9
Mayr, Dobzhansky, and Bush and the Complexities of Sympatric Speciation in Rhagoletis
JEFFREY L. FEDER,* XIANFA XIE,* JUAN RULL,† SEBASTIAN VELEZ,* ANDREW FORBES,* BRIAN LEUNG,*‡ HATTIE DAMBROSKI,*§ KENNETH E. FILCHAK,* AND MARTIN ALUJA†
The Rhagoletis pomonella sibling species complex is a model for sympatric speciation by means of host plant shifting. However, genetic variation aiding the sympatric radiation of the group in the United States may have geographic roots. Inversions on chromosomes 1–3 affecting diapause traits adapting flies to differences in host fruiting phenology appear to exist in the United States because of a series of secondary introgression events from Mexico. Here, we investigate whether these inverted regions of the genome may have subsequently evolved to become more recalcitrant to introgression relative to collinear regions, consistent with new models for chromosomal speciation. As predicted by the models, gene trees for six nuclear loci mapping to chromosomes other than 1–3 tended to have shallower node
depths separating Mexican and U.S. haplotypes relative to an outgroup sequence than nine genes residing on chromosomes 1–3. We discuss the implications of secondary contact and differential introgression with respect to sympatric host race formation and speciation in Rhagoletis, reconciling some of the seemingly dichotomous views of Mayr, Dobzhansky, and Bush concerning modes of divergence.
Ernst Mayr helped to transform speciation into aholistic science. With his influential book Systematics and the Origins of Species, Mayr (1942) integrated and synthesized information from genetics, natural history, biogeography, and phylogenetics into a coherent concept of a biological species and a theory for allopatric speciation. Mayr stressed the critical importance of biogeography and systematics as cornerstones for understanding speciation. Divorced from time, space, and phylogenetic relationship, the analysis of reproductive isolation (the defining characteristic of biological species) loses evolutionary context and meaning. The proper chronological ordering of taxa at various stages of divergence also becomes untenable, prohibiting evaluation of the type, sequence, and importance of ecological, demographic, and genetic factors leading to speciation. Therefore, Mayr (1942, 1963) presented a cogent strategy for studying speciation, clarifying the nature of the question and the critical parameters for investigating the process.
Despite widespread acceptance of Mayr’s general framework for studying speciation, several seemingly dichotomous views and personalities nevertheless have shaped and still greatly influence our understanding of the process. Mayr (1942, 1963) made a forceful argument that geographic isolation (allopatry) is a requisite first step for facilitating divergence in animals. He stressed the coadapted nature of the genome and gene pools, as well as the need for allopatry to break the cohesive chains of gene flow to permit populations to diverge independently. In contrast, Guy Bush (1966, 1969) championed the importance of ecological adaptation in speciation. This view was epitomized in his arguments that certain phytophagous insect specialists speciate sympatrically in the process of shifting and adapting to new host plants. Theodosius Dobzhansky (1937, 1981) pioneered the genetic study of speciation, mapping genetic factors responsible for hybrid sterility and inviability and surveying natural populations to assess levels of genetic variation. He crystallized the view that speciation represents the transformation of within-population variation into between-taxa differences through the evolution of inherent reproductive isolating barriers. Dobzhansky (1981) also was a strong advocate of genetic coadaptation, especially with regard to balanced
polymorphisms in the form of chromosomal inversions. All of these themes are subsumed within the cladistic framework of Hennig and the paleontological perspective of Simpson; speciation represents population bifurcations in which an ancestral population is split into two distinct, descendent daughter lineages on separate evolutionary paths (Wiley, 1978).
However, many of the dichotomies that we envision concerning modes of divergence, the cladistic splitting of taxa, and systematic categories of organisms may blur during species formation. For example, it may not always be the case that geographic isolation is absolute or complete during active stages of species formation. Populations at different stages of divergence may experience periods of spatial isolation interspersed with episodes of contact and differential introgression. Sometimes the new genetic variation introduced by introgression may even open novel environments for populations, facilitating local adaptive divergence (Arnold, 1992). Other times, gene flow will homogenize much of the genome, leaving behind a core of coadapted genes differentially evolved between populations. When these gene complexes persist (because of strong selection and their likely linkage in regions of reduced recombination, for example, in inversions) they form a nucleus from which further divergence can build in sympatry by means of reinforcement or ecological specialization (Rieseberg, 2001), as well as in allopatry if isolation reoccurs. Thus, there has been an increasing realization over the last several decades that fields of recombination often extend beyond taxonomic boundaries (Carson, 1975; Hey, 2001; Templeton, 1989) and introgression can occur between hybridizing species in parts of their genomes but not others (Beltran et al., 2002; Brown et al., 2004; Butlin, 1998; Jiang et al., 2000; Machado et al., 2002; Noor et al., 2001a; Rieseberg et al., 1999; Wu, 2001). Moreover, although bifurcating phylogenies may be constructed for taxonomic groups viewed from the perspective of deep evolutionary time, forcing such a pattern on population divergence may misrepresent a more dynamic and reticulate speciation process (Beltran et al., 2002; Brown et al., 2004; Hewitt, 1989; Machado and Hey, 2003) and, hence, the evolutionary relationships of taxa. Rather than the analogy of a “tree of life,” a “delta of life” composed of many intertangled banks may be more appropriate in several instances.
Here, we investigate the biogeography of the Rhagoletis pomonella sibling species complex, a group of tephritid fruit fly specialists with a potentially reticulate genetics and history (Feder et al., 2003a). Detailed study of the biogeography of R. pomonella flies may seem paradoxical. The four described and several undescribed sibling species constituting the complex are a model for ecological divergence without geographic isolation by sympatric host plant shifts (Bush, 1966, 1969). Moreover, the recent
shift of the species R. pomonella from its ancestral host hawthorn (Crataegus spp.) to introduced, domesticated apple (Malus pumila) within the last 150 years in the eastern United States is often cited as an example of host race formation in action, the hypothesized initial stage of sympatric speciation (Bush, 1966, 1969). However, we have discovered a surprising geographic source of genetic variation contributing to sympatric host shifts (Feder et al., 2003a). Based on gene trees constructed for three anonymous nuclear loci mapping to separate rearrangements on chromosomes 1–3 of the R. pomonella genome, as well as mtDNA, we inferred that an ancestral, hawthorn-infesting fly population became geographically subdivided into Mexican and “Northern” (United States) isolates ≈1.57 million years ago (Mya). Episodes of gene flow from the Altiplano highland fly population in Mexico subsequently infused the Northern population with inversion polymorphism affecting key diapause traits, forming adaptive clines. Later, diapause variation in the latitudinal inversion clines appears to have aided flies in the United States in shifting and adapting to various new plants with different fruiting times. These shifts were mediated by population-level changes in allele (inversion) frequencies, generating premating and postmating reproductive isolation in the process and helping to spawn several new host-specific taxa, including the recently formed apple race. We stress that we are not contending that the R. pomonella complex in the United States evolved in allopatry. Rather, certain raw genetic material contributing to the adaptive radiation of R. pomonella in the United States originated in a different time and place than the proximate ecological host shifts triggering sympatric divergence.
The evidence for past introgression and its contribution to sympatric host shifts could be interpreted as indicating that inversions preferentially flowed from the Mexican Altiplano into the Northern fly population after secondary contact. However, the persistence of latitudinal clines in the United States suggests that environmental factors may have constrained the spread (prevented the fixation) of the inversions relative to other genes. Hawthorns tend to fruit later in southern latitudes (H.D. and J.L.F., unpublished data). Hawthorn-fly populations in the United States track this geographic variation in host phenology, possessing inversion genotypes for chromosomes 1–3 in the “South” that cause them to eclose later in the season (Feder and Filchak, 1999; Feder et al., 2003a; Filchak et al., 2000). The pattern continues into Mexico. In the Altiplano, flies infest their primary hawthorn hosts, Crataegus mexicana and Crataegus rosei var. rosei, from mid-October to late December (J.R., J.L.F., S. Berlocher, and M.A., unpublished data). However, in the United States, R. pomonella infests various different hawthorn species, mainly from mid-August to late October. Mexican flies take significantly longer to eclose than U.S. flies, even those from Texas (H.D., J.L.F., J.R., S. Berlocher, and M.A., unpub-
lished data). Consequently, the positions of the inversion clines represent a balance between diapause selection and migration. In contrast to the inversions, loci mapping to other chromosomal regions generally do not differ in allele frequency between the host races, vary clinally, correlate with the timing of eclosion, nor display high levels of linkage disequilibrium in nature (Berlocher, 2000; Berlocher and McPheron, 1996; Feder et al., 1990, 1993, 2003b). Therefore, these apparently collinear regions of the genome may have introgressed more readily at times in the past between Mexico and the North, homogenizing in frequency because of a lack of differential selection combined with recombination.
The contrasting pattern of genetic differentiation seen for chromosomes 1–3 vs. other genomic regions is consistent with new models of chromosomal speciation (Navarro and Barton, 2003; Noor et al., 2001b; Rieseberg, 2001). In these models, reduced recombination associated with rearrangements facilitates the retention of linked genes conferring adaptation or reproductive isolation between hybridizing taxa. However, collinear portions of the genome tend to introgress because recombination results in weak or no linkage of most genes in these regions to loci causing reproductive isolation. Studies in sunflowers (Rieseberg et al., 1999), the Drosophila pseudoobscura subgroup (Brown et al., 2004; Machado et al., 2002; Noor et al., 2001a), and Anopheles mosquitoes (Besansky et al., 2003; dellaTorre et al., 1997) have found evidence for greater introgression in collinear segments of the genome than inverted segments. If differential introgression is true also for Rhagoletis, then the prediction is that loci mapping outside the inversion carrying chromosomes 1–3 should generally show less genetic divergence between Altiplano and U.S. flies compared with genes within the rearranged chromosomes. Coalescence times for noninverted regions should primarily date to the most recent period of contact and gene flow; rearrangements should display deeper divergence times congruent with the initial separation of Mexican and Northern populations. Thus, the chromosome model is predicated on Rhagoletis inversions having partially introgressed at a distant time in the past. During subsequent periods of geographic isolation between Mexican and Northern populations, these inverted regions accumulated additional host-related, as well as possibly non-host-related, genetic changes. Some of the changes, because of their linkage in rearrangements differing between the populations, reduced the potential for the inversions to introgress between Mexican and U.S. flies.
Here, we examine the applicability of the “rearrangement” model to R. pomonella by means of an expanded DNA sequence analysis of loci encompassing both inverted and likely collinear regions of the genome of the fly. We report a pattern of genetic differentiation that is consistent with the rearrangement hypothesis for differential gene flow; gene trees
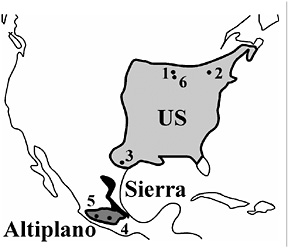
FIGURE 9.1 The current range of R. pomonella in North America. Estimated distributions for the hawthorn-infesting U.S. (light gray) and Mexican Altiplano (dark gray) populations of flies, as well as the recently discovered Sierra Madre Oriental population (black; see text for discussion of “Sierra” flies), are shown. The fly is also distributed patchily in the western United States. Further work is needed to clarify the distribution and origins of these western populations, because they may represent recent introductions. Numbers indicate sampling sites in study (see the Fig. 9.2 legend for site descriptions).
for six nuclear loci mapping to chromosomes other than 1–3 tended to have shallower relative node depths (RNDs) separating Mexican and U.S. sequences than nine genes residing on chromosomes 1–3. We discuss the implications of secondary contact and differential gene flow with respect to sympatric host race formation and speciation in Rhagoletis.
MATERIALS AND METHODS
Fly Populations
Taxa, host plants, collecting sites, and sampling dates for flies are given in Figs. 9.1 and 9.2. Flies were collected as larvae in infested fruit and either (i) dissected from the fruit and frozen for later genetic analysis or (ii) reared to adulthood in the laboratory.
DNA Sequencing
Sequence data were generated for 16 nuclear loci isolated from an R. pomonella EST library, in addition to the three nuclear genes (P220, P2956, and P7) and mtDNA (3′ portion of COI, tRNA-Leu, and COII) analyzed in Feder
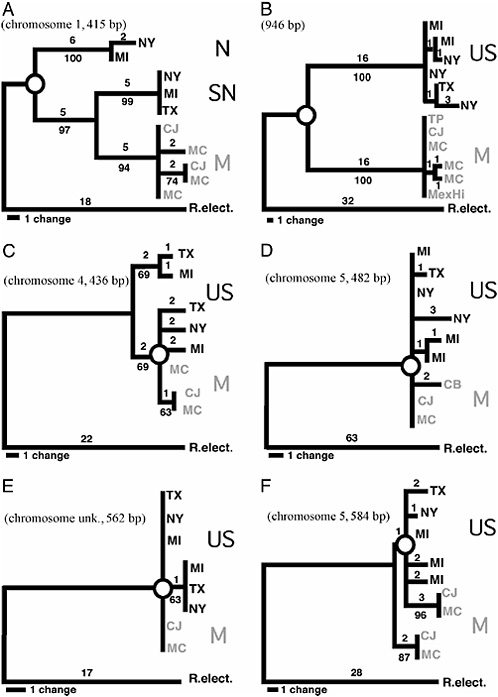
FIGURE 9.2 MP gene trees for P220 (A), mtDNA (B), P661 (C), P309 (D), P3060 (E), and P2620 (F). Trees are scaled so that the longest distance from an allele to the outgroup R. electromorpha (R. elect.) are relatively the same across loci. Chromosome position for loci, bootstrap support for nodes (10,000 replicates), sequence lengths (in bp), and branch lengths (no. of steps) are given. Exact location for P3060 is not known, but the locus does not map to chromosomes 1–3. For locus 220 in A, haplotypes are abbreviated as follows: N, North United States; SN, South/North United States; and M, Mexican (gray). For the other four nuclear loci and mtDNA, U.S. haplotypes are black and Mexican haplotypes are gray. Open circles indicate the contrast between deep and shallow RNDs shown for chromosome 1–3 loci and mtDNA (A and B) vs. genes that do not reside in rearrangements (C–F). Taxa, hosts, location, and collecting dates (month/day/year) are as follows: MI, R. pomonella [apple (M. pumila) and hawthorn (C. mollis)], Grant, MI, 8/15/95; NY, R. pomonella [hawthorn (C. mollis)], Geneva, NY, 9/16/ 00; TX, R. pomonella [hawthorn (C. mollis)], Brazos Bend, TX, 10/6/89; CJ, R. pomonella n.s. [hawthorn (C. mexicana)], Coajomulco, Morelos, Mexico, 11/12/02; MC, R. pomonella n.s. [hawthorn (C. mexicana and C. rosei rosei)], Tancitaro, Michoacan, Mexico, 11/15/02; and R. electromorpha [gray dogwood (Cornus racemosa)], Dowagiac, MI, 9/12/99. Gene trees do not include all of the sequenced alleles for each locus, but subsets that encapsulate the general topological structure for trees. Also, networks incorporating recombinant alleles are not shown for P661 and P3060. Additional alleles and networks are given in supporting information.
et al. (2003a). Nine of the new loci map to the inversion containing chromosomes 1–3, whereas seven genes reside elsewhere in regions that genetic data suggest are not associated with rearrangements (Berlocher, 2000; Berlocher and McPheron, 1996; Feder et al., 1990, 1993, 2003b; Roethele et al., 2001) (Table 9.1). Genomic DNA were PCR amplified for 35 cycles (94°C for 30 sec, 52°C for 1 min, and 72°C for 1.5 min) by using locus-specific primers (Roethele et al., 2001). Products were TA-cloned into pCR II vectors (Invitrogen). PCR amplification products initially were cloned separately for two to three flies from each study site, with from four to six clones sequenced per locus per fly in the 5′ and 3′ directions on an ALF sequencer (Amersham Pharmacia Biotech). To increase sample sizes for certain sites, we also separately amplified genomic DNA for eight flies from the site and TA-cloned the pooled amplification products for sequencing. To avoid analysis of identical alleles from the same individual, sequences generated from the pooled cloning were not included unless they differed from each other.
Gene-Tree Construction
Maximum parsimony (MP) and maximum likelihood (ML) gene trees were constructed by using PAUP* (Version 4.0, Beta 10; Swofford, 2002). For the MP analysis, deletions were treated as a fifth base pair, with indels of identical length and position recoded to count as single mutational steps. Rhagoletis electromorpha, belonging to the sister species group (Rhagoletis tabellaria) to R. pomonella (Bush, 1966), was used as an outgroup.
MP and ML gene trees were very similar, and, thus, only the MP results are presented. Intragenic recombination was tested by using the method of Hudson and Kaplan (1985). Putative recombinant alleles and gene regions were identified, and the alleles were excluded from initial MP gene-tree construction. Recombinant alleles were then added to the trees by hand to generate sequence networks. The molecular clock was tested for each locus for R. pomonella and R. electromorpha sequences by comparing log likelihood scores enforcing vs. relaxing the clock hypothesis for the best supported DNA substitution model identified by using MODELTEST (Posada and Crandall, 1998). To quantify gene-tree topology and genetic divergence, RNDs were calculated between major haplotype classes of alleles segregating in Mexican and U.S. fly populations by dividing the number of substitution differences between a given pair of Mexican and U.S. alleles by the mean number of substitutions between each of these alleles and the R. electromorpha outgroup sequence. Assuming a molecular clock (which none of the nuclear loci or mtDNA violated; Table 9.1), the mean RND for all pairs of Mexican and U.S. alleles between two haplotype classes estimates the age of separation of the haplotypes relative to the divergence of R. electromorpha, given a low to moderate effective size for the ancestral R. pomonella/R. electromorpha population.
RESULTS AND DISCUSSION
Nuclear and mtDNA Gene Trees
Of the 19 total nuclear loci analyzed in the study, 4 were determined to be duplicated loci and excluded from further analysis (P341, P2480, P70, and P2919, mapping to chromosomes 1, 3, 3, and 6, respectively). MP gene trees for the remaining 15 nuclear loci and mtDNA are shown in Fig. 9.2 and supporting information, which is published on the PNAS web site). None of the sequenced genes deviated significantly from a molecular clock (Table 9.1). Nine of the 15 nuclear loci displayed evidence for possible recombination by the method of Hudson and Kaplan (Table 9.1). However, exchange was limited to alleles within identified haplotype classes (i.e., M, S/N, or N) or within geographic populations (Altiplano or United States). The only exceptions were the loci P667, P1700, and P2473, where recombination occurred between major haplotypes within the U.S. population (see supporting information). As would be expected for a nonrecombinant molecule, there was no evidence for exchange among mtDNA sequences.
TABLE 9.1 Loci Sequenced in This Study
Gene Tree Topologies and RND
Gene tree topologies differed significantly between loci mapping to chromosomes 1–3 and those residing elsewhere in the genome (Fig. 9.2). A summary of the differences is shown in Fig. 9.3, where RNDs are plotted between major haplotype classes segregating at loci in Altiplano vs. U.S. flies. RNDs clustered into three groups, corresponding to deep, intermediate, and shallow divergence between Mexican and U.S. haplotypes. Loci tended to fall into different RND categories based on their chromosomal location. Loci mapping to chromosomes 1–3 had significantly greater RNDs than genes on other chromosomes. Six of the nine loci on chromosomes 1–3, as well as mtDNA, had RNDs >0.63 between at least one pair of haplotypes segregating in the United States and Altiplano (Table 9.1). Two of the three loci not displaying deep RNDs (P3072 and P667) showed low levels of disequilibrium, with linked allozymes differentiating the apple and hawthorn host races (standardized disequilib-
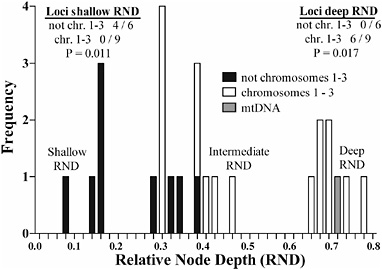
FIGURE 9.3 Distribution of RNDs for nuclear loci not residing on chromosomes 1–3 (black bars), for genes located on chromosomes 1–3 (white bars), and for mtDNA (gray bar). The list on the left gives the number of loci displaying shallow RNDs (<0.16) for the six sequenced genes not mapping to chromosomes 1–3 vs. the nine genes that do. The list on the right gives the number of loci displaying deep RNDs (>0.63). RND values for each locus are given in Table 9.1. P values were determined by two-tailed Fisher’s exact tests.
rium between P3072 and Aat-2, 0.077; P = 0.504; n = 75 scored chromosomes; r value P667/Me, 0.117; P = 0.259; n = 92), suggesting possible weaker associations of these genes with inversions or targets of selection on chromosomes 1 and 2, respectively. In contrast, none of the six loci residing on chromosomes other than 1–3 possessed a deep RND (Table 9.1). Indeed, the deepest RND for any of these six loci was 0.361 (P1700), which was shallower than P22, the third locus on chromosome 3 not possessing a deep RND. Four of the six loci not on chromosomes 1–3 also displayed shallow RNDs of <0.16, not appreciably greater than values found segregating within haplotype classes for these loci within Altiplano and U.S. populations (Fig. 9.2). No locus residing on chromosomes 1–3 possessed a shallow RND (Table 9.1).
Implications of the Gene Trees: Isolation, Contact, and Differential Gene Flow
The tripartite distribution of RNDs for nuclear and mtDNA gene trees is consistent with a hypothesis that Mexican and U.S. fly populations have undergone two cycles of geographic isolation and differential intro-
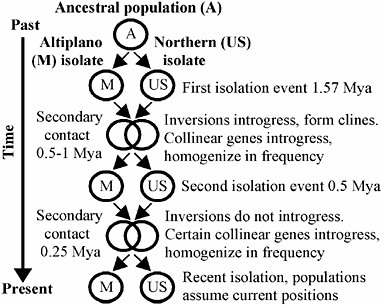
FIGURE 9.4 Biogeographic model depicting two cycles of isolation and differential introgression between Mexican Altiplano and Northern (United States) populations of R. pomonella.
gression (Fig. 9.4). The deep and congruent RNDs for six of the loci on chromosomes 1–3 and mtDNA suggest an initial population subdivision of a Mexican/U.S. common ancestor/1.57 Mya based on an insect mtDNA clock (1.15 × 10−8 substitutions per bp per year) (Brower, 1994). We propose that this initial isolation event was followed by a period of contact from 0.5–1 Mya, during which time gene flow was considerable. Extensive population mixing accounts for the large number of loci displaying intermediate RNDs, as well as for the establishment of adaptive clines for inversions on chromosome 1–3. We do not know the location or extent of the contact zone or clines when they first formed. However, we presume that ecological factors related to host phenology affected the clines in the past in a similar manner as they do currently. Loci residing in other regions of the genome not under selection moved readily between Mexican and Northern populations and recombined, accounting for the lack of deep RNDs for chromosome 4 and 5 loci. In contrast, mtDNA did not introgress during this or any subsequent period of contact.
We hypothesize that the initial period of contact was followed by a second cycle of isolation and introgression (Fig. 9.4). Gene flow was differential during the most recent contact period. Loci residing on chromosomes 4 and 5 tended to move readily between populations, accounting for the shallow RNDs observed for most (4/6; 67%) of these genes (Fig. 9.3
and Table 9.1). In comparison, loci on chromosomes 1–3 did not introgress, resulting in a lack of shallow RNDs. The pattern of gene flow suggests that genetic differences accumulated on chromosomes 1–3 during the second isolation period. To the extent that these changes are defined by inversions (a supposition supported by genetic cross data and population-level linkage disequilibrium values within U.S. populations), they concur with rearrangement models of chromosomal speciation (Rieseberg, 2001; Noor et al., 2001b). Also, the accumulation of additional inversion changes after the hypothesized time when clines were first established suggests that not all diapause-related differences among U.S. flies trace to Mexican origins.
Alternative Hypotheses for the Gene Trees
The pattern of differentiation seen for nuclear loci could potentially also be explained by incomplete linage sorting of balanced inversion polymorphisms present in the ancestral Mexican/U.S. population. In this scenario, Mexican and Northern isolates diverged recently from a common ancestor of modest population size, accounting for the shallow RNDs for loci mapping to chromosomes 4 and 5. In contrast, rearranged regions on chromosomes 1–3 tend to have deeper RNDs because of (i) limited recombination between inversion karyotypes (Mexican and U.S. haplotypes on alternate inversions may often be restricted from coalescing until before the origin of the chromosomal rearrangement separating them in the common ancestor) and (ii) the increased retention time of rearrangements in the ancestral population due to overdominance. At the time of population subdivision, the inversions may have been arrayed in the form of primary clines. Inversions prominent in the South consequently sorted into the Mexican fly population, while a large portion of the polymorphism was retained in the North. As a result, SN haplotypes (alleles that now vary clinally and are found in increasing frequency in southern U.S. fly populations) are genetically more closely related to M haplotypes in Mexico than to alternate N haplotypes segregating in the same host populations (Fig. 9.2A and supporting information).
However, in the absence of a mechanism that coordinately generates inversions throughout the genome, the incomplete lineage-sorting hypothesis has difficulty explaining the clustered distribution of RND values for chromosome 1–3 loci into intermediate and deep categories (Fig. 9.3). Correlated RND values may be expected among loci residing in the same inverted region of a chromosome but not among rearranged regions on different chromosomes, as noted. Moreover, incomplete lineage sorting cannot readily account for the deep RND seen for mtDNA and its congruence with many chromosome 1–3 loci (Figs. 9.2 and 9.3 and Table 9.1).
Given a recent time of separation and modest effective size for the ancestral population, mtDNA should have coalesced quickly and should display minimal differentiation between Mexican and U.S. flies. Last, although inverted regions can be biased toward containing haplotypes with deeper RNDs, unless population splitting was precise, one would still expect to see a subset of inversions shared in common between Mexican and U.S. flies. Haplotypes in the shared inversions should show shallow RNDs, similar to loci on chromosomes 4 and 5. Consequently, the observed gene trees are more consistent with the hypothesis of repeated isolation and secondary contact, with inversions on chromosomes 1–3 becoming increasing more recalcitrant to introgression through time relative to collinear regions of the genome.
Our data could also be explained by a series of gene duplication and deletion events within R. pomonella and the outgroup species R. electromorpha such that many of the haplotype comparisons made in the study were between paralogous rather than orthologous sequences. Four of the original 19 loci amplified in the study were found to be duplicate loci. If similar duplications were accompanied by deletions for many of the other 15 loci, then these duplications/deletions could confound our biogeographic interpretation of the gene trees. However, the deletion scenario, considered alone, suffers the same difficulties as the lineage-sorting hypotheses in explaining the tripartite distribution and deep congruence of chromosome 1–3 nuclear and mtDNA RND values. But it is possible that a composite biogeography/deletion model could account for the pattern. Under this scenario, Mexican and Northern isolates formed ≈1.57 Mya. A period of secondary contact and gene flow followed from 0.5–1 Mya. After this time, Altiplano and Northern populations have remained disjunct. The shallow RNDS observed for loci not on chromosomes 1–3 would be due reciprocal deletions of paralogous genes in R. pomonella and R. electromorpha, resulting in improper comparisons of orthologous Mexican and U.S. haplotypes within R. pomonella to a highly diverged paralogous outgroup sequence for R. electromorpha.
The deletion hypothesis would not negate the contributory roles of allopatry and secondary introgression in facilitating the sympatric radiation of the R. pomonella group by means of host shifting. However, it would call into question whether gene flow was differential for inverted vs. collinear regions of the genome. In essence, there would not have been a second period of recent contact when such a pattern could have been fully generated. Genetic crosses of flies imply that U.S. haplotypes represent allelic variation segregating at single loci (Feder et al., 2003b; Roethele et al., 2001). However, it is difficult to completely rule out the possibility that deletions at very tightly linked duplicated loci generated the observed segregation patterns. Moreover, test cross results for R. pomonella
are not directly germane to resolving the status of R. electromorpha sequences. However, sequence data available for the more distantly related R. cingulata and R. suavis for P661, P309, P2620, and P3060 (loci with shallow RNDs) place R. electromorpha between these two species and R. pomonella. The lack of interspersed clades of sequences containing all or subsets of the four species implies that variation at P661, P309, P2620, and P3060 is allelic and not paralogous.
A Second Mexican Population
Recently, we have discovered a second population of R. pomonella-like flies that infest hawthorns in the Sierra Madre Oriental Mountains of Mexico (Fig. 9.1). The genetics, biogeography, and phenology of the Sierra Oriental population suggest that it may have been a conduit for gene flow between the Altiplano and the North in the past (J.R., J.L.F., X.X., S. Berlocher, and M.A., unpublished data). DNA sequence analysis indicates that Sierra flies are differentiated but, overall, appear to be most closely related to southern U.S. populations (X.X. and J.L.F., unpublished data). The Sierra population abuts the Altiplano population through parts of the states of Veracruz, Puebla, and Hidalgo, Mexico (Fig. 9.1) (J.R., J.L.F., X.X., S. Berlocher, and M.A., unpublished data). We do not know whether the Sierra population contacts U.S. flies. However, if it does, this contact zone is spotty and ephemeral. Hawthorns are rare through the border region but are present in isolated patches in southern New Mexico and, possibly, the Davis Mountains of Texas. The primary hawthorn host for Sierra flies is C. rosei var. parrayana, which is infested from September to early October (J.R., J.L.F., S. Berlocher, and M.A., unpublished data). As is the case for Altiplano and U.S. flies, the diapause characteristics of the Sierra population match host phenology. Sierra flies eclose significantly earlier than Altiplano flies, resulting in potentially substantial allochronic isolation (J.R., J.L.F., X.X., S. Berlocher, and M.A., unpublished data). However, host specificity is not absolute in Mexico. In the transition region between the Altiplano and Sierra, C. mexicana and C. rosei rosei cooccur with C. rosei parrayana and can be found infested by genetically Sierra populations of flies. Here, C. mexicana and C. rosei rosei fruit earlier than they do on the Altiplano and are infested from late September to early November. Thus, host specificity is not as critical a factor isolating Mexican flies as it is for the R. pomonella complex in the United States. However, the spatial and temporal overlap of hawthorns in the transition zone provides a potential bridge for past introgression between the Altiplano and North via the Sierra population.
A Golden Braid
The views of Mayr, Dobzhansky, and Bush may not be as trichotomous as they seem with respect to Rhagoletis. Geographic isolation appears to have established an initial kernel of genetic differentiation that was later expanded on and contributed to sympatric host shifts and new fly taxa. Thus, although geographic context is critical for understanding speciation, allopatry and sympatry should not always be considered as diametrically opposed modes of divergence along an axis of spatial isolation. Differentiation and processes occurring in isolation and contact can interact and compliment each other to accentuate species formation, arguing for a more pluralistic view of modes of speciation (Mallet, 2005). In the case of R. pomonella, the relationship involves a likely sequence of geographic isolation, life-history adaptation, secondary contact, differential introgression, inversion clines, and sympatric host shifts. The evolution of reinforcement can be viewed in an analogous manner, involving non-host-related traits affecting prezygotic isolation rather than ecological adaptation per se. Also, there is no reason to presume that host-related differences that originated in sympatry cannot be solidified by periods of geographic isolation between host-associated populations, although such allopatry is not required to complete the speciation process. Thus, during the time course of differentiation, populations can assume characteristics of both allopatric and sympatric modes of divergence, with phenotypic and genetic elements interacting to further the speciation process.
The connectivity of speciation mode is perhaps best epitomized for R. pomonella if one views the phylogeography of the fly as reflecting sequential adaptation to spatially more finely packaged phenological host niches. At the coarsest level, Altiplano, Sierra Oriental, and Northern R. pomonella populations initially became differentially adapted to temporal and spatial disjunctions in hawthorn fruiting time through a “modular” genetics associated with inversions affecting diapause. After secondary introgression from Mexico, the modular gene blocks became arrayed in the formof broad inversion clines in the North in response to latitudinal variation in hawthorn fruiting time. Last, life-history variation inherent in the clines was extracted on a microgeographic scale [primarily by shifts in allele (inversion) frequencies] to facilitate sympatric shifts and specialization of R. pomonella in the United States to a number of cooccurring host plant species with differing fruiting times. However, host specificity does not appear to be a factor reproductively isolating Altiplano and Sierra flies. Here, geography may act as habitat fidelity does in sympatry, limiting migration and facilitating divergence.
The differences between Altiplano, Sierra, and U.S. populations raise a number of questions. For example, the apparently reduced potential for
rearrangements to introgress implies that these regions of the genome have accumulated additional genetic changes, causing reproductive isolation between Mexican and U.S. flies after their initial establishment in secondary inversion clines in the North. Not all of these changes necessarily reflect host-associated or ecological adaptations. There is no reason that non-host-related differences resulting in prezygotic isolation and hybrid inviability and sterility should not also have accumulated between Mexican and Northern demes during periods of allopatry. Given secondary contact and differential gene flow, the chromosomal speciation models predict that these differences should be concentrated in inversions (Arnold, 1992; Noor et al., 2001b). Therefore, the extent to which intrinsic genomic incompatibilities map to inversion differences between Mexican and U.S. flies needs to be examined. If true, then the inversions would be simultaneously affecting speciation across both allopatric and sympatric scales in R. pomonella. It would be particularly intriguing if any derived differences in the inversions in the U.S. related to latitudinal variation in hawthorn fruiting phenology or interactions between sympatric R. pomonella taxa using different hosts feed back to restrict gene flow between U.S. and Mexican flies, closing the speciation mode braid.
Also, we stress that not all of the host-related changes contributing to sympatric host shifts are diapause-related. Differences in host discrimination (habitat-specific mating) also played a key role in generating the R. pomonella complex. Recently, we demonstrated that host fruit-odor discrimination is an important element of habitat choice for R. pomonella (Linn et al., 2003, 2004). Host choice is important in Rhagoletis because the fly mates only on or near the fruit of its respective host plants (Prokopy et al., 1971, 1972). Thus, variation in host choice translates directly into differences in mate choice and prezygotic isolation. The genetics of fruit-odor discrimination appear to involve loci affecting both preference and avoidance for volatile compounds emitted from the surface of natal and nonnatal fruit (H.D. and J.L.F., unpublished data). Also, F1 hybrids appear to have a reduced ability to orient to host fruit odor in flight-tunnel tests, signifying potentially reduced fitness in the field (Linn et al., 2004). Therefore, the genetics and evolutionary history of host discrimination may prove to be different from diapause traits and not associated with periods of geographic isolation. Because hawthorns are fundamentally similar in Mexico and the United States, there is no reason to expect hawthorn discrimination to be under differential selection or to display a cline.
In conclusion, our study highlights the reticulate nature of speciation at both the population and genomic levels. Students of plant speciation have long embraced this perspective (Anderson, 1949; Arnold, 1992; Grant, 1971; Stebbins, 1959), whereas workers in animal systems are gaining an increased appreciation for the importance of hybridization in meta-
zoan diversity (Arnold, 1992; Coyne and Orr, 2004; Dowling and Secor, 1997; Grant and Grant, 2002; Vollmer and Palumbi, 2002). Many questions remain. Genetic crosses are needed between Altiplano, Sierra, and U.S. flies to assess their taxonomic status and more accurately define the extent of inversion differences separating the populations to strengthen tests for differential introgression. Polytene chromosome spreads are of such poor quality in Rhagoletis that these questions cannot be answered cytologically, but it is nevertheless important to determine whether, for example, SN and M haplotypes now reside on the same or different sets of inversions in U.S. and Mexican populations. Preliminary mating studies indicate that Mexican and U.S. flies are interfertile. However, the relative sterility and viability of F1 and second-generation hybrids remain to be quantified. Moreover, our current understanding of the biogeography of Mexico must be refined, especially in the potential contact zone between Altiplano and Sierra populations, as well as Sierra and U.S. flies, to test for active gene flow. The cause for the lack of mtDNA introgression must also be resolved. Two possibilities are male-mediated gene flow and cytonuclear gene interactions affecting host choice. Last, the paleobiology of Mexico and the Southwest must be further investigated to determine whether the distributions of cooccurring fauna and flora, as well as environmental conditions, are consistent with our historical hypothesis for differential gene flow in R. pomonella. Nevertheless, our results underscore Mayr’s (1942, 1963) emphasis of the critical importance for a fully resolved biogeography and systematics for understanding speciation, even in cases of sympatric divergence in which attention usually is focused on documenting spatial overlap during differentiation. Knowledge of historical information on the biogeography and phylogeography of R. pomonella has helped clarified our understanding of the mechanism of sympatric speciation in these flies by adding a contributory, secondary role for allopatrically evolved inversions in the process.
ACKNOWLEDGMENTS
We thank S. Berlocher, N. Lobo, M. Pale, U. Stolz, and the University of Notre Dame sequencing facility. This work was supported by grants from the National Science Foundation, the United States Department of Agriculture, and the State of Indiana 21st Century Fund (to J.L.F.), and the Mexican Campaña Nacional Contra Moscas de la Fruta and the Instituto de Ecología, Asociación Civil (to J.R. and M.A.).
REFERENCES
Anderson, E. (1949) Introgressive Hybridization (Wiley, New York).
Arnold, M. L. (1992) Natural hybridization as an evolutionary process. Annu. Rev. Ecol. Syst. 23, 237–261.
Beltran, M., Jiggins, C. D., Bull, V., Linares, M., Mallet, J., McMillan, W. O. & Bermingham, E. (2002) Phylogenetic discordance at the species boundary: Comparative gene genealogies among rapidly radiating Heliconius butterflies. Mol. Biol. Evol. 19, 2176–2190.
Berlocher, S. H. (2000) Radiation and divergence in the Rhagoletis pomonella species group: inferences from allozymes. Evolution (Lawrence, Kans.) 54, 543–557.
Berlocher, S. H. & McPheron, B. A. (1996) Population structure of Rhagoletis pomonella, the apple maggot fly. Heredity 77, 83–99.
Besansky, N. J., Krzywinski, J., Lehmann, T., Simard, F., Kern, M., Mukabayire, D., Fontenille, D., Toure, Y. & Sagnon, N.F. (2003) Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: Evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. USA 100, 10818–10823.
Brower, A. V. Z. (1994) Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. USA 91, 6491–6495.
Brown, K. M., Burk, L. M., Henagan, L. M. & Noor, M. A. F. (2004) A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution (Lawrence, Kans.) 58, 1856–1860.
Bush, G. L. (1966) The Taxonomy, Cytology, and Evolution of the Genus Rhagoletis in North America (Mus. Comp. Zool., Cambridge, MA).
Bush, G. L. (1969) Sympatric host race formation and speciation in frugivorous flies of the genus Rhagoletis (Diptera, Tephritidae). Evolution (Lawrence, Kans.) 23, 237–251.
Butlin, R. (1998) What do hybrid zones in general, and the Chorthippus parallelus zone in particular, tell us about speciation? In Endless Forms: Species and Speciation, eds. Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, New York), pp. 367–389.
Carson, H. L. (1975) The genetics of speciation at the diploid level. Am. Nat. 109, 83–92.
Coyne, J. A. & Orr, H. A. (2004) Speciation (Sinauer, Sunderland, MA).
dellaTorre, A., Merzagora, L., Powell, J. R. & Coluzzi, M. (1997) Selective introgression of paracentric inversions between two sibling species of the Anopheles gambiae complex. Genetics 146, 239–244.
Dobzhansky, T. (1937) Genetics and the Origins of Species (Columbia Univ. Press, New York).
Dobzhansky, T. (1981) Dobzhansky’s Genetics of Natural Populations I–XLIII, eds. Lewontin, R. C., Moore, J. A., Provine, W. B. & Wallace, B. (Columbia Univ. Press, New York).
Dowling, T. E. & Secor, C. L. (1997) The role of hybridization and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 28, 593–618.
Feder, J. L. & Filchak, K. E. (1999) It’s about time: The evidence for host plant-mediated selection in the apple maggot fly, Rhagoletis pomonella, and its implications for fitness trade-offs in phytophagous insects. Ent. Exp. Appl. 91, 211–225.
Feder, J. L., Chilcote, C. A. & Bush, G. L. (1990) The geographic pattern of genetic differentiation between host associated populations of Rhagoletis pomonella (Diptera: Tephritidae) in the eastern United States and Canada. Evolution (Lawrence, Kans.) 44, 570–594.
Feder, J. L., Hunt, T. A. & Bush, G. L. (1993) The effects of climate, host plant phenology and host fidelity on the genetics of apple and hawthorn infesting races of Rhagoletis pomonella. Entomol. Exp. App. 69, 117–135.
Feder, J. L., Berlocher, S. H. Roethele, J. B. Smith, J. J., Perry, W. L., Gavrilovic, V., Filchak, K. E. & Aluja, M. (2003a) Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci. USA 100, 10314–10319.
Feder, J. L., Roethele, J. B, Filchak, K., Niedbalski, J. & Romero-Severson, J. (2003b) Evidence for inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella. Genetics 163, 939–953.
Filchak, K. E., Roethele, J. B. & Feder, J. L. (2000) Natural selection and sympatric divergence in the apple maggot Rhagoletis pomonella. Nature 407, 739–742.
Grant, P. R. & Grant, B. R. (2002) Unpredictable evolution in a 30-Year study of Darwin’s finches. Science 296, 707–711.
Grant, V. (1971) Plant Speciation (Columbia Univ. Press, New York).
Hewitt, G. M. (1989) The subdivision of species by hybrid zones. In Speciation and Its Consequences, eds. Otte D. & Endler, J.A. (Sinauer, Sunderland, MA), pp. 85–110.
Hey, J. (2001) Genes, Categories, and Species: The Evolutionary and Cognitive Causes of the Species Problem (Oxford Univ. Press, Oxford).
Hudson, R. R. & Kaplan, N. L. (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111, 147–164.
Jiang, C.-X., Chee, P. W., Draye, X., Morrell, P. L., Smith, C. W. & Paterson, A. H. (2000) Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton). Evolution (Lawrence, Kans.) 54, 798–814.
Linn, C., Jr., Feder, J. L., Nojima, S., Dambroski, H. R., Berlocher, S. H. & Roelofs, W. (2003) Fruit odor discrimination and sympatric host race formation in Rhagoletis. Proc. Natl. Acad. Sci. USA 100, 11490–11493.
Linn, C., Jr., Dambroski, H. R., Feder, J. L., Berlocher, S. H., Nojima, S. & Roelofs, W. (2004) Host specific mating, sympatric speciation and reduced response of hybrid Rhagoletis flies to fruit volatiles. Proc. Natl. Acad. Sci. USA 101, 17753–17758.
Machado, C. A. & Hey, J. (2003) The causes of phylogenetic conflict in a classic Drosophila species group. Proc. R. Soc. London Ser. B 270, 1193–1202.
Machado, C. A., Kliman, R. M., Markert, J. A. & Hey, J. (2002) Inferring the history of speciation from multilocus DNA sequence data: The case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19, 472–488.
Mallet, J. (2005) Speciation in the 21st century. Heredity, in press.
Mayr, E. (1942) Systematics and the Origins of Species (Columbia Univ. Press, New York).
Mayr, E. (1963) Animal Species and Evolution (Belknap, Cambridge, MA).
Navarro, A. & Barton, N. H. (2003) Accumulating postzygotic isolation genes in parapatry: A new twist on chromosomal speciation. Evolution (Lawrence, Kans.) 57, 447–459.
Noor, M. A. F., Grams, K. L., Bertucci, L. A. & Reiland, J. (2001a) The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution (Lawrence, Kans.) 55, 512–521.
Noor, M. A. F., Grams, K. L., Bertucci, L. A. & Reiland, J. (2001b) Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98, 12084–12088.
Posada, D. & Crandall, K. A. (1998) MODELTEST: Testing the model of DNA substitution. Bioinformatics 14, 817–818.
Prokopy, R. J., Bennett, E. W. & Bush, G. L. (1971) Mating behavior in Rhagoletis pomonella (Diptera: Tephritidae). I. Site of assembly. Can. Entomol. 103, 1405–1409.
Prokopy, R. J., Bennett, E. W. & Bush, G. L. (1972) Mating behavior in Rhagoletis pomonella. II. Temporal organization. Can. Entomol. 104, 97–104.
Rieseberg, L. H. (2001) Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358.
Rieseberg, L. H., Whitton, J. & Gardner, K. (1999) Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152, 713–727.




















