7
Cyberinfrastructure and Data Acquisition
7.1 CYBERINFRASTRUCTURE FOR 21ST CENTURY BIOLOGY
Twenty-first century biology seeks to integrate scientific understanding at multiple levels of biological abstraction, and it is holistic in the sense that it seeks an integrated understanding of biological systems through studying the set of interactions between components. Because such an enormous, data-intensive effort is necessarily and inherently distributed over multiple laboratories and investigators, an infrastructure is necessary that facilitates the integration of experimental data, enables collaboration, and promotes communication among the various actors involved.
7.1.1 What Is Cyberinfrastructure?
Cyberinfrastructure for science and engineering is a term coined by the National Science Foundation (NSF) to refer to distributed computer, information, and communication technologies and the associated organizational facilities to support modern scientific and engineering research conducted on a global scale. As articulated by the Atkins panel,1 the technology substrate of cyberinfrastructure involves the following:
-
High-end general-purpose computing centers that provide supercomputing capabilities to the community at large. In the biological context, such capabilities might be used to undertake, for example, calculations to determine the three-dimensional structure of proteins given their genetic sequence. In some cases, these computing capabilities could be provided by local clusters of computers; in other cases, special-purpose hardware; and in still others, computing capabilities on demand from a computing grid environment.
-
Data repositories that are well curated and that store and make available to all researchers large volumes and many types of biological data, both in raw form and as associated derived products. Such repositories must store data, of course, but they must also organize, manage, and document these
|
1 |
”Revolutionizing Science and Engineering Through Cyberinfrastructure: Report of the NSF Blue-Ribbon Advisory Panel on Cyberinfrastructure,” 2003, available at http://www.communitytechnology.org/nsf_ci_report/report.pdf. |
-
datasets dynamically. They must provide robust search capabilities so that researchers can find the datasets they need easily. Also, they are likely to have a major role in ensuring the data interoperability necessary when data collected in one context are made available for use in another.
-
Digital libraries that contain the intellectual legacy of biological researchers and provide mechanisms for sharing, annotating, reviewing, and disseminating knowledge in a collaborative context. Where print journals were once the standard mechanism through which scientific knowledge was validated, modern information technologies allow the circumvention of many of the weaknesses of print. Knowledge can be shared much more broadly, with much shorter lag time between publication and availability. Different forms of information can be conveyed more easily (e.g., multimedia presentations rich in biological imagery). One researcher’s annotations to an article can be disseminated to a broader audience.
-
High-speed networks that connect large-scale, geographically distributed computing resources, data repositories, and digital libraries. Because of the large volumes of data involved in biological datasets, today’s commodity Internet is inadequate for high-end scientific applications, especially where there is a real-time element (e.g., remote instrumentation and collaboration). Network connections ten to a hundred times faster than those generally available today are a lower bound on what will be necessary.
In addition to these components, cyberinfrastructure must provide software and services to the biological community. For example, cyberinfrastructure will involve many software tools, system software components (e.g., for grid computing, compilers and runtime systems, visualization, program development environments, distributed scalable and parallel file systems, human computer interfaces, highly scalable operating systems, system management software, parallelizing compilers for a variety of machine architectures, sophisticated schedulers), and other software building blocks that researchers can use to build their own cyberinfrastructure-enabled applications. Services, such as those needed to maintain software on multiple platforms and provide for authentication and access control, must be supported through the equivalent of help-desk facilities.
From the committee’s perspective, the primary value of cyberinfrastructure resides in what it enables with respect to data management and analysis. Thus, in a biological context, machine-readable terminologies, vocabularies, ontologies, and structured grammars for constructing biological sentences are all necessary higher-level components of cyberinfrastructure as tools to help manage and analyze data (discussed in Section 4.2). High-end computing is useful in specialized applications but, by comparison to tools for data management and analysis, lacks broad applicability across multiple fields of biology.
7.1.2 Why Is Cyberinfrastructure Relevant?
The Atkins panel noted that the lack of a ubiquitous cyberinfrastructure for science and engineering research carries with it some major risks and costs. For example, when coordination is difficult, researchers in different fields and at different sites tend to adopt different formats and representations of key information. As a result, their reconciliation or combination becomes difficult to achieve—and hence disciplinary (or subdisciplinary) boundaries become more difficult to break down. Without systematic archiving and curation of intermediate research results (as well as the polished and reduced publications), useful data and information are often lost. Without common building blocks, research groups build their own application and middleware software, leading to wasted effort and time.
As a field, biology faces all of these costs and risks. Indeed, for much of its history, the organization of biological research could reasonably be regarded as a group of more or less autonomous fiefdoms. Unifying biological research into larger units of aggregation is not a plausible strategy today, and so the federation and loose coordination enabled by cyberinfrastructure seem well suited to provide the major advantages of integration while maintaining a reasonably stable large-scale organizational structure.
Furthermore, well-organized, integrated, synthesized information is increasingly valuable to biological research (Box 7.1). In an era characterized by data-intensive research observations, collecting,
|
Box 7.1 Creating and sustaining a comprehensive cyberinfrastructure (CI; the pervasive applications of all domains of scientific computing and information technology) are as relevant and as required for biology as for any science or intellectual endeavor; in the advances that led to today’s opportunity, the National Science Foundation’s Directorate for Biological Sciences (NSF BIO) made numerous, ad hoc contributions, and now can integrate its efforts to build the complete platforms needed for 21st century biology. Doing so will accelerate progress in extraordinary ways. The time has arrived for creating a CI for all of the sciences, for research and education, and NSF will lead the way. NSF BIO must co-define the extent and fine details of the NSF structure for CI, which will involve major internal NSF partnerships and external partnerships with other agencies, and will be fully international in scope. Only the biological sciences have seen advances as remarkable, sustained, and revolutionary as those in computer and information sciences. Only in the past few years has the world of computing and information technology reached the level of being fully applicable to the wide range of cutting-edge themes characteristic of biological research. Multiplying the exponentials (of continuing advances in computing and bioscience) through deep partnerships will inevitably be exciting beyond any anticipation. The stretch goals for the biological sciences community include both community-level involvement and realization of the complete spectrum of CI, namely, people and training, instrumentation, collaborations, advanced computing and networking, databases and knowledge management; and analytical methods (modeling and simulation). NSF BIO must:
The biology community must decide how it can best interact with the quantitative science community, where and when to intersect with computational sciences and technologies, how to cooperate on and contribute to infrastructure projects, and how NSF BIO should partner administratively. An implementation meeting, as well as briefings to the community through professional societies, will be essential. For NSF BIO to underestimate the importance of cyberinfrastructure for biology, or fail to provide fuel over the entire journey, would severely retard progress and be very damaging for the entire national and international biological sciences community. SOURCE: Adapted from Subcommittee on 21st Century Biology, NSF Directorate for Biological Sciences Advisory Committee, Building a Cyberinfrastructure for the Biological Sciences 2005 and Beyond: A Roadmap for Consolidation and Exponentiation, a workshop report, July 14-15, 2003. |
managing, and connecting data from various modalities and on multiple scales of biological systems, from molecules to ecosystems, are essential to turn that data into information. Each biological subdiscipline also now requires the tools of information technology to probe that information, to interconnect experimental observations and modeling, and to contribute to an enriched understanding or knowledge. The expansion of biology into discovery and synthetic analysis, that is, genome-enabled biology and systems biology as well as the hardening of many biological research tools into high-throughput pipelines, serves also to drive the need for cyberinfrastructure in biology.
Box 7.2 illustrates existing efforts in the development of cyberinfrastructure for biology that are relevant. Note that the examples span a wide range of subfields within biology, including proteomics (PDB), ecology (NEON and LTER), neuroscience (BIRN), and biomedicine (NBCR).
Data repositories and digital libraries are discussed in Chapter 3. The discussion below focuses primarily on computing and networking.
|
Box 7.2 Pacific Rim Application and Grid Middleware Assembly The Pacific Rim Application and Grid Middleware Assembly (PRAGMA) is a collaborative effort of 15 institutions around the Pacific Rim. PRAGMA’s mission is to establish sustained collaborations and advance the use of grid technologies among a community of investigators working with leading institutions around the Pacific Rim. To fulfill this mission, PRAGMA hosts a series of workshop for members to focus on developing applications and on developing a testbed for these applications. Current applications include workflows in biology (protein annotation); linking via Web services climate data (working with some Long-Term Ecological Research [LTER] Network sites in the United States and East Asia Pacific region [ILTER]); running solvation models; and extending telescience application to more institutions. The Protein Data Bank The Protein Data Bank (PDB) was established in 1971 as a computer-based archival resource for macromolecular structures. The purpose of the PDB was to collect, standardize, and distribute atomic coordinates and other data from crystallographic studies. In 1977 the PDB listed atomic coordinates for 47 macromolecules. In 1987, the number began to increase rapidly at a rate of about 10 percent per year due to the development of area detectors and widespread use of synchrotron radiation; by April 1990, atomic coordinate entries existed for 535 macromolecules. Commenting on the state of the art in 1990, Holbrook and colleagues [citation omitted] noted that crystal determination could require one or more man-years. As of 1999, the Biological Macromolecule Crystallization Database (BMCD) of the PDB contain[ed] entries for 2,526 biological macromolecules for which diffraction quality crystals had been obtained. These include proteins, protein-protein complexes, nucleic acids, nucleic acid-nucleic acid complexes, protein-nucleic acid complexes, and viruses. In July 2004, the PDB held information on 26,144 structures (23,676 proteins, peptides, and viruses; 1,338 nucleic acids; 1,112 protein/nucleic acid complexes; and 18 carbohydrates). The National Center for Biotechnology Information The National Center for Biotechnology Information (NCBI), part of NIH’s National Library of Medicine, has been charged with creating automated systems for storing, analyzing, and facilitating the use of knowledge about molecular biology, biochemistry, and genetics. In addition to GenBank, NCBI curates the Online Mendelian Inheritance in Man (OMIM), the Molecular Modeling Database (MMDB) of three-dimensional protein structures, the Unique Human Gene Sequence Collection (UniGene), the Taxonomy Browser, and the Cancer Genome Anatomy Project (CGAP), in collaboration with the National Cancer Institute. NCBI’s retrieval system, Entrez, permits linked searches of the databases, while a variety of tools have been developed for data mining, sequence analysis, and three-dimensional structure display and similarity searching. NCBI’s senior investigators and extended staff collaborate with the external research community to develop novel algorithms and research approaches that have transformed computational biology and will enable further genomic discoveries. |
|
EUROGRID’s Bio GRID Funded by the European Commission, Bio GRID is intended to help biologists and chemists who are not familiar with high-performance computing (HPC) execution systems by developing intuitive user interfaces for selected biomolecular modeling packages and creating compatibility interfaces between the packages and their databases through Bio GRID’s UNICORE platform. The UNICORE system will allow investigators to streamline their work processes, connect to Internet-accessible databases, and run a number of quantum chemistry and molecular dynamics software programs developed as plug-ins by Bio GRID’s staff. The NSF National Ecological Observatory Network (NEON) NEON is a continental-scale research instrument consisting of geographically distributed networked infrastructure, with lab and field instrumentation; site-based experimental infrastructure; natural history archive facilities; and computational, analytical, and modeling capabilities. NEON is intended to transform ecological research by enabling studies on major environmental challenges at regional to continental scales. Scientists and engineers use NEON to conduct real-time ecological studies spanning all levels of biological organization and many temporal and geographical scales. NEON’s synthesis, computation, and visualization infrastructure constitutes a virtual laboratory that enables the development of a predictive understanding of the direct effects and feedbacks between environmental change and biological processes. The NSF Long-Term Ecological Research Network (LTER) Since 1980, NSF has supported the Long-Term Ecological Research (LTER) Network. The LTER program is characterized by long temporal and broad spatial scales of research and fosters ecological comparisons among 26 U.S. sites that illustrate the importance of comprehensive analyses of ecosystems and of distinguishing system features across multiple scales of time and space. Data collected at each site are accessible to other scientists and the general public, and the LTER network works with other research institutions to standardize information management practices to achieve network- and community-wide data integration, facilitating data exchange and advancing data analysis and synthesis. LTER-supported work has included efforts in climate variability and ecosystem response, standardization of protocols for measuring soil properties for long-term ecological research, synthesis of global data on winter ice duration on lakes and rivers, and comparisons of ecosystem productivity, among others. SOURCES: PRAGMA: material adapted from http://www.pragma-grid.net. PDB: material pre-2004 excerpted from T. Lenoir, “Shaping Biomedicine as an Information Science,” pp. 27-45 in Proceedings of the 1998 Conference on the History and Heritage of Science Information Systems, M. Bowden, T. Hahn, and R. Williams, eds., ASIS Monograph Series, Information Today, Inc., Medford, NJ, 1999, available at http://www.stanford.edu/dept/HPST/TimLenoir/Publications/Lenoir_BioAsInfoScience.pdf. Information for 2004 taken from Protein Data Bank Annual Report 2004, available at http://www.rcsb.org/pdb/annual_report04.pdf. NCBI: material adapted from http://www.ncbi.nlm.nih.gov. Bio GRID: material adapted from http://www.eurogrid.org. NEON: material adapted from http://www.nsf.gov/bio/neon/. LTER: material adapted from the LTER brochure, available at http://intranet.lternet.edu/archives/documents/Publications/brochures/lter_brochure.pdf. |
7.1.3 The Role of High-performance Computing
Loosely speaking, processing capability refers to the speed with which a computational solution to a problem can be delivered. High processing capability is generally delivered by computing units operating in parallel and is generally dependent on two factors—the speed with which individual units compute (usually measured in operations per second) and the communications bandwidth between individual units. If a problem can be partitioned so that each subcomponent can be processed independently, then no communication at all is needed between individual computing units. On the other hand, as the dependence of one subcomponent on others increases, so does the amount of communications required between computing units.
Many biological applications must access large amounts of data. Furthermore, because of the combinatorial nature of the exploration required in these applications (i.e., the relationships between different pieces of data is not known in advance and thus all possible combinations are a priori possible), assumptions of locality that can be used to partition problems with relative ease (e.g., in computational fluid dynamics problems) do not apply, and thus the amount of data exchange increases. One estimate of the magnitude of the data-intensive nature of a biological problem is that a comparison of two of the smallest human chromosomes using the best available dynamic programming algorithm allowing for substitutions and gaps would require hundreds of petabytes of memory and hundred-petaflop processors.2
Thus, in supercomputers intended for biological applications, speed in computation and in communication are both necessary—and many of today’s supercomputing architectures are thus inadequate for these applications.3 Note that communications issues deal both with interprocessor communications (e.g., comparing sequences between processors, dividing long sequences among multiple processors) and traditional input-output (e.g., searching large sequence libraries on disk, receiving many requests at a time from the outside world). When problems involve large amounts of data exchange, communications become increasingly important.
Greater processing capability would enable the attack of many biologically significant problems. Today, processing capability is adequate to sequence and assemble data from a known organism. To some extent, it is possible to find genes computationally (as discussed in Chapter 4), but the accuracy of today’s computationally limited techniques is modest. Simulations of interesting biomolecular systems can be carried out routinely for about hundreds of thousands of atoms for tens of nanoseconds. Order-of-magnitude increases (perhaps even two or three orders of magnitude) in processing capability would enable great progress in problem domains such as protein folding (ab initio prediction of three-dimensional structure from one-dimensional sequence information), simulation methods based on quantum mechanics that can provide more accurate predictions of the detailed behavior of interesting biomolecules in solution,4 simulations of large numbers of interacting macromolecules for times of biological interest (i.e., for microseconds and involving millions of atoms), comparative genomics (i.e., finding similar genetic sequences across the genomes of different organisms—the multiple sequence alignment problem), proteomics (i.e., understanding the combinatorially large number of interactions between gene products), predictive and realistic simulations of biological systems ranging from cells to ecosystems), and phylogenetics (the reconstruction of historical relationships between species or individuals). Box 7.3 provides some illustrative applications of high-performance computing in life sciences research.
Any such estimate of the computing power needed to solve a given problem depends on assumptions about how a solution to that problem might be structured. Different ways of structuring a problem
|
2 |
Shankar Subramanian, University of California, San Diego, personal communication, September 24, 2003. |
|
3 |
This discussion of communications issues is based on G.S. Heffelfinger, “Supercomputing and the New Biology,” PowerPoint presentation at the AAAS Annual Meeting, Denver, CO, February 13-18, 2003. |
|
4 |
A typical problem might be the question of enzymes that exhibit high selectivity and high catalytic efficiency, and a detailed simulation might well provide insight into the related problem of designing an enzyme with novel catalytic activity. Simulations based on classical mechanics treat molecules essentially as charged masses on springs. These simulations (so-called molecular dynamics simulations) have had some degree of success, but lead to seriously inaccurate results where ions must interact in water or when the breaking or forming of bonds must be taken into account. Simulations based on quantum mechanics model molecules as collections of nuclei and electrons and entail solving of quantum mechanical equations governing the motion of such particles; these simulations offer the promise of much more accurate simulations of these processes, although at a much higher computational cost. These comments are based on excerpts from a white paper by M. Colvin, “Quantum Mechanical Simulations of Biochemical Processes,” presented at the National Research Council’s Workshop on the Future of Super-computing, Lawrence Livermore National Laboratory, Santa Fe, NM, September 26-28, 2003. See also “Biophysical Simulations Enabled by the Ultrasimulation Facility,” available at http://www.ultrasim.info/doe_docs/Biophysics_Ultrasimulation_White_Paper_4-1-03.pdf. |
|
Box 7.3 The Onset of Cancer It is well known that cancer develops when cells receive inappropriate signals to multiply, but the details of cell signaling are not well understood. For example, activation of the epidermal growth factor signaling pathway is under the control of growth factors that bind to a receptor site on the exterior of a cell. Binding of the receptor initiates a cascade of protein conformational changes through the cell membrane, involving a complex rearrangement of many different proteins, including the Ras enzyme. The Ras enzyme is a molecular switch that can initiate a cascade of protein kinases that in turn transfer the external signal to the cell nucleus where it controls cell proliferation and differentiation. Disruption of this signaling pathway can have dire consequences as illustrated by the finding that mutations of the Ras enzyme have been found in 30 percent of human tumors. Because computer simulations can provide atomic-level detail that is difficult or impossible to obtain from experimental studies, computational studies are essential. However, this requires the modeling of an extremely large complex of biomolecules, including bilayer lipid membranes, transmembrane proteins, and a complex of many intercellular kinases, and thousands of molecules of waters of solvation. Environmental Remediation Microbes may be able to contribute to the cleanup of polluted sites by concentrating waste materials or degrading them into nontoxic form. Understanding the role of gram-negative bacteria in moderating subsurface reduction-oxidation chemistry and the role of such systems in bioremediation technologies requires the study of how cell walls, including many transmembrane protein substituents, interact with extracellular mineral surfaces and solvated atomic and molecular species in the environment. Simulations of these processes requires that many millions of atoms be included. Degradation of Toxic Chemical Weapons Computational approaches can be used for the rational redesign of enzymes to degrade chemical agents. An example is the enzyme phosphotriesterase (PTE), which could be used to degrade nerve gases. Combined experimental and computational efforts can be used to develop a series of highly specific PTE analogues, redesigned for optimum activity at specific temperatures, or for optimum stability and activity in nonaqueous, low-humidity environments or in foams, for improved degradation of warfare neurotoxins. Advanced computations can also facilitate the design of better reactivators of the enzyme acetylcholinesterase (AChE) that can be used as more efficient therapeutic agents against highly toxic phosphoester compounds such as the nerve warfare agents DFP (diisopropyl fluorophosphate), sarin, and soman and insecticides such as paraoxon. AChE is a key protein in the hydrolysis of acetylcholine, and inhibition of AChE through a phosphorylation reaction with such phosphoesters can rapidly lead to severe intoxication and death. Multiscale Physiological Modeling of the Heart The heart has a characteristic volume of around 60 cm3. At a resolution of 0.1 mm, a grid of some 6 × 107 cells is required. If 100 variables are associated with each cell, 10 floating point operations are needed for each time step in a simulation, and the time resolution is around 1 ms (a single heartbeat has a duration around 1 second), a computing throughput of 6 × 1013 floating point operations per second (60 teraflops) is necessary. In addition, a flexible and composable simulation infrastructure is required. For example, for a spatially distributed system, only a representative and relatively small subset of substructures can be represented in the model explicitly, because it is not feasible to model all of them. Contributions of the substructures missing from the model are inferred by an interpolative process. For practical purposes, it will not be known in advance how much and what kinds of detail will be necessary for a useful simulation; the same a priori ignorance also characterizes the nature and extent of the communications required between different levels of the simulation. Thus, the infrastructure must support easy experimentation in which different amounts of detail and different degrees of communication can be explored. SOURCE: The first three examples are adapted with minimal change from D.A. Dixon, T.P. Straatsma, and T. Head-Gordon, “Grand Challenges in Computational Structural and Systems Biology,” available at http://www.ultrasim.info/doe_docs/ESC-response.bio.dad.pdf. |
solution often result in different estimates for the required computing power, and for any complex problem, the “best” structuring may well not be known. (Different ways of structuring a problem may involve different algorithms for its solution, or different assumptions about the nature of the biologically relevant information.) Furthermore, the advantage gained through algorithm advances, conceptual reformulations of the problem, or different notions about the answers being sought is often comparable to advantages from hardware advances, and sometimes greater. On the other hand, for decades computational scientists have been able to count on regular advances in computing power that accrued “for free,” and whether or not scientists are able to develop new ways of looking at a given problem, hardware-based advances in computing are likely to continue.
Three types of computational problem in biology must be distinguished.5 Problems such as protein folding and the simulation of biological systems are similar to other simulation problems that involve substantial amounts of “number crunching.” A second type of problem entails large-scale comparisons or searches in which a very large corpus of data—for example, a genomic sequence or a protein database—is compared against another corpus, such as another genome or a large set of unclassified protein sequences. In this kind of problem, the difficult technical issues involve the lack of good software for broadcast and parallel access disk storage subsystems. The third type of problem involves single instances of large combinatorial problems, for example, finding a particular path in a very large graph. In these problems, computing time is often not an issue if the object can be modeled in the memory of the machine. When memory is too small, the user must write code that allows for efficient random access to a very large object—a task that significantly increases development time and even under the best of circumstances can degrade performance by an order of magnitude.
The latter two types of problem often entail the consideration of large numbers of biological objects (cells, organs, organisms) characterized by high degrees of individuality, contingency, and historicity. Such problems are typically found in investigations involving comparative and functional genomics and proteomics, which generally involve issues such as discrete combinatorial optimization (e.g., the multiple sequence alignment problem) or pattern inference (e.g., finding clusters or other patterns in high-dimensional datasets). Algorithms for discrete optimization and pattern inference are often NP-hard, meaning that the time to find an optimal solution is far too long (e.g., longer than the age of the universe) for a problem of meaningful size, regardless of the computer that might be used or that can be foreseen. Since optimal solutions are not in general possible, heuristic approaches are needed that can come reasonably close to being optimal—and a considerable degree of creativity is involved in developing these approaches.
Historically, another important point has been that the character of biological data is different from that of data in fields such as climate modeling. Many simulations of nonbiological systems can be composed of multiple repeating volume elements (i.e., a mesh that is well suited for finding floating point solutions of partial differential equations that govern the temporal and spatial evolution of various field quantities). By contrast, some important biological data (e.g., genomic sequence data) are characterized by quantities that are better suited to integer representations, and biological simulations are generally composed of heterogeneous objects. However, today, the difference in speed between integer operations and floating point operations is relatively small, and thus the difference between floating point and integer representations is not particularly significant from the standpoint of supercomputer design.
Finally, it is important to realize that many problems in computational biology will never be solved by increased computational capability alone. For example, some problems in systems biology are combinatorial in nature, in the sense that they seek to compare “everything against everything” in a search for previously unknown correlations. Search spaces that are combinatorially large are so large that even
with exponential improvements in computational speed, methods other than exhaustive search must be employed as well to yield useful results in reasonable times.6
The preceding discussion for the life sciences focuses on the large-scale computing needs of the field. Yet these are hardly the only important applications of computing, and rapid innovation is likely to require information technology on many scales. For example, researchers need to be able to explore ideas on local computers, albeit for scaled-down problems. Only after smaller-scale explorations are conducted do researchers have the savvy, the motivation, and the insight needed for meaningful use of high-end cyberinfrastructure. Researchers also need tools that can facilitate quick and dirty tasks, and working knowledge of spreadsheets or Perl programming can be quite helpful. For this reason, biologists working at all scales of problem size will be able to benefit from advances in and knowledge of information technology.
7.1.4 The Role of Networking
As noted in Chapter 3, biological data come in large quantities. High-speed networking (e.g., one or two orders of magnitude faster than that available today) would greatly facilitate the exchange of certain types of biological data such as high-resolution imaging as well as enable real-time remote operation of expensive instrumentation. High-speed networking is critical for life science applications in which large volumes of data change or are created rapidly, such as those involving imaging or remote operation of instrumentation.7
The Internet2 effort also includes the Middleware Initiative (I2-MI), intended to facilitate the creation of interoperable middleware infrastructures among the membership of Internet2 and related communities.8 Middleware generally consists of sets of tools and data that help applications use networked resources and services. The availability of middleware contributes greatly to the interoperability of applications and reduces the expense involved in developing those applications. I2-MI develops middleware to provide services such as identifiers (labels that connect a real-world subject to a set of computerized data); authentication of identity; directories that index elements that applications must access; authorization of services for users; secure multicasting; bandwidth brokering and quality of service; and coscheduling of resources, coupling data, networking, and computing together.
7.1.5 An Example of Using Cyberinfrastructure for Neuroscience Research
The Biomedical Informatics Research Network (BIRN) project is a nationwide effort by National Institutes of Health (NIH)-supported research sites to merge data grid and computer grid cyberinfrastructure into the workflows of biomedical research. The Brain Morphometry BIRN, one of the testbeds driving the development of BIRN, has undertaken a project that uses the new technology by integrating data and analysis methodology drawn from the participating sites. The Multi-site Imaging Research in the Analysis of Depression (MIRIAD) project (Figure 7.1) applies sophisticated image processing of a dataset of magnetic resonance imaging (MRI) scans of a longitudinal study of elderly subjects. The subjects include patients who enroll in the study with symptoms of clinical depressions
|
6 |
Consider the following example. The human genome is estimated to have around 30,000 genes. If the exploration of interest is assumed to be 5 genes operating together, there are approximately 3 × 1020 possible combinations of 30,000 genes in sets of 5. If the assumption is that 6 genes may operate together, there are on the order of 1026 possible combinations (the number of possible combinations of n genes in groups of k is given by n!/(k!(n − k)!), which for large n and small k reduces to nk/k!). |
|
7 |
In the opposite extreme case, in which enormous volumes of data never change, it is convenient rather than essential to use electronic or fiber links to transmit the information—for a small fraction of the cost of high-speed networks, media (or even entire servers!) can be sent by Federal Express more quickly than a high-speed network could transmit the comparable volume of information. See, for example, Jim Gray et al., TeraScale SneakerNet: Using Inexpensive Disks for Backup, Archiving, and Data Exchange, Microsoft Technical Report, MS-TR-02-54, May 2002, available at ftp://ftp.research.microsoft.com/pub/tr/tr-2002-54.pdf. |
|
8 |
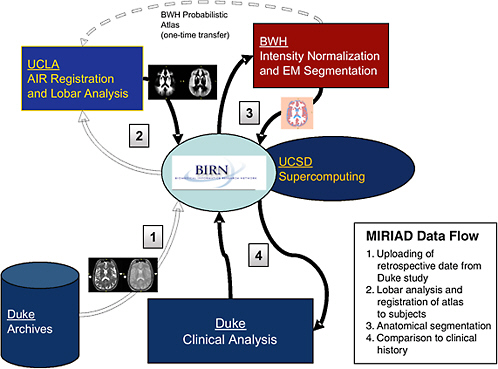
FIGURE 7.1 Steps in data processing in the BIRN MIRIAD project.
1. T2-weighted and proton density (PD) MRI scans from the Duke University longitudinal study are loaded into the BIRN data archive (data grid), accessible by members of the MIRIAD group for analysis using the computer resources at the University of California, San Diego (UCSD) and the San Diego Supercomputer Center (compute grid).
2. The Laboratory of Neuro Imaging at the University of California, Los Angeles (UCLA) performs a nonlinear registration to define the three-dimensional geometric mapping between each subject and a standard brain atlas that encodes the probabilities of each tissue class at each location in the brain.
3. The Surgical Planning Laboratory at Brigham and Women’s Hospital (BWH) then applies an intensity normalization and expectation-maximization algorithm to combine the original image pixel intensities (T2 and PD) and the tissue probabilities to label each point in the images and to calculate the overall volumes of tissue classes.
4. Duke performs statistical tests on the image-processing results to assess the predictive value of the brain morphometry measurements with respect to clinical outcome.
and age-matched controls. Some of the depression patients go on to develop Alzheimer’s disease (AD) and the goal of the MIRIAD project is to measure the changes in brain images, specifically volume changes in cortical and subcortical gray matter, that correlate with clinical outcome.
Of particular significance from the standpoint of cyberinfrastructure, the MIRIAD project is distributed among four separate sites: Duke University Neuropsychiatric Imaging Research Laboratory, Brigham and Women’s Hospital Surgical Planning Laboratory, University of California, Los Angeles Laboratory of Neuro Imaging, and University of California, San Diego BIRN. Each of these sites has responsibility for some substantive part of the work, and the work would not be possible without the BIRN infrastructure to coordinate it.
7.2 DATA ACQUISITION AND LABORATORY AUTOMATION
As noted in Chapter 3, the biology of the 21st century will be data-intensive across a wide range of spatial and temporal scales. Today’s high-throughput data acquisition technologies depend on parallelization rather than on reducing the time needed to take individual data points. These technologies are capable of carrying out global (or nearly global) analyses, and as such they are well suited for the rapid and comprehensive assessment of biological system properties and dynamics. Indeed, in 21st century biology, many questions are asked because relevant data can be obtained to answer them. Whereas earlier researchers automated existing manual techniques, today’s approach is more oriented toward techniques that match existing automation.
7.2.1 Today’s Technologies for Data Acquisition9
Some of today’s data acquisition technologies include the following:10
-
DNA microarrays. Microarray technology enables the simultaneous interrogations of a human genomic sample for complete human transcriptomes, provided that the arrays do not contain only putative protein coding regions. The oligonucleotide microarray can identify single-nucleotide differences and distinguish mRNAs from individual members of multigene families, characterize alternatively spliced genes, and identify and type alternative forms of single-nucleotide polymorphisms. Microarrays are also used to observe in vitro protein-DNA binding events and to do comparative genome hybridization (CGH) studies. Box 7.4 provides a close-up of microarrays.
-
Automated DNA sequencers. Prior to automated sequencing, the sequencing of DNA was performed manually, at many tens (up to a few hundred) of bases per day.11 In the 1970s, the development of restriction enzymes, recombinant DNA techniques, gene cloning techniques, and polymerase chain reaction (PCR) contributed to increasing amounts of data on DNA, RNA, and protein sequences. More than 140,000 genes were cloned and sequenced in the 20 years from 1974 to 1994, many of which were human genes. In 1986, an automated DNA sequencer was first demonstrated that sequenced 250 bases per day.12 By the late 1980s, the NIH GenBank database (release 70) contained more than 74,000 sequences, while the Swiss Protein database (Swiss-Prot) included nearly 23,000 sequences. In addition, protein databases were doubling in size every 12 months. Since 1999, more advanced models of automated DNA sequencer have come into widespread use.13 Today, a state-of-the-art automated sequencer can produce on the order of a million base pairs of raw DNA sequence data per day. (In addition, technologies are available that allow the parallel processing of 16 to 20 residues at a time.14 These enable the determination of complete transcriptomes in individual cell types from organisms whose genome is known.)
-
Mass spectroscopy. Mass spectroscopy (MS) enables the in-quantity identification and quantification of large numbers of proteins.15 Used in conjunction with genomic information, MS information can be used to identify and type single-nucleotide polymorphisms. Some implementations of mass spec-
|
9 |
Section 7.2.1 is adapted from T. Ideker, T. Galitski, and L. Hood, “A New Approach to Decoding Life: Systems Biology,” Annual Review of Genomics and Human Genetics 2:343, 2001. |
|
10 |
Adapted from T. Ideker et al., “A New Approach to Decoding Life,” 2001. |
|
11 |
L. Hood and D.J. Galas, “The Digital Code of DNA,” Nature 421(6921):444-448, 2003. |
|
12 |
L.M. Smith, J.Z. Sanders, R.J. Kaiser, P. Hughes, C. Dodd, C.R. Connell, C. Heiner, et al., “Fluorescence Detection in Automated DNA Sequence Analysis,” Nature 321(6071):674-679, 1986. (Cited in Ideker et al., 2001.) |
|
13 |
L. Rowen, S. Lasky, and L. Hood, “Deciphering Genomes Through Automated Large Scale Sequencing,” Methods in Microbiology, A.G. Craig and J.D. Hoheisel, eds., Academic Press, San Diego, CA, 1999, pp. 155-191. (Cited in Ideker et al., 2001.) |
|
14 |
S. Brenner, M. Johnson, J. Bridgham, G. Golda, D.H. Lloyd, D. Johnson, S. Luo, et al., “Gene Expression Analysis by Massively Parallel Signature Sequencing (MPSS) on Microbead Arrays,” Nature Biotechnology 18(6):630-634, 2000. (Cited in Ideker et al., 2001.) |
|
15 |
J.K. Eng, A.L. McCormack, and J.R.I. Yates, “An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database,” Journal of the American Society for Mass Spectrometry 5:976-989, 1994. (Cited in Ideker et al., 2001.) |
|
Box 7.4 A “classical” microarray typically consists of single-stranded pieces of DNA from virtually an entire genome placed physically in tiny dots on a flat surface and labeled with a fluorescent dye. (Lithographic techniques used to develop semiconductor chips are now used to deposit the DNA on a silicon chip that can later be read optically.) In a microarray experiment, messenger RNA (mRNA) from a cell of interest is extracted and placed in contact with the prepared surface. If the sample contains mRNA corresponding to the DNA on one or more of the dots on the surface, the molecules will bind and the dye will fluoresce. Because the mRNA represents the fraction of genes from the sample that have been transcribed from DNA into mRNA, the resulting fluorescent dots on the surface are a visual indicator of gene expression (or transcription) in the cell’s genome. Different intensities of the dots reflect greater or lesser levels of transcription of particular genes. Obtaining the maximum value from a microarray experiment depends on the ability to correlate the data from a microarray experiment per se with extensive data that identify or classify the genes by other characteristics. In the absence of such data, any given microarray experiment merely points out the fact that some genes are expressed to a greater extent than others in a particular experimental situation. Protein microarrays can identify protein-protein (and protein-drug) interactions among some 10,000 proteins at once.1 As described by Templin,2 [protein] microarray technology allows the simultaneous analysis of thousands of parameters within a single experiment. Microspots of capture molecules are immobilized in rows and columns onto a solid support and exposed to samples containing the corresponding binding molecules. Readout systems based on fluorescence, chemiluminescence, mass spectrometry, radioactivity or electrochemistry can be used to detect complex formation within each microspot. Such miniaturized and parallelized binding assays can be highly sensitive, and the extraordinary power of the method is exemplified by array-based gene expression analysis. In these systems, arrays containing immobilized DNA probes are exposed to complementary targets and the degree of hybridization is measured. Recent developments in the field of protein microarrays show applications for enzyme-substrate, DNA-protein and different types of protein-protein interactions. Here, we discuss theoretical advantages and limitations of any miniaturized capture-molecule-ligand assay system and discuss how the use of protein microarrays will change diagnostic methods and genome and proteome research. NOTE: An overview of microarray technology is available on a private Web site created by Leming Shi: http://www.gene-chips.com/. See also http://www.genome.gov/10000533 and P. Gwynne and G. Page, “Microarray Analysis: The Next Revolution in Molecular Biology,” special advertising supplement, Science 285, August 6, 1999, available at http://www.sciencemag.org/feature/e-market/benchtop/micro.shl. |
-
troscopy today allow 1,000 proteins per day to be analyzed in an automated fashion, and there is hope that a next-generation facility will be able to analyze up to 1 million proteins per day.16
-
Cell sorters. Cell sorters separate different cell types at high speed on the basis of multiple parameters. While microarray experiments provide information on average levels of mRNA or protein within a cell population, the reality is that these levels vary from cell to cell. Knowing the distribution of expression levels across cell types provides important information about the underlying control mechanisms and regulatory network structure. A state-of-the-art cell sorter can separate 30,000 elements per second according to 32 different parameters.17
|
16 |
S.P. Gygi, B. Rist, S.A. Gerber, F. Turecek, M.H. Gelb, and R. Aebersold, “Quantitative Analysis of Complex Protein Mixtures Using Isotope-coded Affinity Tags,” Nature Biotechnology 17(10):994-999, 1999. (Cited in Ideker et al., 2001.) |
|
17 |
See, for example, http://www.systemsbiology.org/Default.aspx?pagename=cellsorting. |
|
Box 7.5 Marine Microorganisms1 Marine microorganisms such as viruses, bacteria, microalgae, and protozoa have a major impact on the ecology of the coastal ocean; present public health issues for coastal human populations as a consequence of the introduction of pathogenic microorganisms into these waters from land runoff, storm drains, and sewage outflow; and have the potential to contaminate drinking water supplies with harmful, pathogenic, or nuisance microbial species. Today, the environmental factors that stimulate the growth of such microorganisms are still poorly understood. To understand these factors, scientists need to correlate environmental conditions with microorganismal abundances at the small spatial and temporal scales that are relevant to these organisms. For a variety of technological and methodological reasons, sampling the environment at the necessary high resolution and identifying microorganisms in situ in near-real time has not been possible in the past. Habitat Sensing2 Understanding in detail the environmental, organismal, and cultural conditions, and the interactions between them, in natural and managed habitats is a problem of considerable biological complexity. Data must be captured and integrated across a wide range of spatial and temporal scales for chemical, physiological, ecological, and environmental purposes. For example, data of interest might include microclimate data; a video of avian behavioral activities related to climate, nesting, and reproduction; and data on soil moisture, nitrate, CO2, temperature, and root-fungi activities in response to weather. |
-
Microfluidic systems. Microfluidic systems, also known as micro-TAS (total analysis system), allow the rapid and precise measurement of sample volumes of picoliter size. These systems put onto a single integrated circuit all stages of chemical analysis, including sample preparation, analyte purification, microliquid handling, analyte detection, and data analysis.18 These “lab-on-a-chip” systems provide portability, higher-quality and higher-quantity data, faster kinetics, automation, and reduction of sample and reagent volumes.
-
Embedded networked sensor (ENS) systems. ENS systems are large-scale, distributed systems, composed of smart sensors embedded in the physical world, that can provide data about the physical world at unprecedented granularity. These systems can monitor and collect large volumes of information at low cost on such diverse subjects as plankton colonies, endangered species, and soil and air contaminants. Across a wide range of large-scale biological applications broadly cast, these systems promise to reveal previously unobservable phenomena. Box 7.5 describes some applications of ENS systems.
Finally, a specialized type of data acquisition technology is the hybrid measurement device that interacts directly with a biological sample to record data from it or to interact with it. As one illustration, contemporary tools for studying neuronal signaling and information processing include implantable probe arrays that record extracellularly or intracellularly from multiple neurons simultaneously.
|
18 |
See, for example, http://www.eurobiochips.com/euro2002/html/agenda.asp. To illustrate the difficulty, consider the handling of liquids. Dilution ratios required for a process may vary by three or four orders of magnitude, and so an early challenge (now largely resolved successfully) is the difficulty of engineering an automated system that can dispense both 0.1-microliter and 1-milliliter volumes with high accuracy and in reasonable time periods. |
Such arrays have been used in moths (Manduca sexta) and sea slugs (Tritonia diomeda) and, when linked directly to the electronic signals of a computer, essentially record and simulate the neural signaling activity occurring in the organism. Box 7.6 describes the dynamic clamp, a hybrid measurement device that has been invaluable in probing the behavior of neurons. Research on this interface will serve both to reveal more about the biological system and to represent that system in a format that can be computed.
|
Box 7.6 The dynamic clamp is a device that mimics the presence of a membrane or synapse proximate to a neuron. That is, the clamp essentially simulates the electrical conductances in the network to which a neuron is ostensibly connected. During clamp operation, the membrane potential of a neuron is continuously measured and fed into a computer. The dynamic clamp program contains a mathematical model of the conductance to be simulated and computes the current that would flow through the conductance as a function of time. This current is injected into the neuron, and the cycle of membrane potential measurement, current computation, and current injection continues. This cycle enables researchers to study the effects of a membrane current or synaptic input in a biological cell (the neuron) by generating a hybrid system in which the artificial conductance interacts with the natural dynamic properties of the neuron. The dynamic clamp can be used to mimic any voltage-dependent conductance that can be expressed in a mathematical model. Depending on the type of conductance, most applications can be grouped in one of the following categories:
The first application of the dynamic clamp involved the stimulation of a gamma-aminobutyric acid (GABA) response in a cultured stomatogastric ganglion neuron. This application illustrated that the dynamic clamp effectively introduces a conductance into the target neuron. Demonstration of an artificial voltage-dependent conductance resulted from simulation of the action of a voltage-dependent proctolin response on a neuron in the intact stomatogastric ganglion, which showed that shifts in the activation curve and the maximal conductance of the response produced different effects on the target neuron. Lastly, the dynamic clamp was used to construct reciprocal inhibitory synapses between two stomatogastric ganglion neurons that were not coupled naturally, illustrating that the dynamic clamp could be used to simulate new networks at will. SOURCE: The description of a dynamic clamp is based heavily on A.A. Prinz, “The Dynamic Clamp a Decade After Its Invention,” Axon Instruments Newsletter 40, February 2004, available at http://www.axon.com/axobits/AxoBits40.pdf. The description of the first application of the dynamic clamp is nearly verbatim from A.A. Sharp, M.B. O’Neil, L.F. Abbott, and E. Marder, “Dynamic Clamp: Computer-generated Conductances in Real Neurons,” Journal of Neurophysiology 69(3):992-995, 1993. |
7.2.2 Examples of Future Technologies
As powerful as these technologies are, new instrumentation and methodology will be needed in the future. These technical advances will have to reduce the cost of data acquisition by several orders of magnitude.
Consider, for example, the promise of genomically individualized medical care, which is based on the notion that treatment and/or prevention strategies for disease can be customized to groups of individuals smaller than the entire population, and perhaps ultimately groups as small as one. Because these groups will be identified in part by particular sets of genomic characteristics, it will be necessary to undertake the genomic sequencing of these individuals. The first complete sequencing of the human genome took 13 years and $2.7 billion. For broad use in the population at large, the cost of assembling and sequencing a human genome must drop to hundreds or thousands of dollars—a reduction in cost of 105 or 106 that would enable the completion of a human genome at such cost in a matter of days.19
Computation per se is expected to continue to drop in cost in accordance with Moore’s law at least over the next decade. But automation of data acquisition will also play an enormous role in facilitating such cost reductions. For example, the laboratory of Richard Mathies at the University of California, Berkeley, has developed a 96-lane microfabricated DNA sequencer capable of sequencing at a rate of 1,700 bases per minute.20 Using this technology, the complete sequencing of an individual 3-billion base genome would take 1,000 sequencer-days. Future versions will incorporate higher degrees of parallelism.
Similar advances in technology will help to reduce the cost of other kinds of biological research as well. A number of biological signatures useful for functional genomics have been susceptible to significantly greater degrees of automation, miniaturization, and multiplexing; these signatures are associated with electrophoresis, molecular microarrays, mass spectrometry, and microscopy.21 Electrophoresis, molecular microarrays, and mass spectrometry provide more opportunities for multiplexed measurement (i.e., the simultaneous measurement of signatures from many molecules from the same source). Such multiplexing can reduce errors due to misalignment of unmultiplexed measures in space and/or time.
In general, the biggest payoffs in laboratory automation are those efforts that can address processes that involve physical material. Much work in biology involves multiple laboratory procedures that each call for multiple fluid transfers, heating and cooling cycles, and mechanical operations such as centrifuging, waiting, and imaging. When these procedures can be undertaken “on-chip,” they reduce the amount of human interaction involved and thus the associated time and cost.
In addition, the feasibility of lab automation is closely tied to the extent to which human craft can be taken out of lab work. That is, because so much lab work must be performed by humans, the skills of the particular individuals involved matter a great deal to the outcomes of the work. A particular individual may be the only one in a laboratory with a “knack” for performing some essential laboratory procedure (e.g., interpretation of certain types of image, preparation or certain types of sample) with high reliability, accuracy, and repeatability.
|
19 |
L.M. Smith, J.Z. Sanders, R.J. Kaiser, P. Hughes, C. Dodd, C.R. Connell, C. Heiner, et al., “Fluorescence Detection in Automated DNA Sequence Analysis,” Nature 321(6071):674-679, 1986; L. Hood and D. Galas, “The Digital Code of DNA,” Nature 421(6921):444-448, 2003. Note that done properly, the second complete sequencing of a human being would be considerably less difficult. The reason is that every member of a biological species has a DNA that is almost identical to that of every other member. In humans, the difference between DNA sequences of different individuals is about one base pair per thousand. (See special issues on the human genome: Science 291(5507) February 16, 2001; Nature 409(6822), February 15, 2001.) So, assuming it is known where to check for every difference, a reduction in effort of at least a factor of 103 is obtainable in principle. |
|
20 |
B.M. Paegel, R.G. Blazej, and R.A. Mathies, “Microfluidic Devices for DNA Sequencing: Sample Preparation and Electrophoretic Analysis,” Current Opinion in Biotechnology 14(1):42-50, 2003, available at http://www.wtec.org/robotics/us_workshop/June22/paper_mathies_microfluidics_sample_prep_2003.pdf. |
|
21 |
G. Church, “Hunger for New Technologies, Metrics, and Spatiotemporal Models in Functional Genomics,” available at http://recomb2001.gmd.de/ABSTRACTS/Church.html. |
While reliance on individuals with specialized technical skills is often a workable strategy for an academic laboratory, it makes much less sense for any organization interested in large-scale production. For large-scale, cost-effective production, process automation is a sine qua non. When a process can be automated, it is generally faster to perform, more free from errors, more accurate, and less expensive.22
Some of the clearest success stories involve genomic technologies. For example, DNA sequencing was a craft at the start of the 1990s—today, automated DNA sequencing is common, with instruments to undertake such sequencing in high volume (a million or more base pairs per day) and even a commercial infrastructure to which sequencing tasks can be outsourced. Nevertheless, a variety of advanced sequencing technologies are being developed, primarily with the intent of lowering the cost of sequencing by another several orders of magnitude.23
An example of such a technology is pyrosequencing, which has also been called “sequencing by synthesis.”24 With pyrosequencing, the DNA to be sequenced is denatured to form a single strand and then placed in solution with a set of selected enzymes. In a cycle of individual steps, the DNA-enzyme solution is mixed with deoxynucleotide triphosphate molecules containing each of the four bases. When the base that is the complement to the next base on the target strand is added, the added base joins a forming complement strand and releases a pyrophosphate molecule. That molecule starts a reaction that ends with luciferin emitting a detectable amount of light. Thus, by monitoring the light output of the reaction (for example, with a CCD camera), it is possible to observe in real time which of the four bases has successfully matched.
454 Life Sciences has applied pyrosequencing to whole-genome analyses by taking advantage of its high parallelizability. Using a PicoTiter plate, a microfluidic system performs pyrosequencing on hundreds of thousands of DNA fragments simultaneously. Custom software analyzes the light emitted and stitches together the complete sequence. This approach has been used successfully to sequence the genome of an adenovirus,25 and the company is expected to produce commercial hardware to perform whole-genome analysis in 2005.
A second success story is microarray technology, which historically has relied heavily on electrophoretic techniques.26 More recently, techniques have been developed that do away entirely with electrophoresis. One approach relies instead on microbeads with different messenger RNAs on their surfaces (serving as probes to which targets bind selectively) and a novel sequencing procedure to
|
22 |
The same can be said for many other aspects of lab work. In 1991, Walter Gilbert noted, “The march of science devises ever newer and more powerful techniques. Widely used techniques begin as breakthroughs in a single laboratory, move to being used by many researchers, then by technicians, then to being taught in undergraduate courses and then to being supplied as purchased services—or, in their turn, superseded…. Fifteen years ago, nobody could work out DNA sequences, today every molecular scientists does so and, five years from now, it will all be purchased from an outside supplier. Just this happened with restriction enzymes. In 1970, each of my graduate students had to make restriction enzymes in order to work with DNA molecules; by 1976 the enzymes were all purchased and today no graduate student knows how to make them. Once one had to synthesize triphosphates to do experiments; still earlier, of course, one blew one’s own glassware.” See W. Gilbert, “Towards a Paradigm Shift in Biology,” Nature 349(6305):99, 1991. |
|
23 |
A review by Shendure et al. classifies emerging ultralow-cost sequencing technologies into one of five groups: microelectrophoretic methods (which extend and incrementally improve today’s mainstream sequencing technologies first developed by Frederick Sanger); sequencing by hybridization; cyclic array sequencing on amplified molecules; cyclic array sequencing on single molecules; and noncyclical, single-molecule, real-time methods. The article notes that most of these technologies are still in the relatively early stages of development, but that they each have great potential. See J. Shendure, R.D. Mitra, C. Varma, and G.M. Church, “Advanced Sequencing Technologies: Methods and Goals,” Nature Reviews: Genetics 5(5):335-344, 2004, available at http://arep.med.harvard.edu/pdf/Shendure04.pdf. Pyrosequencing, provided as an example of one new sequencing technology, is an example of cyclic array sequencing on amplified molecules. |
|
24 |
M. Ronaghi, “Pyrosequencing Sheds Light on DNA Sequencing,” Genome Research 11(1):3-11, 2001. |
|
25 |
A. Strattner, “From Sanger to ‘Sequenator’,” Bio-IT World, October 10, 2003. |
|
26 |
Genes are expressed as proteins, and these proteins have different weights. Electrophoresis is a technique that can be used to determine the extent to which proteins of different weights are present in a sample. |
|
Box 7.7 Optical mapping is a single molecule based physical mapping technology, which creates an ordered restriction map by enumerating the locations of occurrences of a specific “restriction pattern” along a genome. Thus, by locating the same patterns in the sequence reads or contigs, optical maps can detect errors in sequence assembly, and determine the phases (i.e., chromosomal location and orientation) of any set of sequence contigs. Since the input genomic data that can be collected from a single DNA molecule by the best chemical and optical methods (such as those used in Optical Mapping) are badly corrupted by many poorly understood noise processes, this type of technology derives its utility through powerful probabilistic modeling used in experiment design and Bayesian algorithms that can recover from errors by using redundant data. In this way, optical mapping with Gentig, a powerful statistical map-assembly algorithm invented and implemented by the authors, has proven instrumental in completing many microbial genomic maps (Escherichia coli, Yersinia pestis, Plasmodium falciparum, Deinococcus radiodurans, Rhodobacter sphaeroides, etc.) as well as clone maps (DAZ locus of Y chromosome). SOURCE: T. Anantharaman and B. Mishra, Genomics via Optical Mapping (I): 0-1 Laws for Single Molecules, S. Yancopoulos, ed., Oxford University Press, Oxford, England, 2005, in press. |
identify specific microbeads.27 Each bead can be interrogated in parallel, and the abundance of a given messenger RNA is determined by counting the number of beads with that mRNA on their surfaces. In addition to greatly simplifying the sample-handling procedure, this technique has two other important advantages: a direct digital readout of relative abundances (i.e., the bead counts) and throughput increases by more than a factor of 10 compared to other techniques.
A second approach to the elimination of electrophoresis is known as optical mapping or sequencing (Box 7.7). Optical mapping eliminates dependence on ensemble-based methods, focusing on the statistics of individual DNA molecules. Although this technique is fragile and, to date, not replicable in multiple laboratories,28 it may eventually be capable of sequencing entire genomes much more rapidly than is possible today.
A different approach based on magnetic detection of DNA hybridization seeks to lower the cost of performing microarray analysis. Chen et al. have suggested that instead of tagging targets with fluorescent molecules, targets are tagged with microscopic magnetic beads.29 Probes are implanted on a magnetically sensitive surface, such as a floppy disk, after removing the magnetic coating at the probe
|
27 |
S. Brenner, “Gene Expression Analysis by Massively Parallel Signature Sequencing (MPSS) on Microbead Arrays,” Nature Biotechnology 18(6):630-634, 2002. The elimination of electrophoresis (a common laboratory technique for separating biological samples by molecular weight) has many practical benefits. Conceptually, electrophoresis is a straightforward process. A tagged biological sample is inserted into a viscous gel and then subjected to an external electric field for some period of time. The sample differentiates in the electric field because the lighter components move farther under the influence of the electric field than the heavier ones. The tag on the biological sample is, for example, a compound that fluoresces when exposed to ultraviolet light. Measuring the intensity of the fluorescence provides an indication of the relative abundances of components of different molecular weight. However, in practice there are difficulties. The gel must undergo appropriate preparation—no small task. For example, the gel must be homogeneous, with no bubbles to interfere with the natural movement of the sample components. The temperature of the gel-sample combination may be important, because the viscosity of the gel may be temperature-sensitive. While the gel is drying (a process that takes a few hours), it must not be physically disturbed in a way that introduces defects into the gel preparation. |
|
28 |
Bud Mishra, New York University, personal communication, December 2003. |
|
29 |
C.H.W. Chen, V. Golovlev, and S. Allman, “Innovative DNA Microarray Hybridization Detection Technology,” poster abstract presented at Human Genome Meeting 2002, April 14-17, 2004, Shanghai, China; also, “Detection of Polynucleotides on Surface of Magnetic Media, available at http://www.scien-tec.com/news1.htm. |
location, and different probes are attached to different locations. Readout of hybridized probe-target pairs is accomplished through the detection of a magnetic signal at given locations; locations without such pairs provide no signal because the magnetic coating of the floppy disk has been removed from those locations. Also, the location of any given probe-target pair is treated simply as a physical address on the floppy disk. Preliminary data suggest that with the spatial resolution currently achieved, a single floppy diskette can carry up to 45,000 probes, a figure that compares favorably to that of most glass microarrays (of order 10,000 probes or less). Chen et al. argue that this approach has two advantages: greater sensitivity and significantly lower cost. The increased sensitivity is due to the fact that signal strength is controlled by the strength of the beads rather than the amount of hybridizing DNA per se; and so, in principle, this approach could detect even a single hybridization event. Lower costs arguably result from the fact that the most of the components for magnetic detection are mass-produced in quantity for the personal computer industry today.
Laboratory robotics is another area that offers promise of reduced labor costs. For example, the minimization of human intervention is illustrated by the introduction of compact, user-programmable robot arms in the early 1980s.30 One version, patented by the Zymark Corporation, equipped a robot arm with interchangeable hands. This arm was the foundation of robotic laboratory workstations that could be programmed to carry out multistep sample manipulations, thus allowing them to be adapted for different assays and sample-handling approaches.
Building on the promise offered by such robot arms, a testbed laboratory formed in the 1980s by Dr. Masahide Sasaki at the Kochi Medical School in Japan demonstrated the feasibility of a high degree of laboratory automation: robots carried test tube racks, and conveyor belts transported patient samples to various analytical workstations. Automated pipettors drew serum from samples for the required assays. One-armed stationary robots performed pipetting and dispensing steps to accomplish preanalytical processing of higher complexity. The laboratory was able to perform all clinical laboratory testing for a 600-bed hospital with a staff of 19 employees. By comparison, hospitals in the United States of similar size required up to 10 times as many skilled clinical laboratory technologists.
Adoption of the “total laboratory automation” approach was mixed. Many clinical laboratories in particular found that it provided excess capacity whose costs could not be recovered easily. Midsized hospital laboratories had a hard time justifying the purchase of multimillion-dollar systems. By contrast, pharmaceutical firms invested heavily in robotic laboratory automation, and automated facilities to synthesize candidate drugs and to screen their biological effects provided three- to fivefold increases in the number of new compounds screened per unit time.
In recent years, manufacturers have marketed “modular” laboratory automation products, including modules for specimen centrifugation and aliquoting, specimen analysis, and postanalytical storage and retrieval. While such modules can be assembled like building blocks into a system that provides very high degrees of automation, they also enable a laboratory to select the module or modules that best address its needs.
Even mundane but human-intensive tasks are susceptible to some degree of automation. Consider that much of biological experimentation depends on the availability of mice as test subjects. Mice need to be housed and fed, and thus require considerable human labor. The Stowers Institute for Medical Research in Kansas City has approached this problem with the installation of an automated mouse care facility involving two robots, one of which dumps used mouse bedding and feeds it to a conveyor washing machine and the other of which fills the clean cages with bedding and places them on a rack.31 These robots can process 350 cages per hour and reduce the labor needs of cleaning cages by a factor of three (from six technicians to two). At a cost of $860,000, the institute expects to recoup its investment in
6 years, with much of the savings coming from reduced repetitive motion injuries and fewer health problems caused by allergen exposure.
In the future, modularization is likely to continue. In addition, fewer stand-alone robot arms are being used because the robotics necessary for sampling from conveyor belts are often integrated directly into clinical analyzers. Attention is turning from the development of hardware to the design of process control software to control and integrate the various automation components; to manage the transport, storage, and retrieval of specimens; and to support automatic repeat and follow-up testing strategies.
7.2.3 Future Challenges
From a conceptual standpoint, automation for speed depends on two things—speeding up an individual process and processing many samples in parallel. Individual processes can be speeded up to some extent, but because they are limited by physical time constants (e.g., the time needed to mix a solution uniformly, the time needed to dry, the time needed to incubate), the speedups possible are limited—perhaps factors of a few or even ten can be possible. By contrast, parallel processing is a much bigger winner, and it is easy to imagine processing hundreds or even thousands of samples simultaneously.
In addition to quantitative speedups, qualitatively new data acquisition techniques are needed as well. The difficulty of collecting meaningful data from biological systems has often constrained the level of complexity at which to collect data. Biologists often must use indirect or surrogate measures that imply activity. For example, oxygen consumption can be used as a surrogate for breathing.
There is a need to develop new mechanisms to collect data, particularly mechanisms that can form a bridge from the living system to a computer system, in other words, tools that detect and monitor biological events and directly collect and store information about those events for later analysis. Challenges in this area include the connection of cellular material, cells, tissues, and humans to computers for rapid diagnostics and data download, bio-aided computation, laboratory study, or human-computer interactivity, and how to perform “smart” experiments that use models of the biological systems to probe the biology dynamically so that measurements of the spatiotemporal dynamics of living cells at many scales become possible.
A good example of future data acquisition challenges is provided by single-cell assays and single-molecule detection. Traditional assays can involve thousands or tens of thousands of cells and produce datasets that reflect the aggregate behavior of the entire sample. While for many types of experiments this is an appropriate approach, there are current and future biological research issues for which this does not provide sufficient resolution. For example, cells within a population may be in different stages of their life cycle, may be experiencing local variations of environmental conditions, or may be of entirely different types. Alternatively, a probe might not touch the cell type of interest, due to inadequate purification of a sample drawn from a subject that contains many cell types.32 For some biological questions, there is simply not a sufficient supply of cells of interest; for example, certain human nervous system tissue is highly specialized, and a biological inquiry may concern only a few cells. Similarly, in attempts to isolate some diseases, there may be only a few, or even only one, affected cell—for example, in attempts to detect cancerous cells before they develop into a tumor.
Many technologies offer approaches to analyzing and characterizing the behavior of single cells, including the use of mass spectrometry, microdissection, laser-induced fluorescence, and electrophoresis. Ideally, it would be possible to monitor the behavior of a living cell over time with sufficient resolution to determine the functioning of subcellular components at different stages of the life cycle and in response to differing environmental stimuli.
A further challenge in ultrasensitive data acquisition in living cells is that the substances of interest, particularly proteins, occur at a wide range of concentrations (varying by many orders of magnitude). For many important proteins, this may be as few as hundreds of individual molecules. Detection and analysis at such low levels must work even in the face of wide statistical fluctuation, transient modifications, and a wide range of physical and chemical properties.33
At the finest grain, detection and analysis of single molecules could provide further understanding of cellular mechanisms. Again, although there are current techniques to analyze molecular structure (such as nuclear magnetic resonance and X-ray crystallography), these work on large, static samples. To achieve more precise understanding of cellular mechanisms, it is necessary to detect the presence and activity of very small concentrations, even single molecules, dynamically within living cells. Making progress in this field will require advances in chemistry, instrumentation, sensors, and image analysis algorithms.34
Embedded networked sensor (ENS) systems will ride the cost reduction curve that characterizes much of modern electronic systems. Based on microsensors, on-board processing, and wireless communications, ENS systems can monitor phenomena “up close.” Nevertheless, taken as a whole, ENS systems present challenges with respect to longevity, autonomy, scalability, performance, and resilience. For example, off-the-shelf sensors embedded in heterogeneous soil for monitoring soil moisture and nitrate levels raise issues related to calibration when embedded in a previously unknown environment. In addition, the uncertainty in the data they provide must be characterized. Interesting theoretical issues arise with respect to the statistical and information-theoretic foundations for adaptive sampling and data fusion. Also, of course, programming abstractions, common services, and tools for programming the network must be developed.
To illustrate a specific application, consider some of the computing challenges in deploying ENS systems for marine microorganisms. The ultimate goal is to deploy large groups of autonomous, mobile microrobots capable of identifying and tracking microorganisms in real time in the marine environment, while measuring the relevant environmental conditions at the required temporal and spatial scales. Sensors must be mobile to track microorganisms and assess their abundance with a reasonable number of sensors. They must be small, so that they are able to gather information at a spatial scale comparable to the size of the microorganisms and to avoid disturbing them. They must operate in a liquid environment—combined with small sensor size, operation in such an environment raises many difficult issues of mobility, communications, and power, which in turn strongly impact network algorithms and strategies. Also, sensors must be capable of in situ, real-time identification of microorganisms, which requires the development of new sensors with considerable on-board processing capability. Progress in this application—monitoring marine environments and single-cell identification—is expected to be applicable to other liquid environments, such as the circulatory system of higher organisms, including humans.
|
33 |
R.D. Smith et al., “Application of New Technologies for Comprehensive, Quantitative and High Throughput Microbial Proteomics,” abstracts of the Department of Energy’s (DOE) Genomes to Life Systems-Biology Projects on Microbes Sequenced by the U.S. DOE’s Microbial Genome Program, available at http://doegenomestolife.org/pubs/2004abstracts/html/Tech_Dev.shtml#_VPID_289. |
|
34 |
See, for example, the text of the NIH Program Announcement PA-01-049, “Single Molecule Detection and Manipulation,” released February 12, 2001, available at http://grants.nih.gov/grants/guide/pa-files/PA-01-049.html. |




















