3
On the Nature of Biological Data
Twenty-first century biology will be a data-intensive enterprise. Laboratory data will continue to underpin biology’s tradition of being empirical and descriptive. In addition, they will provide confirming or disconfirming evidence for the various theories and models of biological phenomena that researchers build. Also, because 21st century biology will be a collective effort, it is critical that data be widely shareable and interoperable among diverse laboratories and computer systems. This chapter describes the nature of biological data and the requirements that scientists place on data so that they are useful.
3.1 DATA HETEROGENEITY
An immense challenge—one of the most central facing 21st century biology—is that of managing the variety and complexity of data types, the hierarchy of biology, and the inevitable need to acquire data by a wide variety of modalities. Biological data come in many types. For instance, biological data may consist of the following:1
-
Sequences. Sequence data, such as those associated with the DNA of various species, have grown enormously with the development of automated sequencing technology. In addition to the human genome, a variety of other genomes have been collected, covering organisms including bacteria, yeast, chicken, fruit flies, and mice.2 Other projects seek to characterize the genomes of all of the organisms living in a given ecosystem even without knowing all of them beforehand.3 Sequence data generally
|
1 |
This discussion of data types draws heavily on H.V. Jagadish and F. Olken, eds., Data Management for the Biosciences, Report of the NSF/NLM Workshop of Data Management for Molecular and Cell Biology, February 2-3, 2003, Available at http://www.eecs.umich.edu/~jag/wdmbio/wdmb_rpt.pdf. A summary of this report is published as H.V. Jagadish and F. Olken, “Database Management for Life Science Research,” OMICS: A Journal of Integrative Biology 7(1):131-137, 2003. |
|
2 |
|
|
3 |
See, for example, J.C. Venter, K. Remington, J.F. Heidleberg, A.L. Halpern, D. Rusch, J.A. Eisen, D. Wu, et al., “Environmental Genome Shotgun Sequencing of the Sargasso Sea,” Science 304(5667):66-74, 2004. Venter’s team collected microbial populations en masse from seawater samples originating in the Sargasso Sea near Bermuda. The team subsequently identified 1.045 billion base pairs of nonredundant sequence, which they estimated to derive from at least 1,800 genomic species based on sequence relatedness, including 148 previously unknown bacterial phylotypes. They also claimed to have identified more than 1.2 million previously unknown genes represented in these samples. |
-
consist of text strings indicating appropriate bases, but when there are gaps in sequence data, gap lengths (or bounds on gap lengths) must be specified as well.
-
Graphs. Biological data indicating relationships can be captured as graphs, as in the cases of pathway data (e.g., metabolic pathways, signaling pathways, gene regulatory networks), genetic maps, and structured taxonomies. Even laboratory processes can be represented as workflow process model graphs and can be used to support formal representation for use in laboratory information management systems.
-
High-dimensional data. Because systems biology is highly dependent on comparing the behavior of various biological units, data points that might be associated with the behavior of an individual unit must be collected for thousands or tens of thousands of comparable units. For example, gene expression experiments can compare expression profiles of tens of thousands of genes, and since researchers are interested in how expression profiles vary as a function of different experimental conditions (perhaps hundreds or thousands of such conditions), what was one data point associated with the expression of one gene under one set of conditions now becomes 106 to 107 data points to be analyzed.
-
Geometric information. Because a great deal of biological function depends on relative shape (e.g., the “docking” behavior of molecules at a potential binding site depends on the three-dimensional configuration of the molecule and the site), molecular structure data are very important. Graphs are one way of representing three-dimensional structure (e.g., of proteins), but ball-and-stick models of protein backbones provide a more intuitive representation.
-
Scalar and vector fields. Scalar and vector field data are relevant to natural phenomena that vary continuously in space and time. In biology, scalar and vector field properties are associated with chemical concentration and electric charge across the volume of a cell, current fluxes across the surface of a cell or through its volume, and chemical fluxes across cell membranes, as well as data regarding charge, hydrophobicity, and other chemical properties that can be specified over the surface or within the volume of a molecule or a complex.
-
Patterns. Within the genome are patterns that characterize biologically interesting entities. For example, the genome contains patterns associated with genes (i.e., sequences of particular genes) and with regulatory sequences (that determine the extent of a particular gene’s expression). Proteins are characterized by particular genomic sequences. Patterns of sequence data can be represented as regular expressions, hidden Markov models (HMMs), stochastic context-free grammars (for RNA sequences), or other types of grammars. Patterns are also interesting in the exploration of protein structure data, microarray data, pathway data, proteomics data, and metabolomics data.
-
Constraints. Consistency within a database is critical if the data are to be trustworthy, and biological databases are no exception. For example, individual chemical reactions in a biological pathway must locally satisfy the conservation of mass for each element involved. Reaction cycles in thermodynamic databases must satisfy global energy conservation constraints. Other examples of nonlocal constraints include the prohibition of cycles in overlap graphs of DNA sequence reads for linear chromosomes or in the directed graphs of conceptual or biological taxonomies.
-
Images. Imagery, both natural and artificial, is an important part of biological research. Electron and optical microscopes are used to probe cellular and organ function. Radiographic images are used to highlight internal structure within organisms. Fluorescence is used to identify the expressions of genes. Cartoons are often used to simplify and represent complex phenomena. Animations and movies are used to depict the operation of biological mechanisms over time and to provide insight and intuitive understanding that far exceeds what is available from textual descriptions or formal mathematical representations.
-
Spatial information. Real biological entities, from cells to ecosystems, are not spatially homogeneous, and a great deal of interesting science can be found in understanding how one spatial region is different from another. Thus, spatial relationships must be captured in machine-readable form, and other biologically significant data must be overlaid on top of these relationships.
-
Models. As discussed in Section 5.3.4, computational models must be compared and evaluated. As the number of computational models grows, machine-readable data types that describe computational models—both the form and the parameters of the model—are necessary to facilitate comparison among models.
-
Prose. The biological literature itself can be regarded as data to be exploited to find relationships that would otherwise go undiscovered. Biological prose is the basis for annotations, which can be regarded as a form of metadata. Annotations are critical for researchers seeking to assign meaning to biological data. This issue is discussed further in Chapter 4 (automated literature searching).
-
Declarative knowledge such as hypotheses and evidence. As the complexity of various biological systems is unraveled, machine-readable representations of analytic and theoretical results as well as the underlying inferential chains that lead to various hypotheses will be necessary if relationships are to be uncovered in this enormous body of knowledge. This point is discussed further in Section 4.2.8.1.
In many instances, data on some biological entity are associated with many of these types: for example, a protein might have associated with it two-dimensional images, three-dimensional structures, one-dimensional sequences, annotations of these data structures, and so on.
Overlaid on these types of data is a temporal dimension. Temporal aspects of data types such as fields, geometric information, high-dimensional data, and even graphs—important for understanding dynamical behavior—multiply the data that must be managed by a factor equal to the number of time steps of interest (which may number in the thousands or tens of thousands). Examples of phenomena with a temporal dimension include cellular response to environmental changes, pathway regulation, dynamics of gene expression levels, protein structure dynamics, developmental biology, and evolution. As noted by Jagadish and Olken,4 temporal data can be taken absolutely (i.e., measured on an absolute time scale, as might be the case in understanding ecosystem response to climate change) or relatively (i.e., relative to some significant event such as division, organism birth, or environmental insult). Note also that in complex settings such as disease progression, there may be many important events against which time is reckoned. Many traditional problems in signal processing involve the extraction of signal from temporal noise as well, and these problems are often found in investigating biological phenomena.
All of these different types of data are needed to integrate diverse witnesses of cellular behavior into a predictive model of cellular and organism function. Each data source, from high-throughput microarray studies to mass spectroscopy, has characteristic sources of noise and limited visibility into cellular function. By combining multiple witnesses, researchers can bring biological mechanisms into focus, creating models with more coverage that are far more reliable than models created from one source of data alone. Thus, data of diverse types including mRNA expression, observations of in vivo protein-DNA binding, protein-protein interactions, abundance and subcellular localization of small molecules that regulate protein function (e.g., second messengers), posttranslational modifications, and so on will be required under a wide variety of conditions and in varying genetic backgrounds. In addition, DNA sequence from diverse species will be essential to identify conserved portions of the genome that carry meaning.
3.2 DATA IN HIGH VOLUME
Data of all of the types described above contribute to an integrated understanding of multiple levels of a biological organism. Furthermore, since it is generally not known in advance how various components of an organism are connected or how they function, comprehensive datasets from each of these
types are required. In cellular analysis, data comprehensiveness includes three aspects, as noted by Kitano:5
-
Factor comprehensiveness, which reflects the numbers of mRNA transcripts and proteins that can be measured at once;
-
Time-line comprehensiveness, which represents the time frame within which measurements are made (i.e., the importance of high-level temporal resolution); and
-
Item comprehensiveness—the simultaneous measurement of multiple items, such as mRNA and protein concentrations, phosphorylation, localization, and so forth.
For every one of the many proteins in a given cell type, information must be collected about protein identity, abundance, processing, chemical modifications, interactions, turnover time, and so forth. Spatial localization of proteins is particularly critical. To understand cellular function in detail, proteins must be localized on a scale finer than that of cell compartments; moreover, localization of specific protein assemblies to discrete subcellular sites through anchoring and scaffolding proteins is important.
All of these considerations suggest that in addition to being highly heterogeneous, biological data must be voluminous if they are to support comprehensive investigation.
3.3 DATA ACCURACY AND CONSISTENCY
All laboratories must deal with instrument-dependent or protocol-dependent data inconsistencies. For example, measurements must be calibrated against known standards, but calibration methods and procedures may change over time, and data obtained under circumstances of heterogeneous calibration may well not be comparable to each other. Experiments done by multiple independent parties almost always result in inconsistencies in datasets.6 Different experimental runs with different technicians and protocols in different labs inevitably produce data that are not entirely consistent with each other, and such inconsistencies have to be noted and reconciled. Also, the absolute number of data errors that must be reconciled—both within a single dataset and across datasets—increases with the size of the dataset. For such reasons, statistical data analysis becomes particularly important in analyzing data acquired via high-throughput techniques.
To illustrate these difficulties, consider the replication of microarray experiments. Experience with microarrays suggests that such replication can be quite difficult. In principle, a microarray experiment is simple. The raw output of a microarray experiment is a listing of fluorescent intensities associated with spots in an array; apart from complicating factors, the brightness of these spots is an indication of the expression level of the transcript associated with them.
On the other hand, the complicating factors are many, and in some cases ignoring these factors can render one’s interpretation of microarray data completely irrelevant. Consider the impact of the following:
-
Background effects, which are by definition contributions to spot intensity that do not originate with the biological material being examined. For example, an empty microarray might result in some
-
background level of fluorescence and even some variation in background level across the entire surface of the array.
-
Noise dependent on expression levels of the sample. For example, Tu et al. found that hybridization noise is strongly dependent on expression level, and in particular the hybridization noise is mostly Poisson-like for high expression levels but more complex at low expression levels.7
-
Differential binding strengths for different probe-target combinations. The brightness of a spot is determined by the amount of target present at a probe site and the strength of the binding between probe and target. Held et al. found that the strength of binding is affected by the free energy of hybridization, which is itself a function of the specific sequence involved at the site, and they developed a model to account for this finding.8
-
Lack of correlation between mRNA levels and protein levels. The most mature microarray technology measures mRNA levels, while the quantity of interest is often protein level. However, in some cases of interest, the correlation is small even if overall correlations are moderate. One reason for small correlations is likely to be the fact that some proteins are regulated after translation, as noted in Ideker et al.9
-
Lack of uniformity in the underlying glass surface of a microarray slide. Lee et al. found that the specific location of a given probe on the surface affected the expression level recorded.10
Other difficulties arise when the results of different microarray experiments must be compared.11
-
Variations in sample preparation. A lack of standardized procedure across experiments is likely to result in different levels of random noise—and procedures are rarely standardized very well when they are performed by humans in different laboratories. Indeed, sample preparation effects may dominate effects that arise from the biological phenomenon under investigation.12
-
Insufficient spatial resolution. Because multiple cells are sampled in any microarray experiment, tissue inhomogeneities may result in more of a certain kind of cell being present, thus throwing off the final result.
-
Cell-cycle starting times. Identical cells are likely to have more-or-less identical clocks, but there is no assurance that all of the clocks of all of the cells in a sample are started at the same time. Because expression profile varies over time, asynchrony in cell cycles may also throw off the final result.13
To deal with these difficulties, the advice offered by Lee et al. and Novak et al., among others, is fairly straightforward—repeat the experiment (assuming that the experiment is appropriately struc-
|
7 |
Y. Tu, G. Stolovitzky, and U. Klein, “Quantitative Noise Analysis for Gene Expression Microarray Experiments,” Proceedings of the National Academy of Sciences 99(22):14031-14036, 2002. |
|
8 |
G.A. Held, G. Grinstein, and Y. Tu, “Modeling of DNA Microarray Data by Using Physical Properties of Hybridization,” Proceedings of the National Academy of Sciences 100(13):7575-7580, 2003. |
|
9 |
T. Ideker, V. Thornsson, J.A. Ranish, R. Christmas, J. Buhler, J.K. Eng, R. Bumgarner, et al., “Integrated Genomic and Proteomic Analyses of a Systematically Perturbed Metabolic Network,” Science 292(5518):929-934, 2001. (Cited in Rice and Stolovitzky, “Making the Most of It,” 2004, Footnote 11.) |
|
10 |
M.L. Lee, F.C. Kuo, G.A. Whitmore, and J. Sklar, “Importance of Replication in Microarray Gene Expression Studies: Statistical Methods and Evidence from Repetitive cDNA Hybridizations,” Proceedings of the National Academy of Sciences 97(18):9834-9839, 2000. |
|
11 |
J.J. Rice and G. Stolovitzky, “Making the Most of It: Pathway Reconstruction and Integrative Simulation Using the Data at Hand,” Biosilico 2(2):70-77, 2004. |
|
12 |
J.P. Novak, R. Sladek, and T.J. Hudson, “Characterization of Variability in Large-scale Gene Expression Data: Implications for Study Design,” Genomics 79(1):104-113, 2002. |
|
13 |
R.J. Cho, M.J. Campbell, E.A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T.G. Wolfsberg, et al., “A Genome-wide Transcriptional Analysis of the Mitotic Cell Cycle,” Molecular Cell 2(1):65-73, 1998; P.T. Spellman, G. Sherlock, M.Q. Zhang, V.R. Iyer, K. Anders, M.B. Eisen, P.O. Brown, et al., “Comprehensive Identification of Cell Cycle-regulated Genes of the Yeast Saccharomyces cerevisiae by Microarray Hybridization,” Molecular Biology of the Cell 9(12):3273-3297, 1998. (Cited in Rice and Stolovitzky, “Making the Most of It,” 2004, Footnote 11.) |
tured and formulated in the first place). However, the expense of microarrays may be an inhibiting factor in this regard.
3.4 DATA ORGANIZATION
The acquiring of experimental data by some researcher is only the first step in making them useful to the wider biological research community. Data are useless if they are inaccessible or incomprehensible to others, and given the heterogeneity and large volumes of biological data, appropriate data organization is central to extracting useful information from the data. Indeed, it would not be an exaggeration to identify data management and organization issues as a key rate-limiting step in doing science for the small to medium-sized laboratory, where “science” covers the entire intellectual water-front from laboratory experiment to data that are useful to the community at large. This is especially true in laboratories using high-throughput data acquisition technologies.
In recent years, biologists have taken significant steps in coming to terms with the need to think collectively about databases as research tools accessible to the entire community. In the field of molecular biology, the first widely recognized databases were the international archival repositories for DNA and genomic sequence information, including GenBank, the European Molecular Biology Laboratory (EMBL) Nucleotide Sequence Database, and the DNA Databank of Japan (DDJ). Subsequent databases have provided users with information that annotated the genomic sequence data, connecting regions of a genome with genes, identifying proteins associated with those genes, and assigning function to the genes and proteins. There are databases of scientific literature, such as PubMed; databases on single organisms, such as FlyBase (the Drosophila research database); and databases of protein interactions, such as the General Repository for Interaction Datasets (GRID). In their research, investigators typically access multiple databases (from the several hundred Web-accessible biological databases). Table 3.1 provides examples of key database resources in bioinformatics.
Data organization in biology faces significant challenges for the foreseeable future, given the levels of data being produced. Each year, workshops associated with major conferences in computational biology are held to focus on how to apply new techniques from computer science into computational biology. These include the Intelligent Systems for Molecular Biology (ISMB) Conference and the Conference on Research in Computational Biology (RECOMB), which have championed the cause of creating tools for database development and integration.14 The long-term vision for biology is for a decentralized collection of independent and specialized databases that operate as one large, distributed information resource with common controlled vocabularies, related user interfaces, and practices. Much research will be needed to achieve this vision, but in the short term, researchers will have to make do with more specialized tools for the integration of diverse data types as described in Section 4.2.
What is the technological foundation for managing and organizing data? In 1998, Jeff Ullman noted that “the common characteristic of [traditional business databases] is that they have large amounts of data, but the operations to be performed on the data are simple,” and also that under such circumstances, “the modification of the database scheme is very infrequent, compared to the rate at which queries and other data manipulations are performed.”15
The situation in biology is the reverse. Modern information technologies can handle the volumes of data that characterize 21st century biology, but they are generally inadequate to provide a seamless integration of biological data across multiple databases, and commercial database technology has proven to have many limitations in biological applications.16 For example, although relational databases have often been used for biological data management, they are clumsy and awkward to use in many ways.
TABLE 3.1 Examples of Key Database Resources in Bioinformatics
|
Category |
Databases and URLs |
|
Comprehensive data center: broad content including sequence, structure, function, etc. |
NCBI (National Center for Biotechnology and Information): http://www.ncbi.nlm.nih.gov/ |
|
EBI (European Bioinformatics Institute): http://www.ebi.ac.uk/ |
|
|
European Molecular Biology Laboratory (EMBL): http://www.emblheidelberg.de/ |
|
|
TIGR (the Institute of Genome Research): http://www.tigr.org/ |
|
|
Whitehead/Massachusetts Institute of Technology Genome Center: http://www-genome.wi.mit.edu/ |
|
|
DNA or protein sequence |
GenBank: http://www.ncbi.nlm.nih.gov/Genbank |
|
DDBJ (DNA Data Bank of Japan): http://www.ddbj.nig.acjp/ |
|
|
EMBL Nucleotide Sequence Databank: http://www.ebi.ac.uk/embl/index.html |
|
|
PIR (Protein Information Resource): http://pir.georgetown.edu/ |
|
|
Swiss-Prot: http://www.expasy.ch/sprot/sprot-top.html |
|
|
Biomolecular interactions |
BIND (Biomolecular Interaction Network Database): http://www.blueprint.org/bind/bind.php The contents of BIND include high-throughput data submissions and hand-curated information gathered from the scientific literature. |
|
Genomes: complete genome sequences and related information for specific organisms |
Entrez complete genomes: http://www.ncbi.nlm.nih.gov/Entrez/Genome/org.html |
|
Complete genome at EBI: http://www.ebi.ac.uk/genomes/ |
|
|
University of California, Santa Cruz, Human Genome Working Draft: http://genome.ucsc.edu/ |
|
|
MGD (Mouse Genome Database): http://www.informaticsjax.org/ |
|
|
SGD (Saccharomyces Genome Database): http://genomewww.stanford.edu/Saccharomyces/ |
|
|
FlyBase (a database of the Drosophila genome): http://flybase.bio.indiana.edu/ |
|
|
WormBase (the genome and biology of Caenorhabditis elegans): http://www.wormbase.org/ |
|
|
Genetics: gene mapping, mutations, and diseases |
GDB (Genome Database): http://gdbwww.gdb.org/gdb/ |
|
OMIM (Online Mendelian Inheritance in Man): http://www3.ncbi.nlm.nih.gov/Omim/searchomim.html |
|
|
HGMD (Human Gene Mutation Database): http://archive.uwcm.ac.uk/uwcm/mg/hgmdO.html |
|
Category |
Databases and URLs |
|
Gene expression: microarray and cDNA gene expression |
Unigene: http://www.ncbi.nlm.nih.gov/UniGene/ |
|
dbEST (Expression Sequence Tag Database): http://www.ncbi.nlm.nih.gov/dbEST/index.html |
|
|
BodyMap: http://bodymap.ims.u-tokyo.ac.jp/ |
|
|
GEO (Gene Expression Omnibus): http://www.ncbi.nlm.nih.gov/geo/ |
|
|
Structure: three-dimensional structures of small molecules, proteins, nucleic acids (both RNA and DNA) folding predictions |
PDB (Protein Data Bank): http://www.rcsb.org/pdb/index.html |
|
NDB (Nucleic Acid Database): http://ndbserver.irutgers.edu/NDB/ndb.html |
|
|
CSD (Cambridge Structural Database): http://www.ccdc.cam.ac.uk/prods/csd/csd.html |
|
|
Classification of protein family and protein domains |
SCOP (Structure Classification of Proteins): http://scop.mrc-Imb.cam.ac.uk/scop/ |
|
CATH (Protein Structure Classification Database): http://www.biochem.ucl.ac.uk/bsm/cath-new/index.html |
|
|
Pfam: http://pfam.wustl.edu/ |
|
|
PROSITE database for protein family and domains: http://www.expasy.ch/prosite/ |
|
|
BLOCK: http://www.blocks.fhcrc.org/ |
|
|
Protein pathway Protein-protein interactions and metabolic pathway |
KEGG (Kyoto Encyclopedia of Genes and Genomes): http://www.genome.ad.jp/kegg/kegg2.html#pathway |
|
BIND (Biomolecular Interaction Network Database): http://www.binddb.org/ |
|
|
DIP (Database of Interacting Proteins): http:Hdip.doe-mbi.ucla.edu/ |
|
|
EcoCyc (Encyclopedia of Escherichia coli Genes and Metabolism): http://ecocyc.org/ecocyc/ecocyc.html |
|
|
WIT (Metabolic Pathway): http://Hwit.mcs.anl.gov/WIT2/ |
|
|
Proteomics: proteins, protein family |
AFCS (Alliance for Cellular Signaling): http://cellularsignaling.org/ |
|
JCSG (Joint Center for Structure Genomics): http://www.jcsg.org/scripts/prod/home.html |
|
|
PKR (Protein Kinase Resource): http://pkr.sdsc.edu/html/index.shtml |
|
Category |
Databases and URLs |
|
Pharmacogenomics, pharmaco genetics, single nucleotide polymorphism (SNP), genotyping |
PharmGKB (Pharmacogenetics Knowledge Base): http://pharmgkb.org |
|
SNP Consortium: http://snp.cshl.org |
|
|
dbSNP (Single Nucleotide Polymorphism Database): http://www.ncbi.nlm.nih.gov/SNP/ |
|
|
LocusLink: http://www.ncbi.nlm.nih.gov/LocusLink |
|
|
AFRED (Allele Frequency Database): http://alfred.med.yale.edu/alfred/index.asp |
|
|
CEPH Genotype Database: http://www.cephb.fr/cephdb/ |
|
|
Tissues, organs, and organisms |
Visible Human Project Database: http://www.nlm.nih.gov/research/visible/visible-human.html |
|
BRAID (Brain Image Database): http://Hbraid.rad.jhu.edu/interface.html |
|
|
NeuroDB (Neuroscience Federated Database): http://www.npaci.edu/DICE/Neuro/ |
|
|
The Whole Brain Atlas: http://www.med.harvard.edu/AANLIB/home.html |
|
|
Literature reference |
PubMed MEDLINE: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi |
|
USPTO (U.S. Patent and Trademark Office): http://www.uspto.gov/ |
The size of biological objects is often not constant. More importantly, relational databases presume the existence of well-defined and known relationships between data records, whereas the reality of biological research is that relationships are imprecisely known—and this imprecision cannot be reduced to probabilistic measures of relationship that relational databases can handle.
Jagadish and Olken argue that without specialized life sciences enhancements, commercial relational database technology is cumbersome for constructing and managing biological databases, and most approximate sequence matching, graph queries on biopathways, and three-dimensional shape similarity queries have been performed outside of relational data management systems. Moreover, the relational data model is an inadequate abstraction for representing many kinds of biological data (e.g., pedigrees, taxonomies, maps, metabolic networks, food chains). Box 3.1 provides an illustration of how business database technology can be inadequate.
Object-oriented databases have some advantages over relational databases since the natural foci of study are in fact biological objects. Yet Jagadish and Olken note that object-oriented databases have also had limited success in providing efficient or extensible declarative query languages as required for specialized biological applications.
Because commercial database technology is of limited help, research and development of database technology that serves biological needs will be necessary. Jagadish and Olken provide a view of requirements that will necessitate further advances in data management technology, requirements that include
|
Box 3.1 One purpose of database technology is the creation and maintenance of links between items in different databases. Thus, consider the problem in which a primary biological database of genes contains an object (call it A) that subsequent investigation and research reveal to be two objects. For example, what was thought to be a single gene might upon further study turn out to be two closely linked genes (A1 and A2) with a noncoding region in between (A3). Another database (e.g., a database of clones known to hybridize to various genes) may have contained a link to A—call the clone in question C. Research reveals that it is impossible for C to hybridize to both A1 and A2 individually, but that it does hybridize to the set taken collectively (i.e., A1, A2, and A3). How should this relationship now be represented? Before the new discovery, the link was simple: C to A. Now that new knowledge requires that the primary database (or at least the entry for A) be restructured, how should this new knowledge be reflected in the original simple link? That is, what should one do with links connected to the previously single object, now that that single object has been divided into two? The new information in the primary database has three components, A1, A2, and A3. To which of these, if any, should the original link be attached? If the link is discarded entirely, the database loses the fact that C hybridizes to the collection. If the link from C is now attached to all three equally, that link represents information contrary to fact, since experiment shows that C does not hybridize to both A1 and A2. The necessary relationship that must be reflected calls for the clone entry C to link to A1, A2, and A3 simultaneously but also probabilistically. That is, what must be represented is that the probability of the match in the set of three is one and that the probability of match for two or one in the set is zero. As a general rule, such relationships (i.e., one-to-many relationships that are probabilistic) are not supported by business database technology. However, they are required in scientific databases once this kind of splitting operation has occurred on a hypothetical biological object—and such splitting is commonplace in scientific literature. As indicated, it can occur in the splitting of a gene, or in other cases, it can occur in the splitting of a species on the basis of additional findings on the biology of what was believed to be one species. |
a great diversity of data types: sequences, graphs, three-dimensional structures, images; unconventional types of queries: similarity queries, (e.g., sequence similarity), pattern-matching queries, pattern-finding queries; ubiquitous uncertainty (and sometimes even inconsistency) in the data; data curation (data cleaning and annotation); large-scale data integration (hundreds of databases); detailed data provenance; extensive terminology management; rapid schema evolution; temporal data; and management for a variety of mathematical and statistical models of organisms and biological systems.
Data organization and management present major intellectual challenges in integration and presentation, as discussed in Chapter 4.
3.5 DATA SHARING
There is a reasonably broad consensus among scientists in all fields that reproducibility of findings is central to the scientific enterprise. One key component of reproducibility is thus the availability of data for community examination and inspection. In the words of the National Research Council (NRC) Committee on Responsibilities of Authorship in the Biological Sciences, “an author’s obligation is not
only to release data and materials to enable others to verify or replicate published findings but also to provide them in a form on which other scientists can build with further research.”17
However, in practice, this ethos is not uniformly honored. An old joke in the life science research community comments on data mining in biology—“the data are mine, mine, mine.” For a field whose roots are in empirical description, it is not hard to see the origins of such an attitude. For most of its history, the life sciences research community has granted primary intellectual credit to those who have collected data, a stance that has reinforced the sentiment that those that collect the data are its rightful owners. While some fields such as evolutionary biology generally have an ethos of data sharing, the data-sharing ethos is honored with much less uniformity in many other fields of biology. Requests for data associated with publications are sometimes (even often) denied, ignored, or fulfilled only after long delay or with restrictions that limit how the data may be used.18
The reasons for this state of affairs are multiple. The UPSIDE report called attention to the growing role of the for-profit sector (e.g., the pharmaceutical, biotechnology, research-tool, and bioinformatics companies) in basic and applied research over the last two decades, and the resulting circumstance that increasing amounts of data are developed by and held in private hands. These for-profit entities—whose primary responsibilities are to their investors—hope that their data will provide competitive advantages that can be exploited in the marketplace.
Nor are universities and other nonprofit research institutions immune to commercial pressures. An increasing amount of life sciences research in the nonprofit sector is supported directly by funds from the for-profit sector, thus increasing the prospect of potentially conflicting missions that can impede unrestricted data sharing as nonprofit researchers are caught up in commercial concerns. Universities themselves are encouraged as a matter of public law (the Bayh-Dole Act of 1980) to promote the use, commercialization, and public availability of inventions developed through federally funded research by allowing them to own the rights to patents they obtain on these inventions. University researchers also must confront the publish-or-perish issue. In particular, given the academic premiums on being first to publish, researchers are strongly motivated to take steps that will preserve their own ability to publish follow-up papers or the ability of graduate students, postdoctoral fellows, or junior faculty members to do the same.
Another contributing factor is that the nature of the data in question has changed enormously since the rise of the Human Genome Project. In particular, the enormous volumes of data collected are a continuing resource that can be productively “mined” for a long time and yield many papers. Thus, scientists who have collected such data can understandably view relinquishing control of them as a stiff penalty in light of the time, cost, and effort needed to do the research supporting the first publication.19 Although some communities (notably the genomics, structural biology, and clinical trials communities) have established policies and practices to facilitate data sharing, other communities (e.g., those working in brain imaging or gene and protein expression studies) have not yet done so.
|
17 |
National Research Council, Sharing Publication-Related Data and Materials: Responsibilities of Authorship in the Life Sciences, National Academies Press, Washington, DC, 2003. Hereafter referred to as the UPSIDE report. Much of the discussion in Section 3.5 is based on material found in that report. |
|
18 |
For example, a 2002 survey of geneticists and other life scientists at 100 U.S. universities found that of geneticists who had asked other academic faculty for additional information, data, or materials regarding published research, 47 percent reported that at least one of their requests had been denied in the preceding 3 years. Twelve percent of geneticists themselves acknowledged denying a request from another academic researcher. See E.G. Campbell, B.R. Clarridge, M. Gokhale, L. Birenbaum, S. Hilgartner, N.A. Holtzen, and D. Blumenthal, “Data Withholding in Academic Genetics: Evidence from a National Survey,” Journal of the American Medical Association 287(4):473-480, 2002. (Cited in the UPSIDE report; see Footnote 17.) |
|
19 |
Data provenance (the concurrent identification of the source of data along with the data itself as discussed in Section 3.7) has an impact on the social motivation to share data. If data sources are always associated with data, any work based on that data will automatically have a link to the original source; hence proper acknowledgment of intellectual credit will always be possible. Without automated data provenance, it is all too easy for subsequent researchers to lose the connection to the original source. |
Finally, raw biological data are not the only commodities in question. Computational tools and models are increasingly the subject of publication in the life sciences (see Chapters 4 and 5), and it is inevitable that similar pressures will arise (indeed, have arisen) with respect to sharing the software and algorithms that underlie these artifacts. When software is at issue, a common concern is that the release of software—especially if it is released in source code—can enable another party to commercialize that code. Some have also argued that mandatory sharing of source code prevents universities from exercising their legal right to develop commercial products from federally funded research.
Considering these matters, the NRC Committee on Responsibilities of Authorship in the Biological Sciences concluded:
The act of publishing is a quid pro quo in which authors receive credit and acknowledgment in exchange for disclosure of their scientific findings. All members of the scientific community—whether working in academia, government, or a commercial enterprise—have equal responsibility for upholding community standards as participants in the publication system, and all should be equally able to derive benefits from it.
The UPSIDE report also explicated three principles associated with sharing publication-related data and software:20
-
Authors should include in their publications the data, algorithms, or other information that is central or integral to the publication—that is, whatever is necessary to support the major claims of the paper and would enable one skilled in the art to verify or replicate the claims.
-
If central or integral information cannot be included in the publication for practical reasons (for example, because a dataset is too large), it should be made freely (without restriction on its use for research purposes and at no cost) and readily accessible through other means (for example, on line). Moreover, when necessary to enable further research, integral information should be made available in a form that enables it to be manipulated, analyzed, and combined with other scientific data…. [However, m]aking data that is central or integral to a paper freely obtainable does not obligate an author to curate and update it. While the published data should remain freely accessible, an author might make available an improved, curated version of the database that is supported by user fees. Alternatively, a value-added database could be licensed commercially.
-
If publicly accessible repositories for data have been agreed on by a community of researchers and are in general use, the relevant data should be deposited in one of these repositories by the time of publication…. [T]hese repositories help define consistent policies of data format and content, as well as accessibility to the scientific community. The pooling of data into a common format is not only for the purpose of consistency and accessibility. It also allows investigators to manipulate and compare datasets, synthesize new datasets, and gain novel insights that advance science.
When a publication explicitly involves software or algorithms to solve biological problems, the UPSIDE report pointed out that the principle enunciated for data should also apply: software or algorithms that are central or integral to a publication “should be made available in a manner that enables its use for replication, verification, and furtherance of science.” The report also noted that one option is to provide in the publication a detailed description of the algorithm and its parameters. A second option is to make the relevant source code available to investigators who wish to test it, and either option upholds the spirit of the researcher’s obligation.
Since the UPSIDE report was released in 2003, editors at two major life science journals, Science and Nature, have agreed in principle with the idea that publication entails a responsibility to make data freely available to the larger research community.21 Nevertheless, it remains to be seen how widely the UPSIDE principles will be adopted in practice.
As for the technology to facilitate the sharing of data and models, the state of the art today is that even when the will to share is present, data or model exchange between researchers is generally a nontrivial exercise. Data and models from one laboratory or researcher must be accompanied by enough metadata that other researchers can query the data and use the model in meaningful ways without a lot of unproductive overhead in “futzing around doing stupid things.” Technical dimensions of this point are discussed further in Section 4.2.
3.6 DATA INTEGRATION
As noted in Chapter 2, data are the sine qua non of biological science. The ability to share data widely increases the utility of those data to the research community and enables a higher degree of communication between researchers, laboratories, and even different subfields. Data incompatibilities can make data hard to integrate and to relate to information on other variables relevant to the same biological system. Further, when inquiries can be made across large numbers of databases, there is an increased likelihood that meaningful answers can be found. Large-scale data integration also has the salutary virtue that it can uncover inconsistencies and errors in data that are collected in disparate ways.
In digital form, all biological data are represented as bits, which are the underlying electronic representation of data. However, for these data to be useful, they must be interpretable according to some definitions. When there is a single point of responsibility for data management, the definitions are relatively easy to generate. When responsibility is distributed over multiple parties, they must agree on those definitions if the data of one party are to be electronically useful to another party. In other words, merely providing data in digital form does not necessarily mean that they can be shared readily—the semantics of differing data sets must be compatible as well.
Another complicating factor is the fact that nearly all databases—regardless of scale—have their origins in small-scale experimentation. Researchers almost always obtain relatively small amounts of data in their first attempts at experimentation. Small amounts of data can usually be managed in flat files—typically, spreadsheets. Flat files have the major advantage that they are quick and easy to implement and serve small-scale data management needs quite well.
However, flat files are generally impractical for large amounts of data. For example, queries involving multiple search criteria are hard to make when a flat-file database is involved. Relationships between entries are concealed in a flat-file format. Also, flat files are quite poor for handling heterogeneous data types.
There are a number of technologies and approaches, described below, that address such issues. In practice, however, the researcher is faced with the problem of knowing when to abandon the small-scale flat file in favor of a more capable and technically sophisticated arrangement that will inevitably entail higher overhead, at least initially.
The problem of large-scale data integration is extraordinarily complex and difficult to solve. In 2003, Lincoln Stein noted that “life would be much simpler if there was a single biological database, but this would be a poor solution. The diverse databases reflect the expertise and interests of the groups that maintain them. A single database would reflect a series of compromises that would ultimately impoverish the information resources that are available to the scientific community. A better solution would maintain the scientific and political independence of the databases, but allow the information that they contain to be easily integrated to enable cross-database queries. Unfortunately, this is not trivial.”22
Consider, for example, what might be regarded as a straightforward problem—that of keeping straight vocabularies and terminologies and their associated concepts. In reality, when new biological structures, entities, and events have been uncovered in a particular biological context, they are often
described with novel terminology or measurements that do not reveal much about how they might be related to similar entities in other contexts or how they quantitatively function in the contexts in which they exist, for example:
-
Biological concepts may clash as users move from one database to another. Stein discusses several examples:23
-
To some research communities, “a pseudogene is a gene-like structure that contains in-frame stop codons or evidence of reverse transcription. To others, the definition of a pseudogene is expanded to include gene structures that contain full open reading frames (ORFs) but are not transcribed. Some members of the Neisseria gonorrhea research community, meanwhile, use pseudogene to mean a transposable cassette that is rearranged in the course of antigenic variation.”
-
“The human genetics community uses the term allele to refer to any genomic variant, including silent nucleotide polymorphisms that lie outside of genes, whereas members of many model-organism communities prefer to reserve the term allele to refer to variants that change genes.”
-
“Even the concept of the gene itself can mean radically different things to different research communities. Some researchers treat the gene as the transcriptional unit itself, whereas others extend this definition to include up- and downstream regulatory elements, and still others use the classical definitions of cistron and genetic complementation.”
-
-
Evolving scientific understandings may drive changes in terminology. For example, diabetes was once divided into the categories of juvenile and adult onset. As the role of insulin became clearer, the relevant categories evolved into “insulin dependent” and “non-insulin dependent.” The relationship is that almost all juvenile cases of diabetes are insulin dependent, but a significant fraction of adult-onset cases are as well.
-
Names of the same biological object may change across databases. “For example, consider the DNA-damage checkpoint-pathway gene that is named Rad24 in Saccharomyces cerevisiae (budding yeast). [Schizo]saccharomyces pombe (fission yeast) also has a gene named rad24 that is involved in the checkpoint pathway, but it is not the orthologue of the S. cerevisiae Rad24. Instead, the correct S. pombe orthologue is rad17, which is not to be confused with the similarly named Rad17 gene in S. cerevisiae. Meanwhile, the human checkpoint-pathway genes are sometimes named after the S. cerevisiae orthologues, sometimes after the S. pombe orthologues, and sometimes have independently derived names. In C. elegans, there are a series of rad genes, none of which is orthologous to S. cerevisiae Rad17. The closest C. elegans match to Rad17 is, in fact, a DNA-repair gene named mrt-2.”24
-
Implicit meanings can be counterintuitive. For example, the International Classification of Disease (ICD) code for “angina” means “angina occurring in the past.”25 A condition of current angina is indicated by the code for “chest pain not otherwise specified.”
-
Data transformations from one database to another may destroy useful information. For example, a clinical order in a hospital may call for a “PA [posterior-anterior] and lateral chest X-ray.” When that order is reflected in billing, it may be collapsed into “chest X-ray: 2 views.”
-
Metadata may change when databases originally created for different purposes are conceptually joined. For example, MEDLINE was developed to facilitate access to the printed paper literature by
|
23 |
Reprinted by permission from L.D. Stein, “Integrating Biological Databases,” Nature Reviews Genetics 4(5):337-345, 2003. Copyright 2005 Macmillan Magazines Ltd. |
|
24 |
Reprinted by permission from L.D. Stein, “Integrating Biological Databases,” Nature Reviews Genetics 4(5):337-345, 2003. Copyright 2005 Macmillan Magazines Ltd. |
|
25 |
ICD codes refer to a standard international classification of diseases. For more information, see http://www.cdc.gov/nchs/about/otheract/icd9/abticd9.htm. |
-
scientists. The data were assembled in MEDLINE to help users find citations. As a result, authors in MEDLINE were originally treated as text strings, not as people. There was no effort, to identify individual people, so “Smith, J” could be John Smith, Jim Smith, or Joan Smith. However, the name of an individual is not necessarily constant over his or her professional lifetime. Thus, one cannot use MEDLINE to search for all papers authored by an individual who has undergone a name change without independent knowledge of the specifics of that change.
Experience suggests that left to their own devices, designers of individual databases generally make locally optimal decisions about data definitions and formats for entirely rational reasons, and local decisions are almost certain to be incompatible in some ways with other such decisions made in other laboratories by other researchers.26 Nearly 10 years ago, Robbins noted that “a crisis occurred in the [biological] databases in the mid 1980s, when the data flow began to outstrip the ability of the database to keep up. A conceptual change in the relationship of databases to the scientific community, coupled with technical advances, solved the problem…. Now we face a data-integration crisis of the 1990s. Even if the various separate databases each keep up with the flow of data, there will still be a tremendous backlog in the integration of information in them. The implication is similar to that of the 1980s: either a solution will soon emerge or biological databases collectively will experience a massive failure.”27Box 3.2 describes some of the ways in which community-wide use of biological databases continues to be difficult today.
Two examples of research areas requiring a large degree of data integration are cellular modeling and pharmacogenomics. In cellular modeling (discussed further in Section 5.4.2), researchers need to integrate the plethora of data available today about cellular function; such information includes the chemical, electrical, and regulatory features of cells; their internal pathways; mechanisms of cell motility; cell shape changes; and cell division. Box 3.3 provides an example of a cell-oriented database. In pharmacogenomics (the study of how an individual’s genetic makeup affects his or her specific reaction to drugs, discussed in Section 9.7), databases must integrate data on clinical phenotypes (including both pharmacokinetic and pharmacodynamic data) and profiles (e.g., pulmonary, cardiac, and psychological function tests, and cancer chemotherapeutic side effects); DNA sequence data, gene structure, and polymorphisms in sequence (and information to track haploid, diploid, or polyploid alleles, alternative splice sites, and polymorphisms observed as common variants); molecular and cellular phenotype data (e.g., enzyme kinetic measurements); pharmacodynamic assays; cellular drug processing rates; and homology modeling of three-dimensional structures. Box 3.4 illustrates the Pharmacogenetics Research Network and Knowledge Base (PharmGKB), an important database for pharmacogenetics and pharmacogenomics.
3.7 DATA CURATION AND PROVENANCE28
Biological research is a fast-paced, quickly evolving discipline, and data sources evolve with it: new experimental techniques produce more and different types of data, requiring database structures to change accordingly; applications and queries written to access the original version of the schema must
|
26 |
In particular, a scientist working on the cutting edge of a problem almost certainly requires data representations and models with more subtlety and more degrees of resolution in the data relevant to the problem than someone who has only a passing interest in that field. Almost every dataset collected has a lot of subtlety in some areas of the data model and less subtlety elsewhere. Merging these datasets into a common-denominator model risks throwing away the subtlety, where much of the value resides. Yet, merging these datasets into a uniformly data-rich model results in a database so rich that it is not particularly useful for general use. An example—biomedical databases for human beings may well include coding for gender as a variable. However, in a laboratory or medical facility that does a lot of work on transgendered individuals who may have undergone sex-change operations, the notion of gender is not necessarily as simple as “male” or “female.” |
|
27 |
R.J. Robbins, “Comparative Genomics: A New Integrative Biology,” in Integrative Approaches to Molecular Biology, J. Collado-Vides, B. Magasanik, and T.F. Smith, eds., MIT Press, Cambridge, MA, 1996. |
|
28 |
Section 3.7 embeds excerpts from S.Y. Chung and J.C. Wooley, “Challenges Faced in the Integration of Biological Information,” Bioinformatics: Managing Scientific Data, Z. Lacroix and T. Critchlow, eds., Morgan Kaufmann, San Francisco, CA, 2003. |
|
Box 3.2 Biological databases have several characteristics that make them particularly difficult to use by the community at large. Biological databases are
SOURCE: Derived largely from S.Y. Chung and J.C. Wooley, “Challenges Faced in the Integration of Biological Information,” Bioinformatics: Managing Scientific Data, Z. Lacroix and T. Critchlow, eds., Morgan Kaufmann, San Francisco, CA, 2003. |
|
Box 3.3 The Alliance for Cellular Signaling (AfCS), partly supported by the National Institute of General Medical Sciences and partly by large pharmaceutical companies, seeks to build a publicly accessible, comprehensive database on cellular signaling that makes available virtually all significant information about molecules of interest. This database will also be one enabler for pathway analysis and facilitate an understanding of how molecules coordinate with one another during cellular responses. The database seeks to identify all of the proteins that constitute the various signaling systems, assess time-dependent information flow through the systems in both normal and pathological states, and reduce the mass of detailed data into a set of interacting theoretical models that describe cellular signaling. To the maximum extent possible, the information contained in the database is intended to be machine-readable. The complete database is intended to enable researchers to:
The AFCS is motivated by a desire to understand as completely as possible the relationships between sets of inputs and outputs in signaling cells that vary both temporally and spatially. Yet because there are many researchers engaged in signaling research, the cultural challenge faced by the alliance is the fact that information in the database is collected by multiple researchers in different laboratories and from different organizations. Today, it involves more than 50 investigators from 20 academic and industrial institutions. However, as of this writing, it is reported that the NIGMS will reduce funding sharply for the Alliance following a mid-project review in early 2005 (see Z. Merali and J. Giles, “Databases in Peril,” Nature 435:1010-1011, 23 June 2005). |
be rewritten to match the new version. Incremental updates to data warehouses (as opposed to wholesale rebuilding of the warehouse from scratch) are difficult to accomplish efficiently, particularly when complex transformations or aggregations are involved.
A most important point is that most broadly useful databases contain both raw data and data that are either the result of analysis or derived from other databases. In this environment, databases become interdependent. Errors due to data acquisition and handling in one database can be propagated quickly into other databases. Data updated in one database may not be propagated immediately to related databases.
Thus, data curation is essential. Curation is the process through which the community of users can have confidence in the data on which they rely. So that these data can have enduring value, information related to curation must itself be stored within the database; such information is generally categorized as annotation data. Data provenance and data accuracy are central concerns, because the distinctions between primary data generated experimentally, data generated through the application of scientific
|
Box 3.4 Supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health, the Pharmacogenetics Research Network and Knowledge Base (PharmGKB) is intended as a national resource containing high-quality structured data linking genomic information, molecular and cellular phenotype information, and clinical phenotype information. The ultimate aim of this project is to produce a knowledge base that provides a public infrastructure for understanding how variations in the human genome lead to variations in clinical response to medications. Sample inquiries to this database might include the following:
Such queries require a database that can model key elements of the data, acquire data efficiently, provide query tools for analysis, and deliver the resulting system to the scientific community. A central challenge for PharmGKB is that data contained it must be cross-referenced and integrated with a variety of other Web-accessible databases. Thus, PharmGKB provides mechanisms for surveillance of and integration with these databases, allowing users to submit one query with the assurance that other relevant databases are being accessed at the same time. For example, PharmGKB monitors dbSNP, the National Center for BioTechnology Information (NCBI)-supported repository for single nucleotide polymorphisms and short deletion and insertion polymorphisms. These monitoring operations search for new information about the genes of interest to the various research groups associated with the Pharmacogenetics Research Network. In addition, PharmGKB provides users with a tool for comparative genomic analysis between human and mouse that focuses on long-range regulatory elements. Such elements can be difficult to find experimentally, but are often conserved in syntenic regions between mice and humans, and may be useful in focusing polymorphism studies on noncoding areas that are more likely to be associated with detectable phenotypes. Another important issue for the PharmGKB database is that because it contains clinical data derived from individual patients, it must have functionality that enforces the rights of those individuals to privacy and confidentiality. Thus, data flow must be limited both into and out of the knowledge base, based on evolving rules defining what can be stored in PharmGKB and what can be disseminated. No identifying information about an individual patient can be accepted into the knowledge base, and the data must be “massaged” so that patient identity cannot be reconstructed from publicly available data records. |
analysis programs, and data derived from database searches are blurred. Users of databases containing these kinds of data must be concerned about where the data come from and how they are generated. A database may be a potentially rich information resource, but its value is diminished if it fails to keep an adequate description of the provenance of the data it contains.29 Although proponents of online access
|
29 |
P. Buneman, S. Khanna, and W.C. Tan, “Why and Where: A Characterization of Data Provenance,” 8th International Conference on Database Theory (ICDT), pp. 316-330, 2001. Cited in Chung and Wooley, “Challenges Faced in the Integration of Biological Information,” 2003, Footnote 28. |
|
PharmGKB integrates data on clinical phenotypes (including both pharmacokinetic and pharmacodynamic data) and profiles (e.g., pulmonary, cardiac, and psychological function tests; cancer chemotherapeutic side effects), DNA sequence data, gene structure, and polymorphisms in sequence (and information to track haploid, diploid, or polyploid alleles; alternative splice sites; and polymorphisms observed as common variants), molecular and cellular phenotype data (e.g., enzyme kinetic measurements), pharmacodynamic assays, cellular drug processing rates, and homology modeling of three-dimensional structures. Figure 3.4.1 illustrates the complex relationships that are of interest for this knowledge base. 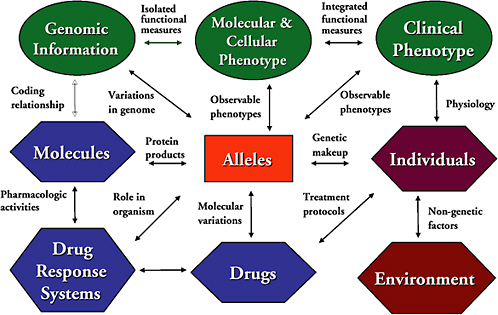 FIGURE 3.4.1 Complexity of relationships in pharmacogenetics. SOURCE: Figure reprinted and text adapted by permission from T.E. Klein, J.T. Chang, M.K. Cho, K.L. Easton, R. Fergerson, M. Hewett, Z. Lin, Y. Liu, S. Liu, D.E. Oliver, D.L. Rubin, F. Shafa, J.M. Stuart, and R.B. Altman, “Integrating Genotype and Phenotype Information: An Overview of the PharmGKB Project,” The Pharmacogenomics Journal 1:167-170, 2001. Copyright 2001 Macmillan Publishers Ltd. |
to databases frequently tout it as an advantage that “the user does not need to know where the data came from or where the data are located,” in fact it is essential for quality assurance reasons that the user be able to ascertain the source of all data accessed in such databases.
Data provenance addresses questions such as the following: Where did the characterization of a given GenBank sequence originate? Has an inaccurate legacy annotation been “transitively” propagated to similar sequences? What is the evidence for this annotation?
A complete record of a datum’s history presents interesting intellectual questions. For example, it is difficult to justify filling a database with errata notices correcting simple errors when the actual entries
can be updated. However, the original data themselves might be important, because subsequent research might have been based on them. One view is that once released, electronic database entries, like the pages of a printed journal, must stand for all time in their original condition, with errors and corrections noted only by the additional publication of errata and commentaries. However, this might quickly lead to a situation in which commentary outweighs original entries severalfold. On the other hand, occasional efforts to “improve” individual entries might inadvertently result in important information being mistakenly expunged. A middle ground might be to require that individual released entries be stable, no matter what the type of error, but that change entries be classified into different types (correction of data entry error, resubmission by original author, correction by different author, etc.), thus allowing the user to set filters to determine whether to retrieve all entries or just the most recent entry of a particular type.
To illustrate the need for provenance, consider that the output of a program used for scientific analysis is often highly sensitive to the parameters used and the specifics of the input datasets. In the case of genomic analysis, a finding that two sequences are “similar” or not may depend on the specific algorithms used and the different cutoff values used to parameterize matching algorithms, in which case other evidence is needed. Furthermore, biological conclusions derived by inference in one database will be propagated and may no longer be reliable after numerous transitive assertions. Repeated transitive assertions inevitably degrade data, whether the assertion is a transitive inference or the result of a simple “join” operation. In the absence of data perfection, additional degradation occurs with each connection.
For a new sequence that does not match any known sequence, gene prediction programs can be used to identify open reading frames, to translate DNA sequence into protein sequence, and to characterize promoter and regulatory sequence motifs. Gene prediction programs are also parameter-dependent, and the specifics of parameter settings must be retained if a future user is to make sense of the results stored in the database.
Neuroscience provides a good example of the need for data provenance. Consider the response of rat cortical cells to various stimuli. In addition to the “primary” data themselves—that is, voltages as a function of time—it is also important to record information about the rat: where the rat came from, how the rat was killed, how the brain was extracted, how the neurological preparation was made, what buffers were present, the temperature of the preparation, how much time elapsed between the sacrifice of the rat and the actual experiment being done, and so on. While all of this “extra” information seems irrelevant to the primary question, neuroscience has not advanced to the point where it is known which of these variables might have an effect on the response of interest—that is, on the evoked cortical potential.
Box 3.5 provides two examples of well-characterized and well-curated data repositories.
Finally, how far curation can be carried is an open question. The point of curation is to provide reliable and trustworthy data—what might be called biological truths. But the meaning of such “truths” may well change as more data is collected and more observations are made—suggesting a growing burden of constant editing to achieve accuracy and internal consistency. Indeed, every new entry in the database would necessarily trigger extensive validity checks of all existing entries individually and perhaps even for entries taken more than one at a time. Moreover, assertions about the real world may be initially believed, then rejected, then accepted again, albeit in a modified form. Catastrophism in geology is an example. Thus, maintaining a database of all biological truths would be an editorial nightmare, if not an outright impossibility—and thus the scope of any single database will necessarily be limited.
A database of biological observations and experimental results provides different challenges. An individual datum or result is a stand-alone contribution. Each datum or result has a recognized party responsible for it, and inclusion in the database means that it has been subject to some form of editorial review, which presumably assures its adherence to current scientific practices (and does not guarantee
|
Box 3.5 GenBank GenBank is a public database of all known nucleotide and protein sequences, distributed by the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine (NLM). As of January 2003, GenBank contained over 20 billion nucleotide bases in sequences from more than 55,000 species—human, mice, rat, nematode, fruit fly, and the model plant Arabidopsis are the most represented. GenBank and its collaborating European (EMBL) and Japanese (JPPL) databases are built with data submitted electronically by individual investigators (using BankIt or Sequin submission programs) and large-scale sequencing centers (using batch procedures). Each submission is reviewed for quality assurance and assigned an accession number; sequence updates are designated as new versions. The database is organized by a sequence-based taxonomy into divisions (e.g., bacteria, viruses, primates) and categories (e.g., expressed sequence tags, genome survey sequences, high-throughput genomic data). GenBank makes available derivative databases, for example of putative new genes, from these data. Investigators use the Entrez retrieval system for cross-database searching of GenBank’s collections of DNA, protein, and genome mapping sequence data, population sets, the NCBI taxonomy, protein structures from the Molecular Modeling Database (MMDB), and MEDLINE references (from the scientific literature). A popular tool is BLAST, the sequence alignment program, for finding GenBank sequences similar to a query sequence. The entire database is available by anonymous FTP in compressed flat-file format, updated every 2 months. NCBI offers its ToolKit to software developers creating their own interfaces and specialized analytical tools. The Research Resource for Complex Physiologic Signals The Research Resource for Complex Physiologic Signals was established by the National Center for Research Resources of the National Institutes of Health to support the study of complex biomedical signals. The creation of this three-part resource (PhysioBank, PhysioToolkit, and PhysioNet) overcomes long-standing barriers to hypothesis-testing research in this field by enabling access to validated, standardized data and software.1 PhysioBank comprises databases of multiparameter, cardiopulmonary, neural, and other biomedical signals from healthy subjects and patients with pathologies such as epilepsy, congestive heart failure, sleep apnea, and sudden cardiac death. In addition to fully characterized, multiply reviewed signal data, PhysioBank provides online access to archival data that underpin results reported in the published literature, significantly extending the contribution of that published work. PhysioBank provides theoreticians and software developers with realistic data with which to test new algorithms. The PhysioToolkit includes software for the detection of physiologically significant events using both classic methods and novel techniques from statistical physics, fractal scaling analysis, and nonlinear dynamics; the analysis of nonstationary processes; interactive display and characterization of signals; the simulation of physiological and other signals; and the quantitative evaluation and comparison of analysis algorithms. PhysioNet is an online forum for the dissemination and exchange of recorded biomedical signals and the software for analyzing such signals; it provides facilities for the cooperative analysis of data and the evaluation of proposed new algorithms. The database is available at http://www.physionet.org/physiobank. |
its absolute truth value). Without the existence of databases with differing editorial policies, some important but iconoclastic data or results might never be published. On the other hand, there is no guarantee of consistency among these data and results, which means that progress at the frontiers will depend on expert judgment in deciding which data and results will constitute the foundation from which to build.
In short, reconciling the tension between truth and diversity—both desirable, but for different reasons—is implicitly a part of the construction of every large-scale database.






















