6
PLENARY SPEAKERS, DAY 2
MODELING AND PANDEMIC PREPAREDNESS,
Professor Neil Ferguson, Professor of Mathematical Biology, Faculty of Medicine, Imperial College of London
I am not going to cover the whole gamut of modeling related to pandemic influenza, as the list of potential modeling questions is long. I will focus principally on whether can we contain pandemics at their source. In this case, the principal threat is an H5N1-based pandemic emanating from Southeast Asia. Models can address many other questions with the right sort of data. A particularly relevant question, given recent experience with SARS, is whether restrictions on people’s movements—such as on international travel—can delay the international spread of a pandemic. I will touch on that topic at the end.
In thinking about preparedness, we might ask how fast a pandemic strain might spread within a country. What will the health care burden be? How can we best use antivirals to reduce mortality and morbidity, protect key personnel, reduce social disruption given significant mortality, and reduce disease attack rates while we prepare a vaccine? More difficult to model than the impact of antivirals is the role of measures to increase “social distance”—such as closing schools or limiting travel within a country—in slowing the spread of disease or reducing attack rates. Models clearly need to capture the logistical constraints and resource limits that always affect such policies. Because we will not know the characteristics of a given influenza strain until a pandemic occurs, we also need real-time techniques for refining model estimates, and potentially even public policies, once a pandemic starts.
What do we need to contain a pandemic at its source? That ambitious goal will require early detection of a pandemic strain along with well-planned and rapidly executed responses. Containment is probably feasible only if we detect the emerging pandemic at its earliest stages – for instance, after a cluster of a few cases has emerged in a village in rural Vietnam or Thailand, for instance..
Containing a potential pandemic at its earliest stages is perhaps made more feasible with the knowledge that transmissibility of a pandemic strain may evolve incrementally, requiring many additional changes after the initial reassortment or mutation event. If the pandemic strain which emerges initially has a value of R0 (the reproduction number) which only slightly exceeds the critical threshold of 1—that is, only one secondary case ensues from each primary case—then spread, while inevitable, will be slow, giving us time to respond.
What are our options? Compared with the past, there are more possibilities. In particular, prophylactic use of antiviral drugs is a potentially key measure to reduce spread. Vaccination will also clearly play a key role if vaccine is available, Other public health measures include those that increase social distance. Here I focus on identifying combinations of control measures
capable of controlling the earliest stages of a pandemic, assessing how sure we can be of the outcome of a containment strategy, and quantifying the resources which would be necessary to deliver these measures.
We have undertaken modeling to address such questions as part of a study called Models of Infectious Disease Agents Study,(MIDAS), funded by the National Institute of General Medical Sciences in NIH. The project includes three basic research groups, one at Virginia Tech led by Dr. Steve Eubank, one at Emory led by Dr. Ira Longini, and one at Johns Hopkins led by Dr. Donald Burke. I am part of the latter study, which includes researchers at Imperial College. I will talk about the work I have been doing with Don's group in modeling pandemic spread in Thailand. Ira Longini has been working on the same topic, and we hope that both these studies will be published in the next few months.
I will talk a bit about the structure of our model and its frailties and assumptions. A key difference between this model and many infectious disease models in the past is its scale. Our approach is to simulate entire countries as realistically as possible, so our computationally intensive simulation models a population of over 85 million. We tried to capture social structure by modeling individuals and households, because—as with many other infectious diseases—households are key location for transmission of influenza. Transmission of influenza also occurs within peer groups at schools and workplaces, and a separate component of the model captures those. We also know that community-based journeys to shops and other locations in the country are key to the longer-range spread of the pathogen. The model captures those by modeling a random contact process between individuals in the population that depends on distance.
We can think of the population as a set of individuals residing within households, in which adults travel to workplaces and children attend schools. An important aspect of this model is data we have collected on how far people in a household typically travel to schools or work. Thus the model tries to capture both the social structure and the real geography of populations—both of which are key to understanding and predicting the spatial and geographic spread of an emergent strain.
How do we simulate the population? We use probably the most detailed dataset available on global population density, called Landscan, which is now being used in Iraq and was used for the tsunami relief effort. This dataset, prepared by Oak Ridge National Laboratory, gives sub-kilometer data on population density globally. We clearly do not know precisely how many people live on every square kilometer of the planet, but the Landscan predicted density figures have been validated using remote-sensing as well as census data. One output of the simulation model we have constructed is maps of population density, which use colour to represent reas of the modeled region in which infection is present or trestment being undertaken..
Capturing age and household structure are also critical for realistically modeling influenza transmission, and the model incorporates data we have collected for Thailand. We chose Thailand not because we felt it was the country of highest risk for emergence of a pandemic, but because it is representative of Southeast Asia, and data on population structure and movements in that country are available. However, we intend to generalize the model to examine Vietnam, among other countries.
The model also incorporates school and workplace demographics, including the distribution of school and worksplace sizes. Dr. Derek Cummings at Johns Hopkins collected these data, which are also important for realistically describing disease transmission.
Most critical is capturing the movement of people. To do so, we use data from the Thai national migration survey, which asked a sample of people the distance they travel to work. The cumulative probability distribution function for those data is typical of many countries: most people travel a very short distance to work, but the distribution includes a fat tail—some people travel tens of kilometers or even 100 kilometers. The distribution of individuals going to school includes a distance cutoff, as students clearly do not travel quite so far to school as adults do to work. Although sample size is limited, the curve is similar to that seen in other countries, so we have some confidence in it. The model also captures the proportion of the population that is working.
The transmissibility of the pathogen will determine the outcome or feasibility of containment policies for pandemic influenza. To investigate this, we are aided by work done by Dr. Mark Lipsitch's group led by Dr. C.E. Mills in 2004, which tried to quantify the transmissibility of pandemic influenza in 1918 and came up with a range of 2 to 3 for R0. Our later work noted that this assumes a serial interval for influenza—the delay between one generation of infections and the next – of about 3.5 days. This value was also assumed by many other past publications, but relatively few data exist with which to back up the figure.
Reanalyzing the available data on the incubation period of influenza and the duration of infectiousness reduces that figure to perhaps 2.5 days, which paradoxically drops estimates of R0. Our revised estimates of R0 for the pandemic waves in the United Kingdom in 1918 and 1919 peak at 1.8, and are considerably less most of the time. However, those adjustments do not mean that pandemic influenza will spread more slowly, because the doubling time of these epidemics remains the same. That is, if we reduce the serial interval and the reproduction number, then the overall rate of spread can remain constant.
The model tries to capture the natural history of influenza as realistically as possible. We are using current H3 and H1 influenza as our principal model. We are well aware that the H5-based pandemic may not show the same natural history parameters, and so are undertaking sensitivity analyses, but we felt it was best to root the model in what we know about human influenza. We are use data published by Moser et al. on the distribution of latent periods of the disease, which average about 1.5 days. The model also incorporates data on the distribution of infectiousness, and we tried to match the model to previous age-dependent attack rates of pandemics.
We base our assumptions on the effect of antiviral drugs on parameters estimated by Dr. Fred Hayden and by Dr. Ira Longini. Our baseline assumptions are that prophylaxis of uninfected individuals reduces their susceptibility by about 30 percent, and the infectiousness of infected people by 60 percent for as long as they are on therapy. A treated person also sees a 65 percent drop in the chance of becoming a clinically diagnosable case.
We assume that only about half of infections result in clinically identifiable disease. That means that should a pandemic strain of H5 emerge through reassortment, its virulence will be less than what we are now seeing. However, if the current human virulence of the avian H5N1 virus remained unchanged for an emergent pandemic H5 strain, severe clinical disease would actually occur in a much higher proportion of cases. Our assumption of the 50 percent clinical case rate is thus quite pessimistic from the perspective of case ascertainment. Our assumption that treatment reduces someone’s chance of becoming a case by 65 percent reduces the average infectiousness of infected individuals still further.
A simulation model can examine almost any policy. We concentrated on a combination of three: treatment of cases and prophylaxis of households; prophylaxis of schools and workplaces; and prophylaxis of everyone within a certain number of kilometers of a diagnosed case. The model can incorporate delays, detection thresholds, and limits on drug use, as well as the impact of measures to increase social distance. Currently I am less satisfied with our estimates of the latter, so I will not present results here.
All these assumptions yield a simulation of uncontrolled pandemic spread in Thailand. We assume that a pandemic will start with a single case in a randomly selected rural area—a reassortment or mutation event in an individual which will increase transmissibility and thus give rise to a growing cluster of cases. At a reproduction number of 1.5, we predict that the epidemic in Thailand will peak within about 120 days. The spatial spread is very fast. After a few hundred cases, we are almost certain to see cases emerging in Bangkok. Once the disease reaches Bangkok, it spreads to the rest of the country very rapidly, because Bangkok is highly connected to all other locations through travel.
The key results from this modeling exercise show the probability of containment as a function of the reproduction number of the pathogen. This analysis does not assume any particular level of transmissibility. Rather, it asks: what is the threshold level of transmissibility beyond which containment will be impossible, or below which containment is feasible? The results indicate that an perfectly implemented policy could achieve containment with a production number of 1.8 or less. As I showed before, that is a feasible range for what we now think the reproduction number of pandemic influenza might be. By perfect implementation, I mean a policy which is started 7 days after the first case of a pandemic strain, and where 90 percent of cases are detected and treated on the day they develop symptoms, 90 percent of their households are treated, 90 percent of their schools and workplaces are treated, and 90 percent of the general population within 10 kilometers of those cases is treated.
If antiviral containment is effective, it reduces the size of the outbreak to a small number of cases—a few hundred at most. The treatment course lasts 5 days, and the prophylaxis course at half the dose lasts 10 days. On average, containment is feasible with an average of under 1 million courses of drug. Even in the worst cases, the pandemic is containable with fewer than 3 million courses. Note that results are averages of a large number of runs of the model. Every run is different, so we have to run it many times to characterize the average behavior.
Perfect implementation is clearly an unrealistic best case, so I will show you briefly what happens when we start relaxing some of our perfect assumptions. One key issue is when we would recognize when a pandemic was potentially starting: that is, when a new cluster of cases in which efficient human-to-human transmission was occurring would be detected. We calculated the probability of containment as a function of whether we have detected the cluster after 0, 10, 20, or 30 cases. What is encouraging is that even if we require a single cluster of 30 cases to realize that the virus has changed, we still see about 95 percent containment under reproduction number (R0) of 1.7, and 100 percent containment under R0 of 1.6.
Some elements are critical to an effective policy. These elements do not include how quickly we identify the first cluster (given that the number of cases is somewhat limited, and that we detect the cluster within a month of the first identified case), but rather that once we start a policy we pursue it effectively. We identify new cases, treat them, and perform prophylaxis rapidly. The model also shows the impact of varying the time required to treat cases and perform
prophylaxis of the case’s contacts. A two-day delay yields considerably less containment, and a four-day delay even less. For four days, we can contain the strain only up to an R0 of about 1.2 (though despite the reduced effectiveness we still only need an average of under 1 million courses of treatment if the policies succeed).That’s because for such a long delay, a whole generation of transmission has been missed, so we are constantly one or two generations of infection behind the pandemic. If we can reduce the delays to two days or less, we are treating people who have just been infected and prophylaxing on their contacts. Remember, I'm assuming a relatively short serial interval for this pandemic strain—about 2.5 days. If the pandemic strain has a much longer incubation and infectious period (as human cases of infection with the avian H5N1 virus currently have), policy effectiveness would be less dependent on very rapid case detection and prophylaxis.
We can also relax many of the assumptions at once and still see a policy that is capable of some degree of containment. One alternative shows a more realistic (but perhaps still optimistic) policy of a 14-day delay to initiation, 75 percent of cases detected, 80 percent of cases treated, 90 percent of households treated, and 90 percent of schools and workplaces treated. Given a two-day delay in all such treatment after a case is detected, we could still contain a pandemic with a reproduction number up to about 1.3 or 1.4.
Remember that I'm modeling purely an antiviral containment strategy. In reality, we would expect to combine that strategy with other measures, especially those that increase social distance. We can improve containment further by adding social distance measures to a prophylaxis-based policy. One example is a quarantine zone. That is, we restrict movement in and out of those areas where we are treating everyone. If we assume that such a policy could reduce traffic in and out of the treatment zone by 80 percent, then we could contain pandemics up to about a R0 of 1.6. Importantly, such a measure makes containment more robust to uncertainty in key model parameters, including the proportion of transmission that occurs in schools and workplaces versus randomly in the community.
If that a containment policy succeeds, it does so by containing the disease in a relatively small geographic area, typically a small rural community in Thailand. We may see the occasional case further afield, but spread is generally quite tightly contained goegraphically. Thus the results from this modeling give some optimism in assessing the feasibility of containing a pandemic.
These results do assume that policies are transnational; i.e. implementation can cross borders. If a pandemic spark occurred at the center of Thailand, that wouldn't necessarily need to be the case for containment to succeed. However, if the original cluster arises in a border region, spread occurs rapidly across the border, meaning a containment policy which is implemented in one country only is doomed to failure. This reinforces the need for a concerted international response—probably with teams on the ground chasing cases.
While a pandemic will spread across land borders, air traffic will be probably be the most important mechanism for international spread in the next pandemic. Compared with 1918, travel capable of transmitting infection over long distances has grown 100-fold to 1,000-fold, and that trend is accelerating. As SARS showed last year, the increasing volume of international air traffic poses clear risks for the rapid spread of novel pathogens. Within a week of the arrival of the index case of SARS at the Metropole Hotel in Hong Kong, the disease had spread to multiple countries around the world.
We have been using a variety of models—both simple and complex—to examine the effectiveness of international restrictions and travel advisories on different pathogens. The conclusions, I have to say, are somewhat pessimistic. For influenza well as SARS, our analysis shows that we would need to reduce traffic by more than 99 percent to have a significant effect on the international spread of an emerging influenza pandemic. This is because when once 100 or 200 cases appear, there is a very high probability that at least one will be exported, owing to the international connectivity in these populations. Even if we reduce that connectivity by 90 percent, we would still need only about 1,000 cases in a source region before being reasonably certain that some export of infection will occur. And an influenza pandemic could entail tens of thousands of cases in the source country if containment is not successful. So while international restrictions might play a small role at the very earliest stages of a pandemic, when only a few dozen cases have appeared, such restrictions become almost irrelevant after a very short period of time. The source country will have so many cases and the world is so connected that we can delay the spread of disease by a week or two at most.
We are using expanded versions of the models of pandemic containment to quantify and predict rates of international spread. We are not quite at the level of global scale simulations, although we are discussing that. Instead, we are using a variety of simpler models to investigate global spread – in particular patch models, which divide the world into small areas and then model traffic between them. People often ask why pandemics emerge in China and Southeast Asia. The Landscan datset map showing areas of high population density offers some insight into this.
Besides quantifying the international spread of influenza, the MIDAS initiative and various projects funded by the European Union are modeling pandemic preparedness options within the continental United States and Europe. These efforts are starting to address some of the questions about the optimal use of antiviral resources to limit mortality and morbidity. We hope preliminary results will be available this summer.
To conclude, our work indicates that a policy of antiviral prophylaxis combined with social distance measures is potentially capable of containing the early stages of a pandemic, but with some caveats. The reproduction number of the virus must be relatively low—less than about 1.8. However, we think that is realistic—particularly if transmissibility evolves incrementally—given our new estimates of the reproduction number of past pandemics.
We need to quickly identify the original cluster where efficient human-to-human transmission occurs. And we need to deliver treatment rapidly to a large proportion of the rural population. That is arguably the area most in need of development. Frankly, I'm skeptical about whether such an approach is possible right now. However, that is not necessarily a reason to rule out containment as a strategy, but rather a spur to improve the public health infrastructure. We also need enough courses of drug—perhaps as many as 3 million. If we end up using more than that, it is likely a containment policy will not succeed in any case, according to this model. We also need excellent case detection and rapid implementation of prophylaxis once the first cluster is identified. That implies that we can ramp up surveillance once a pandemic situation is declared. And we need policies to be transnational.
The policy modeled here entails many logistical hurdles. But given the potentially huge human and economic cost of a pandemic, those hurdles represent challenges to us to make a containment strategy feasible, not reasons to dismiss the strategy outright.
CLINICAL TRIALS OF POTENTIAL PANDEMIC VACCINES: KEY ISSUES
Dr. John Treanor, Associate Professor of Medicine and Microbiology and Immunology, University of Rochester Medical Center
One of the key features of pandemic planning is trying to prepare potentially effective vaccines. I'm going to briefly review what we have learned about evaluating such vaccines.
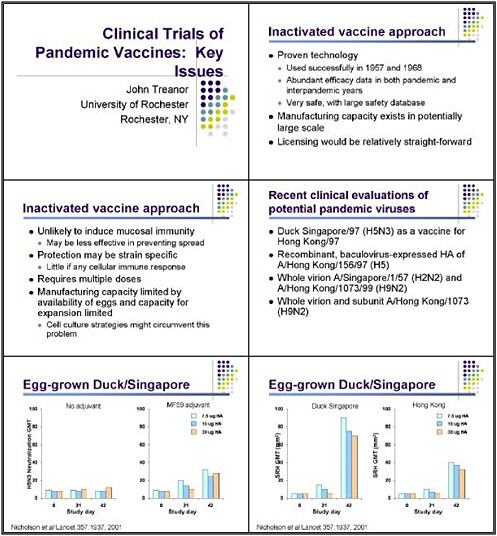
The most straightforward approach for controlling pandemic influenza remains the use of inactivated vaccines, because they have several potential advantages. This is a proven technology that has been used successfully to control pandemic influenza, and abundant efficacy data are already available from both pandemic and inter-pandemic years. A large safety database is also available, as these vaccines have been used in hundreds of millions of people. Thus we have a good idea of what we might expect in the way of side effects if such a vaccine were deployed on a very large scale.
Manufacturing capacity is also already in place. It clearly would not be sufficient to make enough vaccines for everyone in the world, but it is a very large. And, as mentioned yesterday, licensing such a vaccine—using a process already employed for conventional vaccination—would be relatively straightforward compared with some other approaches.
The inactivated vaccine approach does have disadvantages. It is unlikely to induce mucosal immunity, for example, and might therefore not be as effective at preventing transmission as some other strategies. The protection might also be fairly strain specific. Such a vaccine would be likely to induce little, if any, cellular immune responses, and would probably require at least two doses to prime a naive population.
Manufacturing capacity for the current egg-based vaccine strategies is also limited by the availability of eggs. And the facilities that make egg-based vaccines are fairly specialized. Cell-culture technologies would be one way around that, and we clearly need to pursue those vigorously.
Recent clinical evaluations have told us a bit about what future studies of inactivated vaccines should pursue. I'm going to describe them briefly. The strategies used recently have included the Duck Singapore. We have looked at recombinant baculovirus-expressed HAs, and a variety of studies are looking at both whole virus and split virus for H2N2 and H9N2.
Some of the first data came from Karl Nicholson and his group in England, who looked at egg-grown Duck Singapore as a potential vaccine for H5 influenza when the first outbreak was noted in Hong Kong in 1997. Duck Singapore is an antigenically-related H5 virus that does not have the highly cleavable hemagglutinin, and therefore could be used for vaccine production without the need for biocontainment.
One aspect noticed early on was that this vaccine is not particularly immunogenic. So Karl and his group looked at the Duck Singapore formulated as a subunit vaccine in doses of 7.5, 15, or 30 micrograms. They found very little response when looking at a neutralization of the H3N3 virus in microneutralization assay. Two doses of as much as 30 micrograms did not produce any significant response in neutralizing the vaccine virus. The finding that the addition of the adjuvant MF59 did result in significant increases was encouraging, although they are
relatively low titer. And for reasons that are still not completely clear, lower doses seemed to be slightly more immunogenic than higher doses when combined with MF59.
The other interesting finding was that the response was relatively strain specific. Because of containment issues, neutralization tests against the A-Hong Kong H5 were not done directly. Investigators looked instead at the radial hemolysis test. The vaccine did have a tendency—when adjuvanted with MF59—to induce antibody with much greater ability to recognize the Duck Singapore virus than the Human Hong Kong virus, which would be the target of the vaccine program. This is just an example of the observation that inactivated vaccines in an unprimed population are likely to stimulate an immune response that will be fairly specific for the actual antigen in the vaccine.
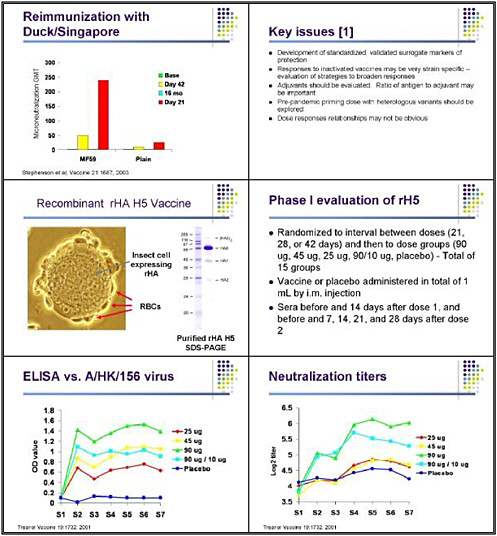
With Karl, Ian Stevenson recently noted a very interesting finding: despite the fact that these responses were relatively minimal and short-lived, significant responses occurred when these individuals were revaccinated 16 months later. The investigators revaccinated everyone in the study who agreed to that with the same vaccine formulation they had received in the first dose. The MF59 group, in particular, saw an increase in neutralizing antibody that was essentially gone 16 months later. But when these individuals received a single additional dose at 16 months, a very significant response occurred by neutralization against the H5N3 virus. This is a very encouraging finding from the point of view of strategies to provide a priming dose before beginning a vaccination campaign.
Those studies taught us about the need to develop standardized and validated surrogate markers of protection. We also learned that the responses to inactivated pandemic vaccines are likely to be strain specific. Adjuvants may play an important role in dose-sparing strategies for an inactivated vaccine. And the use of a pre-pandemic priming dose could generate a more rapid response in the face of an emerging pandemic. Particularly for MF59, the dose-response relationships may not be obvious, and may be more related to the relative ratio of MF59 in antigen than to the total dose of antigen.
At the same time, we did studies looking at recombinant hemagglutinin expressed in a baculovirus system as a vaccine. These were actually the first human trials of a vaccine for H5 in response to the 1997 outbreak in Hong Kong. These studies were possible because of the relative ease of expressing the hemagglutinin—even of a highly pathogenic virus using the baculovirus vector. The great advantage of this kind of approach is the speed with which we can come up with new antigens.
This very complicated study attempted to look at many things at the same time, including the dose of vaccine as well as whether administering the two doses 21, 28, or 42 days apart made any difference. We found that this vaccine was fairly effective in inducing antibody that recognized the baculovirus-expressed hemagglutinin in an ELISA system, although there was a clear-cut dose response, with the highest levels of response seen in individuals who received 90 micrograms. We also looked at as a dose-sparing strategy of giving a large priming dose and then boosting with a smaller dose. That did not work as well as two doses of 90 micrograms in inducing ELISA antibody. Jackie Katz performed these neutralization tests in a containment laboratory—an extremely tedious and difficult way to test lots of sera.
However, none of these vaccines were as effective as we had hoped in generating antibody that could actually neutralize the Hong Kong virus. Two doses of 90 micrograms generated, on average, neutralizing titers on the order of 1 to 32. Those are good but not great
titers, and that is encouraging, but that is a very large amount of antigen. Again, a 90-microgram dose followed by 10 micrograms was somewhat effective, and there was a clear-cut dose relationship in terms of the total amount of hemagglutinin and antigen used—the factor that largely determined the responses.
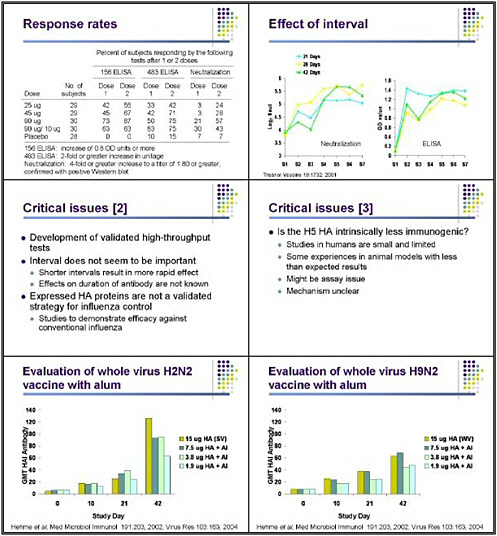
A very complicated algorithm was necessary for trying to decide whether someone had actually responded, because of the difficulties of doing the assay in the first place. This resulted in a very complex definition of a response: a 4-fold or greater increase in titer, to a titer of 1 to 80 or greater by neutralization, confirmed with a positive Western blot. Using those criteria, we found that even two doses of 90 micrograms resulted in a sera response rate of only slightly more than 50 percent of the subjects. The tests did show that immunization is possible but not particularly efficient.
We found that the interval—whether we measured antibody by ELISA or neutralization—did not appear to play an important role in the ultimate response. We saw just as good a response when two doses were separated by 21 days as when they were separated by 28 or 42 days. I can confirm that this vaccine appears to have 100 percent efficacy, because we have not noticed a single case of H5 influenza in any of the vaccine recipients! So that is encouraging news.
But again, this pointed out the technical struggles of doing the assays, and the fact that we have no way of knowing whether these vaccines would have been protective. There is a critical need for any clinical trial to develop well-validated and high-throughput assays, as we anticipate that we will be running many people through trials of candidate H5 vaccines. Getting a better handle on some of the immune responses and how to measure them is one of the critical needs in the field.
We found, however, that interval does not appear to be important, with the proviso that we did not look at the duration of antibody. It is possible that the interval could have had more of an effect on the duration of the antibody response or the development of immune memory. Future trials should probably evaluate those aspects. The expressed hemagglutinin looked like a promising approach, but that is not a validated strategy for influenza control. Pandemic planning should thus confirm whether this approach is actually effective in preventing conventional influenza.
One issue that the Duck Singapore studies and our studies have raised is whether there is something special about the H5 hemagglutinin that makes it intrinsically less immunogenic. I have no idea what the explanation for that would be, but we have noticed poor responses in human trials, and others have reported less-than-expected responses in some scenarios in animals. However, experience so far in humans has been extremely limited, and it is probably premature to conclude that we will not be successful with other vaccine trials with H5 viruses. Still, there might be something about the H5 that makes it intrinsically less immunogenic. I leave it to the immunologists to figure out what that would be, but I think that's an important question to answer.
Norm Hehme and others at GlaxoSmithKline have looked at vaccines for H2N2 viruses, because the next pandemic is likely to be an H2 virus, given previous human history. Those investigators examined two important issues. One is the immunogenicity and safety of a whole virus vaccine—not because it would be more immunogenic, but because it is more efficient to produce, and the yield might therefore be greater without the need for additional purification
steps. Another important issue is whether the addition of alum would help, given that it has been used for influenza vaccines in the distant past and is a readily available and widely used adjuvant for other vaccines.
The study design compared a 15-microgram dose of hemagglutinin in a split product or subunit vaccine formulation against lower doses of hemagglutinin administered with alum. When the researchers used HAI antibody against the H2 virus as the outcome, they found that responses to relatively low doses of hemagglutinin with alum approached but did not equal those seen with the 15-microgram dose. Adjuvant studies need to include truly comparable groups to determine whether the adjuvant itself is increasing the response.
When this group also did a similar study with H9 vaccines, they found that low-dose vaccine with alum appeared to induce responses comparable to those seen with unadjuvanted high-dose vaccine. More recently, Ian Stevenson and his group looked at whole virus and split unit vaccines for H9, and found that whole virus vaccines, particularly at lower doses, might be modestly more immunogenic. The interesting finding in that study was the difference in responses between older and younger individuals, with those over the age of 35 manifesting much better responses—particularly to the first dose of both whole virus and subunit vaccine. Those investigators found a very significant difference in the presence of pre-vaccination antibody against the H9 virus between people born before 1968 and those born after 1968.
Maria Zambon and her group did an extensive series of studies with reassortants to show that this specifically was antibody related to the H2, not a neuraminidase effect. This very interesting phenomenon might be relevant to other potential pandemic viruses.
We learned from this study that dose-sparing roles of adjuvants need to be evaluated by direct comparisons. We also need to consider whether further studies should consider the potential production advantages of whole virus vaccines. And we need to be aware of the potential impact of prior exposure on whether people may respond to a single dose.
The current study that Dr. Fauci alluded to yesterday entails an H5 inactivated vaccine. The goal is to use a virus generated using a process that would be considered a strain change, using a licensed production process. There are two candidate vaccines manufactured using as a seed virus the genetically engineered A-Vietnam strain produced by Dr. Richard Webby and Dr. Robert Webster. The first product is from Sanofi-Avantis and second is from Chiron. Both manufacturers used a production facility to produce subunit vaccines that are very similar in principle to the vaccines that these manufacturers are licensed to produce for conventional influenza.
Our study design is very detailed, because we want to be very clear about the safety of these vaccines before proceeding further. We screen individuals for clinical history and normal laboratory results prior to immunization. They are then randomized to receive either placebo or various doses of the inactivated vaccine, ranging from 7.5 micrograms to 90 micrograms. Although a 90-microgram dose might not be practical for pandemic control, we are hoping that by pushing the dose up, we can root this in the entire dose curve and ensure that at least some people manifest a meaningful response to vaccination.
Serum is obtained before and after vaccination, and clinical evaluation occurs 7 days after vaccine, along with safety labs. That information will be fed to a data safety monitoring board. If
the vaccine meets pre-specified safety rules, the second dose of vaccine will be administered on day 28, with the same type of follow-up in this preliminary group.
An additional cohort of subjects will be enrolled if the vaccine proves safe in the first cohort. The second cohort will be randomized to the same levels of vaccine but will not have laboratory safety. This will give us a total enrollment of 100 subjects in each group at each dose level, plus 50 placebo recipients. The latter are being added mostly to blind the laboratory in assessing the H5 antibody. Based on whether the vaccine is well tolerated, and which doses appear to be effective in generating an immune response, we will do further studies in both elderly subjects and children using the lowest dose that appears to be immunogenic in adults.
Overall, the goal in evaluating an inactivated vaccine is to find the lowest dose that results in a potentially protective immune response in the greatest proportion of people with an acceptable level of safety. Another important goal is to gain experience with the logistical challenges involved in producing a pandemic vaccine. Many of these challenges have in fact come up and have been solved. This learning process has been very important.
Going forward, we need to look at the potential advantages of a whole virus versus subunit vaccines, and to compare egg-grown vaccines with those made using cell culture or other substrates. Evaluating the dose-sparing capacity of adjuvants is critical, and alum and MF59 are the first candidates, because those are the two adjuvants licensed for use with influenza vaccine. We should also look at the schedule and route of administration, including not only inactivated vaccine but also intranasal, transcutaneous, and intradermal administration, again trying to reduce the dose.
We have looked a bit at intradermal vaccines for conventional influenza vaccines, using a clever device made by Becton Dickinson that nurses were able to use very effectively to deliver vaccine intradermally. That study was sponsored by GlaxoSmithKline.
The results—published by Dr. Robert Belshe in the New England Journal of Medicine—showed that an intradermal dose of 6 micrograms of hemagglutinin gave antibody responses in young adults very similar to those seen with an intramuscular dose of 15 micrograms. So essentially half the dose of vaccine given intradermally produced a response in terms of the geometric mean titer (GMT) of H1 antibody to both H1, H3, and type B influenza virus that was very similar to that seen with full-dose vaccine. Actually, the response rate was slightly less in those receiving the intradermal vaccine, and the titer was a little bit less. In individuals over 60, who interestingly did not manifest significant local inflammatory responses to the intradermal vaccine, this difference was more marked, and the intradermal approach did not appear to be as effective.
In interpreting this information, bear in mind that healthy adults respond well to low doses of vaccine administered intramuscularly. So it is premature to conclude that the intradermal approach is necessarily an advantage—that needs to be tested. In healthy adults, intramuscular doses of 7.5 micrograms and 15 micograms do not produce markedly different results. So we need to look at these dose-sparing strategies again using comparable groups.
So, what have we learned in the process of doing these studies? We have learned that the assays for H5 antibody are insensitive, and that we need better assays. It is important to evaluate these vaccines in multiple groups—not just healthy adults but also children and the elderly. And we have to think about an acceptable level of safety, given that this vaccine is designed to protect
against a potentially extremely serious disease but will also be used on a very broad scale. What is an acceptable level of safety for a pandemic vaccine? Should we think about this in the same way as a conventional vaccine, or accept a broader safety profile, particularly in terms of local side effects? Considering local toxicity is important, because although whole virus, alum, and MF59 offer advantages in the studies done so far, all are associated with a significantly increased risk of local side effects, which could factor into the vaccine’s safety profile.
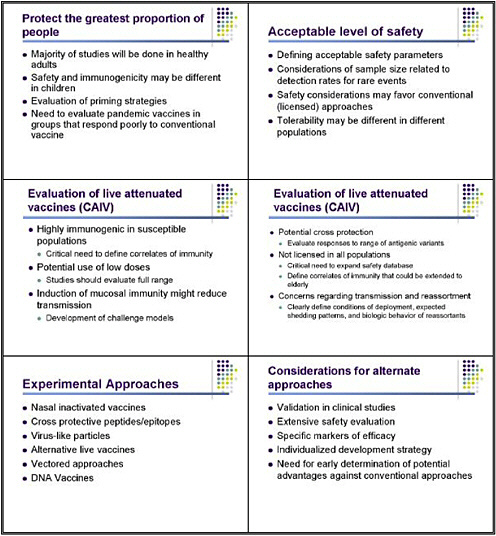
I am going to talk only briefly about other potential approaches. They have not been tested yet in humans, so we have much less experience in designing studies. But we clearly need to consider live vaccines, for several reasons. One is that they are highly immunogenic in susceptible populations. Studies in unprimed, naive children show that these vaccines are extraordinarily immunogenic because individuals are susceptible to infection, and thus those vaccines could be used in highly diluted form. For example, a live vaccine approach could significantly increase the number of doses obtained per egg or per fermenter, because of the potential to use a much lower dose—probably just 10 HID 50. These vaccines are clearly more effective at inducing mucosal immunity, and that might play an important role in their ability to reduce transmission.
What do we need to know? We need to know the correlates of immunity, particularly for live vaccines. We need to look at administering a full range of potential doses with live vaccines. It would be useful to use the live vaccines to develop a form of human challenge model for pandemic influenza, similar to the models that have been so useful for conventional influenza. This type of model would be especially important as we try to get an early signal as to whether candidate vaccines would in fact protect against protection and shedding. The live approach might also induce a broader and more cross-protective response against other antigenic variants within the same subtype—so-called hetereosubtypic immunity. That issue needs to be investigated in both inactivated and live vaccines.
A problem with live vaccines is that they are not now licensed in all populations, for a variety of reasons. Facilitating licensure of attenuated vaccines for conventional influenza in a broader population, particularly children, would be extremely useful. We clearly need to obtain additional safety data in children, and to define correlates of immunity that could be extended to the elderly, given that these vaccines are not now licensed in that age group.
There are some concerns regarding transmission and reassortment. We need to clearly define the conditions of deployment, the expected shedding patterns, and the biological behavior of potential reassortants, to assess whether this is a real or perceived risk of a live vaccine.
Investigators are evaluating a number of more experimental approaches to combating influenza. Some, including universal vaccines and vaccines with cross-protective epitopes, have potential advantages for fighting pandemic viruses. Clinical evaluations should proceed, but we are further from knowing whether those vaccines will work, so I regard them as a long-term goal for pandemic preparedness. They will require extensive safety evaluation, as they are now in phase I: we do not know very much about them. For some, we will need to develop specific markers of efficacy, which may differ from those we use for hemagglutinin-based vaccines. And each will require an individualized development strategy.
Devising a cohesive clinical development plan applicable to every vaccine will thus be difficult. However, it's useful to get early indications of whether these vaccines offer potential advantages over conventional approaches, so we can focus on those that are most worthwhile.
RESEARCH ISSUES IN ANIMAL SURVEILLANCE
Dr. Robert Webster, Professor and Chair, Department of Infectious Diseases, St. Jude's Children's Research Hospital
I would like to thank the organizers for the invitation to speak on this auspicious occasion in honor of John La Montagne. John was my first project officer; he came to Memphis for a site visit when we were preparing the reference antisera to influenza subtypes that are now in the NIAID bank. John held the old goat while I bled it. He was a fine human being.
I want to speak about surveillance. Surveillance in each of the regions with an H5N1 problem is critical. At the time of the meeting, we have recorded 70 humans cases and 47 deaths—33 in Vietnam, 12 in Thailand, and 2 in Cambodia. This is an unprecedented event in the history of influenza, and we must contain this virus.
The difficulty of surveillance lies in obtaining the necessary viruses for analysis. We now know that the outbreaks of H5N1 occurred before 2004 in some of the affected countries, although they were not reported at first. The question of rapid reporting of H5 and H7 must be addressed. One barrier is trade embargoes: if a country reports cases of H5N1, it can hurt its poultry trade. Another factor is national pride: countries want to characterize their own influenza viruses as much as possible. We also need to build surveillance infrastructure in countries such as Lao and Cambodia so that viruses become available for analysis.
The different missions of the Food and Agricultural Organization (FAO), the World Organization for Animal Health (OIE), and WHO also pose challenges. Even today these organizations are not talking to each other in a satisfactory fashion—they are not fully sharing viruses or information. They must sit down together and harmonize their approaches.
H5N1 first occurred in Hong Kong in 1997. That city developed a strategy for dealing with the virus, and it has not seen H5N1 influenza in humans or poultry in 2004 or 2005. The rest of the world largely ignored Hong Kong’s approach, which is part of the problem.
What did Hong Kong do to cope with the virus in 1997? It had limited infrastructure at the time—one small laboratory. The government quickly flew in the brightest young people from all over Asia as well as Laminar flow hoods, and established a temporary isolation laboratory. The staff stayed on the ground to characterize those viruses, and continue to do so today. No infections occurred in any of these staff members. And the agriculture department, the health department, the university, and WHO collaborated successfully to control H5N1 influenza.
After SARS and the laboratory infection of humans, the advice given to countries that lack adequate infrastructure is: do not attempt to isolate viruses. In other words, Hong Kong should not have been isolating H5N1 viruses in 1997 but instead should have been using molecular techniques and reverse transcription- polymerase chain reaction, or sending samples to experts around the world. I think that advice is a mistake. We should have been building infrastructure in these affected countries from the word go.
What did Hong Kong do to achieve the continued absence of H5N1? The country made very simple changes in its agricultural and marketing systems. It banned ducks and geese from live markets, and after Daniel Perez found that quail were capable of replicating every strain of
flu, they were also banned. The country also introduce two clean days per month when all live bird markets were empty and cleaned. They also have instituted the use of inactivated H5 vaccine on poultry farms and have included nonvaccinated sentinel birds in each flock to monitor any virus spread. The result is no H5N1.
Why hasn't this system been copied throughout Asia? The response I usually get is that it's too expensive. Hong Kong can afford to take that approach, but other countries can't. It is too expensive to ban the sale of ducks, geese, quail in these live markets across Asia? Of course it's not. Such stalling indicates a lack of political will, and a reluctance to accept agricultural vaccines. International agencies have failed to promote very simple measures such as keeping ducks, geese, and quail out of live markets. Countries could still take these simple yet profound measures.
Let's turn to agricultural vaccines. We have been talking about vaccines for some time. The problem is that there is a double standard for vaccines. Human vaccines are standardized for antigen content, but agricultural vaccines are not. They are required to induce an HI antibody in poultry, and as a result there are good and bad agricultural vaccines.
Which vaccines are available at the moment, and which have been used in poultry? In Hong Kong, the commercial vaccine in use is based on A/Chicken/Mexico/1994 (H5N1). The homology in the hemagglutinin between this virus and the H5 currently circulated is about 94 percent, so the difference is large, but the vaccine is still effective. China is still using an old virus from 1973, H5N2. Indonesia is using an inactivated highly pathogenic strain, which is a dangerous practice.
Asia is seeing new developments in poultry vaccines. China has developed a fowl pox-based H5, and the United States has developed one as well. These are efficacious. China is developing a reverse genetic H5N1 on an H9N2 backbone, and in the U.S. we have developed a reverse genetic H5N3 on the PR8 backbone.
Good poultry vaccines provide protection despite the antigenic difference. The mechanism of this protection without close homology is not really understood, and we need to pursue this. Is chicken immunology different from human immunology?
These poultry vaccines do not provide sterilizing immunity. Maybe that is because of the lack of antigenic match. However, they can reduce viral load below the level of transmission. In Hong Kong, agricultural researchers have clearly shown that the vaccines now available reduce the load so that transmission does not occur.
Bad vaccines provide protection against disease signs. In other words, the chickens and ducks look fine in the markets, but the birds are shedding high levels or transmissible levels of virus. They thus promote the spread of virus as well as antigenic drift. The lack of regulation is disturbing.
The missing information on the spread of H5N1 in Asia, particulary in Vietnam, is the role of the domestic duck. Let's look at some of the information about ducks in Vietnam. Some 60 million ducks are now raised in Vietnam. Many are free range: these ducks don't go to a home every night; they move from one paddy field to another as rice is harvested, picking up residue grain. The peak numbers of free-range ducks in Vietnam occur in May and October, corresponding to the rice harvest. The plan is to reduce the number of ducks from 60 million to
40 million by banning commercial hatching. However, hatching will likely continue in backyard flocks.
We now know that duck raising increases the risk of H5N1 in both Vietnam and Thailand. Eight percent of households in Vietnam that raised only chickens were infected with serological evidence of H5N1, while 67 percent of households that raised only ducks were infected. Households that raised both ducks and chickens had a 70 percent infection rate. Thus households that raise ducks have infection rates that are eight times higher.
A similar figure has been established in Thailand. However, we should complement the Thais on their great success in containing the second and the third waves of the virus. In October 2004, 39 percent of duck flocks in Thailand tested positive for H5N1, but by February only 2 infected flocks were found. In March, none were detected.
What did Thailand do differently? As we heard yesterday, Thailand could afford to pay compensation to farmers, so it pursued an aggressive program to stamp out the virus. Perhaps the outside world should pay Vietnam to adopt the same strategy.
H5N1 has continued to evolve in ducks in Asia, incrementally increasing its most worrisome biological properties. In November 2002 the virus acquired the ability to kill waterfowl in Hong Kong. All the waterfowl in Kowloon Park in central Hong Kong, including flamingoes and other decorative birds, were susceptible and died of neurological infection.
In 2003 the majority of the H5N1 isolates around Asia were the so-called Z genotype, and they were highly pathogenic in ducks. By 2004, many of the duck isolates were non-pathogenic. Studies by Diane Hulse in my lab in Memphis established some of the key information—which was released immediately—showing that in ducks there is long-term shedding of influenza viruses despite the passage of antibody. Some of the ducks shed virus for up to 17 days. During this period non-pathogenic viruses became dominant in the ducks, but they retained high pathogenicity for chickens; we do not know whether they were pathogenic to humans.
These non-pathogenic H5N1 duck viruses also transmitted naturally to other ducks put into the cage. These properties are very disturbing. The duck is pushing the virus back toward the non-pathogenic state, which naturally occurs with influenza in ducks but these viruses retain high pathogenicity for chickens and presumably humans.
We decided to use Dr. Eric Hoffman’s reverse genetic strategy to generate a vaccine and determine whether it is efficacious in the duck, because we need a vaccine if we want to avoid culling all the ducks. That strategy is the standard eight plasmid system, modified hemagglutinin, that we heard about yesterday. We put on an N3 neuraminidase so we could distinguish between vaccinated and infected birds; these viruses replicate to very high titers.
The strategy was to vaccinate ducks at 2 weeks, boost them at 5 weeks, and challenge them with an H5N1 at 8 weeks—all, of course, in biosafety level 3 containment facilities. The doses of vaccine were surprisingly small: 0.25 to 1.2 micrograms of HA per dose.
We failed to put in a small-enough dose because all the doses of vaccines induced high levels of antibody after one shot, pre-boost HI titers ranged from 500 up to 5,000, and all the ducks were protected from challenge. We are now reducing the dose. So the good news is that the vaccines are efficacious in ducks, and that a very small dose is sufficient to protect them from H5N1 challenge.
What about the lethality of these viruses for ferrets—the models we were talking about yesterday? The human viruses caused severe disease in the ferrets, with central nervous system symptoms and death. The viruses from ducks caused only extremely small numbers of severe cases: one in ferrets, none in chickens. These numbers are too small to mean much, but they indicate that the ferret is an excellent model for determining pathogenic potential in humans.
Duck populations have been increasing rapidly since the early nineties, corresponding to the time when these problems began to occur. The Thais did a back-of-the envelope calculation that Asia is home to some 2 billion domestic ducks—10 to 100 times the number of wild ducks. Is the wild duck—the true migrating duck—involved in spreading this virus? Our view is that the true migrating bird was not responsible for the initial spread in Hong Kong. The virus did spread locally in wild ducks and other wild birds. What is the situation now, after two seasons? Are wild ducks infected? We desperately need to know, because birds that are breeding in Siberia will spread to other areas, including continental Europe.
North Korea has just seen an outbreak of highly infectious disease in poultry. We don't know yet what caused the outbreak—maybe H5N1, maybe not. It was characterized as a non-pathogenic H7N7 virus. Thailand, Japan, and South Korea have stamped out the virus, but what will come across the border from Lao or Cambodia to Thailand? We don't know, nor will we know what will cross the border in China, given the use of vaccine there.
I would like to end with suggestions. The immediate issue is to reduce the likelihood of human-to-human transmission by reducing the viral load in poultry. And we can do that: the international agencies must come aboard to reduce the ducks in live markets and standardize agricultural vaccines.
Stamping out is effective if countries can afford it: Thailand is showing that it can be done. However, the government realizes that it must consider vaccination of the poultry population, including ducks, because this virus will not go away. We desperately need a quality vaccine for poultry right now.
The unresolved issues are many. Why hasn't H5N1 transmitted more freely to mammals including humans? The molecular basis of its pathogenicity still needs to be resolved. Is the Asian human genome special for susceptibility to influenza, given that Asia is the epicenter for these pandemics?
I would like to conclude by acknowledging NIAID, which has supported this program at St. Jude's Children's Research Hospital, in Hong Kong, and in other Asian countries. I would also like to acknowledge the group in Memphis, and particularly the group in Hong Kong, including Malik Peiris and Yi Guan, as well as collaborators in Indonesia, Thailand, and Vietnam.