5
Expanding Our Knowledge of the Limits of Life on Earth
Strategies for minimizing the risk of forward contamination in solar system exploration rely on knowledge about the kinds and numbers of microorganisms carried by a spacecraft. Such descriptions of microbial population structure provide a basis to estimate the likelihood of survival during spaceflight and to model the probability of subsequent growth in an extraterrestrial environment. Implicit in these models is knowing the range of environmental parameters compatible with growth of microbial populations in both nominal and extreme environments1 on Earth. In recent years, the combination of traditional microbiology and new technological advances has identified organisms that can survive and grow under conditions previously thought hostile to life, including extreme temperatures, radiation, high ionic strength, and desiccation. Some of these conditions are similar to those in environments that occur elsewhere in the solar system where terrestrial spacecraft have visited or will visit in the near future. In particular, some of these conditions resemble those likely to occur on Mars, as described in Chapter 4. The current chapter summarizes the changing perspective of microbial life in extreme environments and how technology enables the detection and enumeration of different kinds of organisms in microbial populations. The chapter then explores the limitations, within the context of existing knowledge, to estimating the probability that Earth organisms could grow on Mars.
MODERN VIEWS OF MICROBIAL DIVERSITY
Detection, characterization, and enumeration of single-cell organisms necessarily depend on optical imaging, cultivation, and biochemical or molecular technology. The invention of the microscope in the 1600s was a revolutionary advance that revealed a never-before-seen world of single-cell organisms. Over the ensuing 300 years, biologists relied on microscopy, nutrients required for growth, and biochemical characterization to describe microbial diversity. Differences in morphology, staining characteristics, metabolic capabilities, and physiological properties defined boundaries between different kinds of microorganisms.2 Serial dilution assays3 and colony
formation by individual cells on agar growth medium provided estimates of microbial population sizes. Twenty years ago, microbiologists had described only 5,000 kinds of bacteria and archaea, and biodiversity studies largely ignored the contribution of single-cell organisms to Earth’s global biomass. These modest assessments of microbial diversity and population size were not consistent with a ~3.5 billion-year evolutionary history, during which single-cell organisms have developed an enormous metabolic repertoire to cope with Earth’s dynamic environment. Such underestimates of microbial diversity reflect how difficult it is to identify morphological and biochemical characteristics (phenotypic traits) that are uniquely shared between closely related organisms, as opposed to common ancestral features that persist over the largest of evolutionary timescales.
Discoveries over the last 20 years of microbial life in habitats previously thought to be devoid of life (e.g., hot springs, deep-sea vents, solid ice, subglacial environments), and the ability to detect microorganisms without requiring their cultivation in the laboratory, have greatly expanded our knowledge of habitat range and numbers of microbes in Earth’s biosphere. For example, fluorescent DNA staining of bacterial cells and epifluorescence microscopy revealed that microbial numbers in aquatic environments were 100 to 1,000 times greater than estimates based on cultivation techniques.4 Contemporary culture-independent surveys demonstrate that microbes may be the dominant biomass on Earth (Whitman et al., 1998), and microbes account for most of the genetic and metabolic diversity (Pace, 1997). The number of different kinds of microbes remains unknown, but the use of modern molecular technology indicates that traditional estimates of 5,000 kinds of microbes are low by several orders of magnitude. In addition to high diversity, the number of archaean and bacterial cells on Earth (see Table 5.1) exceeds 3.6 × 1030, while viral and bacteriophage titers are several orders of magnitude higher. These microbial abundances yield a total carbon content that is 50 percent or more of the estimated carbon in all terrestrial plants (Whitman et al., 1998).
MODERN TECHNOLOGY AND MICROBIAL ECOLOGY
The direct interrogation of microbial genomes offers a powerful, culture-independent method for exploring microbial diversity. Every species has a unique collection of gene sequences that make up its genome, and each species can have a very different complement of genes. The collection of gene sequences in a genome specifies the constellation of proteins that orchestrate cellular biochemistry. The aggregate biochemical activity defines the character or phenotype of a particular kind of organism. The DNA sequence of a gene can serve as a proxy for species identification if that gene is present and conserved by evolution in both the unknown and a reference set of well-characterized organisms. The comparison of gene sequences that share a common evolutionary history allows the reconstruction of evolutionary history or phylogeny. When an organism of unknown taxonomic affinity is included in such analyses, computer algorithms can infer its phylogenetic placement in the context of a large molecular database of well-characterized lineages.
|
|
comparison of morphological and physiological traits to differentiate between microbial species. Molecular techniques including DNA:DNA hybridization, determination of GC content of the genome (percentage of the nucleotides containing guanine and cytosine), and phylogenetic inferences based on comparisons of small-subunit ribosomal RNA gene sequences provided more powerful tools for differentiating among microbes. Yet an attempt to define species boundaries according to a defined number of nucleotide differences between homologous genes is arbitrary and in some cases fails to resolve different populations in situ (e.g., Ward et al., 1990). |
TABLE 5.1 Microbial Diversity: Selected Terminology
|
|
Description |
|
|
Type Diversity |
||
|
Prokaryotes |
A general term that encompasses bacteria and archaea, microbial cells that lack a nuclear membrane surrounding their chromosomal DNA, a cytoskeletal matrix, and other membrane-bounded organelles such as mitochondria and chloroplasts |
|
|
Bacteria |
One of the three known domains of life: microbial cells lacking nuclear bound chromosomes with predominantly diacyl glycerol diester membrane lipids |
|
|
Archaea |
One of the three known domains of life: microbial cells lacking nuclear bound chromosomes with predominantly isoprenoid glycerol diethers or diglyceral tetraether membrane lipids |
|
|
Eukarya |
One of the three known domains of life with nuclear bound chromosomes, cytoskelatal organizing matrices, predominantly glycerol fatty acyl diester membrane lipids, and other membrane-bounded organelles such as mitochondria and chloroplasts |
|
|
Psychrophile |
Capable of growth at low temperatures, with an optimal growth temperature below 15°C |
|
|
Psychrotroph |
Capable of growth at low temperatures, with an optimal growth temperature greater than 15°C |
|
|
Mesophile |
Generally defined by optimal temperature for growth, usually between 25°C and 40°C, but often capable of growth from 8°C to 50°C |
|
|
Thermophile |
Optimal temperature for growth is greater than 45°C but not above 80°C |
|
|
Hyperthermophile |
Optimal temperature for growth is 80°C or above |
|
|
Acidophile |
Grows at pH values less than 5 |
|
|
Alkalophile |
Grows at pH values greater than 9 |
|
|
Neutrophile |
Grows with optimal rates near pH 7 |
|
|
Halophile |
Requires high salt concentrations (>2.5 M) for growth |
|
|
Xerophile |
Capable of growing under conditions of low water activity (effective water content) |
|
|
Barophile |
Obligate barophiles are unable to grow at 1 atmosphere of pressure; barotolerant bacteria grow at 1 atmosphere and higher pressures; all barophiles grow optimally under high pressure |
|
|
Physiological Diversity |
||
|
Aerobe |
Capable of using oxygen as terminal electron acceptor; can tolerate levels of oxygen at or greater than 21 percent and has a strictly respiratory-type metabolism |
|
|
Anaerobe |
Grows only in the absence of oxygen; most have fermentative-type metabolism, but some carry out anaerobic respiration using terminal electron acceptors other than oxygen |
|
|
Facultative anaerobe |
Can grow aerobically or anaerobically |
|
|
Microaerophile |
Capable of oxygen-dependent growth at oxygen levels well below 21 percent |
|
|
Autotroph |
Uses carbon dioxide as its sole source of carbon |
|
|
Heterotroph |
Unable to use carbon dioxide as a sole source of carbon and requires one or more organic compounds |
|
|
Chemoorganoheterotroph |
Derives energy from chemical compounds and uses organic compounds as a reductant |
|
|
Chemolithoautotroph |
Relies on reduced chemical compounds as a source of energy and carbon dioxide as a source of carbon; includes hydrogen bacteria, iron bacteria, sulfur bacteria, ammonia oxidizers, nitrite oxidizers obligate methane oxidizers, carbon monoxide oxidizers |
|
|
Mixotroph |
Capable of growing both chemoorganoheterotrophically and chemolithoautotrophically |
|
|
Oligotroph |
Capable of growth on minimal media (1 to 15 µg carbon per liter) |
|
|
Copiotroph |
Requires nutrients at levels 100 times those of oligotrophs |
|
|
SOURCE: Madigan et al. (2002). |
||
There are a variety of methods for analyzing genes from complex, naturally occurring microbial populations. Most rely on the use of gene cloning and/or polymerase chain reaction (PCR)5 to obtain sufficient quantities of a specific gene for DNA sequence analysis. Such culture-independent characterizations of genes that are conserved over evolutionary time have provided many insights about novel metabolic and physiological categories of microbes (Béjà et al., 2000), as well as information about their distribution in nominal and extreme environments. Analyses of ribosomal RNA (rRNA) coding regions have proven particularly informative. These genes are conserved in all known organisms because they are required for the translation of information encoded within genomic DNA into proteins. Current molecular databases contain more than 120,000 reference rRNA sequences (phylotypes) from diverse microbial forms.6 This window on the microbial world has revealed new levels of largely unexplored microbial diversity not represented in laboratory cultures. According to these molecular studies, microbial diversity ranges from 105 to more than 107 kinds of organisms (Pace, 1997). Evidently, traditional microbiology has failed to culture more than 99.9 percent of these newly discovered phylotypes. Much of the newly discovered diversity resides within described lineages, but ongoing investigations continue to reveal deep-branching, basal lineages in all three domains of life (Lopez-Garcia et al., 2001; Moon-van der Staay et al., 2001; Edgcomb et al., 2002; Cifuentes et al., 2000; Dojka et al., 1998, 2000; Dawson and Pace, 2002). Because little is known about the physiology and survivability of many of these phylotypes, their potential impact on forward contamination from spacecraft represents a major concern for planetary protection. For example, even some organisms that do not form heat- and desiccation-resistant endospores7 might survive spaceflight and be capable of colonizing other solar system objects. Also, the large number of undescribed phylotypes may have levels of resistance to such stress that exceed those known for microbes that have been cultured and characterized, possibly including known spore-forming organisms.
Molecular techniques also have demonstrated that microbial lineages that are not easily cultivated sometimes dominate naturally occurring microbial populations. An example of an uncultured group that has been found (by 16S rRNA clone library analysis) to represent a major fraction of the noncultured bacteria in soils, and whose members have vastly different physical and chemical characteristics, is the Acidobacterial Division. Although their function in soils and sediments remains unknown, these bacteria are thought to play important roles in bacterial community function (Felske et al., 2000). Another example is the marine SAR 11 lineage first identified in the early 1990s from samples collected in the Sargasso Sea. These microorganisms were identified qualitatively via gene cloning as belonging to a major group of uncultured bacterioplankton8 in the sea. Yet, scientists have lacked good quantitative information about how this specific group of bacteria contributed to the total oceanic bacterial pool. The use of molecular techniques in combination with microscopy has now shown that SAR 11 makes up as much as 50 percent of the total surface microbial community (from 0 to 140 m below the surface) and 25 percent of the rest of the water column down to the bottom of the sea (Morris et al., 2002; Rappe et al., 2002).
ORGANISMS AT THE LIMITS OF LIFE
In recent years, spectacular discoveries have altered researchers’ ideas about requirements for microbial growth and the ability of microbes to thrive under seemingly inhospitable environmental conditions. Thermophilic
|
5 |
PCR is a molecular technique that can generate many copies of a gene that lies between two regions of known sequence in a genome. For PCR experiments, short oligonucleotide primer pairs that are complementary to conserved domains separated by 1,000 to 2,000 base pairs anneal to single-stranded templates and initiate DNA synthesis with heat-stable DNA polymerase. After completion of the chain-elongation cycle, treatment with heat (90°C) dissociates the new strands to form new single-stranded templates for subsequent rounds of primer reannealing and DNA synthesis. Each cycle of heating, renaturation of primers with new templates, and DNA synthesis (chain elongation) doubles the DNA sequence between the conserved primers, providing an exponential amplification of the DNA over many cycles. This process is illustrated in Box 6.1. |
|
6 |
See, for example, <http://rdp.cme.msu.edu/html/>. |
|
7 |
Endospores (or simply spores) are small, usually single-celled reproductive bodies produced by certain bacteria in response to adverse environmental conditions. They are highly resistant to desiccation and heat, and if viable, are capable of growing into a new organism under favorable conditions. See also footnote 2, Chapter 1. |
|
8 |
“Bacterioplankton” refers to the fraction of life in the water column of the oceans (and lakes) that includes bacteria and archaea. |
microorganisms can thrive at 121°C (Kashefi and Lovely, 2003), whereas cold-adapted lineages (e.g., psychrophiles) are active in icy systems below –15°C (Rivkina et al., 2000; Carpenter et al., 2000; Christner, 2002; Thomas and Dieckmann, 2002). Microbes populate highly acidic (pH ~0.0) and alkaline (pH >12) environments. Some bacteria tolerate very high levels of radiation and desiccating conditions. Chemosynthetic microbial communities near deep-sea hydrothermal vents (Jannasch and Mottl, 1985), bacteria in deep marine sediments (Bale et al., 1997; Parkes et al., 2000), and subglacial microorganisms (Sharp et al., 1999) thrive without direct requirements for solar input. Some subsurface ecosystems may be entirely independent of surface conditions (Chapelle et al., 2002; Priscu et al., 1999b). Bacteria and archaea occupy every imaginable niche and sometimes occur in settings that seem totally incompatible with biological activity. Some microbes survive such conditions by entering into dormant or resting stages (e.g., bacterial spores), whereas extremophiles have adapted physiologies that nurture growth rather than dormancy under these harsh conditions.
Extremophiles are organisms that thrive in what, for most life forms, are intolerably hostile environments.9 The majority of known extremophiles are representatives of the domains Archaea and Bacteria, although certain protists10 have also been found to thrive in these environments. Extremophiles are classified according to the conditions in which they exist, for example as thermophiles (hot), psychrophiles (cold), halophiles (salty), acidophiles (acidic), alkaliphiles (basic), and barophiles (under pressure) (see Table 5.1). These categories are not mutually exclusive; for example, some acidophiles are also thermophiles.
Extremotrophs represent another category of organisms often found in hostile environments. Extremotrophs tolerate the conditions in question but are just barely able to survive them. The only obligate environmental growth constraint for these and all types of microorganisms is a requirement for liquid water. The discovery of extremophiles and extremotrophs points out the extraordinary adaptability of microbial life on Earth and raises the prospect of finding at least microbial life elsewhere in the solar system.
LIFE IN EXTREME ENVIRONMENTS
Cold Environments on Earth and Mars
Earth’s biosphere is cold. Fourteen percent of the biosphere is polar and 90 percent, by volume, of the world’s oceans are below 5°C. More than 70 percent of Earth’s freshwater occurs as ice, and a large portion of the soil ecosystem (~20 percent) exists as permafrost (Priscu and Christner, 2004). Microorganisms in sea ice were noted by early sailors and first studied as a scientific curiosity by Bunt (1964). Despite the global significance of sea ice, which accounts for about 67 percent of Earth’s ice cover, only recently has it been explored for novel microorganisms (Thomas and Dieckmann, 2002; Junge et al., 2002, 2004). Studies of microbial diversity in polar oceans have shown, in addition to psychrophilic bacteria (Wells and Deming, 2003; Huston et al., 2004), a predominance of archaea in the subgroup Crenarchaeota (DeLong et al., 1994). Before that discovery, crenarchaeota were thought to be confined to thermal systems. Detection of microbial life in cold (–5°C) and saline (>5 times saltier than seawater) Antarctic lakes (Priscu et al., 1999a; Takacs and Priscu, 1998; Doran et al., 2002), permanent lake ice (Priscu et al., 1998; Fritsen and Priscu, 1998; Psenner et al., 1999), glacial ice (Christner et al., 2000, 2001; Smith et al., 2004), and polar snow (Carpenter et al., 2000) extends the known boundaries of our biosphere. The recent description of potential bacterial life in Lake Vostok (Priscu et al., 1999b; Karl et al., 1999; Siegert et al., 2001; Bulat et al., 2004) and the discovery of at least 140 other Antarctic subglacial lakes (Priscu et al., 2003) further extend the recognized environmental limits for life on Earth. Yet, the spatial and temporal records for icy systems on Earth are sketchy, and relatively little is known about the psychrophilic or psychrotolerant microorganisms that inhabit them.
Because of the near- or below-freezing temperatures of the martian surface, most microbes will likely exhibit long generation times (the time required to double the number of organisms in a microbial population) that range
from several to many years. Microorganisms capable of growth at subfreezing temperatures with generation times shorter than 1 to 2 years would likely pose the greatest challenge to planetary protection during the next 50 to 100 years.11 There are claims of microbial growth between –17°C and –20°C (e.g., Junge et al., 2004; Carpenter et al., 2000; Priscu and Christner, 2004; Christner, 2002), although their rates are slow owing to thermodynamic constraints (Price and Sowers, 2004).
Under ideal laboratory conditions, psychrophiles capable of growth at low temperature often exhibit generation times that range from hours to weeks (e.g., Ingraham, 1958; Innis, 1975; Ward and Priscu, 1997). Despite these claims, confusion exists about phychrophilic growth at low temperatures: Do psychrophiles, mesophiles, and thermophiles grow at equivalent rates at their respective optimum growth temperatures? This is not a simple question to answer because there is a large variation in the growth rates of different organisms within each group, as well as a large variation in growth rates for an individual microbe, depending on growth parameters such as oxygen supply, nutrient type, nutrient concentration, and pH. Many psychrophiles have generation times in rich liquid culture of a few hours at or close to 0°C (Russell and Hamamoto, 1998). Such values are comparable to those for environmental isolates of mesophiles, although much shorter times have been reported for the latter. It is difficult to extrapolate experimentally measured generation times to growth rates in natural settings, but evidence so far indicates that growth rates in nature are usually lower than those in enriched media (e.g., Ward and Priscu, 1997).
Because of short durations of liquid water12 and the likely absence of organic substrates due to the near-surface oxidizing environment, the growth of psychrophiles in the martian near-surface is likely to be even slower than the growth rates measured in many Earth environments. Chapter 4 describes what is currently known about the distribution and state of water on Mars. Liquid water may be present for short durations near the martian surface, although scientists cannot rule out the longer-term occurrence of liquid water near the surface (e.g., if active hydrothermal systems exist) or, especially, deeper (e.g., at depths where temperatures are always too high for freezing to occur). At low latitudes, the temperature of the top few centimeters of the regolith may exceed 0°C for a few hours of the diurnal cycle, and liquid water may exist at higher latitudes that have low albedo, poorly conductive soils, or large equatorial-facing slopes. Although liquid water may be present for short periods of time in a variety of locations, organisms that experience temperature fluctuations between 0°C and <–20°C must mount stress responses to the damage caused by eutectic freezing and thawing. Under these conditions, there is little time for both recovery and growth over a diurnal cycle where liquid water is present for at most a few hours each day. The generation time for bacteria exposed to temporal freeze-thaw cycles in the permanent ice covers of Antarctic lakes has been estimated to average 2.5 years and can be as long as 9 to 10 years, depending on the annual duration of liquid water (Fritsen and Priscu, 1998).
Increases in cell numbers over time can provide a measure of bacterial growth, but organisms may be metabolically active and increase their total biomass through enlarged cells rather than an increase in cell number. Temperatures below those required for optimal growth generally constrain maximal rates of division but not necessarily productivity (Knoblauch and Jorgensen, 1999).
Recent models predicting that active cells can survive over hundreds of thousands of years at –40°C (Price and Sowers, 2004) have implications for planetary protection policies, if such policies were to span not some decades-
TABLE 5.2 Examples of Cold Environments (and Their Ages) from Which Viable Microorganisms Have Been Recovered
long “period of biological exploration” but rather time periods required for global change. Organisms at these very low temperatures are presumably not increasing in number, but they are capable of producing enough metabolic energy to maintain the integrity of their DNA and essential proteins (Christner, 2002; Napolitano and Shain, 2004) over long periods (see Chapter 8, Box 8.1). Table 5.2 summarizes data for microbial survival in certain cold environments on Earth. Microbes have been show to remain viable for several million years in permafrost environments, and arguments have been made that icy environments provide an excellent repository for maintaining stable DNA (Willerslev et al., 2004a,b). These viability estimates may have implications for potential life in the martian ice caps, which are thought to be ~100 million years old (Fisher et al., 2002).
Permanent Antarctic Lake Ice
The permanently ice-covered lakes of the McMurdo Dry Valleys may be analogs of ancient martian systems (McKay and Stoker, 1989; McKay et al., 2005). Besides an abundance of microbial life in the liquid water columns (e.g., Priscu et al., 1999a), the ice covers themselves have been shown to harbor a microbial consortium of cyanobacteria and bacteria (Priscu et al., 1998; Paerl and Priscu, 1998). These microbes enter the ice on windblown sediments, where they melt into the ice and create liquid water inclusions at a depth approximately 2 m beneath the surface (Priscu et al., 1998) (see Figure 5.1). DNA hybridization studies have shown that the organisms within the ice are indeed seeded from the terrestrial environment rather than from the liquid water column on which the ice floats (Gordon et al., 2000). Once the microbes are bound within the ice cover, they can grow with doubling times averaging 2.5 years, depending on the temporal availability of liquid water within the ice cover (Fritsen and Priscu, 1998). Cyanobacteria supply the consortium with reduced organic carbon and nitrogen via photosynthesis and N2 fixation, and the heterotrophic bacteria recycle carbon and nitrogen back to the cyanobacteria (Priscu et al., 2005). The close spatial and temporal coupling of metabolites within the microbial consortium is essential for the microbes to survive and replicate in what has been characterized as “the edge of life” (Paerl and Priscu, 1998).
Cryoconite Holes
Cryoconite holes form when dark windblown particulate matter accumulates on the surface of a glacier. Solar radiation warms these particles, causing them to melt into the ice, producing a cylindrical basin of liquid water (see Figure 5.2). The cryoconites may (1) remain liquid on warm sunny days, (2) form an ice cover when air temperature drops and solar radiation persists, or (3) freeze completely during winter when solar radiation is low or absent. As with permanent lake ice assemblages, photosynthesis and N2 fixation by algae and cyanobacteria supply sufficient reduced carbon and nutrients to support the development of complex microbial ecosystems (Christner et
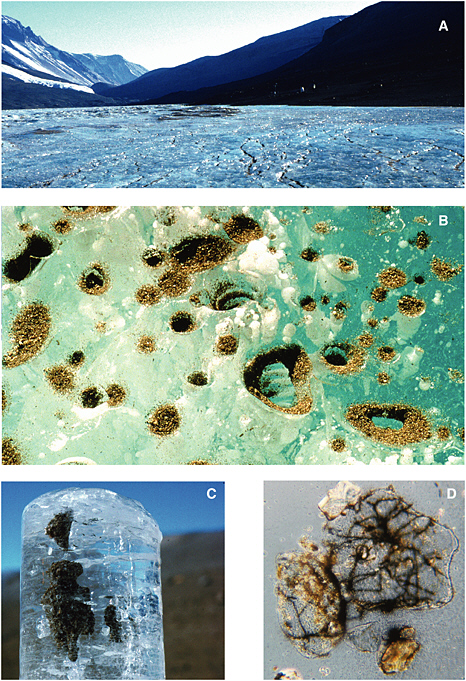
FIGURE 5.1 Sequence of images showing (A) the surface of the Lake Bonney ice cover, (B) a close-up of the surface of the ice cover showing aeolian sediment accumulation, (C) an ice core from 2 m beneath the surface of the ice cover showing sediment accumulation, and (D) microscopic view (1000×) of sediment grains and associated filamentous cyanobacteria. Images courtesy of H. Paerl, University of North Carolina at Chapel Hill, Institute of Marine Sciences.
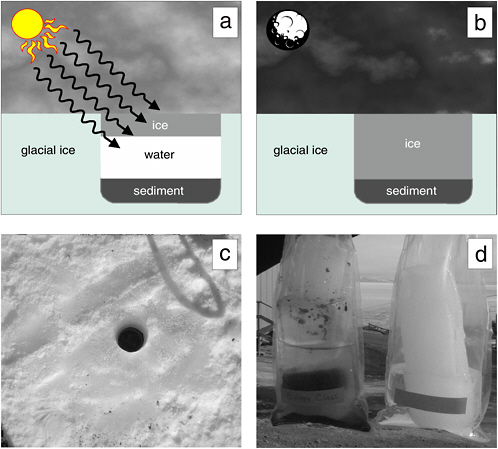
FIGURE 5.2 The cryoconite hole environment in the McMurdo Dry Valleys. (a) In summer, sediment collects on glacial surfaces, and exposure to solar irradiation produces melt pools within the ice, which may subsequently freeze on the surface, and (b) completely freeze during the winter. The 10-cm-diameter cryoconite hole shown in (c) was located on the Canada Glacier and was completely frozen when sampled in January 2001. (d) A comparison of cores retrieved from the cryoconite hole (left) with a core from the adjacent glacial ice. Note the dense layer of sediment and organic material present within the bottom 5 cm of the cryoconite hole core. The diameters of cryoconites on this glacier range from several centimeters to 1 m.
SOURCE: Priscu and Christner (2004), ASM Press, Washington, D.C.
al., 2003). The organic matter produced in the cryoconites can seed the surrounding environment during glacial ablation cycles. Cryoconite hole ecosystems occur globally, being found in Arctic (Mueller et al., 2001), Antarctic (Wharton et al., 1981; Tranter et al., 2004), and in temperate alpine glaciers (Takeuchi et al., 2000). Although dominated by microorganisms, cryoconite holes are one of the few Antarctic terrestrial environments inhabitable by metazoan life. A phylogenetic survey of a cryoconite found in the McMurdo Dry Valleys of Antarctica showed that these ecosystems are inhabited by species quite similar to those in adjacent microbial mat and lake ice
assemblages in this polar desert environment (Christner et al., 2003). The high winds in the Antarctic desert ecosystems disperse organic matter throughout the ecosystem and also provide the biological seed for cryoconite holes and lake habitats. As with the permanent lake ice in the polar deserts of Antarctica, cryoconites serve as biological refuges in an environment that would appear to be inhospitable for life.
Glacial Ice
Earth’s expansive polar ice caps cover ~10 percent of the terrestrial surface with ice and contain ~70 percent of the freshwater on the planet (Patterson, 1994). Archived chronologically within glacial ice are samples of the atmospheric constituents from different times in the past, including biological material such as insects, plant fragments, fungal spores, viruses, and bacteria (Willerslev et al., 1999; Rogers et al., 2005). Studies indicate that ice core samples from nonpolar, high-altitude glaciers contain a greater number and variety of culturable bacterial species than do polar ices (Priscu and Christner, 2004). The difference between polar and nonpolar glaciers results from increased microbial deposition on glaciers contiguous to environments that supply airborne sediments, which serve to transport and protect attached microorganisms. Aerosolized microorganisms can travel large distances on atmospheric currents, often in a viable but dormant state. Remarkably, some atmospheric conditions actually provide a medium for growth, and microbial metabolism has been detected in fog particles (Fuzzi et al., 1997) and super-cooled clouds (Sattler et al., 2001). For an airborne microorganism deposited in glacial ice to retain viability, the stress associated with desiccation, solar irradiation, freezing, an extended period of no growth, and subsequent thawing must not result in a lethal level of unrepairable cellular damage. It is therefore not surprising that many species isolated from glacial ice form spores, structures known to confer resistance to environmental abuses. Many also have thick cell walls or polysaccharide capsules, and they resist repeated cycles of freezing and thawing. Regardless of the ice core’s geographic source, related but not identical species are frequently recovered. Members of the bacterial genera Sphingomonas, Acinetobacter, and Arthrobacter are commonly isolated from glacial samples (Christner et al., 2000, 2001, 2003; Priscu and Christner, 2004) (see Figure 5.3), and they are also the most frequently isolated genera in enrichment surveys of terrestrial subsurface environments (Balkwill et al., 1997). Accordingly, these genera would appear to contain species that can survive for extended times under low-nutrient, nongrowth conditions, and similar survival strategies may be in effect in deep ice and subsurface situations.
Subglacial Lakes
Much attention is currently focused on the possibility that the subglacial environments of Antarctica may harbor microbial ecosystems under thousands of meters of ice, isolated from the atmosphere for as long as the continent has been glaciated (20 million to 30 million years; Naish et al., 2001). The present lake inventory reveals more than 140 Antarctic subglacial lakes (Priscu et al., 2003). Curiosity about the nature of these environments has intensified as a result of the discovery that Lake Vostok, the largest subglacial lake yet known (Studinger et al., 2004), may harbor microbial life (Karl et al., 1999; Priscu et al., 1999b; Bulat et al., 2004). Molecular profiling (16S rDNA) of water from Lake Vostok that has frozen to the bottom of the ice sheet showed close agreement with present-day surface microbiota (Priscu et al., 1999b; Christner et al., 2001). Phylotypes have mapped closely to extant members of the Alpha- and Betaproteobacteria and to the Actinomycetes, the latter of which was also isolated in Vostok glacial ice (Abyzov et al., 1998). These data imply that microbes within Lake Vostok do not represent an evolutionarily distinct subglacial biota (Siegert et al., 2001, 2003). The timescale of isolation within Lake Vostok (~20 million to 25 million years) is not long in terms of microbial evolution compared with its ~3.5 billion-year history on Earth; studies of species divergence of other microorganisms have shown that species-level divergence may take ~100 million years (Lawrence and Ochman, 1998). However, other mechanisms of genetic change (e.g., recombination and mutator genes) could allow more rapid alteration of organism phenotype, allowing for adaptation to conditions within Lake Vostok (Page and Holmes, 1998). Priscu and Christner (2004) used estimates of microbial biomass in subglacial lakes and the polar ice sheets to show that these poorly described systems may contain a previously unrecognized pool of organic carbon similar in magnitude to all of Earth’s surface freshwater ecosystems combined.
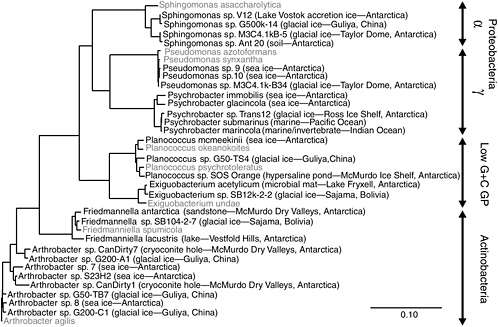
FIGURE 5.3 Phylogenetic analysis of bacteria obtained in microbiological surveys of permanently cold and frozen environments. Isolates from cold habitats are shown in bold, followed by the source environment and geographical location. The 16S rDNA sequences corresponding to nucleotides 27-1492 of the E. coli 16S rDNA were aligned based on secondary structure and used to construct this neighbor-joining tree. The scale bar represents 0.1 fixed substitutions per nucleotide position.
SOURCE: Priscu and Christner (2004), ASM Press, Washington, D.C.
Permafrost
Permafrost is defined as ground that has maintained a temperature lower than 0°C continuously for more than 2 consecutive years. Some permafrost ecosystems are thousands or millions of years old and can serve as “ecological time capsules,” since they may contain micro- and macro-organisms trapped at the time of freezing (Gilichinsky et al., 1992; Tsapin et al., 1999). For example, conspicuous flora and fauna such as bison, horses, and mammoths have been detected by their preserved DNA signatures (Willerslev et al., 2003).
Many of the microorganisms embedded in permafrost soils may remain viable and amenable for cultivation even after hundreds of thousands of years (Vishnivetskaya et al., 2000; see also Table 5.2). This allows for the study of microbial strains that may have been sequestered from ecological interactions with contemporaneous biota for relatively long periods of time. There are multiple physiological adaptations unique to permafrost microbes that can be studied in isolated strains, for example, extremely slow grow rates at –10°C (Bakermans et al., 2003; Rivkina et al., 2000). In addition, other in situ measurements of metabolic activity (e.g., glucose incorporation, methanotrophy) and amino acid chirality support the view that permafrost microbial assemblages are viable and metabolically active in situ.
The combination of low temperatures and relatively high salt content extruded from permafrost soil provides a unique terrestrial hypersaline environment in the form of brine lenses or cryopegs that can reach temperatures of
–15°C and still display metabolic activity in the form of radio-labeled-glucose uptake (Gilichinsky et al., 2003). All these diverse permafrost ecosystems provide terrestrial models for understanding slow rates of microbial growth, low-temperature adaptations, dormancy, and cryoprotection mechanisms that can serve as analogs to other extraterrestrial systems.
Survival in Cold Environments
A phylogenetic comparison of species inhabiting icy environments shows similar phylogenetic 16S rDNA profiles (and therefore the presence of similar kinds of microbes) from Antarctica and other permanently cold nonpolar locales (see Figure 5.3) (Priscu and Christner, 2004). The psychrophilic and psychrotrophic isolates shown in Figure 5.3 originate from locations ranging from aquatic and marine ecosystems to terrestrial soils and glacial ice, with little in common between these environments except that all are permanently cold or frozen. These data argue that clades13 in these bacterial genera evolved under cold conditions and may possess similar strategies to survive freezing and remain active at low temperature. A recent thermodynamic analysis suggested that there is no minimum temperature for metabolism, and the rate at –40°C in ice corresponds to ~10 turnovers of cellular carbon per billion years (Price and Sowers, 2004). Price and Sowers (2004) concluded that microbes in ice and permafrost have metabolic rates similar to those of microbes in water, soil, and sediment at the same temperature. This contention supports the view that, far below the freezing point, liquid water inside ice and permafrost is available for metabolism. As discussed in Chapter 4, such conditions may exist in the martian subsurface. Organisms living in such environments must be able to withstand both the cold conditions and the high salts that often concentrate in the grain boundaries within these systems (Price, 2000). Owing to the difficulty encountered when culturing true psychrophiles and their slow growth rates in the laboratory, relatively little is known of the adaptive mechanisms possessed by this group of microorganisms.
Hot Environments
Many thermophilic organisms (such as the Aquificales, Thermotogales, Sulfolobales, and Archaeoglobales) are representatives of the deep branches of the Archaea and Bacteria, and they are known only from thermal environments. Some of these groups have mesophilic and even psychrotrophic relatives from the gram-positive bacteria14 (including the spore-forming Bacillus), Verrucomicrobiales, and from the methanogens. Heterotrophic organisms such as Geobacillus stearothemophilus and representatives of the Thermotogales are often present in hot environments, but many of these ecosystems are oligotrophic and are dominated by autotrophs that use H2, S, NH4+, and other substrates as electron donors. These ecosystems are often anoxic or contain very low concentrations of oxygen, increasing the importance of alternative terminal electron acceptors such as NO3–, Fe (III), and S (SO4–2, S, and S2O4). Furthermore, some organisms are even capapble of inorganic fermentation (disproportion-ation) using sulfur or sulfite (D’Hondt et al., 2002).
Terrestrial Thermal Environments
Hot springs such as those found in Yellowstone National Park and in Iceland, New Zealand, and Japan have provided scientists with habitats on Earth to study the upper temperature limits of life (Figure 5.4). Brock and other researchers were among the first to describe the physiologies of organisms that could survive and multiply within environments approaching or exceeding boiling temperature (Brock et al., 1972; Brock, 1978). The thermophile Thermus aquaticus has been found in thermal habitats throughout the world. Its temperature range is about 50 to 80°C, and its optimum temperature for growth is around 70°C. Its temperature range overlaps that of the photo-
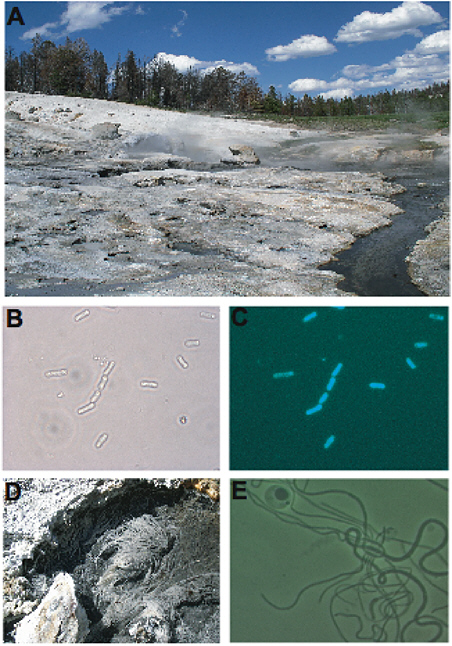
FIGURE 5.4 (A) Coffee Pots, a near-neutral thermal spring in Yellowstone National Park. Resident microbes include phototrophs, chemoautotrophs, and heterotrophs. (B) and (C) are images of the same field of view, except that the cells in Panel C were viewed using epifluorescent microscopy, causing them to autofluoresce and indicating that they contain photosynthetic pigments. (D) Aquificales filaments are macroscopic and resemble tufts of hair attached to thermal spring substrate. (E) Although Thermus sp. have not been detected in this spring, they are found throughout Yellowstone National Park and terrestrial thermal springs worldwide. All micrographs are 1000×.
synthetic bacteria, so that in many hot springs it lives in association with cyanobacteria, ultimately obtaining its energy for growth from the photosynthesis of these organisms. However, it may also be found in lower-diversity environments where temperatures are too high for photosynthesis.
Numerous studies have now described the diversity and productivity of the microbial biota found in geothermal ecosystems. Ecological studies of terrestrial hot-spring microbial communities have reshaped views of microbial biodiversity and of the composition, structure, and function of microbial communities (e.g., Ward et al., 1998). Terrestrial hot-spring microorganisms were among the first extremophiles to be surveyed using molecular technology (Stahl et al., 1985; Ward et al., 1990) and thus were among the first in which the impressive diversity of uncultivated microbial populations in nature was revealed. This has been a typical finding in 16S rRNA gene surveys of microbial diversity in numerous habitats (e.g., Bintrim et al., 1997; Borneman and Triplett, 1997). For example, 16S rRNA studies of hyperthermal hot-spring habitats have led to the discovery of novel uncultivated bacteria (Reysenbach et al., 1994; Pace, 1997) and archaea (Pace, 1997; Barns et al., 1996) that are particularly interesting because they branch near the root of 16S rRNA-derived phylogenetic trees. Because such microorganisms may help researchers to determine characteristics of the most ancestral cells, it is also noteworthy that many of them have been brought into culture (Huber et al., 1995, 1998).
Deep-Sea Hydrothermal Environments
The deep sea was traditionally considered a virtual biological desert. Because phytoplankton, the ocean’s primary producers, are restricted to the upper water column of the sea, the abyssal zone lying at the bottom of the ocean was assumed to be incapable of supporting a significant biological community. However, exploration off the coast of Ecuador in 1977 (Corliss et al., 1979) revealed a diverse community of both prokaryotes and eukaryotes thriving within and surrounding deep-sea hydrothermal vents. Well outside of the ocean’s sunlit photic zone, chemoautotrophic archaea and bacteria, which obtain their energy from the oxidation of inorganic chemicals and fix carbon dioxide for growth, were shown to be the primary producers in deep-sea vent communities, rather than photosynthetic organisms (Karl et al., 1980). The total potential chemosynthetic production for deep-sea hydrothermal ecosystems is estimated to be about 1013 g biomass per year, which represents approximately 0.02 percent of global primary production by photosynthesis in the oceans (McCollom and Shock, 1997).
Deep Subsurface Environments
Early reports of microbes cultivated from Earth’s deep subsurface, primarily from gold mines and oil fields, were met with skepticism because of concerns about contamination. However, improved sampling techniques and the fact that the physiological characteristics of the cultures were consistent with the energy sources and thermal conditions from which they were isolated corroborated the authenticity of their origin (Pedersen, 2000). Early hypotheses suggested that life could extend as much as 10 km deep (Gold, 1999), and preliminary data suggest that microbial communities exist at least 3 to 5 km below Earth’s surface (D’Hondt et al., 2002; Lehman et al., 2001; Fredrickson and Onstott, 1996; Szewzyk et al., 1994; Whitman et al., 1998). Currently, depth is not believed to be the factor limiting subsurface microbial distribution. Rather, it is increased temperature with depth that may restrict life (Pedersen, 2000). The maximum known temperature for microbial growth in the laboratory is 121°C (Kashefi and Lovley, 2003), which is not reached before 5 to 10 km or more in shield rocks, mountains, and deep sediments. The true maximum temperature for life in situ is still unknown, but it is theoretically 150°C (Stetter, 1998). Although surface soil and sediment microbial biomass decrease with depth, some deep subsurface environments harbor microbial abundances as high as 105 to 106 cells cm–3; abundances often increase with increased carbon and energy sources. The vast extent of this biological niche, both below continents and the ocean, results in estimates of subsurface biomass that may account for more than 90 percent of global microbial biomass, 60 to 100 percent of plant biomass, or 3 to 5 × 1017 g of carbon (Whitman et al., 1998; Pedersen, 2000). Although these estimates are based on very preliminary data—and spatially this niche is presumably highly variable—it is clear that deep subsurface biomass is significant and potentially quite large.
In the deep subsurface, a hydrogen-driven biosphere is hypothesized in which hydrogen is formed by either the reaction of gases dissolved in basaltic magma, decomposition of methane (CH4) to C and H at temperatures above 600°C, or radiolysis of water by radioactive isotopes. Under these circumstances, methanogens and acetogens form the basis of the community’s food web, producing methane and acetate, respectively. Claims for the discovery of hydrogen-driven communities within Earth’s deep subsurface have encouraged the search for subsurface life on other planets (Stevens and McKinley, 1995; Chyba and Hand, 2001; Chapelle et al., 2002).
Survival in Hot Environments
Archaea and bacteria dominate hydrothermal ecosystems because temperatures often exceed the habitable limits of eukaryotes (~62°C) and photosynthetic microorganisms (~73°C) (Brock, 1994). The dominance of archaea and bacteria is related in large part to their structural simplicity and metabolic diversity (eukaryotes can be considered structurally complex and metabolically limited). At first, the microbial diversity in hot environments was thought to be relatively low, owing to what was perceived as an extreme environment, a conclusion based primarily on the limited information provided by culturing methods. However, the application of in situ molecular approaches (e.g., characterization of small-subunit rDNA gene diversity) showed that thermal communities consisted of representatives from many of the bacterial and archaeal divisions and led to the discovery of new phylogenetic groups (Barns et al., 1996; Hugenholtz et al., 1998).
The recently discovered hyperthermophile phylum Nanoarchaeota appears to represent yet another deeply branched, independent lineage in the archaean domain. It is unrelated to any cultivated microbial group or environmental sequence.15 Representatives of nanoarcheota were first found in thermal samples from Iceland and were co-cultivated with Ignicoccus, another hyperthermophilic archaean of larger size (Huber et al., 2002, 2003). Preliminary surveys to detect nanoarchaea have shown that this lineage may be much more diverse and also widely distributed in hydrothermal features where other hyperthermophilic archaea such as Ignicoccus exist (Hohn et al., 2002). Recent research further showed that there are at least five lineages clustered by location in thermal sites with temperatures ranging from 70 to 90°C (Chang et al., 2004). Both their genome and cell size are the smallest yet documented, and analyses of their entire genome revealed that many of the genes required for a free-living mode of life are missing, which explains nanoarchaea’s dependence on their host for survival (Waters et al., 2003). Comprehensive environmental studies linked with attempts to cultivate these microbes will provide new insights into the minimal number of genes required for microbial life in symbiotic or parasitic interaction with their hosts. Such studies may shed some light on the range of physiologies and the adaptations required to exist at temperatures close to 100°C and in the presence of S, H2, and CO2.
Desiccating Environments
The Atacama Desert of northern Chile is a high (with most elevations over 2,500 m) and cold desert, with a temperature range of 0 to 25°C. After Antarctica, the Atacama is considered the world’s driest region. Although the Tropic of Capricorn passes through the region, the Atacama lies in the rain shadow of Chile’s Coast Range, which removes the moisture from the atmosphere. (Extremophilic microorganisms that are capable of growing under conditions of low water activity (effective water content expressed as mole fraction) are called xerophiles.) The desert appears completely barren. Most areas receive moisture only from an occasional fog or a shower every few decades. Measurable precipitation rarely occurs, and some places in the area have not had precipitation for hundreds of years. This lack of precipitation places the Atacama environment near the threshold that will support life, as even extremophiles are in low numbers in this environment (Navarro-González et al., 2003). Navarro-González et al. (2003) showed that samples from this Mars-like region had trace levels of total organic matter and
extremely low levels of culturable bacteria. Epifluorescence microscope counts also showed relatively low bacterial abundances of 0.7 × 106 cells per gram and 9.6 × 106 cells per gram for the surface and subsurface Atacama samples, respectively (typical densities in garden soils are 3 to 4 orders of magnitude higher) (Glavin et al., 2004). These epifluorescence counts are much higher than total counts of viable, culturable heterotrophic bacteria (10 to 104 colony-forming units per gram) previously measured by serial dilution plating (Navarro-González et al., 2003), indicating that soil samples from the Atacama Desert contain mostly nonculturable bacteria that are not detected by dilution plating.
Although bacteria in the Atacama soils are near the detection limit of standard analytical techniques, the results of Glavin et al. (2004) imply that the subsurface sample contains a higher total organic content and a higher bacterial cell concentration than do the surface soils. This gradient may result from the presence of a concentration gradient of oxidants with depth. The presence of highly oxidizing conditions within the soils of the Atacama was corroborated by incubation experiments patterned after the Viking labeled-release experiment, but with separate biological and nonbiological isomers. These experiments showed active decomposition of organic species in these soils by nonbiological processes (Navarro-González et al., 2003). The low levels of organic matter in the Atacama Desert in concert with its oxidizing conditions make it an analog for conditions on Mars that offers a potential testing ground for decontamination and life-detection studies.
PROBABILITY OF GROWTH ON MARS
Can we combine our knowledge of the martian environment with our knowledge of extremophilic microorganisms to estimate the probability of growth (Pg) of Earth organisms on Mars? In Chapter 2 (see also Appendix D), it is noted that past estimates of Pg for particular martian environments have ranged as low as 10–10, a value assigned to subpolar regions of Mars within 6 cm of the surface (NRC, 1978). In light of the findings about the martian environment described in Chapter 4 and the growing knowledge of terrestrial psychrophiles and psychrotrophs described in this chapter, the committee suspects that such extremely low values give a false sense of confidence about the risks of microbial contamination of Mars. However, quantifying this suspicion remains very difficult.
Estimates of Pg must account for the physiological tolerances and relative numbers of each kind of microbe on the spacecraft, with special emphasis on extremophiles that might be capable of survival and growth in a specific extraterrestrial environment. As described in Chapter 2, current NASA planetary protection practices for Mars do not consider physiological and genetic diversity, and bioburden estimates fail to differentiate between different kinds of microbes. As described in this chapter, molecular investigations of microbial population structures reveal that many organisms in natural environments are difficult if not impossible to grow in the laboratory. These discoveries have important implications both for knowledge of microbial diversity and inferences about microbes’ ability to survive conditions on Mars, where liquid water might occur.
Estimates of biodiversity based on molecular techniques imply that under controlled nutrient concentrations, salinity, pH, temperature, and so on, microbiologists have successfully cultured fewer than 10 percent and possibly as little as 0.1 percent of the different kinds of organisms from natural settings (Ward et al., 1990). The likelihood of growth in a given environment on Mars may be orders of magnitude lower because experiments involving the accidental delivery of Earth organisms to Mars do not permit the controlled manipulation of environmental conditions that is common during successful attempts to culture microbes on Earth. The probabilities of meeting all conditions necessary to support the growth of an introduced microorganism to the martian surface or subsurface imply that estimates for the probability of growth for a particular kind of microorganism on Mars (![]() , where the superscript i denotes the ith kind of microorganism) may range from 10–3 to much lower values. For these reasons, it seems unlikely that Pg for an arbitrary organism would exceed 10–3, and in fact it could be much lower, given the potential hostility of many martian environments.
, where the superscript i denotes the ith kind of microorganism) may range from 10–3 to much lower values. For these reasons, it seems unlikely that Pg for an arbitrary organism would exceed 10–3, and in fact it could be much lower, given the potential hostility of many martian environments.
Given sufficient resources, molecular techniques could be used to assess microbial diversity Δ on the spacecraft (see Chapter 6). With information about the kinds of organisms present, one could write Equation 2.1 more rigorously, taking into account the varieties of microorganisms i present on the spacecraft:
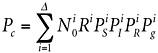
(5.1)
where the variables have the same meaning as in Equation 2.1, except indexed by i to a particular type of microorganism. As before, Equation 5.1 assumes that Pc is small compared with unity. Historically, in the absence of modern DNA phylogenetic methods, there was nearly no knowledge of the varieties i of microorganisms on spacecraft. Instead, because it was believed that spores would be the most likely organisms to survive first interplanetary transport and then the martian surface, Equation 5.1 was collapsed down to culturing sporulating bacteria only, estimating Pc on this basis, provided that Pg for these organisms could be estimated (see Chapter 2).16
To employ Pg to determine bioburden reduction requirements for Mars missions would require knowing enough about Mars and Earth microbes to assign quantitative values to the terms in Equation 5.1. Clearly, research over the past 10 years has broadened researchers’ views on the diversity of extreme habitats in which microorganisms can survive and reproduce on Earth. This advancement is due in part to methodological breakthroughs (primarily in genomic identification and characterization) and a change in research focus by national funding agencies. Knowledge of the capability of Earth microorganisms to survive and propagate on Mars, however, remains quite limited.
Research on extremophiles is still in its infancy, and there is much more to be learned about the physiology and ecology of these organisms. Rapidly changing views on microbial survival and propagation in Earth environments, in concert with considerable uncertainties in knowledge about the martian situation (see Chapter 4), make Pg a scientifically nontractable variable today. Scientists would be hard pressed to accurately compute Pg for microorganisms on Earth; to compute Pg for Earth microorganisms in still poorly understood martian environments currently remains too great a challenge. However, if we were to gain detailed knowledge of the microbial populations on spacecraft (using the techniques described in Chapter 6), and further knowledge of the martian surface and subsurface, most terms in Equation 5.1 might be confidently set to zero, because PiS and Pig may well be extremely close to zero for nearly all kinds i of microorganisms on spacecraft. Were this to prove to be the case, spacecraft bioburden reduction would then have to be applied only to those particular kinds of microorganisms for which PiS and Pig were suspected or known to be appreciable. This might allow an approach to bioburden reduction very different from those employed today. If, for example, future research were to demonstrate that psychrophilic and psychrotrophic extremophiles were universally inactivated (killed) at temperatures low compared with those used in the Viking mission protocols (see Chapter 2), spacecraft might be able to be heat sterilized at much lower temperatures, with fewer consequences for spacecraft material and components.
SUMMARY
Growth and division are the factors required for successful survival in any environment. The fact that microorganisms not only survive but also actually grow in some of the harshest environments on Earth has stimulated scientific curiosity about the evolution and physiology of the organisms that remain viable in this diverse range of temperatures, pH levels, pressures, and salt concentrations. It has also produced a plethora of new research on what have become known as extremophiles. The use of molecular methods to unravel the phylogenies of microorganisms within extreme environments has shown that most of these environments are populated by bacteria, archaea, and unicellar eukarya, which have genetically encoded metabolic plasticity to cope with many divergent environments on Earth. The genomic era has now revealed how extremophiles differ from ordinary microorganisms,
leading to new concepts of microbial evolution and providing impetus to apply genomic methods to research on extremophiles.
The use of molecular methods has revealed that microbial diversity far exceeds what scientists have been able to culture in the laboratory. Genomic tools have made it possible to probe many environments on Earth that were previously thought to be devoid of life. We now know that life occurs in almost every environment on our planet that contains liquid water. Despite that newfound knowledge, the limits to life and its microbial diversity on Earth have not yet been completely defined. Although significant uncertainties remain about the physical and chemical conditions on Mars, it does appear that some subsurface martian conditions are within the known limits of Earth life (see Chapter 4).
The cold temperatures on Mars and the paucity of persistent liquid water would provide selective pressure for the persistence of psychrophilic and psychrotrophic organisms, much as seen within the icy systems on Earth. Earth psychrophiles could survive long-term in the martian environment, but they would not grow rapidly. Data from a variety of cold environments on Earth (see Table 5.2) show that microorganisms can remain immured in ice for many millions of years and remain viable. Willerslev et al. (2004a) concluded that the life-essential nucleic acids DNA and RNA should in theory survive for millions of years in ice and permafrost and remain intact and extractable. At the lowest temperatures of any geologic setting, glacial ice and permafrost are likely to contain the oldest endogenous nucleic acids on Earth.
An understanding of conditions on Mars and the requirements that Earth organisms face for growth in different environments is at the heart of planetary protection and the implementation of planetary protection policy. Chapter 6 describes modern molecular techniques that could be applied to understand the diversity of microbes on spacecraft and methods for eliminating them.
REFERENCES
Abyzov, S.S. 1993. Microorganisms in the Antarctic ice. Pp. 265-295 in Antarctic Microbiology, E.I. Friedmann, ed. Wiley-Liss, Inc., New York.
Abyzov, S.S., I.N. Mitskevich, and M.N. Poglazova. 1998. Microflora of the deep glacier horizons of central Antarctica. Microbiology (Moscow) 67: 66-73.
Bakermans, C., A.I. Tsapin, V. Souza-Egipsy, D.A. Gilichinsky, and K.H. Nealson. 2003. Reproduction and metabolism at –10 degrees C of bacteria isolated from Siberian permafrost. Environ. Microbiol. 5: 321-326.
Bale, S.J., K. Goodman, P.A. Rochelle, J.R. Marchesi, J.C. Fry, A.J. Weightman, and R.J. Parkes. 1997. Desulfovibrio profundus sp. nov., a novel barophilic sulfate-reducing bacterium from deep sediment layers in the Japan Sea. Int. J. Syst. Bacteriol. 47(2): 515-521.
Balkwill, D.L., R.H. Reeves, G.R. Drake, J.Y. Reeves, F.H. Crocker, M.B. King, and D.R. Boone. 1997. Phylogentic characterization of bacteria in the subsurface microbial culture collection. FEMS Microbiol. Rev. 20: 201-216.
Barns, S.M., C.F. Delwiche, J.D. Palmer, and N.R. Pace. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. U.S.A. 93: 9188-9193.
Béjà, O., L. Aravind, E.V. Koonin, M.T. Suzuki, A. Hadd, L.P. Nguyen, S.B. Jovanovich, C.M. Gates, R.A. Feldman, J.L. Spudich, E.N. Spudich, and E.F. DeLong. 2000. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science 289(5486): 1902-1906.
Bintrim, S.B., T.J. Donohue, J. Handelsman, G.P. Roberts, and R.M. Goodman. 1997. Molecular phylogeny of Archaea from soil. Proc. Natl. Acad. Sci. U.S.A. 94: 277-282.
Borneman, J., and E.W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: Evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63: 2647-2653.
Brock, T.D. 1978. Thermophilic Microorganisms and Life at High Temperatures. Springer Verlag, New York.
Brock, T.D. 1994. Life at High Temperatures. Yellowstone Association for Natural Science, History, and Education, Inc., Yellowstone National Park, Wyo. Available at <www.bact.wisc.edu/Bact303/b1>.
Brock, T.D., K.M. Brock, R.T. Belly, and R.L. Weiss. 1972. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Archives fur die Mikrobiologie 84: 54-68.
Bulat, S.A., I.A. Alekhina, M. Blot, J.-R. Petit, M. de Angelis, D. Wagenbach, V. Ya. Lipenkov, L.P. Vasilyeva, D.M. Wloch, D. Raynaud, and V.V. Lukin. 2004. DNA signature of thermophilic bacteria from the aged accretion ice of Lake Vostok, Antarctica: Implications for searching for life in extreme icy environments. International Journal of Astrobiology 3(1): 1-12.
Bunt, J.S. 1964. Primary productivity under sea ice in Antarctic waters 2. Influence of light and other factors on photosynthetic activities of Antarctic marine microalgae. Antarctic Research 1: 27-31.
Carpenter, E.J., S. Lin, and D.G. Capone. 2000. Bacterial activity in South Pole snow. Appl. Environ. Microbiol. 66: 4514-4517.
Chang, C., M. Podar, P. Sammon, E. Mathur, and G. Toledo. 2004. Distribution of the novel phylum Nanoarchaeota in thermal biotopes. Fifth International Conference on Extremophiles, Sept. 19-23, 2004, Cambridge, Md. American Society for Microbiology, Washington, D.C.
Chapelle, F.H., K. O’Neill, P.M. Bradley, B.A. Methe, S.A. Ciufo, L.L. Knobel, and D.R. Lovley. 2002. A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415: 312-315.
Christner, B.C. 2002. Incorporation of DNA and protein precursors into macromolecules by bacteria at –15°C. Appl. Environ. Microbiol. 68: 6435-6438.
Christner, B.C., E. Mosley-Thompson, L.G. Thompson, V. Zagorodnov, K. Sandman, and J.N. Reeve. 2000. Recovery and identification of viable bacteria immured in glacial ice. Icarus 144: 479-485.
Christner, B.C., E. Mosley-Thompson, L.G. Thompson, and J.N. Reeve. 2001. Isolation of bacteria and 16S rDNAs from Lake Vostok accretion ice. Environ. Microbiol. 3: 570-577.
Christner, B.C., E. Mosley-Thompson, L.G. Thompson, and J.N. Reeve. 2003. Bacterial recovery from ancient ice. Environ. Microbiol. 5: 433-436.
Chyba, C.F., and K.P. Hand. 2001. Life without photosynthesis. Science 292: 2026-2027.
Cifuentes, A., J. Anton, S. Benlloch, A. Donnelly, R.A. Herbert, and F. Rodriguez-Valera. 2000. Prokaryotic diversity in Zostera noltiicolonized marine sediments. Appl. Environ. Microbiol. 66(4): 1715-1719.
Corliss, J.B., J. Dymond, L.I. Gordon, J.M. Edmond, R.P.V. Herzen, R.D. Ballard, K. Green, D. Williams, A. Bainbridge, K. Crane, and T.H. van Andel. 1979. Submarine thermal springs on the Galapagos rift. Science 203: 1073-1083.
Dawson, S.C., and N.R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. U.S.A. 99(12): 8324-8329.
DeLong, E.F., K.Y. Wu, B.B. Prezelin, and R.V.M. Jovine. 1994. High abundance of archaea in Antarctic marine picoplankton. Nature 371: 695-697.
D’Hondt, S., S. Rutherford, and A.J. Spivack. 2002. Metabolic activity of subsurface life in deep-sea sediments. Science 295: 2067-2070.
Dojka, M.A., J.K. Harris, and N.R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66(4): 1617-1621.
Dojka, M.A., P. Hugenholtz, S.K. Haack, and N.R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64(10): 3869-3877.
Doran, P.T., C.H. Fritsen, C.P. McKay, J.C. Priscu, and E.E. Adams. 2002. Formation of the 19 m ice cover and associated brine in Lake Vida, Antarctica. Proc. Natl. Acad. Sci. U.S.A. 100: 26-31.
Edgcomb, V.P., D.T. Kysela, A. Teske, A. de Vera Gomez, and M.L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. U.S.A. 99(11): 7658-7662.
Felske A., W.M. de Vos, and A.D.L. Akkermans. 2000. Spatial distribution of 16S rRNA levels from uncultured acidobacteria in soil. Letters in Applied Microbiology 31(2): 118-122.
Fisher, D.A., D.P. Winebrenner, and H. Stern. 2002. Lineations on the “white” accumulation areas of the residual northern ice caps of Mars: Their relation to the “accublation” and ice flow hypothesis. Icarus 159: 36-56.
Fredrickson, J.K., and T.C. Onstott. 1996. Microbes deep inside the Earth. Scientific American 275(4): 68-75.
Fritsen, C.H., and J.C. Priscu. 1998. Cyanobacterial assemblages in permanently ice covers on Antarctic lakes: Distribution, growth rate, and temperature response of photosynthesis. J. Phycol. 34: 587-597.
Fuzzi, G., P. Mandrioli, and A. Perfetto. 1997. Fog droplets—An atmospheric source of secondary biological aerosol particles. Atmos. Environ. 31: 287-290.
Gilichinsky, D., E.A. Vorobyova, L.G. Erokhina, D.G. Fyordorov-Davydov, N.R. Chaikovskaya, and D.G. Fyordorov-Dayvdov. 1992. Long-term preservation of microbial ecosystems in permafrost. Adv. Space Res. 12: 255-263.
Gilichinsky, D., E. Rivkina, V. Shcherbakova, K. Laurinavichuis, and J. Tiedje. 2003. Supercooled water brines within permafrost—An unknown ecological niche for microorganisms: A model for astrobiology. Astrobiol. 3: 331-341.
Glavin, D.P., H.J. Cleaves, M. Schubert, A. Aubrey, and J.L. Bada. 2004. New method for estimating bacterial cell abundances in natural samples by use of sublimation. 2004. Appl. Environ. Microbiol. 70: 5923-5928.
Gold, T. 1999. The Deep Hot Biosphere. Copernicus/Springer-Verlag, New York.
Gordon, D.A., J.C. Priscu, and S. Giovannoni. 2000. Distribution and phylogeny of bacterial communities associated with mineral particles in Antarctic lake ice. Microb. Ecol. 39: 197-202.
Hohn, M.J., B.P. Hedlund, and H. Huber. 2002. Detection of 16S rDNA sequences representing the novel phylum “Nanoarchaeota”: Indication for a wide distribution in high temperature biotopes. Syst. Appl. Microbiol. 25: 551-554.
Huber, H., M.J. Hohn, R. Rachel, T. Fuchs, V.C. Wimmer, and K.O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417: 63-67.
Huber, H., M.J. Hohn, K.O. Stetter, and R. Rachel. 2003. The phylum Nanoarchaeota: Present knowledge and future perspectives of a unique form of life. Res. Microbiol. 154: 165-171.
Huber, R., S. Burggraf, T. Mayer, S.M. Barns, P. Rossnagel, and K.O. Stetter. 1995. Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature 376: 57-58.
Huber, R., W. Eder, S. Heldwein, G. Wanner, H. Huber, R. Rachel, and K.O. Stetter. 1998. Thermocrinis ruber gen. nov., sp. nov., a pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl. Environ. Microbiol. 64: 3576-3583.
Hugenholtz, P., C. Pitulle, K.L. Hershberger, and N.R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180: 366-376.
Huston, A.L., B. Methe, and J.W. Deming. 2004. Purification, characterization and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl. Environ. Microbiol. 70(6): 3321-3328.
Ingraham, J.L. 1958. Psychrophilic and psychrotrophic microorganisms. Ann. Inst. Pasteur Paris 16: 111-118.
Innis, W.E. 1975. Interaction of temperature and psychrophilic microorganisms. Ann. Rev. Microbiol. 29: 445-465.
Jannasch, H.W., and M.J. Mottl. 1985. Geomicrobiology of deep-sea hydrothermal vents. Science 229: 717-725.
Junge, K., F. Imhoff, T. Staley, and J.W. Deming. 2002. Phylogenetic diversity of numerically important Arctic sea-ice bacteria cultured at subzero temperatures. Microb. Ecol. 43: 315-328.
Junge, K., J.W. Deming, and H. Eicken. 2004. Bacterial activity at –2 to –20 degree C in Arctic wintertime sea ice. Appl. Environ. Microbiol. 70: 550-557.
Karl, D.M., C.O. Wirsen, and H.W. Jannasch. 1980. Deep-sea primary production at the Galapagos hydrothermal vents. Science 207: 1345-1347.
Karl, D.M., D.F. Bird, K. Björkman, T. Houlihan, R. Shackelford, and L. Tupas. 1999. Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science 286: 2144-2147.
Kashefi, K., and D.R. Lovley. 2003. Extending the upper temperature limit for life. Science 301: 934.
Knoblauch, C., and B.B. Jorgensen. 1999. Effect of temperature on sulphate reduction, growth rate and growth yield in five psychrophilic sulphate-reducing bacteria from Arctic sediments. Environ. Microbiol. 1: 457-467.
Lawrence, J.G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. U.S.A. 95: 9413-9417.
Lehman, R.M., F.S. Colwell, and G.A. Bala. 2001. Attached and unattached microbial communities in a simulated basalt aquifer under fracture- and porous-flow conditions. App. Environ. Microbiol. 67(6): 2799-2809.
Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Alio, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409(6820): 603-607.
Madigan, M., J.M. Martinko, and J. Parker. 2002. Brock Biology of Microorganisms. 10th Edition. Prentice Hall, Upper Saddle River, N.J.
McCollom, T.M., and E.L. Shock. 1997. Geochemical constraints on chemolithoautotrophic metabolism by microorganisms in seafloor hydrothermal systems. Geochim. Cosmochim. Acta 61: 4375-4391.
McKay, C.P., and C.R. Stoker. 1989. The early environment and its evolution on Mars: Implications for life. Rev. Geophys. 27: 189-214.
McKay, C.P., D.T. Andersen, W.H. Pollard, J.L. Heldmann, P.T. Doran, C.H. Fritsen, and J.C. Priscu. 2005. Polar lakes, streams, and springs as analogs for the hydrological cycle on Mars. Pp. 219-233 in Water on Mars and Life, T. Tokano, ed. Springer-Verlag, Berlin.
Moon-van der Staay, S.Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409(6820): 607-610.
Morris, R.M., M.S. Rappe, S.A. Connon, K.L. Vergin, W.A. Siebold, C.A. Carlson, and S.J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806-810.
Mueller, D.R., W.F. Vincent, W.H. Pollard, and C.H. Fritsen. 2001. Glacial cryoconite ecosystems: A bipolar comparison of algal communities and habitats. Nova Hedwigia 123: 173-197.
Naish, T.R., K.J. Woolfe, P.J. Barrett, G.S. Wilson, C. Atkins, S.M. Bohaty, C.J. Bücker, M. Claps, F.J. Davey, G.B. Dunbar, A.G. Dunn, C.R. Fielding, F. Florindo, M.J. Hannah, D.M. Harwood, S.A. Henrys, L.A. Krissek, M. Lavelle, J. van der Meer, W.C. McIntosh, F. Niessen, S. Passchier, R.D. Powell, A.P. Roberts, L. Sagnotti, R.P. Scherer, C.P. Strong, F. Talarico, K.L. Verosub, G. Villa, D.K. Watkins, P.N. Webb, and T. Wonik. 2001. Orbitally induced oscillations in the East Antarctic ice sheet at the Oligocene/Miocene boundary. Nature 413: 719-723.
Napolitano, M.J., and D.H. Shain. 2004. Four kingdoms on ice: Convergent energetic processes boost energy levels at low physiological temperatures. Proc. R. Soc. Biol. B. Suppl. 271: S273-S276.
Navarro-González, R., F.A. Rainey, P. Molina, D.R. Bagaley, B.J. Hollen, J. de la Rosa, A.M. Small, R.C. Quinn, F.J. Grunthaner, L. Cáceres, B. Gomez-Silva, and C.P. McKay. 2003. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science 302: 1018-1021.
NRC (National Research Council). 1978. Recommendations on Quarantine Policy for Mars, Jupiter, Saturn, Uranus, Neptune, and Titan. National Academy Press, Washington, D.C.
NRC. 2000. Preventing the Forward Contamination of Europa. National Academy Press, Washington, D.C.
Pace, N.R. 1997. A molecular view of microbial diversity and the biosphere. Science 276: 734-740.
Paerl, H.W., and J.C. Priscu. 1998. Microbial phototrophic, heterotrophic, and diazotrophic activities associated with aggregates in the permanent ice cover of Lake Bonney, Antarctica. Microb. Ecol. 36: 221-230.
Page, R.R.M., and E.C. Holmes. 1998. Molecular Evolution: A Phylogenetic Approach. Blackwell Sciences, Oxford, United Kingdom.
Patterson, W.S.B. 1994. The Physics of Glaciers. 3rd edition. Elsevier Science, Inc., Tarrytown, N.Y.
Parkes, R.J., B.A. Cragg, and P. Wellsbury. 2000. Recent studies on bacterial populations and processes in subseafloor sediments: A review. Hydrogeol. J. 8(1): 11-28.
Pedersen, K. 2000. Exploration of deep intraterrestrial microbial life: Current perspectives. FEMS Microbiol. Lett. 185: 9-16.
Price, P.B. 2000. A habitat for psychrophiles in deep Antarctic ice. Proc. Natl. Acad. Sci. U.S.A. 97: 1247-1251.
Price, P.B., and T. Sowers. 2004 Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl. Acad. Sci. U.S.A. 101: 4631-4636.
Priscu, J.C., and B. Christner. 2004. Earth’s icy biosphere. Pp. 130-145 in Microbial Diversity and Prospecting, A.T. Bull, ed. ASM Press, Washington, D.C.
Priscu, J.C., C.H. Fritsen, E.E. Adams, S.J. Giovannoni, H.W. Paerl, C.P. McKay, P.T. Doran, D.A. Gordon, B.D. Lanoil, and J.L. Pinckney. 1998. Perennial Antarctic lake ice: An oasis for life in a polar desert. Science 280: 2095-2098.
Priscu, J.C., C.F. Wolf, C.D. Takacs, C.H. Fritsen, J. Laybourn-Parry, E.C. Roberts, and W.B. Lyons. 1999a. Carbon transformations in the water column of a perennially ice-covered Antarctic Lake. Bioscience 49: 997-1008.
Priscu, J.C., E.E. Adams, W.B. Lyons, M.A. Voytek, D.W. Mogk, R.L. Brown, C.P. McKay, C.D. Takacs, K.A. Welch, C.F. Wolf, J.D. Kirschtein, and R. Avci. 1999b. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286: 2141-2144.
Priscu, J.C., R.E. Bell, S.A. Bulat, C. Ellis-Evans, V.V. Lukin, J.-R. Petit, R.D. Powell, M.J. Siegert, and I. Tabacco. 2003. An international plan for Antarctic subglacial lake exploration. Polar Geography 27(1): 69-83.
Priscu, J.C., E.E. Adams, H.W. Paerl, C.H. Fritsen, J.E. Dore, J.T. Lisle, C.F. Wolf, and J.A. Mikucki. 2005. Perennial Antarctic lake ice: A refuge for cyanobacteria in an extreme environment. Pp. 22-49 in Life in Ancient Ice, J.D. Castello and S.O. Rogers, eds. Princeton University Press, Princeton, N.J.
Psenner, R., B. Sattler, A. Willie, C.H. Fritsen, J.C. Priscu, M. Felip, and J. Catalan. 1999. Lake ice microbial communities in alpine and Antarctic lakes. Pp. 17-31 in Cold-Adapted Organisms, Ecology, Physiology, Enzymology and Molecular Biology, P. Schinner and R. Margesin, eds. Springer-Verlag, Berlin.
Rappe, M.S., S.A. Connan, K.L. Vergin, and S.J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418: 630-631.
Reysenbach A.L., G.S. Wickham, and N.R. Pace. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60: 2113-2119.
Rivkina, E.M., E.I. Friedmann, C.P. McKay, and D.A. Gilichinsky. 2000. Metabolic activity of permafrost bacteria below the freezing point. Appl. Environ. Microbiol. 66: 3230-3233.
Rogers, S.O., W.T. Starmer, and J.D. Castello. 2005. Recycling of organisms and genomes. Life in Ancient Ice, J.D. Castello and S.O. Rogers, eds. Princeton University Press, Princeton, N.J.
Russell, N.J., and T. Hamamoto. 1998. Psychrophiles. Pp. 25-45 in Extremophiles: Microbial Life in Extreme Environments, K. Horikoshi and W.D. Grant, eds. Wiley-Liss, New York.
Sattler, B., H. Puxbaum, and R. Psenner. 2001. Bacterial growth in supercooled cloud droplets. Geophys. Res. Lett. 28: 239-242.
Sharp, M., J. Parkes, B. Cragg, I.J. Fairchild, H. Lamb, and M. Tranter. 1999. Widespread bacterial populations at glacier beds and their relationship to rock weathering and carbon cycling. Geology 27: 107-110.
Sheridan, P.P., V.I. Miteva, and J.E. Brenchley. 2003. Phylogenetic analysis of anaerobic psychrophilic enrichment cultures obtained from a Greenland glacier ice core. Appl. Environ. Microbiol. 69: 2153-2160.
Shi, T., R.H. Reeves, D.A. Gilichinsky, and E.I. Friedmann. 1997. Characterization of viable bacteria in Siberian permafrost by 16S rDNA sequencing. Microb. Ecol. 33: 169-179.
Siegert, M.J., J.C. Ellis-Evans, M. Tranter, C. Mayer, J.-R. Petit, A. Salamatin, and J.C. Priscu. 2001. Physical, chemical and biological processes in Lake Vostok and other Antarctic subglacial lakes. Nature 414:603-609.
Siegert, M.J., M. Tranter, J.C. Ellis-Evans, J.C. Priscu, and W.B. Lyons. 2003. The hydrochemistry of Lake Vostok and the potential for life in Antarctic subglacial lakes. Hydrological Processes 17: 795-814.
Smith, A.W., D.E. Skilling, J.D. Castello, and S.O. Rogers. 2004. Ice as a reservoir for pathogenic human viruses: Specifically, caliciviruses, influenza viruses, and enteroviruses. Medical Hypotheses 63: 560-566.
Stahl, D.A., D.J. Lane, G.J. Olsen, and N.R. Pace. 1985. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl. Environ. Microbiol. 49: 1379-1384.
Stetter, K.O. 1998. Hyperthermophiles: Isolation classification and properties. Pp. 1-24 in Extremophiles: Microbial Life in Extreme Environments, K. Horikoshi and W.D. Grant, eds. Wiley-Liss, New York.
Stevens, T.O., and J.P. McKinley. 1995. Lithoautotrophic microbial exosysems in deep basalt aquifers. Science 270: 450-455.
Studinger, M., R.E. Bell, and A.A. Tikku. 2004. Estimating the depth and shape of subglacial Lake Vostok’s water cavity from aerogravity data. Geophys. Res. Lett. 31: L12401.
Szewzyk, U., R. Szewzyk, and T.A. Stenstrom. 1994. Thermophilic, anaerobic bacteria isolated from a deep borehole in granite in Sweden. Proc. Natl. Acad. Sci. U.S.A. 91: 1810-1813.
Takacs, C.T., and J.C. Priscu. 1998. Bacterioplankton dynamics in the McMurdo Dry Valley lakes: Production and biomass loss over four seasons. Microb. Ecol. 36: 239-250.
Takeuchi, N., S. Kohshima, Y. Yoshimura, K. Seko, and K. Fujita. 2000. Characteristics of cryoconite holes on a Himalayan glacier, Yala Glacier central Nepal. Bull. Glaciol. Res. (Japan) 17: 51-59.
Thomas, D.N., and G.S. Dieckmann. 2002. Antarctic sea ice—A habitat for extremophiles. Science 295: 641-644.
Tranter, M., A. Fountain, C.F. Fritsen, W.B. Lyons, J.C. Priscu, P. Statham, and K. Welch. 2004. Extreme hydrochemical conditions in natural microcosms entombed within Antarctic ice. Hydrological Processes 18: 379-387.
Tsapin, A.I., G.D. McDonald, M. Andrews, R. Bhartial, S. Douglas, and D. Gilichinsky. 1999. Microorganisms from permafrost viable and detectable by 16S RNA analysis: A model for Mars. Fifth International Conference on Mars, July 18-23, 1999. Lunar and Planetary Institute, Pasadena, Calif. Available at <http://marsprogram.jpl.nasa.gov/mgs/sci/fifthconf99/6104.pdf>.
Vishnivetskaya, T., S. Kathariou, J. McGrath, D. Gilichinsky, and J. Tiedje. 2000. Low-temperature recovery strategies for the isolation of bacteria from ancient permafrost sediments. Extremophiles 4: 165-173.
Ward, B.B., and J.C. Priscu. 1997. Detection and characterization of denitrifying bacteria from a permanently ice-covered Antarctic lake. Hydrobiology 347: 57-68.
Ward, D.M., R. Weller, and M.M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 344: 63-65.
Ward, D.M., M.J. Ferris, S.C. Nold, and M.M. Bateson. 1998. A natural view of microbial diversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62: 1353-1370.
Waters, E., M.J. Hohn, I. Ahel, D.E. Graham, M.D. Adams, M. Barnstead, K.Y. Beeson, L. Bibbs, R. Bolanos, M. Keller, K. Kretz, L. Xiaoying, E. Mathur, N. Jingwei, M. Podar, T. Richardson, G.G. Sutton, M. Simon, D. Söll, K.O. Stetter, J.M. Short, and M. Noordewieret. 2003. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. U.S.A. 100: 12984-12988.
Wells, L.E., and J.W. Deming. 2003. Abundance of cytophaga-flavobacterium-bacteriodes and archaea in cold surface-water and nepheloid layers of the Northwest Passage, Canadian Archipelago. Aquat. Microb. Ecol. 31: 19-31.
Wharton, R.A., Jr., W.C. Vinyard, B.C. Parker, G.M. Simmons, Jr., and K.G. Seaburg. 1981. Algae in cryoconite holes on Canada Glacier in southern Victoria Land, Antarctica. Phycologia 20: 208-211.
Whitman, W.B., D.C. Coleman, and W.J. Wiebe. 1998. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95(12): 6578-6583.
Willerslev, E., A.J. Hansen, B. Christensen, J.P. Steffensen, and P. Arctander. 1999. Diversity of Holocene life forms in fossil glacier ice. Proc. Natl. Acad. Sci. U.S.A. 96: 8017-8021.
Willerslev, E., A.J. Hansen, J. Binladen, T.B. Brand, M.P. Gilbert, B. Shapiro, M. Bunce, C. Wiuf, D.A. Gilichinsky, and A. Cooper. 2003. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300: 791-795.
Willerslev, E., A.J. Hansen, and H.N. Poinar. 2004a. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol. Evol. 19: 141-147.
Willerslev, E., A.J. Hansen, T.B. Brand, R. Rønn, I. Barnes, C. Wiuf, D.A. Gilichinsky, D. Mitchell, and A. Cooper. 2004b. Long-term persistence of bacterial DNA. Curr. Biol. 14(1): R9-R10.






















