6
Nucleic Acid Sequence-Based Identification for Detect-to-Warn Applications
Culture-based assays, which typically run for 12 to 24 hours or longer, are normally viewed as an unimpeachable standard for the identification (ID) of microbes. However, nucleic acid-based assays offer a relatively faster response time combined with high sensitivity and specificity, and this has catapulted them to the forefront for laboratory analysis and, in some cases, field analysis. The sequence of nucleic acids has been successfully used in laboratory settings to identify both nonpathogenic1 and pathogenic bacteria.2 Similar studies of microbial diversity have been performed using ribosomal RNA methods.3
It is the view of this committee that sequence-based ID using nucleic acid assays will play a critical role in any defensive architecture against biological agent attack, to confirm whether or not an attack has occurred and if it has, what types of biological agents were used.4 This confirmatory role is essential even if the response time of the assay is relatively slow—on the order of 15-30 minutes. This chapter examines whether DNA or RNA sequence-based detection and ID assays can be accelerated to provide a detect-to-warn capability with a response time on the order of 1 minute starting from aerosolized agent—and if so, how much specificity and sensitivity must be sacrificed in limiting the duration of the process and in automating the overall procedures for unattended field operation.
As discussed in Chapter 1, the sequence of processes that must be included within this response time is as follows:
-
Collecting the sample.
-
Preparing the sample for analysis. This includes removal of assay inhibitors and lysis of the target cells. For dirty samples or for nucleic acid analysis of a spore, these can be significant problems.
-
Performing the assay itself.
-
Analyzing and reporting the results of the assay.
At this writing, the committee is not aware of any existing nucleic acid analysis system that performs the full set of tasks described above for relevant aerosol concentrations of pathogenic organisms within the detect-to-warn time constraint of 1 minute. Presently, this type of multistep analysis is performed manually in laboratory settings by highly trained personnel and requires an hour or more to accomplish. Recent advances in the miniaturization of instrumentation, however, offer the possibility that such nucleic acid-based analysis protocols could be automated and performed in the field by dedicated stand-alone sensors, given much additional development.
This chapter describes representative present-day technologies that perform each of the functions listed above (in sequential order, as a sensor system would operate) and evaluates their potential for use in detect-to-warn applications. Aside from technological feasibility, other factors must also be considered: e.g., sensitivity, specificity, robustness to environmental contaminants, requirements for sample preparation, cost, storage and logistics, ease of implementation into field equipment, and level of multiplexing of the assays. The chapter concludes with the committee's key findings and recommendations.
SAMPLE COLLECTION
As with any other sensor methodology discussed in this report, a nucleic acid sensor would first need to collect an air sample of sufficient volume and pathogen density to be detectable in the assay. For most of the techniques described below, the assay is performed with the organism and target nucleic acid sequence in aqueous solution. Therefore, the air-collection devices (wetted-wall cyclones, air-to-air concentrators, etc.) described in Chapter 4 must convert the air sample to a fluid (hydrosol) and provide it to the assay instrumentation.
For nucleic acid-based detection and identification technologies, one can expect some trade-off between the time required for detection and the starting concentration. Rugged, field-tested aerosol collectors such as the XM-2 have been demonstrated to capture particles in the 1 to 10 μm range with an efficiency of 50 percent or higher, with an effective concentration factor on the order of 5 × 105 (that is, one captured particle per 500 liters of air is concentrated into 1 milliliter of collection fluid.) Bioaerosol collectors of even higher performance have been developed under the guidance of the U.S. Army Edgewood Chem-Bio Center (ECBC) at Aberdeen Proving Grounds.5
From the standpoint of system analysis for the detect-to-warn application, some miniaturization of the overall system dimensions will decrease the time required to transport and prepare the samples for their detection assays, so collection into 1 milliliter is probably less desirable than collection into a 100 microliter or smaller liquid volume. It is important to remember that decreasing the size of the system may lead to more rapid clogging of the fluidics, so that shorter maintenance intervals may be required with the reduction in system size. The same considerations apply to the use of multistage precollection fractionators that could provide concentration factors exceeding 106; improved overall performance is likely to come at the expense of more frequent maintenance.
SAMPLE PREPARATION
Nucleic acid amplification and analysis methods are sensitive to contamination by inhibitors, which can often accompany samples that are collected from the open environment. Furthermore, the nucleic acid of interest typically resides within a cell or spore and must be liberated or made available to the other chemical components of the assay. (While it is true that there is often some detectable, exogenous DNA that was trapped on or within the exosporium during the process of sporulation, assays based solely on this source of DNA may be less sensitive and less reliable.) For reliable assays based on nucleic acid sequences, other than assays that are intracellular, the nucleic acid must be separated from interfering or inhibiting components before the assay is performed.
FIGURE 6.1 Cepheid ultrasonic fluidic component for lysing spores to get access to DNA.
A number of effective methods have been developed over the years to lyse cells and remove contaminants from samples. Cell lysis using chaotropic salts (e.g., guanidinium isothiocyanate) is one effective method and is often followed by capture of the DNA (and not the dirt) on some silica-based substrate under high-salt conditions.6 Once the dirt is washed away, the DNA is then eluted from the silica in a low-salt buffer and then analyzed. This and similar methods are available commercially, and they are often performed in plastic tubes using a centrifuge7 or vacuum or pressure to transport and process the samples. These commercial techniques, while effective, require between 30 minutes and 2 hours and are therefore not suitable, in the committee's view, for consideration for the detect-to-warn application.
Recently, the chemistry that is described in the patent by W. Boom has been implemented using microfabricated silica surfaces within microfluidic systems.8 The committee is not aware, however, of any studies on the rates or kinetics involved in the use of such microfluidic silica surfaces for the overall process of trapping, washing, and releasing nucleic acids from lysed bacteria. Using the chemistry of Boom, Ness and Belgrader have demonstrated the polymerase chain reaction (PCR) of F. tularensis spiked into raw sewage water, which normally inhibits any PCR.9
One rapid method to lyse cells, including spores—which are difficult to disrupt—is ultrasonication with glass or composite microbeads (Figure 6.1). This method has been shown to open spores in as little as 10 seconds10 and has been integrated into an existing commercial DNA analysis instrument (Figure 6.2).11 This continues to be an area of

FIGURE 6.2 Arrays of microfabricated pillars with silica surfaces.
|
6 |
W.R. Boom, H.M.A. Adriaanse, T. Kievits, and P.F. Lens. August 10, 1993. Process for isolating nucleic acid. U.S. Patent 5,234,809. |
|
7 |
Qiagen at http://www.Qiagen.com and MoBio at http://www.Mobio.com. |
|
8 |
D.D. Hansmann, J.P. Grace, M.G. Lower, G.M. Oosta, N.W. Loomis, E.B. Shain, and T.G. Schapira. Devices and methods utilizing arrays of structure for analyte capture. January 13, 1998. U.S. Patent 5,707,799. D.D. Hansmann, J.P. Grace, M.G. Lower, G.M. Oosta, N.W. Loomis, E.B. Shain, and T.G. Schapira. Devices and methods utilizing arrays of structure for analyte capture. September 14, 1999. U.S. Patent 5,952,173. |
|
9 |
K. Ness and P. Belgrader, Lawrence Livermore National Laboratory. Unpublished data communicated to the committee in 2003. |
|
10 |
P. Belgrader, W. Benett, D. Hadley, J. Richards, P. Stratton, R. Mariella, Jr., and F. Milanovich. 1999. PCR detection of bacteria in seven minutes. Science 284:449-450. P. Belgrader, D. Hansford, G. Kovacs, K. Ventkateswaran, R. Mariella, Jr., F. Milanovich, S. Nasarabadi, M. Okuzumi, F. Pourahamadi, and M. Northrup. 1999. A minisonicator to rapidly disrupt bacterial spores for DNA analysis. Anal. Chem. 71:4232-4236. |
|
11 |
P. Belgrader, M. Okuzumi, F. Pourahamdi, D.A. Borkholder, and M.A. Northrup. 2000. A microfluidic cartridge to prepare spores |
active research. For the purposes of this report, the committee expects that it will be possible to implement an ultrasonication procedure with a duration of 10 seconds. There are no data regarding the percent of DNA or RNA that will be released by this process, but a reasonable guess might be that roughly 30 percent of the nucleic acids from spores or vegetative bacteria could be released within a sample volume of 100 microliters or less (see Box 6.1).
In other research, a flow-through high-frequency lysis module has been demonstrated to release DNA from spores without the addition of microbeads or chemicals.12 Processing without additives and in a flow-through manner will simplify integration with other sample processing modules and decrease cost due to consumables, which is important for detect-to-warn applications. The residence time of the continuously moving sample in the flow-through lysis module was not optimized; it ranged from about 10 to 60 seconds in the initial studies. The use of standing waves of ultrasound within a microfluidic channel has also been demonstrated to separate and concentrate particles that are suspended in an aqueous solution.13 This could be employed as a processing step within a detect-to-warn system.14
Methods for using electric fields to attract, focus, or separate bacteria, cells, and DNA in solution have been demonstrated and may work within time scales that have benefit for this application as well. The electrophoretic concentration and fractionation of bacteria are well documented in the literature. Typical electrophoretic mobility values for bacteria are on the order of 2 × 10-4 square centimeters per volt-second.15 A field strength of 100 volts per centimeter can result in an average velocity of 0.2 millimeter per second, which may be fast enough to be useful for detect-to-warn applications.16
The majority of biological particles are amphoteric; that is, they can be net negative, net positive, or net neutral, depending on the local pH. The pH at which a particle is neutral is called the isoelectric point (pI)—at this pH the particle experiences no net force when an electric field is applied. By generating a suitable pH gradient parallel to an electric field, particles in the field will migrate to their pI and remain fixed at that position. This technique has been used successfully to concentrate bacteria in flowing conditions.17
Finally, biological particles also become polarized when placed in an electric field. These induced dipoles can lead to net migration in an applied, nonuniform field, which is referred to as dielectrophoresis.18 The net velocity of a particle within such a nonuniform field depends on its dielectric
|
|
for PCR analysis. Biosensors and Bioelectronics 14:849-852. M.T. Taylor, P. Belgrader, B.J. Furman, F. Pourahamdi, G.T.A. Kovacs, and M.A. Northrup. 2001. Lysing bacterial spores by sonication through a flexible interface in a microfluidic system. Anal. Chem. 73:492-496. |
|
Box 6.1 Below, the committee outlines a detection system that is only slightly beyond what has already been demonstrated in controlled laboratory settings. The committee is relatively confident that a system could be produced that would be able to perform a single-target (selectable), real-time PCR assay within 3 minutes, starting with the collection of an aerosol sample. There are no data that would allow an extrapolation of such a system to the use of a deeply multiplexed (15 target organisms) PCR assay with any confidence. The assay proceeds through the following steps, with a time budget of 180 seconds: sample collection, sample preparation, performance of an assay, and analysis and reporting.
|
function relative to that of the surrounding media as well as on the frequency and strength of the applied electric field. In batch mode, cells can travel at velocities up to 0.1 millimeter per second.19 Early prototypes of dielectrophoresis-based devices can be rather small and simple in appearance (Figure 6.3).
|
|
Y. Huang, J. Yang, X.B. Wang, F.F. Becker, and P.R. Gascoyne. 1999. The removal of human breast cancer cells from hematopoietic CD34+ stem cells by dielectrophoretic field-flow-fractionation. J. Hemat. Stem Cell Res. 8:481-490. T. Schnelle, T. Muller, R. Hagedorn, A. Voight, and G. Fuhr. 1999. Single micro electrode dielectrophoretic tweezers for manipulation of suspended cells and particles. Biochem. Biophys. Acta 1428(1):99-105. H. Morgan, M.P. Hughs, and N.G. Green. 1999. Separation of submicron bioparticles by dielectrophoresis. Biophysical Journal 77:516-525. T. Schnelle, T. Muller, G. Gradl, S.G. Shirley, and G. Fuhr. 2000. Dielectrophoretic manipulation of suspended submicron particles. Electrophoresis 21:66-73. R. Miles, P. Belgrader, K. Bettencourt, J. Hamilton, and S. Nasarabadi. 1999. Dielectrophoretic manipulation of particles for use in microfluidic devices. J. Microelectromechanical Systems 1:497-501. |
NUCLEIC ACID ASSAYS
Assays based on nucleic acid sequencing can be divided into two main groups:
-
Group I: techniques that use amplification of one or more target sequences of the nucleic acid (RNA and DNA require different protocols) and
-
Group II: techniques that do not use amplification of the nucleic acid.
The following sections describe assays in each category, with specific comments on the potential for detect-to-warn applications. For each of the techniques described, there may be multiple methods to detect the presence of the target sequence(s). These may be based on use of fluorescent labels, chemiluminescent reporters, colorimetric labels, mass-spectrometric tags, or other sensing means. The committee tried to consider and describe the tagging or sensing methods that seem most useful for rapid detection and to identify areas in which further improvements in speed and sensitivity are required in order to enhance the potential of the technology for detect-to-warn applications.

FIGURE 6.3 Photograph of hybrid microfluidic circuit that contains a commercial micropump (Mainz, Germany), a dielectrophoresis chip, load and waste wells, and fluidic interconnects, potted in silicone rubber.
Both Group I and Group II techniques can use hybridization (binding) of single-stranded target DNA of unknown sequence to arrays of potentially complementary sequences of single-stranded DNA.20 The concept of hybridization to DNA arrays fabricated on flat substrates such as plastic or glass is illustrated in Figure 6.4. Short strands of single-stranded DNA, so-called oligonucleotides ("oligos") are synthesized in known sequences and attached to spots in the array in a known and predetermined way. Detection of the spots that contain hybridized target DNA after the hybridization event allows one to infer the sequence of DNA in the unknown target. The utility of this technique, in combination with other DNA and RNA manipulation techniques, is discussed in greater detail in the remainder of this chapter. RNA readily
FIGURE 6.4 Concept of the detection of the sequence of unknown target DNA via hybridization to oligonucleotides in an array. This has become a popular method to detect DNA sequences, identify organisms, and study gene expression.
hybridizes to form double-stranded structures to its complementary sequence (A-T, C-G, G-C, U-A). Thus, arrays of single-stranded DNA can be used to detect RNA via hybridization.
Work has been published21 that describes a large acceleration of the process of hybridization of nucleic acids to surface-immobilized DNA using electrophoretic transport of the nucleic acid in solution to specific sites on the array (Figure 6.4).
Group I: Assays That Use Amplification Techniques
The majority of nucleic acid-sequence-based assays that are performed today rely on amplification of target sequences at some point in their procedures. The reason for this is that a number of amplification techniques have been demonstrated to perform, under reproducible conditions, selective amplification and copying of target sequences of DNA or RNA by factors of 109 or higher. This ability to generate such high numbers of the selected sequence permits the detection of the sequence of interest using relatively easy methods, even when few copies of the sequence of interest were originally present. Single-copy amplification and detection have been reported routinely, although the risk of false negative results due to Poissonian sampling errors normally compels the practitioners to avoid working at such low starting numbers.22
All of the amplification techniques employ enzymes or engineered fragments of enzymes as critical, consumable reagents. Also, all of these techniques consume nucleotides A, C, G, and T or U as part of the chemical reactions that copy the sequence of interest. The PCR method (see below) requires both
short oligomers, known as primers, as well as the individual nucleotides, while the ligase chain reaction (LCR),23 for example, requires oligomers, but does not require individual nucleotides. One could contemplate the reclamation and reuse of some reagents, such as the enzymes, salts, and nucleotides, but repeated operation of any of the amplification techniques tends to generate spurious products, even in the absence of the official target sequence.
Single-target assays have been performed using well-characterized samples (known to have no inhibitors), wherein results have been observed within 5 to 7 minutes of the time of starting the assay.24 As performed in typical laboratory settings, however, the overall process of collecting a sample, preparing it, performing the assay and detection process, and analyzing the results requires at least 1 hour.
Polymerase Chain Reaction Method
The vast influence of PCR is reflected by the fact that the PubMed search engine found more than 30,000 publications in 2001 and 2002 (as of August 2002) with the word PCR in their titles. PCR is an enzyme-based chemical reaction25 that manufactures copies of one or more selected regions of double-stranded DNA sequences, known as target sequences, on samples of DNA, referred to as substrates. Not surprisingly, the enzyme that is the heart of PCR is polymerase, which is found in various forms in all bacteria and higher organisms. PCR per se does not work with RNA.26
Numerous commercial versions of the polymerase enzyme or its engineered fragment are available today. The one feature that all commercial polymerases used in PCR have in common is that they are thermally stable, so they are not destroyed when heated to 96°C or 97°C for a few seconds per cycle. They can perform their copying and amplification process over a range of temperatures from 50°C to above 70°C. The process of copying the DNA sequence of interest, known as extension, may proceed at a rate of roughly 100 bases per second, depending upon the conditions. In linear amplification, only one strand of a double-stranded Watson-Crick pair is copied. However, in typical PCR, both complementary sequences of a Watson-Crick pair are copied during each cycle of the PCR. Thus, after N cycles, assuming that the process has worked efficiently (high productivity), for each copy of target DNA sequence that was present when the reaction process began, the reaction will have manufactured (2N−1) copies. If the productivity of the system is low, however, one may find little or no amplification after N cycles.27
Cycling times of 15 to 20 seconds have been reported with high productivity for a single-target PCR assay using 25-microliter reaction volumes in a polypropylene sample tube, starting with 50 through 5,000 bacteria.28 If the appropriate reagents are used so that more than one sequence and its complement are
|
23 |
D.Y. Wu. 1989. The ligation amplification reaction (LAR)—Amplification of specific DNA sequences using sequential rounds of template-dependent ligation. Genomics 4:560-569. K. Backman. 1992. Ligase chain reaction: Diagnostic technology for the 1990s and beyond. Clinical Chemistry 38(3) 457-458. |
|
24 |
Belgrader et al., 1999. See note 10 above. |
|
25 |
K. Mullis, F. Faloona, S. Scharf, R. Saiki, G. Horn, and H. Erlich. 1986. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harbor Symposia on Quantitative Biology 51:263-273. K.B. Mullis, H.A. Erlich, N. Arnheim, G.T. Horn, R.K. Saiki, and S.J. Scharf, Cetus Corporation. July 28, 1987. Process for amplifying, detecting, and/or cloning nucleic acid sequences. U.S. Patent 4,683,195. R.K. Saiki, D.H. Gelfand, S. Stoffel, S.J. Scarf, R. Higuchi, G.T. Horn, K.B. Mullis, and H.A. Ehrlich. 1988. Primer-directed enzymatic amplification of DNA with thermostable DNA polymerase. Science 239:487-491. |
|
26 |
Roche Molecular Systems has developed an RNA assay, AMPLICOR, which detects HIV, an RNA virus, using the reverse transcriptase activity of one polymerase enzyme in a single-tube assay. There are also single-tube assays that use a separate reverse transcriptase enzyme to convert RNA into DNA. Once this step is performed, the reaction mixture can be heated to 96°C, at which point the PCR may begin, and the reverse transcriptase enzyme is permanently denatured. |
|
27 |
In this publication, even starting with 108 copies of target DNA in a well-characterized sample, running only a single-target assay showed a rapid decrease in detectable product when 20 cycles were performed with a total duration below 3 to 4 minutes; See, for example, M.U. Kopp, A.J. Mello, and A. Manz. 1998. Chemical amplification: Continuous flow PCR on a chip. Science 280:1046-1048. |
|
28 |
Belgrader et al., 1999. See note 10 above. |
copied, the process is referred to as multiplex PCR. It is commonly found that multiplex PCR requires longer cycling times.29
There are several methods of monitoring the products of PCR. One of the most accurate is the use of fluorogenic probes. Examples are Taqman30 or Molecular Beacons.31 These reactions are sometimes referred to as real-time PCR, since it is possible to monitor the reaction yield after every cycle of the PCR.32 When compared with the original PCR procedure that required electrophoresis to determine the presence of product(s), real-time PCR has provided increased sensitivity, specificity, and rapidity of the assay.33 When the samples are well characterized, it is possible both to detect and to quantify the number of starting copies of target DN A in the PCR.
When PCR is used without fluorogenic probes, the product is typically detected using gel or capillary electrophoresis or via hybridization against an array or microarray of probes (see discussion below on microarrays and HySeq patents and Affymax patents). The overwhelming majority of PCR that is performed around the world today is not real-time PCR.
Reliable PCR has several requirements. First, the reagents and cycling temperatures that are used to perform the assay must have been thoroughly tested and optimized. The instrument that is used must perform as required in terms of cycling times and temperatures. Finally, the sample must have been thoroughly evaluated by experts,34 who then guide the necessary sample preparation35 prior to the PCR.
One exciting recent development has been the use of PCR with other techniques in order to increase the power of the overall procedure. For example, in situ PCR is being used with flow cytometry.36 Also, PCR has been integrated with electrophoresis to increase the throughput of the assays.37 Similarly, the use of mass spectroscopy has enabled rapid throughput of PCR analyses.38
|
29 |
S. Nasarabadi, Lawrence Livermore National Laboratory. Discussions with the committee in 2002. |
|
30 |
R. Higuchi, C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions. Biotechnology 11:1026-1030. M.S. Ibrahim, R.S. Lofts, P.B. Jahrling, E.A. Henchal, V.W. Weedn, M.A. Northrup, and P. Belgrader. 1998. Real-time microchip PCR for detecting single-base differences in viral and human DNA. Anal. Chem. 70:2013-2017. Belgrader et al., 1999. See note 10 above. P. Belgrader, W. Benett, D. Hadley, G. Long, R. Mariella, Jr., F. Milanovich, S. Nasarabadi, W. Nelson, J. Richards, and P. Stratton. 1998. Rapid pathogen detection using a microchip PCR array instrument. Clinical Chemistry 44:2191-2194. |
|
31 |
S. Tyagi and F.R. Kramer. 1996. Molecular beacons: Probes that fluoresce upon hybridization. Nature Biotechnology 14:303-308. |
|
32 |
One of the most powerful Taqman instruments in terms of spectroscopic analyses and depth of possible multiplexing does not provide real-time data after each cycle. One must wait until all of the preprogrammed thermal cycles have run and the analysis software has performed its functions before one may view the data. Total elapsed time has typically been 2 to 3 hours. |
|
33 |
Higuchi et al., 1993. See note 30 above. Belgrader et al., 1999. See note 10 above. Belgrader et al., 1998. See note 30 above. |
|
34 |
T. Moretti, B. Koons, and B. Budowle. 1998. Enhancement of PCR amplification yield and specificity using ampliTaq Gold DNA polymerase. Biotechniques 25:716-722. |
|
35 |
P. Belgrader, D. Hansford, G. Kovacs, K. Ventkateswaran, R. Mariella, Jr., F. Milanovich, S. Nasarabadi, M. Okuzumi, F. Pourahamadi, and M. Northrup. 1999. A minisonicator to rapidly disrupt bacterial spores for DNA analysis. Anal. Chem. 71:4232-4236. |
|
36 |
B.K. Patterson, C. Goolsby, V. Hodara, K.L. Lohman, and S.M. Wolinsky. 1995. Detection of CD4+ T cells harboring human immunodeficiency virus type 1 DNA by flow cytometry using simultaneous immunphenotyping and PCR-driven in situ hybridization: Evidence of epitope masking of the CD4 cell surface molecule in vivo. Journal of Virology 69:4316-4322. D.E. Gibellini, M.C. Re, G. Furlini, and M. La Placa. 1997. Flow cytometry analysis of an in situ PCR for the detection of human immunodeficiency virus type-1 (HIV-1) proviral DNA. Methods in Molecular Biology 71:113-122. E.M. Gaynor, M.L. Mirsky, and H.A. Lewin. 1996. Use of flow cytometry and RT-PCR for detecting gene expression by single cells. Biotechniques 21:286-291. |
|
37 |
J. Cheng, E.L. Shelton, L.Wu, A. Uribe, L.O. Gerrue, J. Carrino, M.J. Heller, and J.P. O’Connell. 1998. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nature Biotechnology 16:541-546. |
|
38 |
N.H. Chiu, K. Tang, P. Yip, A. Braun, H. Koster, and C.R. Cantor. 2000. Mass spectrometry of single-stranded restriction fragments captured by an undigested complementery sequence. Nucleic Acids Research 28:e31. |
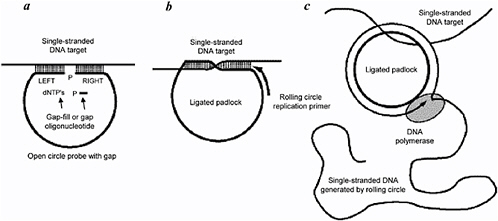
FIGURE 6.5 Rolling-circle amplification (RCA) technique: (a) a single-stranded DNA oligonucleotide probe with recognition sequences on each end binds to (recognizes) complementary target sequences in a single-stranded DNA target. A small gap-fill oligonucleotide fills the gap in the probe and completes a double-strand section to make a so-called ligated padlock, or circle of DNA hybridized to the target; (b) and (c) a DNA polymerase replication primer is added that continuously runs around the ligated padlock circle, replicating a long strand of DNA that contains many repeats of sequence complementary to that of the circle. SOURCE: P.M. Lizardi, X. Huang, Z. Zhu, P. Bray-Ward, D.C. Thomas, and D.C. Ward. 1998. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genetics 19:225-232.
Techniques Based on Amplification That Do Not Use PCR
There are a number of techniques that use amplification of the nucleic acid target sequence that do not employ what is strictly defined as classical PCR. For example, as shown in Figure 6.5, rolling circle amplification (RCA) uses a relatively small circular template of single-stranded DNA in a mixture of polymerase and appropriate reagents to generate long strands of complementary DNA (cDNA) that contain many repeats of the sequence in the circular template.39 The small circular template can be used as a probe to bind to and thereby identify unknown target DNA. After the RCA is completed, the target DNA ends up being attached to a long strand of cDNA. RCA has been demonstrated to work with intracellular RNA as the template (in situ detection), but this requires the use of multiple enzymes and does not proceed quickly enough to be applicable in the detect-to-warn application.40
Copy strands containing hundreds of repeats can be generated in as few as 2 minutes,41 although the reaction presently requires 1 to 2 hours of preparation and ligation time. An important advantage of RCA over PCR is that it does not require thermal cycling—the copying reaction can proceed isothermally once it begins—which simplifies the instrumentation. RCA is a linear amplification process, while PCR is an exponential amplification process. The fact that RCA produces a substantial amount of cDNA (thereby making the detection easier) in a few minutes gives it some far-term promise as a technology for fast DNA
|
39 |
P.M. Lizardi, X. Huang, Z. Zhu, P. Bray-Ward, D.C. Thomas, and D.C. Ward. 1998. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genetics 19:225-232. B. Schweitzer, S. Wiltshire, J. Lambert, S. O'Malley, K. Kukanskis, Z. Zhu, S.F. Kingsmore, P. M. Lizardi, and D.C. Ward. 2000. Immunoassays with rolling circle DNA amplification: A versatile platform for ultrasensitive antigen detection. Proc. Natl. Acad. Sci. 97:10113-10119. |
|
40 |
A.T. Christian, M.S. Pattee, C.M. Attix, B.E. Reed, K.J. Sorensen, and J.D. Tucker. 2001. Single-base and mRNA detection by rolling circle amplification in individual cells. Proc. Natl. Acad. Sci. 98:14238-14243. |
|
41 |
Lizardi et al., 1998. See note 39 above. |
detection, provided there is substantial additional development. If one started with a relatively high number of copies of the sequence of interest, RCA might produce a detectable amount of product before PCR did.
Strand-displacement amplification (SDA) uses a recognition sequence such as 5'-GTTGAC-3' that is hybridized to the end of the target DNA via a primer. This recognition sequence is nicked in each cycle and a fragment of an appropriate polymerase binds to this site, replicates the complementary 3'-5' target strand, and the enzyme displaces the original 5'-3' strand, freeing it to diffuse away in solution (see Figure 6.6).42 After an initial heating step to denature the original target DNA, this process proceeds isothermally without heating cycles and can produce 1012 copies of a single target DNA sequence in 30 minutes at 37°C.43 An in situ hybridization version of the technique has been used to detect as few as one gene copy of human immunodeficiency virus (HIV) DNA in individual cells.44 Though reasonably fast in its amplification and very sensitive, this technique so far requires more than 2 hours of sample and reagent preparation time.
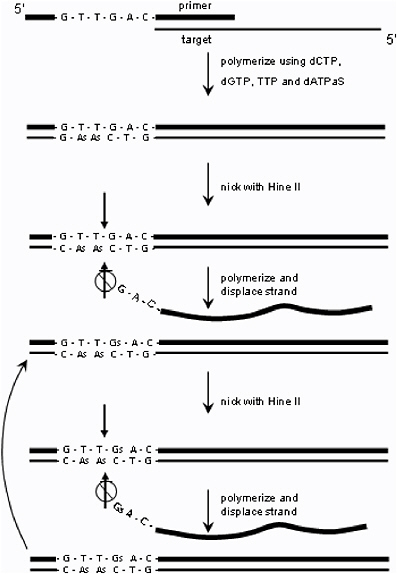
FIGURE 6.6 Strand-displacement amplification (SDA).
As shown in Figures 6.7 and 6.8, a hybrid of RCA and SDA can be achieved by first replicating cDNA from a circular template and then using SDA on the resulting cDNA strands to create dendritic trees of cDNA (so-called branched DNA or bDNA), all bound to a central core.45 Such techniques may merit further monitoring and consideration, simply because of their ability to produce copious quantities of cDNA in a few minutes.
Another technique that can amplify DNA or RNA is nucleic acid-sequence-based amplification (NASBA).46 The process uses a collection of enzymes (T7 RNA polymerase, Ribonuclease (RNase) H,
|
42 |
T.G. Walker. 1992. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc. Natl. Acad. Sci. 89:392-396. T.G. Walker. 1992. Strand displacement amplification—An isothermal, in vitro DNA amplification technique. Nucleic Acids Research 20(7):1691-1696. |
|
43 |
G. Nuovo. 2000. In-situ strand displacement amplification: An improved technique for the detection of low copy nucleic acids. Diagnostic Molecular Pathology 9(4):195-202. |
|
44 |
Walker, 1992. See note 42 above. |
|
45 |
Lizardi et al., 1998. See note 39 above. F.B. Dean, J.R. Nelson, T.L. Giesler, and R.S. Laskin. 2001. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11:1095-1099. |
|
46 |
J. Compton. 1991. Nucleic acid sequence-based amplification. Nature 350:91-92. L. Malek, S. Darash, C. Davey, G. Henderson, M. Howes, P. Lens, and R. Sooknanan. 1992. Application of NASBA isothermal nucleic-acid amplification method to the diagnosis of HIV-1. Clinical Chemistry 38:458. |
alfalfa mosaic virus (AMV) reverse transcriptase, and other reagents) to copy RNA or DNA isothermally at room temperature. Amplification factors of 108 can be obtained in 60 minutes. Because it can replicate messenger RNA (mRNA), which is present typically in viable organisms only, nucleic acid sequence-based amplification (NASBA) has been used not only for detection and identification but also for the detection, identification, and discrimination of viable organisms via the detection of mRNA expressed due to a heat-shock step.47 The fact that this technique is isothermal, can replicate RNA as well as DNA, and can be used as a viability test might allow it to be used as a second confirmation step following a detect-to-warn stage.
Another non-PCR amplification method considered is the ligase amplification reaction (LAR), also known as the ligase chain reaction (LCR).48 This method, mentioned briefly above, is similar to PCR in its need for thermal cycling but uses two oligonucleotide probes that bind to adjacent sites on the target DNA at the lower temperature of the thermal cycle. The two oligonucleotides then ligate and are driven off (denatured) as the temperature cycles to the peak. This process is repeated to create many copies of the ligated oligo
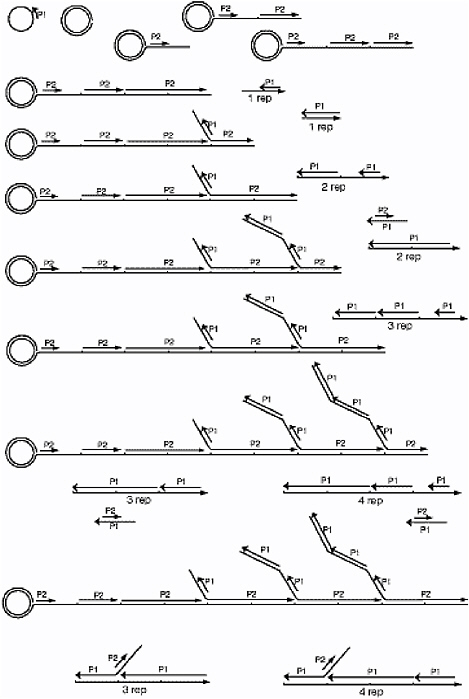
FIGURE 6.7 A combination of RCA and SDA.
|
|
H. Revets, D. Marissens, S. de Wit, P. Lacor, N. Clumeck, S. Lauwers, and G. Zissis. 1996. Comparative evaluation of NASBA HIV-1 RNA QT, AMPLIcor-HIV monitor, and QUANTIPLEX HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 34:1058-1064. |

FIGURE 6.8 Another combination of RCA and SDA.
molecules and is claimed to have the ability for single-base-mutation discrimination (i.e., the oligos do not ligate if there is a mutation at the juncture site). Since this method requires thermal cycling and the accurate hybridization and ligation of relatively long oligomers of DNA for each cycle, the committee does not believe it is a contender for use in a 1-minute detection system.
Finally, rapid doubling times for RNA have been reported with the use of Qß replicase. For example, the MDV fragment has been copied using Qß replicase in 12 seconds. The conditions for which this was achieved were limited, and the majority of RNA fragments that were studied in this research did not display such rapid doubling times.49 Until this technique is shown to work with RNA from lysed cells and in a multiplex format, the committee cannot include it as a likely component of a 1-minute detection system.
The idea of producing an integrated system that does rapid identification using PCR or other target-amplification assay in under 5 minutes is already being pursued at the research stage around the world. Although this research is still ongoing, the committee judges that the probability of a first prototype demonstration being realized within 2 years is reasonably high.
Detection of Amplified Target Sequences Using Array Technologies
Since the late 1980s, DNA microarray (so-called "DNA chip") technology has been developed to provide a method for the parallel analysis of target DNA strands.50 The technique builds upon the work of Southern51 and typically deposits an array of synthesized oligos in spots on a two-dimensional surface of silica or various polymers (Figure 6.4). The coding sequence of oligos in each site is the same and is known, and it differs from site to site in a known and designed way.
Target DNA strands that are washed onto the array and allowed to settle will hybridize to oligos having an exactly complementary sequence but will only weakly bind to oligos that do not have an exact match if the chemistry of the solution is properly adjusted. The hybridized target DNA is usually labeled in some manner such as with a fluorescent dye molecule so that the pattern of hybridization across the array can be detected. The sequence of the original target DNA can then be inferred using a computer algorithm, provided that the coding sequence of the oligos in the array was properly designed and that the target DNA strands are not too long and do not have large repeats of sequence. Much work has been put into the development of this technology over the past 10 years, and it is now used ubiquitously in gene-expression studies, drug development, bioagent analysis and detection, and many other fields. The concept of using such an array of oligos to determine the sequence of a sample is known as sequencing by hybridization.52
|
49 |
J.L. Burg, A.M. Juffras, Y.Wu, L. Blomquist, and Y. Du. 1996. Single molecule detection of RNA reporter probes by amplification with Qß replicase. Mol. and Cellular Probes 10:357-271. J.S. Shah, J. Liu, J. Smith, S. Popoff, G. Radcliffe, W. J. O’Brien, G. Serpe, D.M. Olive, and W. King. 1994. Novel, ultrasensitive, Q-beta replicase amplified hybridization assay for detection of Chlamydia trachomatis. J. Clin. Microbiol. 32:2718-2724. S. Paillasson, S.M. Van De Corput, R.W. Dirks, H.J. Tanke, M. Robert-Nicoud, and X. Ronot. 1997. In-situ hybridization in living cells: Detection of RNA molecules. Experimental Cell Research 231:226-233. |
|
50 |
Eggers et al., 1993. See note 20 above. J.B. Lamature, K.L. Beattie, B.E. Burke, M.D. Eggers, D.J. Ehrlich, R. Fowler, M.A. Hollis, B.B. Kosicki, R.K. Reich, S.R. Smith, R.S. Varma, and M.E. Hogan. 1994. Direct detection of nucleic acid hybridization on the surface of a charge coupled device. Nucleic Acids Research 22(11):2121-2125. S.P.A. Fodor. 1997. DNA sequencing—Massively parallel genomics. Science 277(5324):393. |
|
51 |
Southern, 1982. See note 20 above. |
|
52 |
M.C. Pirrung, J.L. Read, S.P.A. Fodor, and L. Stryer, Affymax Technologies. September 1, 1992. Large scale photolithographic solid phase synthesis of polypeptides and receptor binding screening thereof. U.S. Patent No. 5,143,854. |
One of the drawbacks of conventional DNA microarray technology for the detect-to-warn application is that the conventional hybridization step and subsequent rinses typically can take several hours. Another drawback is that nonspecific binding of target DNA to oligos can occur, and one typically needs well-controlled experiments and good imaging and computer algorithms to correct for this. One of the most important drawbacks is that the technology typically uses PCR to amplify the amount of target DNA before applying it to the array to maximize the signal. These factors all combine to make the conventional DNA microarray technology unsuitable for detect-to-warn applications.
To address these drawbacks, workers have improved the speed of hybridization to times of less than 5 minutes by using electrophoretic transport to draw the target DNA to the oligo sites quickly, as discussed with Figure 6.4.53 This particular approach has still required the use of PCR-amplified and -labeled target DNA, and the amplification steps take time. Others have developed alternative labeling systems that use electrically or optically sensed labels to try to improve the speed and sensitivity of detection,54 but these methods do not meet the time requirements for detect-to-warn and still require PCR-amplified DNA for trace-level detection. To try to eliminate the effort and time required for the labeling of target DNA, workers have also developed a hybridization-detection method using permittivity sensing of changes in the electrochemical boundary layer on electrodes in the test sites to which the oligos are attached.55 The relative signal change sensed by this method using even PCR-amplified target DNA was relatively small (less than 20 percent), and it does not seem to be a sufficiently robust candidate to improve microarray performance. Surface-plasmon resonance has also been reported in the literature as a technique that can detect hybridization without the use of labels.56 Direct, unlabeled detection of 16S rRNA on DNA microarrays has also been reported on a notional detector, as described in Box 6.2.
Techniques based on mass spectrometry have also been developed to detect nucleic acid sequences.57 While these work quite well in the laboratory, they do require about 108 molecules for detection and thus PCR amplification, which slows them down, as well as vacuum pumps and sophisticated instrumentation. In addition, they require significant sample preparation and cleanup for contaminated samples and have difficulty deconvolving the signals from mixtures of DNA from different organisms that may be present in environmental samples if the selectivity of the PCR amplification is insufficient. It is difficult to see how these mass-spectrometry techniques could be implemented conveniently and inexpensively in field- or building-monitoring situations for detect-to-warn requirements.
|
|
W.J. Dower, S.E. Cwirla, and R.W. Barrett, Affymax Technologies. July 11, 1995. Peptide library and screening systems. U.S. Patent No. 5,432,018. M.C. Pirrung, J.L. Read, S.P.A. Fodor, and L. Stryer, Affymax Technologies.April 11, 1995. Large scale photolithographic solid phase synthesis of an array of polymers. U.S. Patent No. 5,405,783. D. Campbell, Affymax Technologies. Novem ber 8, 1994. Chiral synthesis of alpha-aminophosponic acids. U.S. Patent 5,362,899. |
|
Box 6.2 Below, the committee describes an identification system that does not exist but that might, conceivably, be created to function within the 1-minute time limit that is desired for the detect-to-warn (DTW) application. An estimate is made of the performance requirement of a conceptual DTW system against a hypothetical aerosol challenge. The committee first estimates the physiologically relevant concentration of this agent in air, looking at the required sensitivity that this DTW system must have to detect the minimum concentration of agent that would present a hazard if it is breathed for 5 minutes. It is assumed that the individuals involved would take protective action within a few minutes of being warned. The lower limit of sensitivity is examined, because a system with poor performance in terms of sensitivity would be relatively prone to false negatives, even for hazardous levels of agent. The committee starts by estimating the physiologically relevant concentration of an aerosolized pathogenic agent inside of a building, for a person not particularly exerting herself or himself. The exchange of air within the alveoli (the innermost surface of the lungs) is approximately 15 liters per minute. It is also assumed that the agent is present in its spore form, with a physiologically hazardous (LD50) dose of roughly 8,000 spores. For a worst case analysis, it is assumed that 100 percent of the spores are viable and 100 percent of the spores that are breathed into the alveoli are captured and can begin their infective actions. Based on these assumptions, the aerosol concentration of these spores that would present an LD50 dose within 5 minutes of breathing would be 100 spores per liter of air.a If the capture efficiency in the lungs is lower or if the percent of viable spores is lower, then the corresponding aerosol concentration that would present a hazard within a time frame of 5 minutes would be higher. The assay proceeds through the following steps, with a time budget of 60 seconds: sample collection, sample preparation, performance of an assay, and analysis and reporting.
|
|
Group II: Sequence-Based Assays That Do Not Use Amplification Techniques
Although there have been musings about directly reading the sequence of DNA by using a mass spectrometer or a scanning-probe microscope or a nanometer orifice, the techniques that are used today to determine the sequence of RNA or DNA are based upon hybridization.58 These techniques include sequencing by hybridization, such as array-based techniques.59 Multiple signal-transduction mechanisms are also possible, including signal amplification, which the committee views as different from sequence amplification. One example of a signal amplification technique is the use of a label that chemiluminesces or that can undergo repeated electrochemical oxidation-reduction reactions. Thus, if a single hybridization event occurs with DNA that is carrying this label, the label can emit many photons via chemiluminescence—horseradish peroxidase/substrate reactions, used in enzyme-linked immunosorbent assay (ELISA) or the Origen biodetector—or can participate in cyclic voltammetry using ruthenium complexes,60 ferrocene complexes,61 or osmium tetraoxide-2,2'-bipyridine.62
|
58 |
The nucleic acid sequencing work in the Human Genome Project relies primarily on capillary electrophoresis. However, all that is necessary for identification of the agents considered here is the detection of small fragments of virulent genes or plasmids. This is commonly done via hybridization. |
|
59 |
W.J. Wilson, C.L. Strout, T.Z. DeSantis, J.L. Stilwell, A.V. Carrano, and G.L. Andersen. 2002. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell Probes 16:119-127. |
|
60 |
J. Miller, N. Frank, and T.J. Mead. 2001. Investigations of 5’-labeled ruthenium nucleotides as electron acceptor complexes. Abstr. Papers Am. Chem. Soc. 221:150. |
|
61 |
Yu et al., 2001. See note 54 above. |
|
62 |
E. Paleek, M. Fojta, and F. Jelen. 2002. New approaches in the development of DNA sensors: Hybridization and electrochemical detection of DNA and RNA at two different surfaces. Bioelectrochemistry 56:85-90. |
An alternative to the sample preparation strategy described in the previous section (that of lysing the cells or spores and freeing the DNA from them) is to try to insert or diffuse the appropriate amplification/detection reagents into cells that are still intact or have had their outer membranes permeabilized. This method may avoid some of the time required for sample preparation. This so-called in situ detection (also known as in situ hybridization if it uses labeled single-stranded DNA probes) has been tried successfully with modifications of the RCA technique described above.63 A similar method has also been demonstrated using a technique known as catalyzed signal amplification (CSA). This technique employs biotinylated DNA probes and a colorimetric detection process based on peroxidase-conjugated streptavidin, which is activated by application of the chromogenic substrate diaminobenzidine.64 In this latter method the DNA is not amplified; instead, many copies of a marker dye are generated via the peroxidase/diaminobenzidine reaction. Very sensitive single-gene-copy detection within a cell has been demonstrated, though the cell preparation time is far too long (assay time of more than 2 hours in these cases) for the detect-to-warn application,65 showing that single molecules can be probed and imaged inside single cells, though much work remains to develop this technique further.
In a more positive vein, molecular beacons (Figure 6.9) are molecules that exhibit relatively little fluorescence when unbound to target DNA but fluoresce when bound to their complementary sequence.66 Molecular beacons could be attractive for the detect-to-warn application if means could be found to bring them together with the DNA or RNA in cells quickly, perhaps using electric-field effects or other methods to overcome the time lag due to diffusion as described above. Trace-level detection could be enhanced for the molecular beacons via development of quenchable Stokes-shift dyes (similar to Cy3 or Cy5) to minimize signal background and/or via development of more strongly emitting dyes.
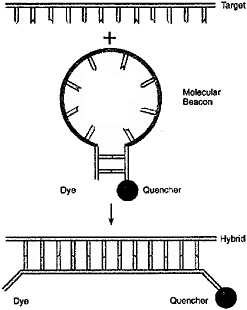
FIGURE 6.9 Concept of a molecular beacon.
In an idea similar to molecular beacons and depicted schematically in Figure 6.10, Nie and coworkers have demonstrated the use of gold and silver nanoparticles as both attachment substrates and quenchers for fluorescently labeled oligonucleotides.67 They report enhanced discrimination of single-base mismatches between oligo probes and the target DNA, although the hybridization kinetics are slow for both the nanoparticle-based probes and molecular beacons (50 percent hybridization is achieved in about 10 minutes). The slow hybridization is explained by a strong stability of the fluor-oligo-nanoparticle (or quencher) complex, limiting the tendency of the oligo to unfold and bind with the target. These are attractive areas and might merit further consideration, but they will need much additional work to improve the speed of the assay.
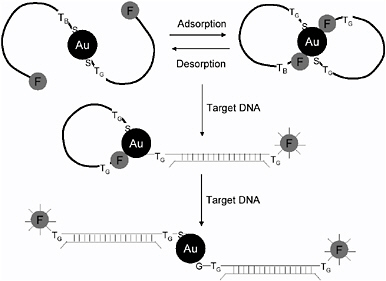
FIGURE 6.10 Nanoparticles used for DNA labeling and detection. In this method, oligos are tethered on one end to Au or Ag nanoparticles (dark circles) and are conjugated to fluorophores on their other end (grey circles). The fluorophores have some tendency to adhere to the nanoparticles under quiescent conditions, and their fluorescence is quenched in this mode. When a short single strand of target DNA hybridizes to the oligo, however, a rigid double-stranded section of DNA is formed that pulls the fluorophore from the surface of the nanoparticle, allowing it to emit light again.
Nie and coworkers have also shown that 20-nanometer fluorescent nanoparticles can be conjugated to DNA-binding proteins such as the restriction enzyme EcoRI, bound to specific sites on target DNA strands, and imaged once bound.68 This is interesting work, but so far the experimental preparation times far exceed that required for detect-to-warn.
Another approach to signal amplification would be to use a probe with a complementary sequence for rolling circle amplification. If such a probe hybridized to a surface-immobilized sequence but maintained its RCA target sequence as unhybridized, one could, after washing the array, add a circular template that was complementary to the unhybridized portion and run RCA to generate long strands of linear sequence, complementary to the circular probe. If the circular probe possesses a sequence that is complementary to a Molecular Beacon probe, then these could attach at every corresponding complementary sequence on the growing linear strand, producing hundreds of fluorescent labels for each original hybridization event.
Detection of the hybridization of DNA is the heart of both the Taqman and Molecular Beacon probes. There has been a report of the use of surface-bound Molecular Beacon probes that detect the hybridization of unlabeled DNA. The surface-bound probes exhibit reduced levels of fluorescence until the hybridization event makes them fluorescent.69 This has potential for the direct detection of ribosomal RNA (see Box 6.2 on a notional detector).
DETECTION, IDENTIFICATION, ANALYSIS, AND REPORTING
The specific labeling and detection methods appropriate to each technique were discussed as each technique was presented. The assay signal that is detected is either an electronic signal or a photonic
|
68 |
Taylor et al., 2001. See note 10 above. |
|
69 |
H. Wang, J. Li, H. Liu, Q. Liu, Q. Mei, Y. Wang, J. Zhu, N. He, and Z. Lu. 2002. Label-free hybridization detection of a single nucleotide mismatch by immobilization of molecular beacons on an agarose film. Molecular and Cellular Probes 16:119-127. |
one that is then converted into an electrical signal. Compared with the difficulty of the assays themselves, the analysis and reporting of any of these signals are relatively straightforward with present instrumentation and computers and, in the committee's view, do not pose a significant technical challenge for detect-to-warn applications, even in a miniaturized format.
Nucleic acid sequence detection technology provides a powerful set of tools to advance science and medicine. However, the concept of detect-to-warn operation in a biosensor is an extremely challenging goal for this type of technology, primarily due to the demand for speed. The committee cannot say with any confidence that a nucleic acid sequence-based technology will be able to play a role in a 1-minute detection/identification system. However, if existing technologies can be pushed beyond what has yet been done, and if they can be integrated into a system, then one can at least imagine a system that could include a nucleic acid-based detection/identification assay. For example, a sequence-based technology that could provide confirmation of an attack and identify the organisms involved could play a vital role in resolving alarms from faster, nonspecific detectors, even if the sequence-based technology had a response time on the order of several minutes. Box 6.1 describes a hypothetical 3-minute detection/identification system based on PCR.
The committee notes several promising developments that could help to reduce the response times of sequence-based detector/identifier systems. The basic process that leads to spore disruption and cell lysis during ultrasonication with beads is becoming better understood—beads may not be necessary at all; nano- or microbubbles may be all that are needed—and it may be possible to design a sample-processing front end to extract ribosomal RNA rapidly. Also, the implementation of the chemistry of Willem Boom in a microfluidic format may be customized to extract, purify, concentrate, and release RNA in a microfluidic system in seconds. Using electrophoresis, rapid transport of nucleic acids for stringent hybridization can be achieved. Significant progress has also been made using DNA arrays70 to detect and identify organisms using ribosomal RNA. Finally, several label-free technologies could conceivably provide a readout of hybridization in seconds: surface-plasmon resonance, immobilized fluorescence-resonant-energy-transfer probes, and holographic films. It may be that research into one or more of these technologies will ultimately break the challenging kinetic barrier that currently prevents detection/identification systems based on these technologies from achieving anything close to a 1-minute overall performance. Of course, a label-free method would only be effective if it had a high detection sensitivity; otherwise, although the readout time might be reduced, the overall analysis time might be increased, since a larger volume of sample would need to be collected to provide sufficient target organisms for detection.
If the nucleic acid-based assay is to run unattended, the stability of reagents becomes a major issue. Many reagents for nucleic acid-based chemistries need to be refrigerated until use and lose their activity within hours. Reagents with poor stability would lead to signals that change over time even when detecting the same levels of targeted organisms. Highly stable reagents would not only increase the time a detector could operate unattended, but would also improve the overall reproducibility/reliability of the system.
STRAWMAN CONCEPT FOR A FAST RNA DETECTION/IDENTIFICATION SYSTEM
If any detection/identification system based on a nucleic acid sequence assay can come close to meeting a 1-minute detect-to-warn requirement, it may be one based on the sequence determination of unamplified ribosomal RNA. One such notional detection and identification system is described in Box 6.2. The basic technique has been used for the identification and categorization of bacteria,71 but the proposed methods for accelerating the assay in Box 6.2 are hypothetical and as yet untried.
The ribosomes are organelles, found in all bacteria (prokaryotes) as well as in organisms with nuclei (eukaryotes). Typically, there are about 10,000 ribosomes per bacterium.72 The RNA that occurs in the ribosomes (rRNA) is single-stranded and has three characteristic subunits consisting of roughly 120 nucleotides, 1,540 nucleotides, and 3,000 nucleotides, respectively. By contrast, the genomic DNA in a bacterium typically has about 4 million base pairs. Due to the much greater length of the bacterial genomic DNA, the DNA affords better identification of both species and strain of a bacterium than does the rRNA, and an rRNA assay alone may be more appropriate as a rapid screening tool than as a tool for the precise identification of the bioagent.73 However, the committee suggests a notional detector that uses a hybridization assay with a sequence of rRNA as its identifier because the presence of 10,000 copies of the rRNA in each bacterium may enable detection without resorting to the more time-consuming step of amplification of the nucleic acid.
FINDINGS AND RECOMMENDATIONS
Detection and identification of organisms by assays based on nucleic acid sequencing are accepted worldwide as offering the greatest information content and sensitivity of any single technology. In the committee's view, the detect-to-warn biosensor application is extremely challenging for such detection technologies, primarily because they are presently too slow. Substantial basic research on nucleic acid detection has already been conducted, funded by many agencies, companies, and governments, but there has never before been an urgent reason to develop techniques that could respond in less than 1 minute. The committee's major findings and recommendations are as follows:
Finding 6-1: Sample preparation, including sample handling, transport, and system integration, represents the single most important challenge to be faced in the production of a detect-to-warn (DTW) system that performs an identification assay.
Recommendation 6-1: Support research on sample handling, transport, and system integration for assays that are compatible with DTW system requirements. This research may include, but is not limited to, the following:
-
Fabrication and interconnection of miniature components and fluidics that serve DTW.
-
Pressure-driven and electrophoretic transport.
-
Acoustics or ultrasonics for sample handling and preparation.
-
Dielectrophoresis for transport and separations.
-
Surface-chemistry-based techniques for rapid cleanup and concentration of nucleic acids (such as described by the Boom patent).
-
Manufacture and use of hybridization arrays for identification within a few seconds.
Finding 6-2: Neither the front end (sample collectors) nor the back end (fluorescent labels, diode lasers, and detection hardware) of nucleic acid sequence detection systems present fundamental obstacles to the development of detect-to-warn systems. However, even if it proves to be technically feasible to perform the detect-to-warn function in an autonomous fashion, the additional difficulty of manufacturing a reliable, fieldable autonomous system poses an important obstacle.
Recommendation 6-2: Support R&D on collection and detection systems that is consistent with overall system requirements for a detect-to-warn system. Special attention should be paid to the interfacing of modern, complex collector technologies with the sample handling system, avoidance of sampling errors
(e.g., those that result from having low numbers of target sequences in the sample volume), miniaturization of components, and prevention of clogging in any system that would continuously sample the environment.
Finding 6-3: Highly stable reagents not only improve on the time a particular system may operate unattended but also may improve the overall reproducibility and reliability of the detection as well as the logistics and storage requirements to support the detector.
Recommendation 6-3: R&D should be conducted to develop reagents for nucleic acid assays with improved chemical stability.
Finding 6-4: Although a detect-to-warn system has its highest impact if it can initiate responses within approximately 1 minute of an attack, technologies that provide confirmation of the attack and identify the organisms involved will serve a vital function in the overall defensive architecture, even if their response times are several minutes.
Recommendation 6-4: R&D should be conducted to develop an integrated, fully automated PCR system, including sample collection, preparation, and analysis, initially with a 15-minute or so overall confirmation time and later with a 5-minute or so confirmation time.
Finding 6-5: Of the technologies considered, the determination of the sequence of unamplified target ribosomal RNA appears to offer the best potential for a 1-minute DTW system. However, cost and maintenance factors associated with required reagents would still be important issues.
Recommendation 6-5: Support research on an integrated assay for rapid sequence determination of ribosomal RNA (rRNA). This would include the selection of appropriate processing steps, their proper order, and minimum acceptable duration.





















