessary actions to enhance infectious disease reporting by medical health care and veterinary health care providers. Innovative strategies to improve communication between health care providers and public health authorities should be developed by working with other public health agencies (e.g., the Food and Drug Administration [FDA], the Health Resources and Services Administration [HRSA], USDA, the Department of Veterans Affairs [VA], state and local health departments), health
TABLE 4-1 Selected Sentinel Surveillance Systems for Monitoring Infectious Diseases
|
EMERGEncy ID NET |
EMERGEncy ID NET is an interdisciplinary, multicenter, emergency department-based network based at 11 university-affiliated, urban hospital emergency departments with more than 900,000 combined annual patient visits. Research projects include investigation of bloody diarrhea; prevalence of Shiga toxin-producing Escherichia coli; rabies postexposure prophylaxis practices, and nosocomial emergency department M. tuberculosis transmission. |
|
Foodborne Diseases Active Surveillance Network (FoodNet) |
FoodNet is a collaborative project among the CDC, the 9 Emerging Infections Program sites (EIPs), the U.S. Department of Agriculture (USDA), and the U.S. Food and Drug Administration (FDA). FoodNet consists of active surveillance of laboratories, physicians, and the general population for foodborne diseases and related epidemiologic studies designed to help public health officials better understand the epidemiology of foodborne diseases in the United States. |
|
Gonococcal Isolate Surveillance Project (GISP) |
GISP is a collaborative project to monitor antimicrobial resistance in Neisseria gonorrhoeae in the United States. Participants of GISP include the CDC, five regional laboratories, and selected local STD clinics. |
|
National Molecular Subtyping Network for Foodborne Disease Surveillance (PulseNet) |
PulseNet is a national network of local public health laboratories that performs DNA “fingerprinting” on pathogens that may be foodborne. The network permits rapid comparison of these “fingerprint” patterns through an electronic database at the CDC. |
|
National Nosocomial Infections Surveillance (NNIS) System |
The NNIS system is conducted by the Hospital Infections Program to collect high-quality nosocomial infection surveillance data that can be aggregated into a national database. NNIS is a cooperative effort between the CDC and acute care general hospitals that volunteer to participate in this surveillance system. |
|
Unexplained Deaths and Critical Illnesses Surveillance System |
Active population-based surveillance through coroners and medical examiners is conducted in 4 Emerging Infections Program sites (EIPs) with a total population of 7.7 million 1- to 49-year-olds. Surveillance is passive for clusters of unexplained deaths and illnesses. |
|
United States Influenza Sentinel Physicians Surveillance Network |
Approximately 260 physicians around the country report each week to the CDC the total number of patients seen and the number of those patients with influenza-like illness by age group. |
|
SOURCE: CDC. |
|
|
BOX 4-5 Nationally Notifiable Infectious Diseases in the United States, 2003 Acquired immunodeficiency syndrome (AIDS) Anthrax Botulism Botulism, foodborne Botulism, infant Botulism, other (wound and unspecified) Brucellosis Chancroid Chlamydia trachomatis, genital infections Cholera Coccidioidomycosis Cryptosporidiosis Cyclosporiasis Diphtheria Ehrlichiosis Ehrlichiosis, human granulocytic Ehrlichiosis, human monocytic Ehrlichiosis, human, other or unspecified agent Encephalitis/meningitis, Arboviral Encephalitis/meningitis, California serogroup viral Encephalitis/meningitis, eastern equine Encephalitis/meningitis, Powassan Encephalitis/meningitis, St. Louis Encephalitis/meningitis, western equine Encephalitis/meningitis, West Nile Enterohemorrhagic Escherichia coli Enterohemorrhagic Escherichia coli, O157:H7 Enterohemorrhagic Escherichia coli, shiga toxin positive, serogroup non-O157 Enterohemorrhagic Escherichia coli shiga toxin+ (not serogrouped) Giardiasis Gonorrhea Haemophilus influenzae, invasive disease Hansen disease (leprosy) Hantavirus pulmonary syndrome Hemolytic uremic syndrome, post-diarrheal Hepatitis, viral, acute Hepatitis A, acute Hepatitis B, acute Hepatitis B virus, perinatal infection Hepatitis C, acute Hepatitis, viral, chronic Chronic hepatitis B Hepatitis C virus infection (past or present) HIV infection HIV infection, adult(≥13 years) HIV infection, pediatric (<13 years) Legionellosis Listeriosis Lyme disease Malaria Measles Meningococcal disease Mumps Pertussis Plague Poliomyelitis, paralytic Psittacosis Q fever Rabies Rabies, animal Rabies, human Rocky Mountain spotted fever Rubella Rubella, congenital syndrome Salmonellosis Shigellosis Streptococcal disease, invasive, Group A Streptococcal toxic-shock syndrome Streptococcus pneumoniae, drug resistant, invasive disease Streptococcus pneumoniae, invasive in children <5 years Syphilis Syphilis, primary Syphilis, secondary Syphilis, latent Syphilis, early latent Syphilis, late latent Syphilis, latent unknown duration Neurosyphilis Syphilis, late, non-neurological Syphilis, congenital Syphilitic stillbirth Tetanus Toxic-shock syndrome Trichinosis Tuberculosis Tularemia Typhoid fever Varicella (morbidity) Varicella (deaths only) Yellow fever SOURCE: CDC. |
sciences educational programs, and professional medical organizations (e.g., the American Medical Association, the American Society for Microbiology, the American Nurses Association, the American Veterinary Medical Association, the Association for Professionals in Infection Control and Epidemiology, the Association of Teachers of Preventive Medicine).
In addition to improving disease reporting by health care providers, efforts are needed to expand disease reporting from clinical laboratories. Automated laboratory reporting of notifiable infectious diseases from private clinical laboratories has been shown to improve dramatically the timeliness and quality of disease reporting for many notifiable infectious diseases, such as foodborne bacterial diseases (Effler et al., 1999; Overhage et al., 1997; Panackal et al., 2002). CDC has developed the standards and security measures needed for automated reporting of notifiable infectious diseases, having achieved consensus on critical issues. As of June 2002, however, relatively few states had implemented automated reporting of infectious diseases from major clinical laboratories using these standards.
CDC should expeditiously implement automated electronic laboratory reporting of notifiable infectious diseases from all relevant major clinical laboratories (e.g., microbiology, pathology) to their respective state health departments as part of a national electronic infectious disease reporting system. The inclusion of antimicrobial resistance patterns of pathogens in the application of automated electronic laboratory reporting would assist in the surveillance and control of antimicrobial resistance.
EXPLORING INNOVATIVE SYSTEMS OF SURVEILLANCE
Advances in information technology that allow automated reporting from laboratories may also be helpful in the development of other new systems, such as those incorporating remote sensing, as well as automated systems of syndromic surveillance.1 In some sites, data describing patient illnesses before definitive diagnosis (e.g., fever, cough) are being transmitted electronically by health care providers and monitored centrally by those responsible for disease surveillance.
Syndromic surveillance is not new, although advances in information technology may improve its potential usefulness (see Appendix B for a more detailed discussion of syndromic surveillance). Historically, syndromic sur
veillance has proven quite useful in limited circumstances. For example, cruise ships that dock in U.S. ports are required to notify the U.S. Public Health Service when the number of visits to the ship’s clinic reaches a threshold; public health investigation of and response to these threshold events has led to a marked reduction in the frequency of bacterial food- and waterborne illnesses among passengers in the past two decades.
It has been argued that surveillance of presenting symptoms of illness in emergency departments or clinics could be used to detect a mass release of a biological agent earlier than would be possible through more traditional surveillance. The influx of resources for enhancing recognition of bioterrorism events has resulted in numerous attempts to automate potentially relevant data and provide these data to a central entity responsible for epidemic detection. However, reporting of data from many clinics and hospitals is currently difficult to accomplish in real time in much of the civilian sector because of the number of incompatible systems in operation.
One of the more advanced and efficient systems of encounter-level data is the Electronic Surveillance for Early Notification of Community-based Epidemics (ESSENCE), developed by DOD, which provides syndromic surveillance in military treatment facilities using a grouping of International Classification of Disease (ICD) codes. Evaluation of the usefulness of this system for the timely detection of epidemics is ongoing (see Box 4-6).
In 2001, the Department of Health and Mental Hygiene in New York City established a surveillance system for detection of bioterrorism events. The technical setup and daily statistical analyses, as well as any disease investigations, if needed, occur at the Department of Health. The system was labor-intensive in its first few weeks, in the immediate aftermath of September 11, 2001, when staff were placed at every participating hospital to ensure that the medical providers completed daily forms and to conduct real-time data entry. Beginning in October 2001, the system was transitioned to a completely electronic system for data transfer so that staff on site were no longer required, and existing data systems were used so as not to require additional work by the hospital staff. The system was the first to detect the start of widespread influenza activity in the New York City, as well as the first indicator of norovirus activity in the area—well before the increased reports of institutional and cruise ship outbreaks. As one of the major purposes of this system is to ensure rapid detection of disease syndromes that might indicate the prodrome of a bioterrorist event, the analysts err on the side of increased sensitivity and are required to investigate several false alarms in the process.
Another approach is reflected in the Rapid Syndrome Validation System, developed by Sandia National Laboratories. This system enables health care providers in emergency departments to enter clinical and demographic
|
BOX 4-6 The Electronic Surveillance System for Early Notification of Community-Based Epidemics The Electronic Surveillance System for Early Notification of Community-Based Epidemics (ESSENCE) is a syndromic surveillance system for the detection of infectious disease outbreaks at military treatment facilities worldwide. ESSENCE was initially developed by DOD’s Global Emerging Infections System (GEIS) to serve 104 primary and emergency care clinics in the National Capital Region. Since the September 11, 2001, terrorist attack, it has been expanded to include the entire Military Health System (121 Army, 110 Navy, 80 Air Force, and 2 Coast Guard installations worldwide). ESSENCE uses data from the Ambulatory Data System, which contains diagnoses of DOD health care beneficiaries and is located at all military treatment facilities. The data are captured at the military treatment facilities and are then sent to a centralized server in Denver, which feeds information directly into the secure server located at the Walter Reed Army Institute of Research, the Central Hub of GEIS. Data are captured daily; however, there is a lag time in the transfer of data of 1 to 4 days from the time of the initial patient visit. Data are classified according to seven syndrome groups that have been identified based on International Classification of Diseases, 9th Revision (ICD-9) codes. These groups are as follows:
|
data on patients with infectious disease syndromes and to report directly to the health department (Sandia National Laboratories, 2002).
The resource requirements for automated reporting of syndromic data from most hospitals, clinics, or emergency departments are currently high, but these costs may be reduced over time with standardization of software. The resources required may also be reduced if the surveillance system uses data that are already being collected, and data transfer can occur automatically without requiring staff resources on either end. The primary resource requirements for such systems are analytic staff to evaluate the data and disease investigators to respond to any potential outbreaks detected. The most critical need for these systems is to ensure effective links to local and state public health agencies that would need to respond in the event of an alarm. The central role of public health agencies in the effectiveness of any such system is crucial, since they have both the authority and the expertise for investigations needed to respond should an attack be detected.
The use of other existing health databases, including 911 calls and pharmacy records, is also being explored. A clinical validation study of the
Graphs are created and historical data are used in baseline comparisons of the data to monitor the defined syndromes for trends that could signify an event due to an emerging infectious disease. A geographic information system (GIS) is used to perform data visualization, as well as to determine the geographic component of an outbreak. The central hub of ESSENCE can provide data in terms of syndrome, age, gender, clinic, location, and health care provider. The graphs are made available daily to public health officials on a secure website so they can review and analyze any potential emerging infection outbreak scenarios. GEIS was recently awarded a 4-year, $12 million grant from the Defense Advanced Research Projects Agency (DARPA) for the creation of ESSENCE II. This effort involves collaboration with the Johns Hopkins School of Public Health, the George Washington University School of Public Health, Carnegie Mellon University, IBM, and Cycorp. These partners will work together to create a surveillance system for detecting a potential biological attack on the U.S. military. Plans are for the system to actively obtain data on the following: health maintenance organization (HMO) billing, over-the-counter drug sales, school absenteeism, and military pharmacy, laboratory, and radiology orders. ESSENCE II will track these data continually to detect abnormalities, and will transmit alerts and notifications when an abnormal situation appears. In addition to the partners listed above, a team of epidemiologists and computer researchers will be involved in the development of ESSENCE II. SOURCE: Department of Defense–Global Emerging Infections System, 2002. |
Emergency Management Services 911 syndromic surveillance system showed sensitivity in detecting illness suggestive of influenza, although the system had poor specificity (Greenko et al., 2002). Surveillance of antimicrobial and over-the-counter drugs is being explored for its usefulness in early detection of an epidemic in a community.
Some evidence indicates that these systems may be used to detect epidemics of influenza and some gastrointestinal illnesses earlier than would otherwise be the case (J. Duchin, Public Health, Seattle and King county, personal communication, January 30, 2002; J. Pavlin, GEIS, personal communication, January 28, 2002). Data from these systems have also been used in tabletop exercises for a bioterrorism event and have filled a critical need for rapid and continuous assessment of health care utilization for particular problems; this is likely to be the case as well for naturally occurring epidemics, such as influenza pandemics and other crises.
The potential usefulness of these systems for early detection of individual class A biological agent infections is unclear. The assumptions on which these systems are based require closer examination with regard to
their usefulness in the early detection of bioterrorism. A key issue for syndromic surveillance systems is determining statistical thresholds for response that are sufficiently sensitive and specific to detect severe illnesses (such as anthrax) earlier than would be possible through traditional methods without overtaxing the public health system with false alarms. This determination is difficult for a rare event or an event causing severe illness or death. Numerous false signals from these systems have to date resulted in the diversion of limited public health resources, and some areas are raising the threshold for which they may investigate a “signal” at the expense of loss of sensitivity and timeliness. Geospatial coding may be useful in this regard.
In summary, syndromic surveillance is likely to be increasingly helpful in the detection and monitoring of epidemics, as well as the evaluation of health care utilization for infectious diseases. At the same time, the potential exists for syndromic surveillance to draw resources away from other systems that have proven to be robust in the detection of microbial threats. Although novel approaches utilizing nonspecific data may prove useful, particularly for conditions for which empirical diagnosis and treatment represent the standard of care, a balance should be sought between strengthening what is known to be helpful (e.g., diagnosis of patients with infectious illness, strengthening of the liaison between clinical care providers and health departments) and the exploration and evaluation of new approaches.
Research on innovative systems of surveillance that capitalize on advances in information technology should be supported. Before widespread implementation, these systems should be carefully evaluated for their usefulness in detection of infectious disease epidemics, including their potential for detection of the major biothreat agents, their ability to monitor the spread of epidemics, and their cost-effectiveness. Research on syndromic surveillance systems should continue to assess such factors as the capacity to transmit existing data electronically, to standardize chief complaint or other coded data, and to explore the usefulness of geospatial coding; CDC should provide leadership in such evaluations. In addition, promising approaches will need to be coordinated nationally so that data can be shared and analyzed across jurisdictions.
DEVELOPING AND USING DIAGNOSTICS
Etiologic diagnosis—identifying the microbial cause of an infectious disease—is the cornerstone of effective disease control and prevention efforts, including surveillance. The first recognized case of inhalational anthrax in the 2001 bioterrorist attack was diagnosed by examination of a
Gram stain of the patient’s cerebrospinal fluid. Yet for various reasons, including restrictions imposed by managed care, laboratory regulations (e.g., CLIA2), and the increasing use of empirical therapy, etiologic diagnosis has declined significantly over the past decade; as a result, the quality of clinical care, surveillance, and training has been compromised. In addition, a dangerous consequence of decreased etiologic diagnosis has been an increase in the inappropriate use of broad-spectrum antibiotics and the emergence of antimicrobial resistance. A specific diagnosis, including results of antimicrobial resistance testing, allows for more appropriate treatment, avoids the inappropriate use of antibiotics, and also informs public health actions.
The dramatic rise in the number of unexplained causes of community acquired pneumonia in adults is testament to the current crisis in etiologic diagnosis. For example, prospective studies evaluating the causes of community acquired pneumonia in adults have failed to identify the cause in 40 to 60 percent of cases (Bartlett et al., 2000). In virtually all studies of community acquired pneumonia when diagnoses have been made, Streptococcus pneumoniae has accounted for two-thirds of all bacteremic cases. It has been suggested that sputum cultures in the large percentage of undiagnosed cases have failed to yield S. pneumoniae mainly because of inadequate specimen collection; delays in seeding cultures; use of less sensitive techniques for recovering the organism (Bartlett et al., 1998); and barriers imposed by meeting requirements of individual health care plans, such as the requirement to send specimens to a specified laboratory regardless of geographic location or availability of tests of highest quality. Permitting these unacceptable practices to persist is causing problems for the U.S. capacity to respond to microbial threats.
Current diagnostic approaches for collection of environmental samples or clinical specimens involve primarily hands-on, ad hoc procedures using a variety of devices and instruments. In the clinical arena, these procedures result in specimens of variable quantity and quality. The process is relatively laborious and nonstandardized. A few common methods for specimen disruption are applied to each specimen type without particular regard for the possible diversity of pathogens and their requirements, nor are special precautions taken in a uniform manner to minimize degradation of pathogens. Recovery of fastidious microbes from clinical specimens has almost certainly suffered as a result of the time demands and resource
constraints typical of today’s clinical workplaces, as well as recent laboratory downsizing (Bartlett et al., 1998). Technology developments, such as the use of microsonicators for efficient rapid microbial lysis (Belgrader et al., 1999a; Taylor et al., 2001b), are likely to improve the current situation. Problems with lack of standardization and nonuniformity of procedures are even more prevalent in environmental microbial detection; air and water are among the environmental specimen types that are collected and processed most frequently.
Disincentives to conduct careful laboratory diagnoses include the costs of additional tests, particularly comprehensive ones, and claims that the test results return too late to be of use in clinical management. However, a precise diagnosis obtained from skilled laboratory analysis can be invaluable and cost-effective, even if specific therapy is not available for the condition diagnosed, and is important for public health surveillance. This is particularly true for acute respiratory infections such as bronchitis and pharyngitis, illnesses with a low percentage of bacterial infection that are nonetheless often treated with antibiotics (Gonzales et al., 2001). Treating such illnesses with antibiotics is costly and represents the type of inappropriate antibiotic use that fosters drug resistance and exposes patients to the risks associated with antibacterial drug consumption (see the discussion later in this chapter). Progress has been made in etiologic diagnosis of pediatric patients in some communities. In Finland, for example, etiologic diagnosis is made in 90 percent of cases of severe respiratory disease in children (Nohynek et al., 1991). This success rate is attributed to an emphasis on etiologic diagnosis, coupled with a situation in which highly skilled microbiologists work within the relatively compact health care system that characterizes the Scandinavian medical community. In the United States, however, the underuse of etiologic diagnosis and the overuse of antibiotics remain significant health care problems.
Clinical microbiology continues to rely heavily upon cultivation-based methods and should continue to do so. Cultivation methods have improved considerably over the past several decades, with advances being made in the scope and diversity of media components, control of environmental conditions, use of heterologous host cells, and use of growth-promoting factors (Mukamolova et al., 1998). A number of recently recognized and newly described microbial pathogens, including spirochetes, rickettsia, actinomycetes, and a variety of viruses, have been cultivated successfully in the laboratory. Cultivation is the most widely used approach in laboratories, clinics, and health care facilities throughout the world, especially in developing countries, and hence is currently the most common microbial detection platform for international surveillance. It is important to note that cultivation, despite being slow, limited in sensitivity for some clinically relevant microbes, and the least technologically sophisticated approach,
nevertheless provides the most ready assessment of complex microbial phenotypes (behaviors), such as drug resistance. Cultivation allows for the “fingerprinting” of an organism, important for the understanding of microbes and how they spread.
Diagnostic Pathology
The ability of diagnostic pathology to help in recognizing and understanding diseases—both old and emerging, both in humans and in animals—is often overlooked. Basic anatomic pathology involves analyzing tissues from dead specimens, making observations, interpreting the findings, and following up with histopathology studies of samples under a microscope. In recent years, the advent of molecular pathology has heightened the power of diagnostic pathology. With such tools as immunohistochemistry, in situ hybridization, and polymerase chain reaction (PCR) assays, pathologists can now identify etiology more rapidly than ever before and, in many cases, where doing so would previously have been impossible (see Appendix C for a more detailed discussion of advanced diagnostic methods). Diagnostic immunohistochemistry was key to confirming both the West Nile virus and the anthrax outbreaks. However, current disease surveillance systems, for human diseases and zoonoses alike, fail to make adequate use of diagnostic pathology. No infectious disease pathology program is currently supported, in contrast with other specialty pathology training programs, such as those in cardiovascular disease. In the medical community today, many unexplained deaths evade rigorous pathological studies. Overall, the number of autopsies being performed has declined, and the system is highly capricious as to the determination of when an autopsy is necessary. In addition, there is overarching concern about training and skill levels with respect to ensuring that the latest technologies in diagnostic pathology are being applied (see Box 4-7).
The availability of archived biological samples can facilitate the understanding of new pathogens and the response to outbreaks. More countries should be encouraged to maintain (publicly aknowledged) registries of blood and tissue specimens, whether from zoonotic or human events, to make it feasible to search for specific susceptibility factors. For example, in 1998 an outbreak of viral encephalitis occurred in Malaysia. Analysis of archival collections of 1995 serum samples from pigs showed that some of those samples contained antibodies to the previously unrecognized Nipah virus. This finding allowed investigators to conclude that the virus had been circulating in the population for some time. A large and invaluable resource of archived samples is DOD’s Triservice Serum Repository, begun in 1985, which contains 25 million specimens collected from military personnel. This resource serves many research needs; it provided important informa-
|
BOX 4-7 The Value of Autopsies The records of medical examiners and coroners provide vital information about patterns and trends of mortality in the United States and are excellent sources of data for public health studies and surveillance. For example, during the outbreak of West Nile encephalitis in New York, autopsies were performed under the jurisdiction of New York City’s Office of the Chief Medical Examiner because of the obvious public health implications (Shieh et al., 2000). Other notable cases in which autopsies contributed to the discovery or increased understanding of emerging infectious diseases include investigations of hantavirus pulmonary syndrome, Ebola hemorrhagic fever, leptospirosis associated with pulmonary hemorrhage, new variant Creutzfeldt-Jakob disease, and Nipah virus encephalitis. Autopsy rates are decreasing in the United States (Shieh et al., 2000; Sinard, 2001). National statistics reveal that the performance of autopsies declined from 41 percent of hospital deaths in 1961 to 5 to 10 percent in the mid-1990s (CDC, 2001n; Hasson and Schneiderman, 1995; Burton and Nemetz, 2000). A survey of 244 hospitals conducted by the College of American Pathologists in 1994 showed that half of U.S. hospitals had autopsy rates at or below 8.5 percent, and three-quarters had autopsy rates below 13.5 percent. Accreditation programs in internal medicine recommend maintaining an autopsy rate of at least 15 percent (CDC, 2001n). Autopsy rates have fallen for a variety of reasons. First is a lack of insurance reimbursement, so that hospital financial managers and pathology departments must absorb the autopsy costs. Second is lack of incentive. Managed care has resulted in fewer pathologists who have to work longer and harder, imposing workforce burdens with little incentive to take on the added workload imposed by increasing the rate of autopsies. Third is a decreased emphasis on pathology in medical schools. Many medical students graduate with no training in autopsy procedure or little opportunity to view an autopsy in progress. Fourth is the misconception that other diagnostic modalities, such as computed tomography and magnetic resonance imaging, have replaced the need for autopsies. And finally, some pathologists have become concerned about the increased risk of occupational exposure to potentially fatal pathogens (Hanzlick and Baker, 1998). |
tion on hantavirus from samples that had been obtained from military recruits from the southwestern United States.
Microbiological Diagnosis and Development of New Diagnostic Tools
At the same time that traditional, available diagnostic tools are not being utilized, newer, improved etiologic diagnostic tools are needed. The capability for etiologic diagnosis could be significantly improved if inexpensive, rapid, sensitive, and specific tests were available to differentiate not only between viruses and bacteria, but also among different types of viruses and bacteria. The sensitivity of detection methods for cultivation-
amenable microorganisms is currently suboptimal. When traditional diagnostic methods are rigorously applied to syndromes of suspected infectious etiology, such as pneumonia, encephalitis, lymphocyte-predominant meningitis, pericarditis, acute diarrhea, and sepsis, only a minority of cases can be explained microbiologically. In addition, a long list of chronic inflammatory diseases with features of infection remain poorly understood and inadequately explained from a microbiological perspective.
Newer technologies can succeed where methods for pathogen identification through serology or cultivation have failed in the past because of the absence of specific reagents or fastidious requirements for agent replication. The newly available technologies include methods based on the analysis of microbial nucleic acid sequences (e.g., DNA microarrays, PCR), analysis of microbial protein sequences (e.g., mass spectrophotometry), immunological systems for microbe detection (e.g., expression libraries), and host response profiling. Over the past decade, the use of molecular pathogen discovery methods has resulted in the identification of several novel agents, including Borna disease virus, hepatitis C virus, Sin Nombre virus, HHV-6, HHV-8, Bartonella henselae, and Tropheryma whippelii. The advent of molecular pathology has heightened the power of diagnostic pathology. Despite these achievements, however, sensitive and specific rapid diagnostic tools that can be used to diagnose common diseases in the office, clinic, and emergency room simply are not being used or are unavailable.
For example, although many different microbe-specific PCR assays have been described, only a small proportion has actually entered into routine clinical practice. Examples include assays for N. gonorrheae, C. trachomatis, herpes simplex virus, and HIV. A modest number of recent studies have confirmed that the use of these molecular diagnostic tests can reduce patient-care costs and favorably impact patient management (Dumler and Valsamakis, 1999; Ramers et al., 2000). In particular, the development of rapid, real-time (semiquantitative) PCR with point-of-care microbial detection within 30 minutes (Belgrader et al., 1998, 1999b) could potentially alter the use of antibiotics on a widespread scale and reduce antibiotic resistance (Bergeron and Ouellette, 1998). Some of the factors that may have limited more widespread use of these theoretically appealing molecular approaches include difficulty in obtaining specimens and transporting them to the laboratory, a paucity of studies that address clinical validation, and the need for specialized expertise.
CDC and NIH should work with FDA, other government agencies (e.g., DOD, USDA, the national laboratories), and industry on the development, assessment, and validation of rapid, inexpensive and cost-effective, sensitive, and specific etiologic diagnostic tests for microbial threats of public health importance.
Strategies for developing and deploying inexpensive, sensitive, and specific rapid diagnostics will need to be developed for several different scenarios, including ambulatory and bedside, as well as use in developing countries. Ambulatory diagnostics should target primarily childhood respiratory diseases; these tests need to be as inexpensive as possible and widely available for use in both offices and clinics to ensure timely results. For hospitalized patients or those with more serious conditions, a single platform for diagnosis of multiple agents is needed. For example, if pneumonia is the issue, a single PCR assay that can detect common bacterial and viral pathogens should be able to provide an etiologic diagnosis and allow specific therapy. Hospitals could adopt a single platform as an interim standard that would allow tests to be developed at multiple sites with some assurance that they could be implemented at most sites. Using the same platform for bioterrorist threats would stimulate the more rapid availability of local testing for bioterrorist agents and would not preclude competing test platforms or research on better approaches. Overseas diagnostics should be rugged, simple, inexpensive, and tailored to diseases and zoonotic agents of local importance.
Public health agencies and professional organizations (e.g., those concerned with patient care, health education, and microbiological issues) should promulgate and publicize guidelines that call for the intensive application of existing diagnostic modalities and new modalities as they are established. Such guidelines should be incorporated into continuing education programs, board examinations, and accreditation practices. Payers for health care should cover diagnostic tests for infectious diseases to increase specific diagnoses and thereby inform both public health and medical care, including monitoring of inappropriate use of antimicrobials.
As diagnostic tests become more sensitive and increasingly capable of detecting the presence of and differentiating among various types of microorganisms, it is imperative that health care practitioners be able to make clinical sense of the results. Again, a renewed emphasis on laboratory and etiologic diagnostic training should be a regular part of medical education. A positive laboratory test, coupled with corresponding clinical signs and symptoms of disease, can assist decision-making regarding appropriate therapy; treatment decisions should not rest purely on diagnostic test results, however, regardless of their sensitivity and specificity. As necessary as improved diagnostic tests are, enthusiasm for them must not override their meaning in the clinical presentation of illness.
EDUCATING AND TRAINING THE MICROBIAL THREAT WORKFORCE
The number of qualified individuals in the workforce required for microbial threat preparedness is dangerously low. For example, in 2001 the need for at least 600 new epidemiologists in public health departments across the United States was identified because of the requirements for bioterrorism preparedness alone. Yet only 1,076 students graduated with a degree in epidemiology in the year 2000 and are potentially seeking employment in government, academia, or private industry, and the largest percentage are trained in chronic disease, not infectious disease epidemiology. According to the National Association of City and County Health Officers, the most needed occupations between 1999 and 2000 were public health nurses, environmental scientists and specialists, epidemiologists, health educators, and administrative staff.
The real-world information and skills needed for confronting microbial threats must be better integrated into the training of all health care professionals to ensure a prompt and effective response to any and all infectious disease threats, whether naturally occurring or maliciously introduced. It is vital that on-the-job training opportunities, especially for students and new health professionals, be further developed to ensure that professionals responsible for infectious disease control are well trained in real-life situations, and to expose health professions students to career paths in infectious disease prevention and control. Academic health centers are the intellectual hub of these types of professional training programs, and as such serve as the ideal recipients for increased investment in microbial threat education and outreach. Training programs are also urgently needed for health care workers who are currently in mid- and upper-level management positions.
Training programs in applied epidemiology are critical for the development of a strong microbial threat workforce. Currently, some applied epidemiology training programs are in place both domestically and internationally. These programs may provide a solid foundation on which present programs can be expanded and future epidemiology training programs modeled. One example of such a program is the Epidemic Intelligence Service (EIS), a 2-year postgraduate program at CDC that currently enrolls about 70 new officers each year. A typical class comprises primarily medical doctors, but also includes nurses, veterinarians, dentists, and doctoral graduates in epidemiology and the social and behavioral sciences. The majority of EIS officers (also known as “disease detectives”) train at CDC headquarters, and the remainder go to either a state or a large local health department. The latter officers are trained in broad, front-line public health experience; officers deal with surveillance using this information to focus
investigations and to implement policy. The officers at CDC headquarters are trained in specialized, disease- or problem-specific areas.
The initial EIS program, which started over 50 years ago, was focused on communicable disease, but over the years has expanded to include officers trained in environmental health, chronic disease, injuries, and maternal and child health. EIS officers provide international outbreak assistance through investigations in all regions of the world. The success of the program has led to the development of similar training offices in 20 countries worldwide.
EIS officers have played a pivotal role in the success of many investigations, including polio eradication in Africa and Asia, hantavirus outbreak in the southwestern United States, West Nile virus outbreak in the northeastern and southeastern United States, Ebola outbreaks in Uganda and Zaire, and bioterrorism preparedness. Still, there is room for improvement. In particular, EIS needs to be expanded to include more officers who spend full- or part-time assignments in infectious disease control programs at state and local public health departments. In addition, it should be recognized that this is a training program. Thus, while EIS officers often provide an invaluable service during their training, their true value lies in their development as a much-needed cadre of professionals for the future.
For more than 20 years, CDC has collaborated with ministries of health around the world to help establish field epidemiology training programs (FETPs). The goal of these programs is to provide service to the sponsoring government or agency while also training public health workers in epidemiology and outbreak investigation. During training, staff members, trainers, and trainees (or fellows) work with ministries of health and national governments to provide and enhance core public health functions, including disease prevention and control, surveillance, and the supplying of information needed to support informed policy and legislation. In most countries, applied training programs also serve as the backbone for the development and implementation of health information and surveillance systems, bulletins that give program managers and decision makers timely information, and supervision and training of other health workers in the health care system.
The FETPs have trained more than 900 international public health leaders in epidemiology and outbreak investigation, and approximately 420 more are currently in training (CDC, 2002r). Many of these programs have contributed to efforts in infectious disease control and prevention. The FETPs would be enhanced if all programs were assured of laboratory support in the diagnosis of infectious diseases, as is the case with the Thailand FETP.
The Association of Public Health Laboratories’ Emerging Infectious Diseases Laboratory Fellowship Program trains qualified bachelor’s and
|
BOX 4-8 Training in Foreign Animal Disease Control Plum Island (located 1 1/2 miles off the northeastern end of Long Island, New York) under the administration of the U.S. Department of Agriculture’s Agricultural Research Service (ARS) and the Animal and Plant Health Inspection Source (APHIS) is one of the principal locations in the United States where infectious foreign animal disease agents are studied. Scientists on Plum Island have the laboratory capability to diagnose more than 35 exotic animal diseases, and they perform thousands of diagnostic tests each year to detect the presence of foreign animal disease agents. The tissue and blood samples that are tested are submitted by veterinarians suspecting an exotic disease in domestic livestock or by animal import centers testing quarantined animals for foreign diseases. Samples are also submitted by animal health professionals in other countries who need help with a diagnosis. An integral part of the laboratory’s mission is training animal health professionals in the recognition of foreign animal diseases. Staff present several courses each year at Plum Island to give veterinarians, scientists, professors, and veterinary students the opportunity to study the clinical signs and pathological changes caused by foreign animal diseases. SOURCE: Animal and Plant Health Inspection Service, 1992. |
master’s candidates in laboratory science so they can understand the role and importance of public health laboratory services, and it trains doctoral candidates (microbiologists in particular) to conduct high-priority infectious disease research in public health laboratories. The program is highly competitive and limited to a small number of trainees each year. This program should be evaluated to assess how well it is meeting the purpose for which it was initiated and to provide recommendations for further expansion and enhancement. Increased training in the recognition and diagnosis of animal diseases that can indirectly affect human health is also needed (see Box 4-8).
CDC, DOD, and NIH should develop new and expand upon current intramural and extramural programs that train health professionals in applied epidemiology and field-based research and training in the United States and abroad. Research and training should combine field and laboratory approaches to infectious disease prevention and control. Federal agencies should develop these programs in close collaboration with academic centers or other potential training sites. Domestic training programs should include an
educational, hands-on experience at state and local public health departments to expose future and current health professionals to new career options, such as public health.
VACCINE DEVELOPMENT AND PRODUCTION
Vaccines are an essential element in the success of modern medical science. Vaccines have played a central role in providing people around the world with longer and better lives. Indeed, it is difficult to exaggerate the impact of vaccination on the health of the world’s population. With the exception of safe water, no other modality, not even antibiotics, has had such a large effect on mortality reduction and population growth (Plotkin and Mortimer, 1994; Szucs, 2000). As we enter the twenty-first century, however, vaccine development and production are dependent on a complex set of issues, including the translation of basic research into the development of effective vaccines, regulatory requirements, liability concerns, market forces, and competing priorities that have led to periodic shortages of routine vaccines, as well as a lack of vaccines for diseases that affect predominantly developing regions of the world. Added to this complex situation is the need for vaccines to protect against microbial agents that could be used in a bioterrorist attack.
Despite these challenges, opportunities for major breakthroughs in vaccine development are more promising than ever before as the front across which the basic sciences are advancing broadens. In the past decade, scientists have made great strides forward in biotechnology, genomics, and understanding of the molecular basis of pathogenesis. Advances in immunology include the development of tetramer technology that permits functional analysis of T cells (Klenerman et al., 2002); understanding of the role of dendritic cells in antigen processing and presentation (Guermonprez et al., 2002), of antigen-receptor signaling (Myung et al., 2000), and of the role of heat-shock proteins in antigen presentations (Srivastava, 2002); and demonstration of the induction of both antibody-mediated and cell-mediated immune responses to a wide range of infectious disease agents. Success in using virus-like particles may soon lead to the development of vectors that simultaneously carry several different antigens (Moss, 1996).
The development of new delivery systems is likely to continue to be an important area of innovation in vaccines. The improved fundamental knowledge of helper T cells offers opportunities to create new means of encouraging cell-mediated immunity. Mucosal immunity is being studied extensively, as are new types of adjuvants. Opportunities also exist to follow up the success with hepatitis B vaccine by developing other recombinant vaccines (IOM, 2001d). Advances in viral pathogenesis reveal that disease agents such as vaccinia have multiple strategies to circumvent both
antibody- and cell-mediated responses (Moss and Shisler, 2001). New knowledge about the role of cytokines in immune responsiveness and function (Hunter and Reiner, 2000) offers hope for adjuvants for immune responses. Characterization of the antigenic domains on multiple organisms and the ability to express them on a single organism provides the opportunity to vaccinate against multiple disease agents (Henderson and Moss, 1999). And recent advances in reverse genetics for negative-stranded RNA viruses now permit influenza virus to be made to order, permitting rapid development of new vaccine strains (Neumann and Kawaoka, 2001). On all of these fronts, additional, well-funded basic research is likely to have significant social benefits (Kurstak, 1993).
Meeting the Need for Translating Basic Research
Despite the remarkable advances in basic knowledge of immunology and microbiology, proportional translation of these findings to new vaccine development has not occurred. Reflecting on the HIV pandemic and the terrible toll it has taken throughout both the developed and, in particular, the developing world, no clearer need for an effective vaccine exists. After two decades of substantial progress in the field of HIV/AIDS, however, we are still short of the target of having a vaccine to prevent the spread of infection.
An increased commitment to research is necessary to ensure the development of vaccines for the numerous infectious diseases that threaten the world’s human population. Vaccines against acute respiratory infections, diarrheal diseases, malaria, tuberculosis, STDs, and dengue are desperately needed. Vaccines are in the pipeline for many diseases that burden the developing world—Chagas’ disease, onchocerciasis and lymphatic filariasis, leishmaniasis, schistosomiasis, and malaria—but they are in the predevelopment stage and are still far from ready for use in humans (WHO, 1999a). Efficacious vaccines for malaria and dengue, arguably two of the most important arthropod-borne diseases, are not available despite extensive developmental efforts. The problems associated with the development of malaria vaccines have been the subject of many scientific and lay publications, and an efficacious vaccine is not likely to be available soon. Development of a dengue vaccine has been complicated by the issue of the potential for immune enhancement. Four serotypes of dengue virus exist; infection with one serotype typically provides lifelong immunity against reinfection with the homologous serotype, but does not provide immunity against infection with a heterologous serotype. Such secondary heterologous infections are an important risk factor for dengue hemorrhagic fever and shock syndrome (Beaty, 2000). Thus, development of a vaccine strategy that will result in balanced immunity to all four serotypes is a necessity;
however, achieving this has proven to be very difficult, and a number of new strategies are now undergoing investigation.
Responding to Marketplace and Policy Issues
Numerous economic and public health policy issues complicate efforts in vaccine development, production, and deployment. In the last 40 years, few pharmaceutical manufacturers have considered vaccines an attractive business opportunity because of the low return on investment and the exposure to legal liability (Rappuoli et al., 2002). Companies perceive little market incentive to develop vaccines for diseases that occur sporadically and affect the poorest populations—vaccines that may also have little chance of being employed effectively. This has certainly been seen in the area of tropical diseases, especially arthropod- and rodent-borne diseases. Even if inexpensive and efficacious vaccines or drugs are developed, their use by those most in need may be hampered by a lack of public health infrastructure. For example, a safe and effective vaccine for yellow fever has existed for decades. Yet this disease continues to be a pathogen of significant importance in humans in Africa and South America, and its emergence remains a constant threat in urban areas of the tropics and potentially even in Asia, where it would represent a public health catastrophe (Monath, 2001).
The infrastructure for the manufacture of vaccines is steadily deteriorating, and shortages currently exist even in the availability of certain routine vaccines. For example, the difficulty of producing sufficient influenza vaccine in extremely mild interpandemic years, such as 2000 and 2001, signals a potential disaster during a pandemic year. In 2000, supplies of the tetanus and diptheria booster fell short. By the fall of 2001, CDC was reporting shortages of five vaccines, some of which are combination vaccines that protect against eight childhood diseases. Of the eight recommended routine childhood vaccines, five are produced by a single major manufacturer; consequently, if supply is interrupted or a manufacturer ceases production, there may be few or no alternative sources of vaccine (GAO, 2002). The reality is that the infrastructure does not exist to produce even a sufficient supply of currently licensed vaccines, let alone to develop new vaccines against emerging microbial threats. Thus in the event of an outbreak—whether naturally occurring or an act of bioterrorism— the United States will not have the capacity to produce sufficient vaccines to safeguard the population.
The anthrax attacks of 2001 in the United States generated widespread public awareness of infectious agents that had previously not been regarded as worthy of much public attention or substantial federal research funding. Before September 2001, several infectious agents had been identified as
being of concern for possible use in a biological attack. As a result, early efforts were undertaken to explore new avenues for the development of needed vaccines. For example, DOD proposed that government-owned, contractor-operated (GOCO) facilities investigate, develop, and produce vaccines against potential biological agents such as tularemia, plague, and anthrax. Recognizing that neither the government nor private industry alone could develop protection against these threats, DOD initiated a set of new partnerships and contracting strategies to prevent a national catastrophe.
To reap the advantages of scientific advances, private, industrial sources of innovation in vaccines must be strengthened and protected. Here the base is clearly too narrow. In contrast to the vast complex of institutions and individuals involved in basic medical research, only four leading companies worldwide have been responsible for developing new vaccines during the past two decades. It was not mergers and acquisitions that concentrated responsibility for vaccine innovation in the hands of four multinational firms; rather, the economic forces that drove firms out of the industry were the rising costs of innovation, production, and distribution and the shrinking margins allowed by monopsony, or the concentration of buying power in the hands of a relatively small number of public agencies. In the United States, several large companies ceased vaccine production because the total world market for vaccines was so much smaller than that for pharmaceuticals, government purchases allowed only narrow profit margins, and liability continued to be an issue (Galambos and Sewell, 1995; IOM, 2001d).
The current economic situation surrounding vaccine innovation has not improved, and thus will not encourage the entry or reentry of large pharmaceutical companies with extensive resources that could be dedicated to vaccine innovation. If anything, the economic situation has deteriorated further. The increasing costs associated with discovery have become a major barrier. In 1976, it cost an estimated $54 million to develop a new chemical entity (Hansen, 1979). By 1987 that figure had increased to $231 million (DiMasi et al., 1991). Today it costs an estimated $500 million to devise a new drug or vaccine, and that figure includes only the cost of discovery. If one adds the charges for development and clinical evaluation, the total cost increases to $800 million to $1 billion. Moreover, before any of that investment can be recovered, a new vaccine must pass through a complex and lengthy regulatory process. Without lowering standards, the United States has made progress in streamlining the regulatory process, yet cooperative improvements in regulatory systems must continue to be examined and addressed.
Incremental changes along these lines will be important, but improvements with global—not national or even regional—distribution will also be essential. A major development that could have a significant impact on the economic problems outlined above would be international harmonization
of the industry’s technical requirements and regulatory processes. The European Union (EU) has demonstrated how to change on a regional basis; now the move must be made to an international level of standardization and regulation. Cooperative measures short of thoroughgoing harmonization would help. Clinical trials and designs could be standardized, and testing authorities could begin to accept lot release test results without major structural changes on a national basis. By working with increased vigor toward the development of international standards and an international compact that would finally eliminate redundancy in a costly and time-consuming process, it may be possible to improve substantially the economic conditions that have in the past sharply reduced the number of firms contributing to vaccine innovation.
The pricing problem with vaccines is a critical issue. To prevent further defections from the industry and encourage healthy entry into the market, this problem must be addressed head-on. To some extent, a price-controlled market for vaccines exists in the United States, and to a greater extent, price control through government purchasing exists in much of the developed and developing worlds. Half of the vaccines in the United States are purchased by the Vaccines for Children Fund and other government-controlled programs. The creation of these funds was a major step forward. So, too, was the development of vaccine production capabilities in the developing world, where 75 percent of the world’s supply of vaccines is now manufactured. India, China, Pakistan, Egypt, Nigeria, Brazil, and other developing nations produce diphtheria, tetanus, and pertussis (DTP) vaccine for use in their countries. In Europe and Japan, government-controlled markets have long prevailed. Under political pressures to hold down costs, many government agencies place downward pressure on margins, which makes it difficult to sustain the needed level of innovation and production.
Only the developed nations can afford a pricing system that favors innovation. The manner in which vaccines are priced in the U.S. market cannot be applied to markets in Africa, Latin America, and Southeast Asia. The European market is also likely to have a different level of pricing, although that situation should change as the EU makes further progress toward consolidation and growth. With a consolidated market comparable to that of the United States, the EU should, at some point, be able to make a contribution to sustaining innovation similar to that of the United States. This development would have the enormous advantage of alleviating some of the political pressures that are mounting in the United States against differential pricing.
With the necessary political will and effective leadership from the developed world, a new economic environment for vaccine innovation can be created. By developing and deploying new vaccines, it will be possible to build on the tremendous accomplishments of the WHO campaigns; deal
effectively with the massive problems created by rotavirus, malaria, dengue, tuberculosis, and reemerging organisms; and perhaps even cope with the threat of biological terrorism. Without changes in the economics of vaccine innovation, however, significant progress on these fronts cannot be expected.
A State of Crisis
Our nation—and the world—faces a serious crisis with respect to vaccine development, production, and deployment. Concern has increased over the inadequacy of vaccine research and development efforts, periodic shortages of existing vaccines, and the lack of vaccines to prevent diseases that affect persons in developing countries disproportionately. Yet, too little has been done to resolve these issues. The evolving threat of intentional biological attacks makes the need for focused attention and action even more critical.
The challenges associated with vaccine innovation, production, and deployment are many and complex. Solutions will require a novel, coordinated approach among government agencies, academia, and industry. Issues that must be examined and addressed in a more meaningful and systematic fashion include the identification of priorities for research, the determination of effective incentive strategies for developers and manufacturers, liability concerns, and streamlining of the regulatory process. Currently, the federal government is neither addressing all of these challenges at a sufficiently high level nor providing adequate resources. Leadership, empowerment, and accountability are urgently needed at the cabinet level to ensure a comprehensive, integrated vaccine strategy that will address the following critical elements:
The U.S. Secretary of Health and Human Services should ensure the formulation and implementation of a national vaccine strategy for protecting the U.S. population from endemic and emerging microbial threats. Only by focusing leadership, authority, and accountability at the cabinet level can the federal government meet its national responsibility for ensuring an innovative and adequately funded research base for existing and emerging infectious diseases and the development of an ample supply of routinely recommended vaccines. The U.S. Secretary of Health and Human Services should work closely with other relevant federal agencies (e.g., DOD, the Department of Homeland Security, VA), Congress, industry, academia, and the public health community to carry out this responsibility.
The U.S. Secretary of Defense, the U.S. Secretary of Health and Human Services, and the U.S. Secretary of Homeland Security should work closely with industry and academia to ensure the rapid development and deployment of vaccines for naturally occurring or intentionally introduced microbial threats to national security. The federal government should explore innovative mechanisms, such as cooperative agreements between government and industry or consortia of government, industry, and academia, to accelerate these efforts.
The Administrator of USAID, the U.S. Secretary of Health and Human Services, and the U.S. Secretary of State should work in cooperation with public and private partners (e.g., leaders of foundations and other donor agencies, industry, WHO, UNICEF, the Global Alliance for Vaccines and Immunization) to ensure the development and distribution of vaccines for diseases that affect populations in developing countries disproportionately.
NEED FOR NEW ANTIMICROBIAL DRUGS
Antibiotics
Unfortunately, complacency toward infectious diseases in the 1960s, overconfidence in existing antibiotics, and competition from highly profitable opportunities for pharmaceutical development and sale in other fields of medicine resulted in a lag in the production of new classes of antibiotics. This occurred despite significant advances in the fundamental science that has fueled pharmaceutical innovation in many other areas.
As a result of the looming crisis previously discussed, public pressure, and apparent scientific opportunities, many companies intensified their efforts in antibiotic drug discovery in the early 1990s. The complete sequencing of all major bacterial pathogens affecting humans, development of high-throughput screening, combinatorial chemistry, and microarray assays promised a golden age of antibiotic drug discovery. Indeed, at first glance, the situation with respect to antibiotics currently in clinical development looks encouraging. Several new antibiotic variations are in the first three phases of clinical development, with billions of dollars having been invested in their development (see Table 4-2). Not one new class of antibiotics, however, is in development. Rather, these “new” antibiotics belong to existing classes, including macrolides and quinolones, that have been used to treat humans for years. The absence of new classes in the pipeline and the fact that, even for compounds in Phase I, an additional 8 years is required
on average before launch, is alarming when one considers the ever-increasing number of antibiotic-resistant organisms.
It has become apparent that the discovery of new antibiotics is not as easy as was once believed. Despite a plethora of highly validated targets, these targets are not feasible unless a chemical entity can be found that penetrates the cell wall and inhibits growth. In addition, the chemical compound must not be highly toxic and preferably should have good oral bioavailability. These technical hurdles, coupled with competition for resources within pharmaceutical companies from other significant medical needs with larger market opportunities, have led to reduced investment in or, in the case of most companies, elimination of antibiotic drug discovery programs. In fact, today there is only one large pharmaceutical company with a robust antibiotic research program.
Recent discoveries have linked many chronic diseases to infectious etiologies, and it remains to be seen whether this will inspire a renewed interest in antibiotic development for these conditions. Nevertheless, the development of an antibiotic is an expensive and risky process; no guarantee can be made that the antibiotic will remain effective and the investment will be regained before the patent period has ended. Bacterial threats associated with bioterrorism raise additional concerns. In an initial bacterial bioterrorist attack, the level of antibiotic susceptibility will not be known. More than one bacterial agent may be released simultaneously, and bacterial weapons resistant to even the newest antibiotics (e.g., quinolones) can be generated. Thus, new classes of broad-spectrum antibiotics are urgently needed.
Antivirals
Expanding knowledge in the fields of genomics, cell biology, structural biology, and combinatorial chemistry has resulted in the rapid development of some new antivirals. This is exemplified by the dramatic increase in antiviral drugs targeting HIV and, to a much lesser extent, influenza viruses. Yet only a few broad-spectrum antiviral agents are on the market, and the availability of specific antivirals to the majority of RNA and DNA viruses is largely lacking. This situation is exacerbated by the fact that many viruses evolve to circumvent the action of antivirals and the immune response through the development of resistance.
Despite the rapid advances in technology, antivirals for only HIV, hepatitis B and C, herpes viruses, and influenza have been targeted for development. As with vaccines, this situation is due, in large part, to marketing considerations, the absence of a profit margin large enough to repay the costs of development, and the limited potential for sufficient net profit. The problem is well illustrated by the case of influenza. Four companies were
TABLE 4-2 Antibiotics and Antivirals in Development
|
Product Name |
Company |
Indication |
Development Status |
|
Antibiotics |
|||
|
antibiotic (topical) |
Antex Biologics Gaithersburg, MD |
skin and soft tissue infections |
Phase I |
|
Augmentin SR beta lactam antibiotic (modified release formulation) |
GlaxoSmithKline Philadelphia, PA Rsch. Triangle Park, NC |
respiratory tract infections, including penicillin-resistant Streptococcus pneumoniae |
application submitted |
|
Cidecin© daptomycin for injection |
Cubist Pharmaceuticals Lexington, MA |
complicated skin and soft tissue infections, community-acquired pneumonia and certain resistant infections |
Phase III |
|
Cleocin© clindamycin XR |
Pharmacia Peapack, NJ |
acute bacterial sinusitis, dental infections, streptococcal pharyngitis/ tonsillitis, uncomplicated skin and skin structure infections |
Phase I |
|
dalbavancin |
Versicor Fremont, CA |
skin and soft tissue infections |
Phase II |
|
daptomycin |
Cubist Pharmaceuticals Lexington, MA |
complicated skin and soft tissue infections |
Phase III completed |
|
bacteremia in endocarditis, community-acquired pneumonia requiring hospitalization, VRE infections |
Phase III |
||
|
complicated urinary tract infections |
Phase II completed |
||
|
E1010 (carbapanem antibiotic) |
Elsai Teaneck, NJ |
broad spectrum antibiotic |
Phase I |
|
Factive broad-spectrum fluoroquinolone antibiotic (IV formulation) |
GlaxoSmithKline Philadelphia, PA Rsch. Triangle Park, NC |
respiratory tract infections |
Phase III |
|
Factive broad-spectrum fluoroquinolone antibiotic (oral formulation) |
GlaxoSmithKline Philadelphia, PA Rsch. Triangle Park, NC |
respiratory and urinary tract infections |
application submitted |
|
Helicide© bismuth subutrate, tetracycline and metronidozole |
Axcan Pharma Mont St.-Hiliare, Quebec |
eradication of Helicobacter pylori |
application submitted |
|
Iseganan HCl oral solution |
IntraBiotics Pharmaceuticals Mountain View, CA |
prevention of oral mucositis caused by radiotherapy and chemotherapy |
Phase III |
|
prevention of ventilator-associated pneumonia |
Phase II |
||
|
Iseganan HCl solution for inhalation |
IntraBiotics Pharmaceuticals Mountain View, CA |
treatment of respiratory infections in cystic fibrosis patients |
Phase I/II |
|
ISV-401 |
InSite Vision Alameda, CA |
bacterial conjunctival infections |
Phase II |
|
Product Name |
Company |
Indication |
Development Status |
|
Ketek© ketolide |
Aventis Pharmaceuticals Bridgewater, NJ |
first-line therapy for respiratory tract infections in adults |
application submitted |
|
first-line therapy for respiratory tract infections in children |
Phase I/II |
||
|
Levaquin™ levofloxacin |
Janssen Research Foundation Titusville, NJ R.W. Johnson P.R.I. Raritan, NJ |
nosocomial pneumonia |
application submitted |
|
community-acquired pneumonia (short-course therapy), prostatitis |
Phase III |
||
|
Lumenax™ rifaximin |
Salix Pharmaceuticals Raleigh, NC |
traveler’s diarrhea |
application submitted |
|
MB1 594AN |
Micrologix Biotech Vancouver, British Columbia |
Propionibacterium acnes |
Phase II |
|
MB1 594AN |
Micrologix Biotech Vancouver, British Columbia |
Propionibacterium acnes |
Phase II |
|
oritavancin |
InterMune Pharmaceuticals Brisbane, CA |
treatment of gram-positive bacterial infections |
Phase III |
|
ramoplanin |
Genome Therapeutics Waltham, MA |
prevention of bloodstream infections caused by the gram-positive bacteria vancomycin resistant enterococci (VRE) |
Phase III |
|
Tequin™ gatifloxacin |
Bristol-Myers Squibb Princeton, NJ |
prostatitis |
Phase III |
|
otitis media, pediatric meningitis |
Phase II |
||
|
Ligecycline |
Wyeth Pharmaceuticals Philadelphia, PA |
infection caused by antibiotic-resistant bacteria |
Phase III |
|
ABT-492 (quinolone) |
Abbott Laboratories Abbott Park, IL |
respiratory and urinary tract infections |
Phase II |
|
ABT-773 |
Abbott Laboratories Abbott Park, IL |
respiratory infections; ketolide |
Phase III |
|
Antivirals |
|||
|
ACH-126,443 (beta-L-Fd4C) |
Achillion Pharmaceuticals New Haven, CT |
chronic hepatitis B |
Phase II |
|
ACH-126,445 (L-Oddu) |
Achillion Pharmaceuticals New Haven, CT |
Epstein-Barr virus |
Preclinical |
|
adefovir dipivoxil |
Gilead Sciences Foster City, CA |
chronic hepatitis B |
Phase III |
|
adenovirus antiviral |
Barr Laboratories Pomona, NY |
undetermined |
Phase I |
|
amdoxovir (DAPD) |
Triangle Pharmaceuticals Durham, NC |
chronic hepatitis B |
Phase II |
|
clevudine (L-FMAU) |
Triangle Pharmaceuticals Durham, NC |
chronic hepatitis B |
Phase II |
|
Product Name |
Company |
Indication |
Development Status |
|
Coviracil© emtricitabine |
Triangle Pharmaceuticals Durham, NC |
chronic hepatitis B |
Phase II |
|
EHT899 |
Enzo Biochem Farmingdale, NY |
chronic active hepatitis associated with hepatitis B |
Phase II |
|
MIV-210 |
Medivir Huddinge, Sweden |
hepatitis B |
Phase I |
|
MIV-606 |
Medivir Huddinge, Sweden Reliant Pharmaceuticals Liberty Corner, NJ |
herpes zoster |
Phase II |
|
Picovir™ pleconaril |
Aventis Pharmaceuticals Bridgewater, NJ ViroPharma Exton, PA |
viral respiratory infection (adult) |
application submitted disapproved by FDA, terminated |
|
PNU-243672 |
Pharmacia Peapack, NJ |
prevention and treatment of infections caused by herpes viruses in immunocompromised patients |
Phase I |
|
relbivudine (LdT) |
Novirio Pharmaceuticals Cambridge, MA |
treatment of chronic hepatitis B |
Phase II |
|
torcitabine (LdC) |
Novirio Pharmaceuticals Cambridge, MA |
treatment of chronic hepatitisB |
Phase II |
|
Valtrex© valacyclovir |
GlaxoSmithKline Philadelphia, PA Rsch. Triangle Park, NC |
cold sores, HSV suppression in immunocompromised patients, prevention of HSV transmission |
Phase III |
|
XTL-001 in combination with lamivudine |
XTL Biopharmaceuticals Rehovot, Israel New Ipswich, NH |
treatment of chronic hepatitis B |
Phase II |
|
XTL-002 |
XTL Biopharmaceuticals Rehovot, Israel New Ipswich, NH |
treatment of hepatitis C |
Phase I |
|
NOTE: The content of this survey was obtained through government and industry sources based on the latest information. Survey current as of March 1, 2002 (http://www.phrma.org). A list of medicines approved and in development for HIV infection and AIDS can be found in PhRMA’s report, New Medicines in Development for AIDS. SOURCE: Adapted from PhRMA, 2002. |
|||
initially developing antineuraminidase inhibitors. After two consecutive years of mild influenza, two of the companies ceased their development of the drugs, and the fact that only two remaining companies were producing them raised concern regarding the market’s capacity to support continued production. This situation could have catastrophic results during the next emergence of pandemic influenza. Stockpiling of these drugs is critical, and strategies that have been proven effective in maintaining an adequate supply of them must be implemented.
The development of new, improved therapies for influenza and other viruses is essential. However, incentives may be necessary to foster the development of antivirals for those viruses that do not represent large market opportunities but have high morbidity and mortality. The threat of certain viruses being used as agents of biological terrorism emphasizes the increased need for the development of new antivirals, as well as broadspectrum antivirals and immunomodulators, especially for those agents for which there are no vaccines, such as Ebola and Marburg. The possible targets for antiviral development include each step in the replication cycle, from virus attachment to release (see Table 4-3).
Antivirals to Human Immunodeficiency Virus (HIV)
An effective vaccine to prevent HIV infection has not yet been developed. The remarkable advances in the treatment of HIV have resulted largely from advances in antiviral chemotherapy. Combination chemotherapy that suppresses the replication of HIV results in a pronounced reduction in illness and death. Multidrug treatment of patients with indinavir, zidovudine, and lamivudine can reduce the serum levels of HIV to less than 50 copies per ml (see Figure 4-2). Although these treatments reduce replication, however, they do not completely suppress viral replication, and it is probable that a smoldering virus replication is present and difficult to detect. Nonetheless, the immune function of both CD4 and CD8 cells is regenerated, and persistent opportunistic infections are often resolved. It is this restoration of the immune function that has transformed the natural history of AIDS (Richman, 2001). The dramatic restoration of immune function comes at a cost, however—the expense, inconvenience, and toxicity of antiretroviral therapy.
Approximately 10 billion (1010) HIV virus particles are generated daily in an infected host (Perelson et al., 1996). With a mutation rate of about 10–5 nucleotides per replication cycle and no proofreading mechanism for reverse transcription, approximately one mutation is generated for each new genome of 92,000 nucleotides (Mansky and Temin, 1995). Thus, genomes with a mutation in any gene, as well as many with double mutations, could be generated daily. As a result, drug-resistant mutants can develop
TABLE 4-3 Stages of Virus Replication and Possible Targets of Action of Antiviral Agents
|
Stage of Replication |
Classes of Selective Inhibitors |
Viruses |
|
Cell Entry Attachment Penetration |
Soluble receptor decoys, antireceptor antibodies, fusion protein inhibitors Soluble ICAM,B anti ICAM |
GeneralA Retroviruses (HIV)R PicornavirusesR |
|
Uncoating Release of viral genome |
Ion channel blockers, capsid stablizers |
Influenza AR PicornavirusesR |
|
Transcription of viral genome* Transcription of viral Messenger RNA Replication of viral genome |
Inhibitors of viral DNA polymerase, RNA polymerase, reverse transcriptase, helicase, primase, or integrase Ribavirin |
Retroviruses (HIV)R, HerpesvirusesR, Hepatitis BR, Hepatitis CR Respiratory Syncitical Virus (RSV), Arenaviruses (Lassa), Hanta viruses |
|
Translation of viral proteins Regulatory proteins (early) Structural proteins (late) |
Interferons, antisense oligonucleotides, Ribozymes Inhibitors of regulatory proteins |
GeneralA Papillomaviruses GeneralR |
|
Posttranslational modifications Proteolytic cleavage Myristoylation, glycosylation |
Protease inhibitors |
Retroviruses (HIV)R PicornavirusesR |
|
Assembly of virion components |
Interferons, assembly protein inhibitors |
GeneralA |
|
Release Budding, cell lysis |
Neuraminidase inhibitors, antiviral antibodies, cytotoxic lymphocytes |
GeneralA |
|
*Depends on specific replication strategy of virus, but virus-specified enzyme required for part of process AGeneral, potentially applicable to all viruses. BIntercellular adhesion molecule. REmergence of resistance. SOURCE: Modified from Hayden, 2001. |
||
TABLE 4-4 Approved Antiretroviral Drugs
|
Nucleoside reverse-transcriptase inhibitors |
|
|
Abacavir (ABC) |
Stavudine (d4T) |
|
Didanosine (ddI) |
Zalcitabine (ddC) |
|
Lamivudine (3TC) |
Zidovudine (AZT) |
|
Non-nucleoside reverse-transcriptase inhibitors |
|
|
Efavirenz |
Nevirapine |
|
Delavirdine |
|
|
Protease inhibitors |
|
|
Amprenavir* |
Nelfinavir |
|
Indinavir* |
Ritonavir |
|
Lopinavir* |
Saquinavir* |
|
*Often or usually used with low-dose ritonavir for pharmacological enhancement. Reprinted with permission from Nature (Richman, 2001) copyright (2001) Macmillan Publishers Ltd. |
|
rapidly and are highly transmissible, leading to treatment failure. The emergence of multidrug-resistant HIV means that new drugs are needed, and drugs against additional targets are in fact under development (see Table 4-4). Yet new drugs will face problems of bioavailability and the inevitable emergence of resistance. The other difficulty is the high cost of the drugs, making optimal chemotherapy for HIV available only to persons in the upper socioeconomic groups, even in the developed world.
Two approaches to augmenting HIV-specific immunity are under investigation: therapeutic vaccination and strategic treatment interruption (STI). Therapeutic vaccination may induce new cellular and humoral immune responses in HIV-infected persons. STI is based on the hypothesis that after the arrest of progressive disease and the partial recovery of the immune system through potent antiretroviral therapy, the temporary interruption of therapy will release HIV antigen. This “autoimmunization,” followed by reprotection of the immune system with reintroduced chemotherapy, can be performed in cycles until augmented immunity can change the natural history of infection. The combined use of chemotherapy and immunological approaches is in its infancy and is an important topic for further research. Insight gained into combination chemo- and immunotherapy will also be used in addressing the extensive viral epidemics of chronic hepatitis B and C.
Antivirals to Influenza Viruses
The first option for the control of influenza is efficacious, economically beneficial vaccines. However, it takes at least 6 months to prepare a new
influenza virus vaccine. In the face of an emerging pandemic strain, a specific vaccine would not be immediately available, and less-specific anti-virals would be the only option. Two families of antivirals are available for influenza viruses: (1) two derivatives of adamantine—amantadine (Symmetrel) and rimantadine (Flumadine), and (2) two neuraminidase inhibitors—zanamivir (Relenza) and oseltamivir (Tamiflu). Amantadine and rimantadine target primarily the M2 ion channel, with the hemagglutinin as a secondary target; they are efficacious for influenza A viruses but do not inhibit influenza B viruses (Hay et al., 1985). The two neuraminidase inhibitors block the enzymatic site on both influenza A and B viruses and are efficacious on all subtypes of the viruses, including the H5N1 and H9N2 viruses that were transmitted to humans in Hong Kong (Gubareva et al., 2000). Therapeutic benefits from each of these antineuraminidase influenza drugs are dependent on early initiation of therapy; they include shortening of the infection by 1.5 days and reduction in the sequelae of influenza, such as middle-ear infection (Gubareva et al., 2000; Hayden et al., 1999; Walker et al., 1997).
Both families of anti-influenza drugs are approved for prophylaxis, and the degree of protection provided is essentially complete. Unfortunately, therapeutic use of amantadine and rimantadine results in the rapid emergence of resistant strains, which are shed and transmitted to treated patients (Hayden and Hay, 1992). However, resistant strains are rarely detected in naturally circulating influenza A viruses. Resistance to the neuraminidase inhibitors has also been recognized, but is much more difficult to achieve either experimentally or in the field. Mutations are usually detected first in the hemagglutinin near the receptor binding sites, resulting in a reduction in the affinity of binding and easier release of virus by the enzyme activity of the neuraminidase. Under continued drug pressure from neuraminidase inhibitors, mutations occur in the neuraminidase enzyme active center, leading to resistance. This resistance comes at a price to the virus, including compromised enzyme stability, a change in optimal pH (McKimm-Breschkin et al., 1996; Gubareva et al., 1997), and reduced transmissibility of the resistant virus in animals (Herlocher et al., 2002). The importance of immune surveillance in combination with chemotherapy became apparent for immunocompromised patients when prolonged use of antineuraminidase inhibitors in an influenza-infected transplant patient resulted in selection of drug-resistant mutants (Gubareva et al., 1998). Thus, future developments in antiviral research must address the emergence of resistant mutants and include strategies that incorporate immunostimulation with vaccines and/ or cytokines.
Other Antiviral Agents
Approved efficacious antiviral agents for herpes simplex virus (HSV) and varicella virus, including a number of nucleoside analogues (e.g., adenine arabinoside [Ara-A], acyclovir [Zovirax], valacyclovir [Valtrex]; see Field and Laughlin, 1999), are widely used for therapy and prophylaxis. If patients have normal immune response to HSV, the development of resistance to nucleoside analogues is not a significant problem, but the current antivirals are not curative. Antiviral agents for hepatitis B and C (interferons plus antivirals) are approved or in development. Ribavirin, a triazole nucleoside analogue, inhibits RNA polymerases and transcription and is broadly reactive against respiratory syncitial virus, papillomaviruses, and arenaviruses (see Table 4-3).
As a result of increasing antibiotic resistance, drug options for treatment of some bacterial infections (e.g., vancomycin-resistant enterococci and vancomycin-resistant staphylococci) are increasingly limited. Although defining the precise public health risk of emergent antibiotic resistance is not simple, the problem is global in scope and very serious. Many generic but essential antibiotics are in short supply (Strausbaugh, 2001), and the development of new antibiotics has been severely curtailed. Antivirals are available for only a limited number of viruses, and few are in development. When one considers the bacterial and viral threats of bioterrorism and the ability to generate antibiotic-resistant bacterial weapons, the urgent need for new antimicrobials becomes clear. Thus, many of the issues raised regarding vaccine production apply also to antibiotics and antivirals.
The U.S. Secretary of Health and Human Services should ensure the formulation and implementation of a national strategy for developing new antimicrobials, as well as producing an adequate supply of approved antimicrobials. The U.S. Secretary of Health and Human Services should work closely with other relevant federal agencies (e.g., DOD, the Department of Homeland Security), Congress, industry, academia, and the public health community to carry out this responsibility.
Need for Stockpiling
The 2001 anthrax outbreak clearly demonstrated the need to have access to a stockpile of effective antimicrobial agents for immediate use during the aftermath of a bioterrorist attack. In the absence of vaccines, antimicrobials are the only effective population-based preventive measure for use against a bioterrorist attack once exposure has occurred, provided, of course, that an effective drug exists. Fortunately, ciprofloxacin was de-
termined to be the appropriate drug in this situation, and an ample supply was readily available. The same may not be true when the next influenza pandemic eventually occurs, likely resulting in tens of thousands of deaths. Although stockpiling of antiviral drugs for influenza is a component of the pandemic plan developed by the United States and WHO (WHO, 1999e), we have yet to begin stockpiling antivirals effective against influenza. The time has come to move forward with this plan and determine which drugs are needed; the quantity required; the costs of production, storage, and distribution; and the authority under which the drugs will be used.
The U.S. Secretary of Health and Human Services and the U.S. Secretary of Homeland Security should protect our national security by ensuring the stockpiling and distribution of antibiotics, antivirals (e.g., for influenza), and antitoxins for naturally occurring or intentionally introduced microbial threats. The federal government should explore innovative mechanisms, such as cooperative agreements between government and industry or consortia of government, industry, and academia, to accelerate these efforts.
INAPPROPRIATE USE OF ANTIMICROBIALS
For a variety of reasons previously discussed, the pharmaceutical industry is developing fewer new antimicrobials than in previous years. Whereas it appeared at one time that an endless supply of effective new drugs to treat resistant infections would exist, such is no longer the case. Therefore, immediate action must be taken to preserve the effectiveness of available drugs.
Factors leading to the increasing problem of antimicrobial resistance are well known and understood. Many genes for resistance occur on cassettes that can move between organisms, across species boundaries (Leverstein-van Hall et al., 2002), and between chromosomes and plasmids. Resistance genes in bacteria are commonly grouped together on the same mobile genetic elements, with the crucial practical consequence that the use of any single drug may select for resistance to a wide group of drugs. Thus, an antimicrobial employed in food and animal production that has never before been used to treat infection in humans can select for resistance to other drugs used to treat humans.
Resistant bacteria often persist in vivo even in the absence of continued selection by antibiotics, although in some cases resistance gradually diminishes once antibiotic pressures have been reduced. One explanation for continued resistance involves the lethal effect of the loss of certain plasmids when bacteria divide. Some resistant microbes are less fit, but resistant strains arising in a clinical context are generally virulent and can often
persist for extended periods of time once established. Therefore, it is imperative to actively pursue and address the problem; it will be too late to effect useful change once most microbes have become resistant to the available drugs.
Antibiotic resistance resulting from the inappropriate overuse of antibiotics is not a new problem. A number of expert committees and professional organizations have studied the problem, issued reports, and made recommendations (Alliance for the Prudent Use of Antibiotics, 2001; CDC, 2001o; FDA, 2000; GAO, 1999; Center for Science in the Public Interest, 1998; NRC, 1999). Unfortunately, little has been done to change the situation, especially in the United States. Resistance due to the inappropriate use of antibiotics compromises the efficacy of many classic and highly effective antibiotics, such as penicillin for pneumococci and vancomycin for enterococci, as well as that of some newer antibiotics, such as ciprofloxacin and other types of fluorinated quinolones for gonococci, Salmonella, and Campylobacter. The recent discovery of an enterococcal gene for vancomycin resistance in S. aureus was alarming even though it had been predicted on the basis of the ability of the genes to transfer across species boundaries during mixed culture (CDC, 2002d). In the case of enterococcal and staphylococcal infection, alternative therapies have been introduced, but resistance to these new drugs has already been documented (Tsiodras et al., 2001; Herrero et al., 2002). The specter of untreatable infections—a regression to the pre-antibiotic era—is looming just around the corner.
Preventing the overuse of antimicrobials is not an easy task because of the revolutionary effects the drugs have had on human and animal health. Because antimicrobials are highly effective, there is an understandable tendency to use them in any situation in which they might be helpful. These effective drugs are relatively inexpensive compared with other medical interventions. Patients demand the drugs when they have an illness they imagine to be treatable with antibiotics. Doctors prescribe antibiotics for that same reason, often in the absence of diagnostic tests to determine the etiology of infection, and also because patients want and expect to be treated with them. In many areas of the world where little money is available for health care, antimicrobials are readily available without a doctor’s prescription, and as a result are often taken unnecessarily or inadequately. Many problems associated with antimicrobial resistance have arisen in poor and developing areas of the world, and have subsequently spread globally.
In addition to avoiding the inappropriate use of antibiotics to treat viral disease, prudence dictates use of the appropriate antimicrobial when an etiologic diagnosis is made. For example, the rapid rise in drug-resistant malaria has led to the development of newer, generally more expensive therapies for the disease. This in turn has resulted in an increase in the
prescribing of these newer drugs, even in areas where there is no demonstrated resistance to first-line therapies. The use of first-line therapies must be continued in areas where resistance has not been documented, and newer therapies should be used only when first-line therapies are ineffective or in areas of resistance. To this end, it is essential to monitor resistance patterns around the world.
Decreasing Inappropriate Use of Antimicrobials in Human Medicine
Decreasing the inappropriate use of antimicrobials in human medicine is a complex task that requires a multipronged effort fueled by a sense of urgency. The inappropriate use of antibiotics for treatment of viral diseases can be averted by the increased use of available diagnostic tests and the development of better point-of-care, inexpensive, rapid, sensitive, and specific diagnostic tests, which would enable the rational use of new antivirals as they become available (see the earlier discussion of the development of diagnostics). The decreased use of antibacterials for viral respiratory infections and other syndromes should lessen selective pressures for the emergence of resistant bacteria. FDA has recently included this message on label inserts of antibiotics.
If this important objective is to be achieved, the general public and health care providers must be better educated and informed about the importance of administering antimicrobial therapy properly. The need is urgent to both educate and monitor all categories of practitioners and drug dispensers in developing countries where medicines are sold directly to the public over the counter and dispensed by private practitioners in an ad hoc manner. More attention needs to be given to improving practitioner education and compliance. Patient care would be improved by the development and dissemination of better evidence-based treatment guidelines. More research is needed on methods for treating infections to minimize the emergence of resistance without a loss of efficacy. Infection control programs must be supported in hospitals in an effort to decrease the transmission of resistance both within the hospitals and in the community. Surveillance for patterns of resistance in hospitals and in the community must be continued and expanded; this will require a coordinated effort among public health organizations, private medicine, and industry. Because resistant microbes arise throughout the world and travel broadly to all regions, the needs and problems of the economically and health care disadvantaged regions of the world must be considered.
The world is facing an imminent crisis in the control of infectious diseases as the result of a gradual but steady increase in the resistance of a number of microbial agents to available therapeutic drugs. Although defining the precise public health risk of emergent antimicrobial resistance is not
a simple task, there is no doubt that the problem is of global concern and is creating dilemmas for the treatment of infections in both hospitals and community health care settings.
CDC, FDA, professional health organizations, academia, health care delivery systems, and industry should expand efforts to decrease the inappropriate use of antimicrobials in human medicine through (1) expanded outreach and better education of health care providers, drug dispensers, and the general public on the inherent dangers associated with the inappropriate use of antimicrobials, and (2) the increased use of diagnostic tests, as well as the development and use of rapid diagnostic tests, to determine the etiology of infection and thereby ensure the more appropriate use of antimicrobials.
Decreasing Inappropriate Overuse of Antimicrobials in Animal Husbandry and Agriculture
Clearly, a decrease in the inappropriate use of antimicrobials in human medicine alone is not enough. Substantial efforts must be made to decrease inappropriate overuse of antimicrobials in animals and agriculture as well.
Although estimates vary widely, the total amount of antimicrobials used in Europe and the United States in animal husbandry and agriculture far outweighs the total used in humans (McEwen and Fedorka-Cray, 2002). The majority of this use is for growth promotion or preventive therapy in healthy animals. Mounting evidence suggests a relationship between antimicrobial use in animal husbandry and an increase in bacterial resistance in humans (Alliance for the Prudent Use of Antibiotics, 2002), a view supported by an IOM committee that reviewed the use of drugs in food animals (IOM, 1999b). The use of antimicrobials in food animals leads to antibiotic resistance, which can then be transmitted to humans through the food supply (Swartz, 2002; Fey et al., 2000; Smith et al., 2002; White et al., 2001).
A study published in 2001 found that 20 percent of ground meat samples obtained from supermarkets in the Washington, D.C., metropolitan area were contaminated with Salmonella. Of these bacteria, 84 percent were resistant to at least one antibiotic and 53 percent to at least three antibiotics (White et al., 2001). This study supports previous findings that foods of animal origin are potential sources of ceftriaxone-resistant Salmonella infections in humans. Similarly, researchers found that between 17 and 87 percent of chickens obtained in supermarkets in four states contained strains of Enterococcus faecium that were resistant to quinupristin–
dalfopristin, an approved antimicrobial for use in humans (McDonald et al., 2001). The researchers believed that the use of virginiamycin, an antibiotic of the streptogramin group, in farm animals had created a reservoir of streptogramin–resistant E. faecium in the food supply, which could contribute to foodborne dissemination of resistance as the clinical use of quinupristin–dalfopristin increases.
Substantial evidence supports that certain types of resistant organisms, such as vancomycin-resistant enterococci, emerged initially in animals because of the use of similar drugs for growth promotion or prophylaxis (O’Brien, 2002). Consideration of this association led to a ban on the use of avoparacin, a vancomycin analogue, in Europe (Wegener et al., 1999). The decreased use of antimicrobials for growth promotion or prophylaxis in many European countries has been associated with a subsequent stabilization in resistance or a gradually decreasing resistance in animal flora (Aarestrup et al., 2001). WHO has called for all antimicrobials used for disease control in food animals to be prescribed by veterinary health care providers, and for termination or rapid phase-out of antimicrobials used for growth promotion if they are used for human treatment (WHO, 2000f). Various other groups have suggested that because of the increasing risk of antimicrobial resistance, the subtherapeutic use of antibiotics for growth promotion should be banned (some would include use for prophylaxis in the ban as well) if they are also used in humans (Union of Concerned Scientists, 2002; Alliance for the Prudent Use of Antimicrobials, 2002).
The main argument against a ban is the potential economic hardships to livestock and poultry producers, which would result in higher costs for consumers. According to the IOM Committee on the Use of Drugs in Food Animals, such a ban would increase the price of meat by an estimated 0.013 to 0.06 cents per pound; this translates to $4.84 to $9.72 per person each year, depending on the meat and the cut (IOM, 1999b). Yet, evidence suggests that animals can be raised efficiently without the use of growth-promoting antimicrobials (Emborg et al., 2001; Wierup, 2001).
Critics of the ban also argue that it would result in poorer production efficiency and an increased incidence of infectious disease in animals. However, it has been noted that subtherapeutic antibiotics are most effective in animals under the stress of inadequate nutrition and suboptimal sanitary conditions (Braude et al., 1953); therefore, improved hygiene and changes in animal husbandry practices to control disease could potentially eliminate the need for growth promoters (Emborg et al., 2001). In Denmark, the elimination of antimicrobial growth promoters from broiler chicken feed did not result in a change in death rates or a decrease in kilograms of broilers produced per square meter. Danish scientists also reported that the decreased use of virginiamycin and avilamycin in animals was followed by decreases in resistance to these drugs (Aarestrup et al., 2001).
FDA should ban the use of antimicrobials for growth promotion in animals if those classes of antimicrobials are also used in humans.
The committee endorses the Public Health Action Plan to Combat Antimicrobial Resistance developed by the Interagency Task Force on Antimicrobial Resistance and the recommendations of the WHO Global Strategy for the Containment of Antimicrobial Resistance (see Boxes 4-9 and 4-10). Although the broad scope of these recommendations defies easy implementation, we must seize the opportunity immediately to do as much as we can while organizing the resources and plans needed to carry out other initiatives. To do nothing is, in effect, to allow the continued evolution of antimicrobial-resistant microbes, which poses serious near- and long-term threats to global health. The total burden of human illness due to resistant bacteria that have been transferred from animals to humans is unknown, but the guiding principle should be that we must do what the available evidence suggests will help stem the tide of increasing resistance before it is too late. By endorsing these recommendations, we will join belatedly much of the rest of the developed world, which already has made similar recommendations and, in many cases, implemented them. These changes should be accompanied by substantial outcomes research on the effects on animal health, resistance prevalence in animals and humans, and the economics of food production.
VECTOR-BORNE AND ZOONOTIC DISEASE CONTROL
The majority of emerging infectious diseases are zoonoses (i.e., diseases transmitted from animals to humans under natural conditions). Vector-borne and rodent-borne diseases are especially notable in this regard, remaining major causes of morbidity and mortality in humans in the tropical world and representing a large proportion of newly emerged diseases (see the discussion in Chapter 3). Exacerbating the situation is the potential for many of these agents to be weaponized and used by bioterrorists. Because of their resurging public health importance and their exceptional ability to cause epidemics, vector-borne and zoonotic diseases will undoubtedly continue to pose significant risks to human health in the future.
Unfortunately, the national and international capacity to address these diseases is limited. The many reasons for this include (1) the lack of efficacious vaccines for many of these pathogens; (2) decreased support for and deterioration of the public health surveillance and control infrastructure for vector-borne and zoonotic diseases; (3) erosion in the numbers of scientists trained in relevant fields, including medical entomology, vector ecology, zoonoses, and tropical medicine; (4) the development of resistance to drugs
|
BOX 4-9 WHO Global Strategy for Antimicrobial Resistance In response to the growing problem of antibiotic resistance, WHO has worked with many partners, including the American Society for Microbiology and the Alliance for the Prudent Use of Antibiotic (APUA), to develop the WHO Global Strategy for Containment for Antimicrobial Resistance. The seven key recommendations emanating from the 25 expert reports used to formulate the strategy are summarized below. Increase Awareness of the Antibiotic Resistance Problem International organizations: Obtain worldwide commitments to establish prudent antibiotic use policies National and municipal organizations: Publicize the outcomes of programs from other countries Educate the general public Promote communication Evaluate the curricula of universities Health care institutions: Use effective teaching methods for education prescribers Health care workers: Educate the general public Improve Surveillance of Antibiotic Resistance National and municipal organizations: Coordinate local surveillance networks Recruit leaders for surveillance networks Support a reference laboratory Share results of surveillance with international organizations Monitor resistance in food animals Monitor sentinel human populations Health care institutions: Develop local surveillance network Maintain a laboratory with adequate quality assurance and trained technicians Health care workers: Initiate a local surveillance network Pharmaceutical companies: Undertake postmarking surveillance to detect the emergence of resistance to new antibiotics Support surveillance networks Improve Antibiotic Use in People National and municipal organizations: Enforce the prudent use of antibiotics Create national and regional guidelines Update guidelines based on surveillance data Eliminate financial incentives that promote the misuse of antibiotics Monitor advertising Consider the impact of new drugs on resistance during the drug approval process Limit general access to new drugs Establish postmarking surveillance accords Health care institutions: Establish an Infection Control Committee Establish a Drugs and Therapeutics Committee Establish guidelines for appropriate antibiotic use Appoint an antimicrobial resistance monitor Reduce the spread of infection Create pharmacy reports |
|
Establish and disseminate list of essential drugs Educate employees Maintain a laboratory Health care workers: Prescribe antibiotics prudently Improve hygiene Improve Antibiotic Use in Animals National and municipal organizations: Increase awareness of the antibiotic resistance problem Regulate antibiotic prescriptions for animals Restrict growth promoter use in animals Regulate antibiotic use in animals Set a risk standard for resistance Consider human and nonhuman uses simultaneously Monitor advertising Veterinarians: Promote a prudent use of antibiotics in animals Develop local guidelines for antibiotic use Food animal producers: Improve farm hygiene Reduce the use of antibiotics as growth promoters Improve animal husbandry Researchers: Perform risk–benefit analysis of growth promoter use Assess environmental impact Examine food processing and distribution methods Encourage New Product Development National and municipal organizations: Provide incentives to industry Protect intellectual property rights Facilitate networking Pharmaceutical companies: Increase research and development in several areas Increase Resources to Curb Antibiotic Resistance in the Developing World International organizations: Share results of surveillance internationally Secure technical and financial support for developing countries Invest in a worldwide vaccine strategy to reduce antibiotics Ensure the availability of vaccines and quality drugs Facilitate communication among the countries of the world Safeguard privacy and human rights Promote appropriate international laws National and municipal organizations: Decrease the risk of infectious disease Ensure antibiotic availability Share resources with other countries Increase Funding for Surveillance, Research, and Education National and municipal organizations: Increase funding for a surveillance network Increase funding for research Increase funding for education SOURCE: World Health Organization, 2001i. |
|
BOX 4-10 Public Health Action Plan to Combat Antimicrobial Resistance A Public Health Action Plan to Combat Antimicrobial Resistance was developed by an Interagency Task Force on Antimicrobial Resistance that was created in 1999. This plan reflects a broad-based consensus of federal agencies on actions needed to address antimicrobial resistance. Part I focuses on domestic is-sues, Part II, yet to be developed, will identify actions that address international concerns more specifically. The following is a summary of the Task Force’s recommendations on the four domestic focus areas. 1. Surveillance Develop and implement a coordinated national plan for antimicrobial resistance Ensure the availability of reliable drug susceptibility data for surveillance Monitor patterns of antimicrobial drug use Monitor antimicrobial resistance in agricultural settings to protect the public’s health by ensuring a safe food supply, as well as animal and plant health 2. Prevention and Control Extend the useful life of antimicrobial drugs through appropriate use policies that discourage overuse and misuse Improve diagnostic testing practices Prevent infection transmission through improved infection control methods and use of vaccines Prevent and control emerging antimicrobial resistance problems in agriculture and human and veterinary medicine Ensure that comprehensive programs to prevent and control antimicrobial resistance involve a wide variety of nonfederal partners and the public so these programs become a part of routine practice nationwide 3. Research Increase understanding of microbial physiology, ecology, genetics, and mechanisms of resistance Augment the existing research infrastructure to support a critical mass of researchers in antimicrobial resistance and related fields Translate research findings into clinically useful products, such as novel approaches to detecting, preventing, and treating antimicrobial-resistant infections. 4. Product Development Ensure that researchers and drug manufacturers are informed of current and projected gaps in the arsenal of antimicrobial drugs, vaccines, and diagnostics and of potential markets for these products Stimulate the development of priority antimicrobial products for which market incentives are inadequate while fostering their appropriate use Optimize the development and use of veterinary drugs and related agricultural products that reduce the transfer of resistance to pathogens that can infect humans SOURCE: CDC, 2001o. |
in pathogens and to chemical pesticides in arthropods; (5) population growth associated with rampant and unplanned urbanization in the tropics and increased juxtapositions of humans, animal reservoirs, and vectors; (6) increased trafficking of pathogens, vectors, and animal reservoirs; and (7) societal and behavioral changes and practices that contribute to greatly increased disease incidence.
To deal with the many needs and issues involving vector-borne and zoonotic diseases is an overwhelming task. Nonetheless, a number of key issues must be addressed promptly to provide the expertise, infrastructure, resources, and tools required to develop the capacity to respond to the threats posed by vector-borne and zoonotic diseases.
Human Resource Capacity
The critical national needs for specialists in vector-borne and zoonotic diseases (e.g., medical entomologists, vector biologists, vector ecologists, mammologists, ornithologists) were illustrated dramatically by the recent emergences of Sin Nombre virus and West Nile virus in the United States. The loss of medical entomologists, for example, was recognized as a national problem in the 1980s (NRC, 1983). However, attitudes changed little in academic departments across the nation as a result. Medical entomology positions were invariably lost when occupants retired or moved. This situation resulted in a dramatic reduction overall in the number of medical entomologists/vector biologists. For example, the United States is facing a critical shortage of morphological systematists who are also field biologists capable of collecting arthropod vectors within the context of their specific environments. As newer molecular taxonomic approaches become more established in monitoring vectors, traditional systematists are needed to guide and verify the application of such approaches. Specialists in vector ecology are also increasingly in short supply, as are academic institutions capable of the relevant training (Spielman, 1994). Experts in the ecology of both vectors and reservoirs are needed to monitor the distribution, abundance, composition, and density of species, and the prevalence of infections that are relevant to human health.
With the emergence of Lyme disease, human erlichiosis, and West Nile encephalitis in the United States and the resurgence of vector-borne diseases throughout the world (Gratz, 1999; Gubler, 1998), some progress has been made as organizations such as CDC, NIH, WHO, and foundations have begun to promote research in molecular vector biology. Increased funding and program development opportunities, especially in molecular vector biology, have led to greater numbers of scientists entering the field, many of whom are now assuming faculty positions in universities. Perhaps as the
public health importance of vector-borne and zoonotic disease control is better recognized, student demand will grow, and previously lost positions in much-needed related areas of science will be regained. The end result would be a renewed stream of students to replenish the depleted ranks of medical entomologists, ecologists, and vector biologists at all levels.
The momentum to address national and international needs in medical entomology/vector biology could be leveraged in numerous ways. For example, CDC could establish a medical entomology/vector biology Epidemic Intelligence Service (EIS) program, and its entomologic EIS officers could provide support to national and international jurisdictions requesting entomologic expertise. In addition, regional centers of excellence in medical entomology/vector biology could be established to provide needed services, training, and research. These centers would preferably be incorporated into larger interdisciplinary infectious disease centers (see the later discussion), which would serve as resources to regions and nations in addressing issues involving vector-borne and other emerging diseases and bioterrorism.
Efficient training strategies are necessary to address the human resource needs at all levels, from understanding disturbances in the enviroment that impact on the abundance and distribution of vectors and animal reservoirs, to identification and processing of species, to gene identification and characterization, to development of geographic information systems (GIS) and other approaches for control of vector-borne and zoonotic diseases. The emergence of West Nile virus resulted in an intensive effort by CDC to train individuals in mosquito identification, processing, and control, and the emergence of Sin Nombre virus led to similar efforts by CDC to train individuals in rodent identification, surveillance, and control. WHO provides workshops devoted to specific important issues concerning vector-borne diseases in disease-endemic areas. Web-based training programs, courses, and texts might be used to address critical short term-needs. However, alternative approaches will be necessary to develop a new generation of leaders and trainers in the field.
The Biology of Disease Vectors course is notable in this regard. This intensive 2-week course, which is supported by the MacArthur Foundation, the WHO Special Program for Research and Training in Tropical Diseases, and the Howard Hughes Medical Institute, provides learning and networking opportunities for vector biologists. The course alternates annually between the United States and a disease-endemic country. Students participating in the course come from both developed and developing countries, and internationally recognized faculty provide invaluable networking and career opportunities. Such courses, strategically targeted to an area of national need, not only expedite the development of new leaders, but can also advance fields scientifically.
In efforts to rebuild the human resource capacity needed to address vector-borne and zoonotic diseases, it will be critical to increase training and research opportunities in applied, field-oriented vector biology and zoonotic disease research. CDC, DOD, NIH, and other federal agencies should continue to encourage research projects and programs that investigate the biological, behavioral, entomological, and environmental determinants of pathogen emergence, and that incorporate modern and robust molecular and quantitative tools into these investigations. Notable successes in this area include CDC’s partnering with state and university scientists to address newly emergent vector-borne and rodent-borne diseases, such as Lyme disease, West Nile fever, and hantavirus pulmonary syndrome. These efforts leveraged CDC funds and talents to address emerging disease issues, supported applied epidemiological research and training in disease-endemic sites, and enhanced communication and partnering between CDC and state and local institutions.
The continued presence of long-term, sustainable laboratories in selected disease-endemic countries is critical. Such laboratories are invaluable for research, training, and surveillance for tropical and emerging infectious diseases. Historically, Naval Medical Research Units, United States Army, and WHO laboratories have provided opportunities for trainees to obtain tropical disease research experience. More recent programs, such the NIH International Collaborations in Infectious Disease Research (ICIDR) and Tropical Disease Research Units, provide training and research opportunities in field-oriented vector biology and control. The NIH ICIDR grants, together with the companion Actions for Building Capacity Program at the Fogarty Center, emphasize epidemiological research and training in disease-endemic areas in the context of NIH-funded research. Such programs also initiate interactions with collaborators in tropical regions, and yield long-term benefits in terms of establishing public health infrastructure, training and research opportunities, and listening posts in areas of the world where many pathogens emerge.
CDC, DOD, NIH, and USDA should work with academia, private organizations, and foundations to support efforts at rebuilding the human resource capacity at both academic centers and public health agencies in the relevant sciences—such as medical entomology, vector and reservoir biology, vector and reservoir ecology, and zoonoses—necessary to control vector-borne and zoonotic diseases.
Need to Increase the Armamentarium for Vector Control
Expanded research into the biological and ecological determinants of vector maintenance and transmission of pathogens, together with the explosion of information that will occur in the mosquito post-genomics era (Holt et al., 2002) are likely to result in new and unforeseen approaches and targets to control vector-borne diseases. Examples of research areas with the potential to increase the armamentarium for control of vector-borne diseases and to augment currently available control approaches are described below.
For the foreseeable future, traditional approaches to reducing vector populations or repelling vectors will remain the first lines of defense against emerging and resurging vector-borne diseases. Clearly, the development of new, environmentally acceptable pesticides will be critical to mitigate the potential for dramatic increases in such diseases (Sina and Aultman, 2001).
Improved Pesticides
Discontinuance of DDT usage has exacerbated the burden of vector-borne diseases in many parts of the world (Attaran et al., 2000). The resurgence of vector-borne diseases and resistance to alternative pesticides, therefore, has forced some countries to resume DDT usage. Since domicile treatment with DDT has not been associated with major adverse environmental consequences, this practice should be allowed for vector control in public health emergencies until equally effective and inexpensive substitutes for DDT are developed. DDT may help control vector-borne diseases, such as dengue and malaria, not only by killing vectors, but also by repelling them (Roberts et al., 2000). Residual DDT in homes may repel mosquitoes, thereby disrupting the close association between the human host and anthropophilic and endophilic vectors and dramatically reducing opportunities for pathogen transmission. At the same time, care will be needed to ensure that the availability of DDT for public health uses does not result in its use in agricultural applications. The development of efficacious and environmentally sensitive alternatives to DDT needs to become a major research objective.
Novel Strategies to Prolong Pesticide Usage
Pesticide usage in integrated pest management programs, which incorporate established agricultural practices for mitigating the evolution of resistance (e.g., rotation of pesticides used, inclusion of refugia with no pesticide applications), would extend the useful life of existing pesticides. Incorporating new molecular tools for diagnosing pesticide resistance into
control programs could also result in more effective and efficient pesticide usage. Moreover, the development of novel strategies for prolonging pesticide efficacy, such as negative cross-resistance, should be possible in this era of high-throughput screening (Pittendrigh and Gaffney, 2001). More information on the effect of the prevalence of resistance to pesticides on the control of vector-borne diseases would be of great value for risk assessment.
New Repellents
As noted, repellents remain a first line of defense against emerging or resurging vector-borne diseases. DEET is the most efficacious repellent currently available commercially; however, its relicensing has been problematic because of adverse effects associated with its overuse in children, presumably due to its lipophilic nature (Qiu et al., 1998). Modern high-throughput and genomic approaches may permit the identification of new molecules with repellent activity similar to that of DEET (and DDT), but without adverse effects. Understanding of the molecular basis of vector olfaction and host seeking (Hill et al., 2002) could lead to the development of new repellents and attractants to control vectors (Day et al., 2001).
New Biopesticides and Biocontrol Agents to Augment Chemical Pesticides
The increase in pesticide resistance necessitates new investigations into biocontrol agents, such as viruses and bacteria, that could be incorporated into integrated pest management approaches for vector control. New formulations of Bacillus thruringiensis and Bacillus sphaericus show promise for control of vectors, even in tropical regions (Thiery et al., 1997; Regis et al., 2001). Baculoviruses from mosquitoes may be useful for vector control (Afonso et al., 2001). Other biopesticide agents could be improved using molecular genetic approaches to make them more efficacious control agents. For example, viruses could be used to transduce effector molecules in order to enhance vector knockdown or manipulate vector phenotypes.
Novel Strategies to Interrupt Pathogen Transmission
Strategies for vector-borne disease control remain focused on approaches that involve immunizing humans, using pesticides to reduce vector populations, or repellents to reduce contact with vectors. If pesticide resistance and parasite resistance to drugs continue to increase, if public health infrastructure cannot be rebuilt, and if mortality rates from vector-borne diseases persist or increase, novel approaches now emerging from
investigations of vector molecular biology and pathogen–vector–host interactions may be necessary to control these diseases.
Insights into the molecular basis of pathogen–vector–host interactions suggest new strategies for controlling vector-borne diseases (Foy et al., in press; Willadsen, 2001). Immunizing hosts to vector-specific determinants of pathogen transmission (e.g., salivary effector proteins that enhance pathogen infection; see Titus and Ribeiro, 1988) could provide broad-spectrum protection against multiple pathogens or strains (Kamhawi et al., 2000, Valenzuela et al., 2001). Other critical determinants of pathogen infection of and transmission by vectors (e.g., vector proteolytic enzymes, which process arbovirus proteins and condition vector infection) could be targeted for transmission-blocking vaccines (Carter, 2001). Immunizing vertebrate hosts to immunologically privileged antigens of vectors could kill or impair blood-feeding mosquitoes, a strategy that works for ticks (Willadsen and Billingsley, 1996) and may also be useful against mosquito vectors, which frequently feed on humans (Foy et al., in press). Theoretically, these vectors would feed on other hosts (zooprophylaxis), thereby reducing pathogen transmission.
Genetic approaches in which vector populations are manipulated to become incompetent vectors are being investigated for their potential to interrupt pathogen transmission. Such approaches could minimize potential environmental issues associated with pesticide usage and prevent an ecological vacuum that other vectors could occupy. The vector population could theoretically be genetically immunized to make it nonpermissive to pathogen transmission. The “immunogens” could be driven into vector populations by harnessing naturally occurring arthropod systems, such as transposable elements, symbionts, or transducing viruses, which would be vector-specific (Beaty, 2000). RNAi, which was recently documented in vectors (Adelman et al., 2001), could be exploited in such programs. Proof of principle has been provided that vectors can be molecularly manipulated to make them refractory to arboviruses and trypanosome and malaria parasites (Beard et al., 2002; Olson et al., 1996, Ito et al., 2002). Recent progress in vector molecular biology suggests that continued research in these areas may provide new approaches for the control of vector-borne diseases, although the success of genetic manipulation in control programs is by no means certain (Boete and Koella, 2003). Research utilizing genetically manipulated vectors would require ecological studies (Scott et al., 2002) to determine the feasibility of such an approach to vector control and would require addressing the benefits, risks, and social and political issues associated with such a control strategy (Alphey et al., 2002).
DOD and NIH should develop new and expand upon current research efforts to enhance the armamentarium for vector control. The development of safe and effective pesticides and repellents, as well as novel strategies for prolonging the use of existing pesticides by mitigating the evolution of resistance, is paramount in the absence of vaccines to prevent most vector-borne diseases. In addition, newer methods of vector control—such as biopesticides and biocontrol agents to augment chemical pesticides, and novel strategies for interrupting vector-borne pathogen transmission to humans—should be developed and evaluated for effectiveness.
Geographic Information Systems and Robust Models for Predicting and Preventing Vector-borne and Zoonotic Diseases
Also complicating the control of vector-borne and zoonotic diseases has been a lack of knowledge of fundamental epidemiologic, genetic, biologic, and environmental determinants that condition potential increased transmission to humans by the respective nonhuman vectors or reservoirs. Because biological and ecological factors condition the transmission of pathogens by vectors and from animal reservoirs to humans, GIS and robust models (see Appendix E for a discussion of modeling) offer the potential to provide predictive capability for the emergence of vector-borne and zoonotic diseases. The ecological and quantitative capabilities of GIS make it possible to identify some of the determinants of endemicity and emergence. The developing hantavirus GIS models are promising in this regard (Boone et al., 1998; Glass et al., 2000; Hjelle and Glass, 2000; Yates et al., 2002b). The inclusion of genetic information concerning vector competence and rodent permissiveness, as well as other epidemiologically important information, such as gene flow in vector and reservoir populations (e.g., Black et al., 2001; Gorrochotequi-Escalante et al., 2002), may improve surveillance and risk assessment strategies for zoonoses and enhance the predictive capability of GIS and model systems. New GIS and robust models could revolutionize surveillance, risk assessment, and prevention strategies for zoonoses and permit the focusing of resources and talent on prevention efforts in areas of greatest risk, an especially important capability in resource-limited environments.
CDC, DOD, and NIH should work with state and local public health agencies and academia to expand efforts to exploit geographic information systems (GIS) and robust models for predicting and preventing the emergence of vector-borne and zoonotic diseases.
COMPREHENSIVE INFECTIOUS DISEASE RESEARCH AGENDA
Research remains an essential underpinning of the capacity to prevent and control infectious diseases. Despite recommendations made in the 1992 IOM report Emerging Infections: Microbial Threats to Health in the United States, calling for increased research on factors underlying the emergence of infectious diseases and an extramural grant program for research on surveillance and applied control methods, significant gaps remain in the overall infectious disease research agenda of the United States. To ensure that the nation is strategically poised to protect itself against the threat of infectious diseases and to maximize its assistance to developing countries in their efforts to combat these diseases, further investments must be made to support a diverse array of multidisciplinary research domains. These new investments must be part of an overall strategy for improved public health preparedness and protection against infectious disease threats, and a comprehensive system of accountability must be in place to ensure that no critical areas are neglected.
The considerable amount of new resources now becoming available for biodefense research makes this a critical time to develop a comprehensive research agenda. The most effective use of these new funds will involve integration of the evolving threat posed by the intentional use of biological agents as weapons into the broader context of infectious disease research. As previously noted, bioterrorism represents but the extreme end of a continuum of serious infectious disease threats, including the emergence of new infectious diseases, the resurgence of old ones, the appearance of new antimicrobial-resistant forms of old diseases, recognition of the infectious etiology of chronic diseases, and the creation of bioengineered organisms that produce disease in unforeseen ways. For such an integrated agenda to be effective, it must address both long- and short-term needs, involve both basic science and applied public health research, be multidisciplinary in nature, and utilize modern and robust molecular and quantitative tools.
Scientific research can yield a greater understanding of the biology and pathogenesis of organisms that cause disease, the biology of disease-spreading vectors, and the ways in which the human immune system responds to infection and disease. Several factors beyond these traditional foci of infectious disease research, however, play significant roles in the emergence of infectious disease threats (see Chapter 3). For example, malnutrition has long been known to play a role in susceptibility to death from diarrhea, respiratory infection, and malaria. Not as well understood are the roles of famine, war, crowding, urbanization, and population growth. Risky behaviors, such as illicit drug use and unprotected sex, are closely linked to several emerging infectious diseases. Ecological factors surrounding a lack of clean water and poor sanitation have also been linked to diseases such as
cholera and plague. Additional ecological factors (e.g., deforestation or other forms of land use change) are associated with many emerging vector-borne and zoonotic diseases, such as dengue, malaria, yellow fever, Lassa fever, Lyme disease, and West Nile encephalitis, but remain poorly understood. New grounds for mosquito breeding have developed in waste dumps, threatening vector control efforts that have traditionally focused on vector breeding in swamps and marshes. As previously noted, agricultural practices have been closely linked to the spread of antibiotic resistance, influenza outbreaks, and diseases of food crops and animals. Migration, travel, and commerce have been associated with several microbial threats to health.
Human development and large-scale social phenomena are closely connected to infectious disease threats at a global level. National security and an enlightened self-interest require that countries recognize the direct impact of social, economic, political, and ecological factors, especially in developing countries. In additional to technical and financial support, a research program focused on the global social and ecological factors affecting infectious disease emergence should be established. Only recently have studies been conducted within the traditional biomedical, social epidemiology, and medical anthropology research arenas to begin to address these factors and the interventions necessary to combat them.
Inferences about the etiology of disease are typically drawn through statistical association of natural observations or experiments. Recognizing, however, that the emergence of infectious disease is usually not attributable to any single factor, but the result of complex interactions among numerous and often unknown physical, biological, ecological, and socioeconomic variables, it is clear that multidisciplinary studies, including dynamic analyses of such interactions, are needed.
NIH should develop a comprehensive research agenda for infectious disease prevention and control in collaboration with other federal research institutions and laboratories (e.g., CDC, DOD, the U.S. Department of Energy, the National Science Foundation), academia, and industry. This agenda should be designed to investigate the role of genetic, biological, social, economic, political, ecological, and physical environmental factors in the emergence of infectious diseases in the United States and worldwide. This agenda should also include the development and assessment of public health measures to address microbial threats. A sustained commitment to a robust research agenda must be a high priority if the United States is to dramatically reduce the threat of naturally occurring infectious diseases and intentional uses of biological agents. The research agenda should be flexible to permit rapid assessment of new and emerging threats, and should be rigorously reevaluated
on a 5-year basis to ensure that it is addressing areas of highest priority.
Successfully carrying out such an integrated, comprehensive research agenda will entail collaboration among multiple government agencies, academia, and the private sector. Collaborations between the academic research and public health communities are essential to ensure that priority research areas are addressed in a timely manner and that findings can be readily applied. Research driven by grants in the infectious disease arena rarely includes the kind of practical research, such as evaluations of programs and interventions, that is of value to public health workers in the field. Even within schools of public health, applied public health research has not been a priority. This problem can be addressed, in part, by creating faculty positions that are accountable to both academic centers and health departments. A coordinated approach must extend from fundamental laboratory science through operation, evaluation, and intervention research. In addition, the full support and engagement of a range of professional disciplines, including such often-overlooked fields as entomology, ecology, and anthropology, are needed.
INTERDISCIPLINARY INFECTIOUS DISEASE CENTERS
As previously noted, addressing the highly complex nature of infectious disease emergence requires the involvement of experts from a broad range of disciplines and health sectors. Collaborative links within and between universities and among international, federal, and state governments currently exist. The present structure of academic and public health institutions, however, requires that most of these arenas operate independently of each other. Opportunities for convergence and synergism are often lost unless experts convene under the same roof (or on the same campus) to discuss a problem. Not only are opportunities lost for collaboration, but there are often unnecessary redundancies of effort and expense. Furthermore, the absence of an interdisciplinary collaborative approach results in failure to adequately train the workforce needed to address the emerging microbial threats facing the world today. While federally proposed Research Centers of Excellence in Biodefense and Emerging Infections may provide some of the infrastructure needed to address specific emerging disease threats (e.g., basic research, vaccine and antimicrobial drug development), they do not meet the need for such an approach.
Many types of infectious disease problems—including the search for infectious triggers of chronic disease, the emergence of antibiotic resistance, nosocomial infections, and zoonotic infections—could be addressed through an interdisciplinary approach. The majority of emerging diseases that
threaten humans are of zoonotic origin. In the past 10 years, the world has had to respond to Sin Nombre virus and other hantaviruses from rodents, Nipah virus from bats via pigs, influenza viruses from aquatic birds, and West Nile virus from birds via mosquitoes. Zoonotic diseases have also emerged when domestic animals have served as reservoirs. As discussed earlier, antimicrobial-resistant organisms have emerged in part as a result of the agricultural use of antimicrobials for disease prevention and growth promotion in chickens, pigs, cattle, and even fish and shellfish. Indeed anthrax, used as an agent of bioterrorism in 2001, is a naturally occurring zoonotic disease. The vulnerability of the United States and the developed world to agroterrorism attack using agents such as foot and mouth disease virus and the high socioeconomic cost of diseases of livestock could be better addressed through an interdisciplinary approach to microbial threats. It is imperative that those in the human, animal, agricultural, and environmental sciences come together to examine such threats.
Our understanding of many recent emerging disease threats has come mostly from a cadre of scientists who are at home in the laboratory, in the clinic, or in the field. These scientists have usually been formally trained in one medical/biomedical/veterinary discipline, but have gained additional training and experience in other disciplines pertinent to disease prevention and control. The disciplinary base of this cadre of scientists has been remarkably diverse, including clinical medicine, veterinary medicine, microbiology, virology, molecular biology, pathology, immunology, toxicology, epidemiology, public health, mammalogy, wildlife biology, medical entomology, and ecology. Some of these scientists have had valuable tertiary expertise as well, in such areas as epidemiologic modeling, GIS and remote sensing technologies, health education, administration and management, and public policy. Other relevant disciplines include economics, anthropology, and ethics.
This cadre of scientists has provided invaluable knowledge and skills to address the multidisciplinary nature of infectious disease control. Younger counterparts rely increasingly on molecular approaches and computer-based tools, and often lack training and experience in the basic disciplines most pertinent to infectious disease prevention and control, such as epidemiologic field observations and investigations. Unfortunately, today there are too few scientists who can bring to bear all the various tools and approaches that may be of use in the detection, diagnosis, investigation, prevention, and control of emerging infectious diseases.
These problems are not unique to infectious diseases. In the past, one solution has been to create centers of interdisciplinary excellence, perhaps best exemplified by the cancer research centers that have proven invaluable to advances in cancer prevention ansd treatment. Denmark has met the need for an interdisciplinary perspective in addressing emerging infectious dis-
eases by developing a zoonosis center as an element of its national public health institution, the Statens Serum Institut, uniting veterinary and human health professionals. The committee believes much could be gained if the United States were to create similar interdisciplinary infectious disease centers for research, education, training, and public service. Given the nation’s lack of infrastructure in this area, such centers would have to be established with bricks and mortar, not as completely virtual centers.
Interdisciplinary infectious disease centers should be developed to promote a multidisciplinary approach to addressing microbial threats to health. These centers should be based within academic institutions and link (both physically and virtually) the relevant disciplines necessary to support such an approach. They would collaborate with the larger network of public agencies addressing emerging infectious diseases (e.g., local and state health agencies, CDC, DOD, the U.S. Department of Energy, FDA, the Food Safety and Inspection Service, NIH, the National Science Foundation, USAID, USDA), interested foundations, private organizations, and industry. The training, education, and research that these centers would provide are a much-needed resource not only for the United States, but also for the entire world.
The proposed centers would provide space to bring people together so that their proximity would generate work across intellectual discipline– driven boundaries on a research agenda that requires a cross-disciplinary approach. This is exactly what comprehensive cancer centers have done so well, bringing together clinicians (pediatricians, internists, oncologists, radiologists, and surgeons), basic scientists, epidemiologists, pharmacologists, immunologists, virologists, cell biologists, structural biologists, radiation biologists, and radiation therapists. Interdisciplinary work requires that those involved have not only good will toward and awareness of each other, but also a means of actually talking to each other frequently, often casually—contacts that in time lead to new kinds of work that bridge multiple disciplines. Seminars on various arenas of work given regularly in a center help bring people and ideas together. Economists, sociologists, medical anthropologists, epidemiologists, medical geographers, and others might need to pool their talents with those of immunologists, vaccine developers, and infectious disease clinicians and health care workers to solve persistent problems of community- or regionally-based outbreaks of infection. A center would help unite faculty of schools of public health, medical school basic and clinical faculty, and local and state public health officials.
Nowhere is the opportunity for interdisciplinary work greater than in the global infectious diseases arena. To make such work a reality, we need to create space and support for people from multiple disciplines to work
|
A Vision for Interdisciplinary Infectious Disease Centers Interdisciplinary, multidisciplinary research projects. The centers would foster interactions among university faculty in public health, clinical medicine, veterinary medicine, and the relevant basic and social sciences, as well as among those working in local, state, and national public health systems. The centers would focus on high-priority public health problems and leverage expertise across multiple organizations. A training venue. The aim would be to develop future scientists and leaders with the kind of broad perspective needed to work across boundaries in their efforts to control new and reemerging microbial threats to health. A venue providing a point of entry or primary exposure for young scientists might take the form of summer fellowship programs, internships, and other training opportunities. A link to primary and reference laboratory diagnostic systems. No common ground for developing a proper national reference diagnostic system and communal repository of infectious agent stocks and other diagnostic reagents currently exists. The centers would also provide an ideal venue for creating such a system and for training in laboratory technologies. Information/database systems. Databases would provide pertinent epidemiological, diagnostic, and other important information for all members of the scientific community. Some aspects of this information/database system might be made available via the Internet, and an e-mail network (perhaps as a subunit of the highly successful Pro-Med system) might also be provided. |
together. Emerging infectious diseases and persistent infections create an urgent need for such centers. In addition to helping to meet national and even international needs in addressing emerging infectious diseases, the proposed centers would play a critical role in improving our national capacity to deal with nosocomial infections, drug resistance, socioeconomic issues in the emergence and transmission of infection, and the infectious etiologies of cancer and chronic inflammatory and degenerative diseases. Individual centers would be expected to have different foci of interest, so as to provide the nation with a broad-based ability to deal with infections of all kinds.
A case in point to suggest the value of such centers is the importance of addressing the zoonotic aspects of influenza. While WHO oversees global surveillance of human influenza through centers in London, Atlanta, Tokyo, and Melbourne, there is little interaction between this program and animal influenza surveillance programs. Influenza viruses that have been
transmitted from wild aquatic bird reservoirs through domestic poultry and pigs and then on to humans represent the most significant threat of influenza to humans. Although the Office International des Epizooties (OIE) deals with the reportable diseases of animals, only certain influenza virus types that are highly pathogenic for poultry are of concern—those viruses that are found to be rather nonpathogenic in poultry, but for which a potential threat to humans exists, are ignored. Given the remarkable mutability of influenza viruses, it might be expected that there would be strong links between WHO and its human influenza tracking system and OIE and its still-primitive animal influenza tracking system. It can be argued that such linkage will be effected only through the development of several interdisciplinary infectious disease centers focused on zoonoses.