3
Inter-Locus Antagonistic Coevolution as an Engine of Speciation: Assessment with Hemiclonal Analysis
WILLIAM R. RICE,* JODELL E. LINDER,* URBAN FRIBERG,*† TIMOTHY A. LEW,* EDWARD H. MORROW,* AND ANDREW D. STEWART*
One of Ernst Mayr’s legacies is the consensus that the allopatry model is the predominant mode of speciation in most sexually reproducing lineages. In this model, reproductive isolation develops as a pleiotropic byproduct of the genetic divergence that develops among physically isolated populations. Presently, there is no consensus concerning which, if any, evolutionary process is primarily responsible for driving the specific genetic divergence that leads to reproductive isolation. Here, we focus on the hypothesis that inter-locus antagonistic coevolution drives rapid genetic divergence among allopatric populations and thereby acts as an important “engine” of speciation. We assert that only data from studies of experimental evolution, rather than descriptive patterns of molecular evolution, can provide definitive evidence for this hypothesis. We describe and use an experimental approach, called hemiclonal analysis, that can be used in the laboratory model system to simultaneously screen nearly the entire genome for both standing genetic variation within a population and the net-selection gradient acting on the variation. Hemiclonal analysis has four stages: (i) creation of a laboratory “island population”; (ii) cytogenetic cloning of nearly genomewide haplotypes to construct hemiclones; (iii) measurement of additive genetic
|
* |
Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, CA 93106; and |
|
† |
Department of Ecology and Environmental Science, Umeå University, 901 87 Umeå, Sweden. |
variation among hemiclones; and (iv) measurement of the selection gradient acting on phenotypic variation among hemiclones. We apply hemiclonal analysis to test the hypothesis that there is ongoing antagonistic coevolution between the sexes in the D. melanogaster laboratory model system and then discuss the relevance of this analysis to natural systems.
There is widespread agreement among evolutionary biologists that the allopatry model is responsible for generating much of the species diversity presently found within sexually reproducing lineages (Coyne and Orr, 2004; Dobzhansky, 1937; Futuyma, 1986; Gavrilets, 2004; Mayr, 1942; Muller, 1942; White, 1978). Although substantial empirical evidence supports the operational steps of the basic allopatry model (Rice and Hostert, 1993), there is no general consensus regarding the relative importance of alternative evolutionary processes that drive the specific genetic divergence that leads to reproductive isolation. Because allopatry necessitates that populations be physically separated, there can be no direct selection for reproductive isolation and therefore it must develop as a pleiotropic byproduct of the genetic differences that accrue due to the independent evolution of populations.
At a minimum, recurrent mutation and random genetic drift of neutral variation lead to genetic differentiation among allopatric populations. However, natural selection can greatly accelerate the rate of genetic divergence. Van Valen (1973) used paleontological evidence to conclude that adaptation to the physical environment is asymptotic (declining in rate with time). Rapid adaptation occurs when a population initially experiences a new physical environment, but the rate of evolution slows as the population becomes progressively more adapted to the prevailing physical conditions. The rate of adaptation diminishes with time as the lag-load (the reduction in mean population fitness due to the average trait of a population differing from its optimal value) of the population decreases. In contrast, adaptation to the biotic environment is expected to be nonasymptotic when enemies (for example, predator and prey, host and pathogen, or resource competitors) become locked in a perpetual arms race of adaptation and counteradaptation. In this case of interspecific antagonistic coevolution, the lag-load of a species does not diminish with time because adaptive progress is continually eroded due to counter-evolution by enemy species.
An analogous cycle of antagonistic coevolution can take place between genes that reside within the genome of a single species (inter-locus antagonistic coevolution). In this case, adaptive allelic replacement at one
locus increases the lag-load at a second locus by generating selection for a new optimal allele, and the resulting adaptive allelic replacement at the second locus increases the lag-load at the first locus, thereby stimulating a new round of the antagonistic cycle of adaptation and counteradaptation. The genetic conflict that drives inter-locus antagonistic coevolution is termed “intragenomic” when conflict occurs between genes that reside in a single individual [for example, genetic conflict associated with genomic imprinting (Haig, 2002), meiotic drive (Jaenike, 2001), and/or cytonuclear conflict (Werren, 1997)] and “intergenomic” when gene products from different loci mediate conflicts of interest between different individuals of the same species (Rice and Holland, 1997).
Inter-locus antagonistic coevolution (intragenomic or intergenomic) can potentially drive rapid genetic divergence among allopatric populations because the antagonistic cycle of adaptation and counteradaptation maintains a persistent lag-load at each interacting locus and thereby drives perpetual evolutionary change. In this article, we focus on intergenomic conflict because two of its forms are predicted to contribute to reproductive isolation by causing genetic divergence among allopatric populations (Parker and Partridge, 1998; Rice, 1996, 1998; Rice and Holland, 1997). Intergenomic conflict can occur both within and between the sexes. An example of intrasexual conflict occurs between gene loci that mediate the male “offense” and “defense” phenotypes in the context of male–male competition to fertilize eggs. A male that mates a virgin female is selected for a defense phenotype, that is, to prevent the female from mating with other males and to prevent his stored sperm from being displaced by a secondary male if the female remates. A male encountering a previously mated female is selected for an offense phenotype, that is, to induce the female to mate with him even if she has ample stored sperm from another male, and then to replace the stored sperm from the previous male with his own. To the extent that the male offense and defense phenotypes are controlled, at least in part, by gene products derived from different gene loci, these interacting genes can antagonistically coevolve in a perpetual cycle of adaptation and counteradaptation.
Persistent male courtship, insemination, and seminal fluid proteins are known to be harmful to females in many species because they lower the female’s survival, fecundity, and short-term fertility (Arnqvist, 1989; Burpee and Sakaluk, 1993; Chapman et al., 1993, 1995; Cohet and David, 1976; Crudgington and Siva-Jothy, 2000; Dean, 1981; Fowler and Partridge, 1989; Friberg and Arnqvist, 2003; Kasule, 1986; McKinney et al., 1983; Moore et al., 2001; Partridge and Fowler, 1990; Pitnick and Garcia-Gonzalez, 2002; Prout and Clark, 2000). Available evidence indicates that the harm to females is an incidental byproduct of adaptations that increase male fertilization success in promiscuous species (Hosken et al., 2001; Morrow et al., 2003). Genes
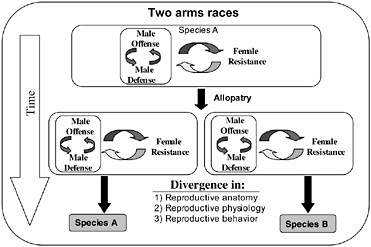
FIGURE 3.1 Inter-locus antagonistic coevolution within and between the sexes can promote the rapid evolution of reproductive traits that are expected to contribute to reproductive isolation (prezygotic mating isolation and postzygotic hybrid infertility) among allopatric populations.
expressed in females are expected to evolve to reduce the harmful effects of interacting with males. If the counteradaptations by female-expressed genes reduce the efficacy of male–male competition to fertilize eggs, then an inter-locus intersexual arms race is expected to ensue between genes that promote female resistance to male-induced harm and the genes that code for male offense and defense (Parker and Partridge, 1998; Rice, 1996, 1998; Rice and Holland, 1997).
The arms races between male offense and defense, and both of these processes with female resistance, would be expected to occur independently in allopatric populations and drive rapid genetic divergence among them (Fig. 3.1). Because, in principle, these arms races can be expected to cause changes in phenotypes influencing reproductive traits (mating behavior and reproductive physiology and anatomy), the genetic divergence produce by inter-locus antagonistic coevolution is expected to cause reproductive isolation by means of pleiotropy in the context of prezygotic reproductive isolation and hybrid infertility, and be an important engine of speciation (Fig. 3.1).
There are two major lines of evidence supporting the hypothesis that inter-locus arms races within and between the sexes drive rapid genetic divergence among allopatric populations: studies of molecular evolution (Swanson and Vacquier, 2002) and studies of experimental evolution (Holland and Rice, 1999; Hosken et al., 2001; Martin and Hosken, 2003; Rice, 1996; Wigby and Chapman, 2004). Molecular studies have demonstrated
that reproductive proteins evolve more rapidly than most other types of proteins (Swanson and Vacquier, 2002). For example, in abalone (genus Haliotis) the gene lysin codes for a protein that dissolves a tunnel through the glycoprotein coat surrounding the egg, and the gene verl codes for the glycoprotein. From the sperm’s perspective, a lysin gene product that more rapidly dissolves a tunnel through the glycoprotein coat is favored because it helps the sperm win in sperm competition. But, from the egg’s perspective, slower penetration of the sperm through the glycoprotein coat is expected to be favored because it provides more time for a block to polyspermy (polyspermy is fatal to the egg) when many sperm compete to fertilize the same egg. This opposing selection on sperm penetration rate sets up a potential arms race between the verl and lysin genes (Frank, 2000; Rice, 1998; Rice and Holland, 1997; Swanson and Vacquier, 2002; Vacquier et al., 1997). In support of this arms race, studies of molecular evolution have demonstrated that both verl and lysin are evolving rapidly due to positive Darwinian selection (Swanson and Vacquier, 2002). As the arms race progresses independently among allopatric populations, the capacity for fertilization to occur between sperm and eggs derived from different populations would be expected to diminish due to coevolution between lysin and verl following different evolutionary trajectories in separated populations.
However, the pattern observed in the studies of molecular evolution has an alternative explanation. The verl gene may be antagonistically evolving due to an interspecific arms race with one or more pathogens that gain entry to the egg by transgressing the glycoprotein coat (Rice, 1998; Rice and Holland, 1997; Vacquier et al., 1997). In this case, the evolution at the lysin locus does not create a lag-load at the verl locus, but instead it evolves to track the evolution in verl that occurs in response to an interspecific arms race between host and pathogen. This alternative explanation for the same molecular data illustrates the problem with using descriptive studies of molecular evolution to test the hypothesis that inter-locus arms races are driving genetic divergence among populations. Because the data are correlative and do not directly measure selection, descriptive studies of molecular evolution can provide supporting evidence for inter-locus arms races but cannot provide definitive evidence.
Studies of experimental evolution in the laboratory are capable of measuring simultaneously standing genetic variance, selection on this variation, and response to the selection at a level of detail that cannot be achieved in natural populations. As a consequence, these studies can be used to provide a direct assessment of the potential for inter-locus antagonistic coevolution within and between the sexes. In the following section, we describe an experimental approach (hemiclonal analysis) to screen nearly the complete genome of Drosophila melanogaster for both genetic
variation (expressed under outbred conditions) and the net-fitness selection gradient on this variation. Because the flies are assayed in the outbred state and in an environment to which they have adapted at large size for hundreds of generations, we were able to obtain direct experimental evidence for the inter-locus arms race between the sexes that can drive the genetic divergence that leads to reproductive isolation. The technique that we describe can be applied only to laboratory populations, but we assert that laboratory populations can be constructed in such a way that the study of their evolution provides an essential complement to studies of natural populations.
HEMICLONAL ANALYSIS
The ability to predict the direction of evolutionary change requires that one establish that there is (i) standing additive genetic variation for a trait, and (ii) a non-zero net-fitness selection gradient on the trait in the prescribed direction. Because of the recent, rapid advance of DNA-based molecular tools, our capacity to measure certain types of standing genetic variation in natural populations is no longer a limiting factor, for example, in genes with highly conserved regions that permit PCR primers to be readily constructed. However, measuring heritable standing genetic variation for fitness-related traits, and especially net-fitness itself, in natural populations remains a daunting task, although these traits can be approximated in special circumstances (see, for example, Kruuk et al., 2000). Although it is possible in the laboratory to accurately measure genetic variation and fitness components associated with many traits, the relevance of these measurements to net-fitness and heritability in nature is dubious. As a consequence, although we can presently measure the standing genetic variation for certain types of traits (such as microsatellites) of natural populations with a high degree of accuracy, levels of heritable phenotypic variation, and the selection gradients on this variation, are unknown for most traits (Fig. 3.2). To circumvent this problem we have focused on the analysis of a locally adapted laboratory population of D. melanogaster. In this context, by cloning nearly entire genomes and amplifying them to large numbers, we can measure accurately both standing genetic variation for a trait and net-selection acting on this variation. Our purpose in this laboratory analysis is not to duplicate the natural history of wild populations, but to study the evolution of laboratory-adapted populations in their own right to deduce basic principles of evolution. Many laboratories throughout the world study laboratory populations; however, we describe here a particular form of evolutionary investigation that we call “hemiclonal analysis” that specifically applies
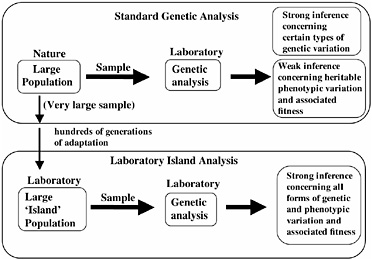
FIGURE 3.2 Genetic analysis of natural and laboratory-adapted populations. Laboratory analysis of samples of organisms taken from wild populations permits strong inference concerning levels of certain types of genetic variation (for example, microsatellites), but only weak inference concerning levels of heritable variation for phenotypic traits, and selection on this variation. When laboratory island populations are analyzed, strong inference is possible for all forms of genetic and phenotypic variation, and its adaptive significance, because the organisms are measured in the same environment to which they have adapted for hundreds of generations.
to the D. melanogaster model system. Our approach has four stages that we describe and discuss in the sections below.
STAGE 1: CREATE A LABORATORY ISLAND POPULATION
The study of island populations has played an important role in the study of evolution, beginning with the pioneering studies of Charles Darwin and Alfred Wallace. It is our view that one of the major reasons that island populations have been particularly informative is that they are much simpler then continental populations (in the context of both biotic and abiotic factors) and therefore easier to understand. Laboratory populations represent island populations that conveniently reside within the laboratory where joint measurements of fitness and genetic variation are more feasible. They are far simpler than natural island populations, but this simplicity provides tractability in the context of evolutionary analysis. By comparison, ecologists have gained important insights into the extinction and colonization process by studying very small island populations of vertebrates in nature, despite the fact that these “islands” are
little more than large protruding rocks, 1–16 m on a side (see, for example, Schoener et al., 2004).
The utility of using laboratory populations to study evolution depends upon how much their evolution is regulated by the same principles that control the evolution of natural populations. Most past laboratory studies of D. melanogaster have used highly inbred stocks such as Oregon-R, Laussan-S, and Canton-S, or genetic samples that have been recently derived from nature and therefore have not adapted to the laboratory environment. In the latter case, the flies are tested in a novel environment so measures such as heritability and selection on the standing, heritable phenotypic variance are difficult to interpret. Inbred laboratory stocks lack these problems, but they have been bottlenecked many times, their extreme and uncontrolled crowding interferes with many forms of behavioral interactions that have historically been important in the species, and their laboratory culture varies among laboratories and stock centers. As a consequence, these inbred laboratory populations have evolved under crowded, uncontrolled conditions that preclude the importance of the rich repertoire of behavioral interactions within and between the sexes, and they have undoubtedly fixed for large numbers of deleterious alleles. To avoid these undesirable aspects of standard laboratory stocks, our laboratory, like others (e.g., the laboratories of Brian Charlesworth, Linda Partridge, and Michael Rose), has started a new laboratory population. The large outbred laboratory population that we study (LHM) was founded by Larry Harshman (now at the University of Nebraska) from 400 inseminated females collected in an orchard near Modesto, California in 1991. Since then, it has been maintained at a large effective population size (>1,800 breeding adults). In 1995 the population density of juveniles and adults was reduced so that juvenile density was consistently maintained at between 150 and 200 individuals per vial, and adult density was reduced further to only 16 pairs per vial (placed on its side to allow more horizontal space for the flies to spread out). The low adult density increased the potential for behavioral interactions to contribute the adult fitness of both sexes.
The population of flies pass through three sets of 56 10-dram vials during their 2-week generation cycle. On day 1, the eggs that were laid at the end of the previous generation are randomly reduced to 150–200 per vial to prevent the extreme crowding that occurs in most mass-transfer laboratory cultures. The flies remain in these “juvenile competition” vials for 12 days during which larval competition, pupation, and the early adult stages occur. On day 12, the flies are mixed among vials and, after being randomly culled to 16 pairs per vial, are transferred to new “adult competition” vials where they reside for 48 h. Live yeast (10 mg) is applied to the top of the 10 ml of killed yeast medium in each vial. There is
a steep linear relationship between the amount of live yeast applied and average female fecundity, indicating that live yeast is the major factor limiting female fecundity (Linder and Rice, 2005). In the adult competition vials, females compete intensely for the limited supply of live yeast, and males compete to inseminate females and fertilize their eggs. Eighteen hours before the end of the 2-week generation cycle, the flies are transferred to “oviposition vials” (with no live yeast added), and the eggs laid at this time are used to begin the next generation. As a consequence, egg production in the oviposition vials represents the lifetime offspring production of both sexes. Put another way, the flies are selected to be “big-bang” reproducers, analogous to semelparous salmon. During the 2-week generation cycle, adults live for at most 6 days, and there is virtually no adult mortality during this time; however, larval mortality does occur in the juvenile competition vials at a rate of ≈10% (Chippindale et al., 2001). In the following sections, all measurements of lifetime fitness and phenotypic traits are taken under conditions that closely match those of the routine culture of the LHM base population.
For the purpose of assaying the flies for lifetime fitness and other traits such as sperm displacement, we also have backcrossed genetic markers [brown eye (bw), brown-eye-dominant (bwD), and nubbin wings (nub2)] and a compound-X [C(1)DX y f] into the LHM base population. Each marker or chromosome has been backcrossed a minimum of 10 times through the LHM base population. Only the brown-eyed (LHM-bw) and compound-X(LHM-DX) replicas of the LHM base population were used in the assays described below.
At the time of this writing, the LHM population has adapted to the laboratory, at continuous large effective size, for 350 generations. With such a long period to adapt to the laboratory environment, most polygenic quantitative traits should have had sufficient time to at least approach their new optima. The capacity for rapid evolution to a new environment is manifest in newly derived Drosophila populations (for example, see Frankham and Loebel, 1992) and is also illustrated by the transplantation experiments of Reznick et al. (1997) in which natural populations of guppies were shown to rapidly adapt to new environments under field conditions. Because the LHM population has such a long history of adaptation to a prescribed laboratory environment, we are able to measure both its genetic and fitness variation in the environment to which it is adapted, and thereby assess antagonistic coevolution within and between the sexes. Because the model organism is D. melanogaster, we can apply the powerful set of genetic tools available only in this species to take full advantage of the laboratory island population.
STAGE 2: CYTOGENETIC CLONING AND CONSTRUCTION OF HEMICLONES
There are many quantitative genetic techniques available to test for additive genetic variation for a trait and to measure net-selection on this variation. One of the most powerful techniques available with the D. melanogaster model system is the isolation and amplification of individual chromosomes through the use of balancers (which suppress recombination between homologous chromosomes). With this technique, many different chromosomes can be randomly sampled, and genetic variation can then be measured among different isochromosomal lines. Because the individual chromosomes can be amplified to many copies, high statistical power can be achieved.
A limitation with the use of balancers to construct isochromosome lines is that balancers are effective in suppressing recombination only when used on single chromosomes. Therefore the entire genome cannot be screened simultaneously with this technique. To eliminate this problem, we (in collaboration with my former postdoctoral associate, A. Chippindale, and former graduate student J. Gibson) have devised a method that relies on the lack of homologous recombination in male Drosophila, rather than balancers, to clonally amplify chromosomes, as originally described in Rice (1996). The advantage of this technique is that a haplotype spanning nearly the entire genome (99.5%, including the X and both major autosomes, but excluding the dot fourth chromosome) can be clonally amplified simultaneously. As a consequence, nearly the entire genome of D. melanogaster can be screened for genetic variation (and selection, see below) simultaneously.
The protocol for cytogenetic cloning has been described in detail (Chippindale et al., 2001), and it is schematically outlined in Fig. 3.3. A random male is drawn from the base population and mated to a “clone-generator” female. These females carry a random Y chromosome from the LHM base population, an attached X (both X chromosomes in females cosegregate as a single linkage unit), and a translocation of the two major autosomes (which, in heterozygotes, causes the two autosomal chromosomes to cosegregate as a single linkage unit, among the living offspring). A single son from this cross (carrying a random genomic haplotype from the base population, which includes the X and the two major autosomes) is randomly selected and mated to many clone-generator females. The sons from these crosses that are retained (half of all sons are retained; the discarded half are homozygous for the translocation and are identifiable by the recessive markers on the translocation) carry the same genomic haplotype as their father and are next crossed to many clone-generator
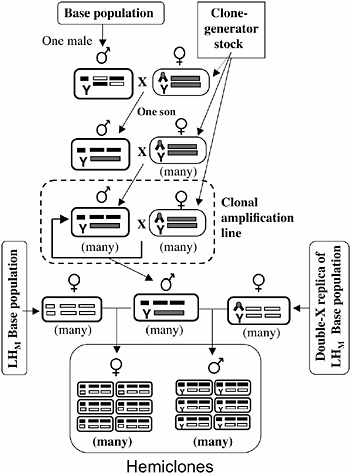
FIGURE 3.3 A schematic of the cytogenetic cloning protocol and the construction of male and female hemiclones. Ellipses depict individuals, and rectangles within ellipses depict chromosomes (X/Y at left position, autosome-2 at middle position, and autosome-3 at right position). The compound X chromosome (chevron) is C(1)DX y f, and the autosomal translocation (rectangle spanning the two autosomal chromosome positions) is T(2;3) rdgC st pp bwD.
females to produce a clonal amplification line that perpetuates the genomic haplotype (Fig. 3.3).
Males from a clonal amplification line are next crossed to one of two types of females: wild-type females from the LHM base population or females from an attached-X replica of the LHM base population that is continuously backcrossed to the LHM population (Fig. 3.3). Half of the males or females from these crosses are completely wild type (the other half express a dominant genetic marker, bwD, and are discarded) and constitute a hemiclone. Members of a hemiclone share in common one
nearly complete genomic haplotype, each expressed in a different random genetic background. A hemiclone is equivalent to the offspring that would be produced by randomly picking a group of eggs from the base population and then fertilizing each egg with a cloned copy of the same sperm.
STAGE 3: MEASURING GENETIC VARIATION
To measure genetic variation for an arbitrary trait in the base population, multiple genomic haplotypes are independently sampled, cytogenetically cloned, and used to construct clonal amplification lines (Fig. 3.3). Next, hemiclones are constructed independently two or more times from each clonal amplification line, and the phenotypic value of each individual in each hemiclone is measured. Finally, random-effects analysis of variance is used to partition phenotypic variation among and between hemiclones to estimate additive genetic variance among hemiclones (Chippindale et al., 2001). Individuals within a hemiclone share half of their genetic variation in common, so that two times the additive genetic variation among hemiclones divided by the total phenotypic variation approximates the heritability of the trait in the base population.
The additive genetic variation among hemiclones contains no nonadditive dominance variation, nor epistatic variation between alleles that reside in the genomic haplotype of a hemiclone and those in its genetic background. It does, however, potentially contain nonadditive epistatic variation between nonallelic genes that reside in the same genomic haplotype. Epistasis can occur between nonallelic genes that reside in genomic haplotypes inherited from (i) the father, (ii) the mother, or (iii) a mixture of these two. Only epistasis between genes that both reside in the paternal haplotype are included in the measure of additive genetic variation among hemiclones (a quarter of the four possible pair-wise types). The inclusion of some epistatic variation in the estimate of additive genetic variation is not unique to hemiclonal analysis. In fact, because of the lack of recombination in male Drosophila, it is a confound that is shared in common with most forms of quantitative genetic analysis with Drosophila. For example, a paternal half-sib design to estimate additive genetic variance includes epistatic variance among alleles that reside on the same chromosome because lack of recombination in male Drosophila keeps these alleles together during meiosis. Similarly, the well-known North Carolina II breeding design (Comstock and Robinson, 1952) also confounds additive and epistatic variation, when applied to D. melanogaster, because the protocol uses balancer chromosomes that cause whole chromosomes to segregate like single giant supergenes (e.g., see Hughes, 1997).
In effect the hemiclonal analysis technique of estimating additive ge-
netic variation among genomic haplotypes treats the genome as if it were a single highly pleiotropic locus in males (but not in females). In the more conventional procedures of paternal half-sib analysis of variance and off-spring-sire regression in D. melanogaster, the male genome segregates as if it were three gene loci (corresponding to the X and the two major autosomes, and ignoring the 0.5% of the genome found on the dot fourth chromosome and the small number if genes residing on the Y sex chromosome). Recombination occurs in females in paternal half-sib analysis of variance and offspring-sire regression designs (as it does in hemiclonal analysis), but these recombined chromosomes do not contribute to the covariance used to estimate heritability. The lack of recombination in males causes each male chromosome to be transmitted intact from father to offspring; hence, each pair of homologous chromosomes in males behaves like a pair of alleles residing at a single highly pleiotropic locus. The only way that we see to disentangle epistatic variation from estimates of additive genetic variance in D. melanogaster that are free from confounding maternal effects would be to carry out analysis of more distant paternal relatives. Because our measures of additive genetic variation among hemiclones include a limited amount (25%) of the potential epistatic variation, they represent an upper bound for the level of additive genetic variation among diploid individuals.
STAGE 4: MEASURING THE NET SELECTION GRADIENT ON STANDING PHENOTYPIC VARIATION
To measure the net selection gradient on phenotypic variation, we needed to first measure the average net-fitness associated with each hemiclone. To estimate this value, a prescribed number of eggs from a hemiclone are placed in a vial with genetically marked (LHM-bw) competitors (we have specifically used a 1:2 ratio of hemiclonal eggs to competitor eggs in previous assays; see Chippindale et al., 2001). Next, the vials of eggs are put through the same culturing protocol as the LHM base population, and then the numbers of offspring produced by the hemiclonal individuals are measured. In this way, each fitness component is weighted by its relative contribution to total fitness, and a measure of net-fitness is obtained. This net-fitness measure of each hemiclone is averaged across many different genetic backgrounds and thus appropriately weighted for the influence of different genetic backgrounds. Lastly, once the average net-fitness of a random set of hemiclones is measured, a bivariate plot of average net-fitness vs. average trait value is constructed, and the slope (selection gradient) is estimated with reduced major axis regression.
APPLICATION OF HEMICLONAL ANALYSIS TO INTER-LOCUS ANTAGONISTIC COEVOLUTION
To use hemiclonal analysis to assess the potential for inter-locus sexual conflict to drive genetic divergence among allopatric populations, we review results from two recent studies in our laboratory. In the first study, a random set of 35 genomic haplotypes was sampled from the LHM base population and used to construct five replicated sets of the 35 female hemiclones (Linder and Rice, 2005). The number of females in each replicate was 20 per hemiclone. The females were collected as virgins and then mass mated by brief (90 min) exposure to a 3:1 ratio of males to females from the LMM base population. Once mated, the females were randomly divided into two experimental treatments during the 2-day adult competition phase of their 2-week generation cycle. The treatments were (i) a “male-protected” environment in which the 10 hemiclonal females competed with 6 unrelated females for the resource that limited their lifetime fecundity (10 mg of live yeast, applied to the surface of the killed-yeast medium) in the absence of persistent courtship from males (no males present), and (ii) a “male-exposed” environment that was identical to the former treatment except that females competed in the presence of males at a 1:1: sex ratio of males to female, i.e., they competed under the social environment experienced during the normal propagation of the LHM base population. The environmental conditions under which the flies were assayed (for example, timing of events, food levels, and densities and ages of flies) closely matched those to which the LHM base population had adapted for >300 generations. Finally, the lifetime fecundity of the females was compared between the two treatments by measuring egg production during the last 18 h of their 2-week generation cycle (which is equivalent to egg production in the oviposition vials during the normal propagation of the LHM base population). Any reduction in fecundity in the male-exposed compared with the male-protected treatments estimated the total cost of interacting with males, that is, the reduction in fecundity owing to resources spent by females when they decamped, kicked, and otherwise responded to persistent interactions with males. Control experiments demonstrated that males did not compete with females for their limiting resource (live yeast) and that differences in fly density did not contribute to the difference in fecundity between treatments (Linder and Rice, 2005).
All 35 hemiclones experienced a decline in lifetime fecundity due to their interactions with males (Fig. 3.4, modified from Linder and Rice, 2005). On average, lifetime fecundity was reduced by 15.4% in the male-exposed treatment relative to the male-protected treatment. However, some hemiclones were harmed more than others (Fig. 3.4), and this het-
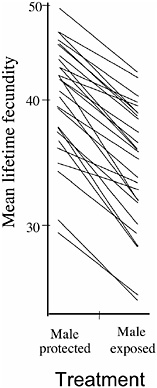
FIGURE 3.4 Aninteraction plot of the average lifetime fecundity of 35 hemiclones when reared in adult competition vials with and without males (modified from Linder and Rice, 2005).
erogeneity in the degree to which lifetime fecundity was reduced by interactions with males measured variation for female resistance to male-induced harm. Analysis of variance was used to test for and estimate heritable variation among hemiclones for the degree of resistance to males, and highly significant genetic variation was observed (P < 0.0001; Linder and Rice, 2005). Heritable variation among hemiclones was estimated to contribute only 2.4% of the total phenotypic variation among individuals, indicating that standing heritable variation was low, as would be expected for a polygenic trait subject to strong directional selection. However, 17% of the total genetic variation among hemiclones for lifetime fecundity was due to variation in female resistance to male-induced harm, indicating that this trait contributed substantially to total genetic fitness variation among females.
Because males harm females through their seminal fluid (Chapman et al., 1995), the rate of secondary mating with different males (i.e., remating rate) was a candidate phenotype contributing to female resistance. We
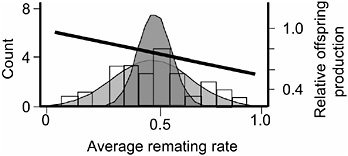
FIGURE 3.5 The distribution of average remating rate of the 35 hemiclones when expressed in females, and the selection gradient on this variation. The lightly stippled curve is a normal distribution fit to the data, and it depicts phenotypic variation among hemiclones. The darker curve depicts genetic variation: it is a normal distribution centered at the sample mean but with variance set equal to the estimated genetic variation among hemiclones (modified from Linder and Rice, 2005).
measured this trait as the percentage of females that remated at least once during their 2 days in the adult competition vials. We found significant heritable variation among hemiclones for remating rate (heritability = 18.2%, additive coefficient of variation = 14.4%, P < 0.0001, Fig. 3.5, modified from Linder and Rice, 2005). Female hemiclones that remated at higher average rate experienced a higher proportional reduction in their lifetime fecundity (P < 0.0001), indicating that reluctance to remate contributed substantially to the female resistance phenotype (Linder and Rice, 2005). We also found a significant negative correlation between our measure of female remating rate and female lifetime fecundity (P = 0.0269; Fig. 3.5), indicating that there was a negative selection gradient on female propensity to remate. Because (i) there was virtually no adult mortality during the 2-week generation cycle of the LHM base population (Chippindale et al., 2001), and (ii) there is no measurable correlation between adult and juvenile fitness (Chippindale et al., 2001), this negative correlation indicated a negative net-selection gradient on female remating rate. At this time, we have not completed an independent assay of lifetime fitness for the 35 hemiclones, so we were unable to directly test for a net-fitness selection gradient on female remating rate. However, an independent assay of a separate, smaller set of 16 hemiclones for which we have completed both a total fitness assay (Rice and Chippindale, 2001) and an assay for remating rate (T.A.L., E.H.M., and W.R.R., unpublished results) corroborated a negative net selection gradient on remating rate.
To test for inter-locus antagonistic coevolution between the sexes, the same 35 hemiclones were expressed in males and assayed for remating
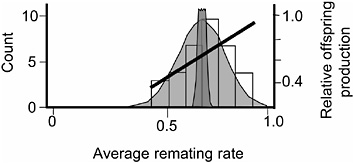
FIGURE 3.6 The distribution of average remating rate of the 35 hemiclones when expressed in males, and the selection gradient on this variation. Normal curves are defined as in Fig. 3.5.
rate. The logic underlying this second assay was to determine whether there was additive genetic variation for remating rate in males, whether this variation was at least partially nonoverlapping with that controlling remating in females, and whether the net-selection gradient on remating rate in males was positive.
The protocol for measuring remating rate in males followed that of the male-exposed treatment of the female resistance assay except that the hemiclones were expressed as males and the females expressed random genotypes drawn from the base population (LHM-bw). We found significant additive variation among male hemiclones for remating rate (proportion of nonvirgin females that remated at least once with the hemiclonal males during their 2 days in the adult competition vials (heritability = 1.4%, additive coefficient of variation = 8.7%, P < 0.05; Fig. 3.6 and Friberg et al., 2005). We also found a significant positive correlation between male remating rate and male adult fitness measured during the assay (P = 0.0081, Fig. 3.6). Because we found (i) no negative correlations between male remating rate and any other adult male fitness components, and (ii) past work in our laboratory found no measurable correlation between adult and juvenile fitness (Chippindale et al., 2001) the positive correlation between male adult offspring production and male remating rate indicated a positive net-selection gradient on this character in males. Again, because we have not yet completed an independent assay of net fitness variation among the 35 male hemiclones, we were unable to directly test for a positive net-selection gradient on male remating rate. However, a smaller assay of 17 different hemiclones was available from a related study that tested for both remate rate (Friberg et al., 2005) and lifetime fitness (Rice and Chippindale, 2001). Here, a positive correlation was found between male remating rate and male lifetime
fitness (P = 0.029), corroborating the significant positive net-selection gradient on this trait deduced from the measures of male adult lifetime offspring production from the larger sample of 35 hemiclones.
In sum, we found significant additive genetic variation among hemiclones for remating rate in both males and females, but the net-selection gradient on this trait was positive in males and negative in females. To look for independent genetic variation for remating rate in males and females, we constructed a bivariate plot of remating rate of the same hemiclones when expressed in males vs. females. No significant correlation was found (r = −0.175, P = 0.316), indicating that remating rate in the two sexes is controlled by different genetic variation. Because there is independent genetic variation for remating rate in the two sexes, and because it is selected in opposite directions in each sex, we conclude that this trait presently is evolving in opposite directions in the two sexes and therefore that sexually antagonistic coevolution for mating rate is currently in evidence in this laboratory island population.
INTERPRETATION OF RESULTS FROM HEMICLONAL ANALYSIS
In the above section, we used hemiclonal analysis to provide evidence that (i) females have genetic variation for resistance to male-induced harm, (ii) resistance contributes substantially to total genetic variation for net fitness, (iii) propensity to remating strongly influences the degree of female resistance, and (iv) there is unique genetic variation for remating rate in males and females that is selected and evolving in opposite directions in the two sexes. These data provide support for the hypothesis that perpetual inter-locus, intersexual arms races contribute to rapid genetic divergence among allopatric populations, and owing to the phenotypes that coevolve (reproductive behavior, physiology, and anatomy) are likely to be contributing to the specific genetic divergence that leads to reproductive isolation and speciation.
The data that we described, however, came from a laboratory island population rather than directly from nature. Some might argue that such populations are too artificial and hence tell us nothing about evolution in nature. We disagree. We cannot statistically extrapolate from our laboratory island population to natural populations of D. melanogaster because our laboratory population is not a random sample from the natural environment. We can, however, use laboratory island populations to make inferences about the fundamental principles of evolution and then use logic to extrapolate to the process of evolution in nature. Just as Darwin (1859) used his study of island tortoise populations to deduce general evolutionary principles (rather than extrapolate to specific continental
populations of tortoises), we used a laboratory island population to assess the evolutionary principles that underlie inter-locus antagonistic coevolution between the sexes. Our finding, that after hundreds of generations of coevolution we can detect an ongoing arms race between the sexes, supports the conclusion that perpetual arms races occur in nature and contribute substantially to the genetic divergence that leads to reproductive isolation and speciation.
A study such as ours could not feasibly be carried out in nature and therefore is possible only in the context of laboratory island populations. For example, the fact that we were able to detect heritabilities among hemiclones of only 2.4% for female resistance illustrates the substantial statistical power of this approach. The genetic measurements that we obtained for standing genetic variance and heritability took advantage of a broad array of genetic tools that are available only in laboratory populations of D. melanogaster. So, in general, we see two options: (i) study only natural populations and wait for technology to advance to the point that experiments such as ours are possible in situ, or (ii) study laboratory island populations where these experiments are possible today. We see a clear advantage to the second option.
We believe that the process of biotic evolution has basic underlying principles that apply to manmade microcosms just as they do to natural ecosystems. If we want to estimate the current or historical trajectory of a natural population, then we need to study that population in situ. But, if we want to understand the evolutionary principles that underlie evolution in nature, rather than specific evolutionary histories, then we can study them just as effectively, and in general more so because of reduced technical constraint, in the context of laboratory island populations. There is the danger that newly constructed laboratory populations will display misleading transients, and, as a result, caution is needed when interpreting results from laboratory populations that have not coevolved over a protracted number of generations (Sgrò and Partridge, 2000). Nonetheless, laboratory island populations make possible evolutionary analysis that cannot be achieved in nature and thereby provide an essential complement to direct studies of populations in nature.
CONCLUSIONS
The allopatric model of speciation, as originally articulated by Dobzhansy (1937) and Mayr (1942), requires genetic divergence among physically isolated populations. Although sequence data are available for only a small number of genes that cause reproductive isolation, the available data indicate that these genes evolve rapidly under positive Darwinian selection [see article by H. A. Orr, “The Genetic Basis of Reproductive
Isolation: Insights from Drosophila” (Chapter 2)]. A fundamental question that remains is the identification of the selective process that drives the rapid divergence of the genes that lead to speciation. In this article, we show that experimental evolution, and more specifically hemiclonal analysis, provides support for the hypothesis that inter-locus antagonistic coevolution promotes rapid genetic divergence among allopatric populations.
ACKNOWLEDGMENT
This work was supported by National Science Foundation Grants DEB99-96164, DEB-0128780, and DEB-0410112 (to W.R.R.).
REFERENCES
Arnqvist, G. (1989) Multiple mating in a water strider—Mutual benefits or intersexual conflict. Anim. Behav. 38, 749–756.
Burpee, D. M. & Sakaluk, S. K. (1993) Repeated matings offset costs of reproduction in female crickets. Evol. Ecol. 7, 240–250.
Chapman, T., Hutchings, J. & Partridge, L. (1993) No reduction in the cost of mating for Drosophila-melanogaster females mating with spermless males. Proc. R. Soc. London Ser. B 253, 211–217.
Chapman, T., Liddle, L. F., Kalb, J. M., Wolfner, M. F. & Partridge L. (1995) Cost of mating in Drosophila-melanogaster females is mediated by male accessory-gland products. Nature 373, 241–244.
Chippindale, A. K., Gibson, J. R. & Rice, W. R. (2001) Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc. Natl. Acad. Sci. USA 98, 1671–1675.
Cohet, Y. A. & David, J. R. (1976) Deleterious effects of copulation in Drosophila females as a function of growth temperature of both sexes. Experientia 32, 696–697.
Comstock, R. E. & Robinson, H. F. (1952) In Heterosis, ed. Gowen, J. W. (Iowa State College Press, Ames, IA), pp. 494–516.
Coyne, J. A. & Orr, H. A. (2004) Speciation (Sinauer, Sunderland, MA).
Crudgington, H. S. & Siva-Jothy, M. T. (2000) Genital damage, kicking and early death—The battle of the sexes takes a sinister turn in the bean weevil. Nature 407, 855–856.
Darwin, C. (1859) On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life (John Murray, London).
Dean, J. M. (1981) The relationship between lifespan and reproduction in the grasshopper Melanoplus. Oecologia 49, 385–388.
Dobzhansky, T. (1937) Genetics and the Origin of Species (Columbia Univ. Press, New York).
Fowler, K. & Partridge, L. (1989) A cost of mating in female fruit-flies. Nature 338, 760–761.
Frank, S. A. (2000) Sperm competition and female avoidance of polyspermy mediated by sperm-egg biochemistry. Evol. Ecol. Res. 2, 613–625.
Frankham, R. & Loebel, D. A. (1992) Modeling problems in conservation genetics using captive drosophila populations—Rapid genetic adaptation to captivity. Zool. Biol. 11, 333–342.
Friberg, U. & Arnqvist, G. (2003) Fitness effects of female mate choice: preferred males are detrimental for Drosophila melanogaster females. J. Evol. Biol. 16, 797–811.
Friberg, U., Lew, T. A., Byrne, P. G. & Rice, W. R. (2005) Assessing the potential for an ongoing arms race within and between the sexes: Selection and heritable variation. Evolution, in press.
Futuyma, D. J. (1986) Evolutionary Biology (Sinauer, Sunderland, MA).
Gavrilets, S. (2004) Fitness Landscapes and the Origin of Species (Princeton Univ. Press, Princeton, NJ).
Haig, D. (2002) Genomic Imprinting and Kinship (Rutgers Univ. Press, New Brunswick, NJ).
Holland, B. & Rice, W. R. (1999) Experimental removal of sexual selection reverses intersexual antagonistic coevolution and removes a reproductive load. Proc. Natl. Acad. Sci. USA 96, 5083–5088.
Hosken D. J., Garner T. W. J. & Ward, P. I. (2001) Sexual conflict selects for male and female reproductive characters. Curr. Biol. 11, 489–493.
Hughes, K. A. (1997) Quantitative genetics of sperm precedence in Drosophila melanogaster. Genetics 145, 139–151.
Jaenike, J. (2001) Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst. 32, 25–49.
Kasule, F. K. (1986) Repetitive mating and female fitness in Dysdercus-cardinalis (Hemiptera, Pyrrhocoridae). Zool. J. Linn. Soc. 88, 191–199.
Kruuk, L. E. B., Clutton-Brock, T. H., Slate, J., Pemberton, J. M., Brotherstone, S. & Guinness, F. E. (2000) Heritability of fitness in a wild mammal population. Proc. Natl. Acad. Sci. USA 97, 698–703.
Linder, J. E. & Rice, W. R. (2005) Natural selection and genetic variation for female resistance to harm from males. J. Evol. Biol. 18, 568–575.
Martin, O. Y. & Hoskin, D. J. (2003) The evolution of reproductive isolation through sexual conflict. Nature 423, 979–982.
Mayr, E. (1942) Systematics and the Origin of Species (Columbia Univ. Press, New York).
McKinney, F., Derrickson, S. R. & Mineau, P. (1983) Forced copulation in waterfowl. Behavior 86, 250–294.
Moore, A. J., Gowaty, P. A., Wallin, W. G. & Moore, P. J. (2001) Sexual conflict and the evolution of female mate choice and male social dominance. Proc. R. Soc. London Ser. B 268, 517–523.
Morrow, E. H., Arnqvist, G. & Pitnick, S. (2003) Adaptation versus pleiotropy: Why do males harm their mates? Behav. Ecol. 14, 802–806.
Muller, H. J. (1942) Isolating mechanisms, evolution and temperature. Biol. Symp. 6, 71–125.
Parker, G. A. & Partridge, L. (1998) Sexual conflict and speciation. Philos. Trans. R. Soc. London B 353, 261–274.
Partridge, L. & Fowler, K. (1990) Nonmating costs of exposure to males in female Drosophila-melanogaster. J. Insect Physiol. 36, 419–425.
Pitnick, S. & Garcia-Gonzalez, F. (2002) Harm to females increases with male body size in Drosophila melanogaster. Proc. R. Soc. London Ser. B Biol. 269, 1821–1828.
Prout, T. & Clark, A. G. (2000) Seminal fluid causes temporarily reduced egg hatch in previously mated females. Proc. R. Soc. London Ser. B 267, 201–203.
Reznick, D. N., Shaw, F. H., Rodd, F. H. & Shaw, R. G. (1997) Evaluation of the rate of evolution in natural populations of guppies (Poecilia reticulata). Science 275, 1934–1937.
Rice, W. R. (1996) Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381, 232–334.
Rice, W. R. (1998) In Endless Forms: Species and Speciation, eds., Howard, D. J. & Berlocher, S. H. (Oxford Univ. Press, New York), pp. 261–270.
Rice, W. R. & Chippindale, A. K. (2001) Sexual recombination and the power of natural selection. Science 294, 555–559.
Rice, W. R. & Holland, B. (1997) The enemies within: intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav. Ecol. Sociobiol. 41, 1–7.
Rice, W. R. & Hostert, E. E. (1993) Laboratory experiments on speciation—What have we learned in 40 years? Evolution 47, 1637–1653.
Schoener, T. W., Spiller, D. A. & Losos, J. B. (2004) Variable ecological effects of hurricanes: The importance of seasonal timing for survival of lizards on Bahamian islands. Proc. Natl. Acad. Sci. USA 101, 177–181.
Sgrò, C. M. & Partridge, L. (2000) Evolutionary responses of the life history of wild-caught Drosophila melanogaster to two standard methods of laboratory culture. Am. Nat. 156, 341–353.
Swanson W. J. & Vacquier, V. D. (2002) The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144.
Vacquier, V. D., Swanson, W. J. & Lee, Y. H. (1997) Positive Darwinian selection on two homologous fertilization proteins: What is the selective pressure driving their divergence? J. Mol. Evol. 44, S15–22.
Van Valen, L. (1973) A new evolutionary law. Evol. Theory 1, 1–30.
Werren, J. H. (1997) Biology of Wolbachia. Annu. Rev. Entomol. 42, 587–609.
White, M. J. D. (1978) Modes of Speciation (Freeman, San Francisco).
Wigby, S. & Chapman, T. (2004) Female resistance to male harm evolves in response to manipulation of sexual conflict. Evolution 58, 1028–1037.






















