3
Advances in Technologies with Relevance to Biology: The Future Landscape
This chapter provides an overview and a perspective on the breadth and types of technologies that may have an impact on the life sciences enterprise of the future, with the understanding that there are inherent difficulties in anticipating or predicting how any of these technologies alone or in combination will alter the nature of the future threat “landscape.”
Rather than attempt to cover the technology landscape in a comprehensive manner, this chapter (1) highlights technologies likely to have obvious or high-impact near-term consequences; (2) illustrates the general principles by which technological growth alters the nature of future biological threats; and, (3) highlights how and why some technologies are complementary or synergistic in bolstering defense against future threats while also enhancing or altering the nature of future threats.
There is immense diversity and rapid evolution of technologies with relevance to (or impact on) the life sciences enterprise. Their impact(s) may be beneficial or detrimental depending on how these tools and technologies are applied. Some may be seen as “coming out of left field”; that is, these technologies may have very different applications from those originally intended, or may be combined in unexpected, nontraditional configurations. The combination of nanotechnology and biotechnology is one such example of a synergistic combination.
Many of the technologies discussed in this chapter create novel opportunities for scientists (and others) to explore aspects of biological and chemical diversity that cannot be accessed through natural mechanisms
or processes. Given the unpredictable nature of technological change, it is difficult if not impossible to describe in definite terms what the global technology landscape will look like in 5 to 10 years, both with regard to the emergence of technologies with dual-use applications and the global geography of future breakthroughs. New, unexpected discoveries and technological applications in RNAi and synthetic biology arose even during the course of deliberations by this committee. If this report, with the same charge, were prepared even a year or two in the future, many of the details presented in this chapter would likely be different.
A CLASSIFICATION SCHEME FOR BIOLOGICAL TECHNOLOGIES
Despite the seemingly disparate and scattered goals of recent advances in life sciences technologies, the committee concluded that there are classes or categories of advances that share important features. These shared characteristics are based on common purposes, common conceptual underpinnings, and common technical enabling platforms. Thus, the technologies outlined in this chapter are categorized according to a classification scheme devised by the committee and organized around four groupings:
-
Acquisition of novel biological or molecular diversity. These are technologies driven by efforts to acquire or synthesize novel biological or molecular diversity, or a greater range of specificity, so that the user can then select what is useful from the large, newly-acquired diversity pool. The goal is to create collections of molecules with greater breadth of diversity than found so far in nature, as well as with types of diversity that may not exist in nature. The kinds of molecules that might be generated include, for example, enzymes with enhanced or altered activities, as well as molecules composed of “unnatural” amino acids. Technologies in this category include those dedicated toward DNA synthesis; the generation of new chemical diversity (i.e., through combinatorial chemistry); those that create novel DNA molecules (from genes to genomes) using directed in vitro molecular evolution (e.g., “DNA shuffling”1); and those that amplify or simply collect previously uncharacterized sequences (genomes) directly from nature (i.e., bioprospecting). All of these technologies require a subsequent selection step, such that molecules, macromolecular complexes, or even microbes with the desired properties can be identified and isolated from a large and very diverse pool of possibilities. Toward this end, new high-throughput screening (including the use of robotics and advanced information management systems) have become critical enabling technologies.
-
Directed design. These are technologies that involve deliberate efforts to generate novel but predetermined and specific biological or molecular diversity. The use of these technologies begins with a more defined, preexisting understanding of the desired endproduct and its molecular features. One then synthesizes or re-engineers the desired product or its components. Examples include but are not limited to rational, structure-aided design of small-molecule ligands; the genetic engineering of viruses or microbes; and, the emerging field of “synthetic biology.”
-
Understanding and manipulation of biological systems. These are technologies driven by efforts to gain a more complete understanding of complex biological systems and an ability to manipulate such systems. Examples include “systems biology”; gene silencing (e.g., RNA interference); the generation of novel binding (affinity) reagents; technologies focused on developmental programs (e.g., embryonic stem cells); genomics and genomic medicine; the study of modulators of homeostatic systems; bioinformatics; and, advanced network theory.
-
Production, delivery, and “packaging.” These are technologies driven by efforts in the pharmaceutical, agriculture, and healthcare sectors to improve capabilities for producing, reengineering, or delivering biological or biology-derived products and miniaturizing these processes. Examples include the use of transgenic plants as production platforms, aerosol technology, microencapsulation, microfluidics/microfabrication; nanotechnology; and, gene therapy technology. [Some of these technologies are related to the manipulation of biological systems—e.g., nanotechnology—and may also be applied to the generation (category 1) or design (category 2) of novel biological diversity or to the analysis of complex biological systems (category 3).]
The classification scheme serves several important purposes. It:
-
highlights commonalities among technologies and, by so doing, draws attention to critical enabling features;
-
provides insight into some of the technical drivers behind biology-related technology;
-
facilitates predictions about future emerging technologies; and,
-
lends insight into the basis for complementarities or synergies among technologies and, as such, facilitates the analysis of interactions that lead to either beneficial or potentially malevolent ends.
Limitations of the classification scheme include the fact that it is based on a relatively small number of relevant technologies (i.e., the committee’s
list of technologies may be biased and is inevitably incomplete) and the acknowledgment that there are many ways to categorize these technologies. As a reflection of the latter dilemma, the committee found that some of the technologies discussed in this chapter could have been classified in more than one category. The category assignment in these cases was guided by the nature of the particular applications that the committee had in mind when considering each of the relevant technologies.
The examples below serve as a finite set of future technologies that represent and illustrate each of the four categories. For each example the following issues are addressed: the purpose of the technology, its current state of the art, and future applications. The coverage of these issues for each of the technologies is not intended to be exhaustive. The technologies covered in this chapter include not only those that open up new possibilities for the creation of novel or enhanced biological agents but also those that expose new vulnerabilities (i.e., targets for biological attack). Details are limited to those necessary for a clear explanation of the plausibility of use.
1. ACQUISITION OF NOVEL BIOLOGICAL OR MOLECULAR DIVERSITY
Given the clear capability of at least some microbes and viruses to evolve quickly, acquire new genes, and alter their behavior, it might seem reasonable that over hundreds of thousands of years all conceivable biological agents have been “built” and “tested” and that the agents seen today are the most “successful” of these. Thus, is there any reason to think that it might be possible to create a more successful biological agent? Possibly not, but it is important to understand that “successful” in this context means the most able to survive within, on, or near human populations over time. “Success” does not necessarily equate with virulence or pathogenicity, the ability to cause disease or injury.
The kinds of basic biological diversity found in nature today, or those that have potentially evolved in the natural world and been tested for fitness over time, may have been (and are still) limited by certain natural constraints, including available building blocks—nucleotides and amino acids; natural mechanisms for generating genetic diversity; and, the strength and nature of selective pressures over time. Nor has there been enough time over the history of the earth for nature to have explored more than a tiny fraction of the diversity that is possible.2 The technologies described in this section are those that seek to create a much wider and deeper set of diverse biological molecules, many of which may never have been generated or given a fair chance for succeeding in nature (although success may be defined in different ways).3
Techniques have been developed to expand both the diversity of nucleotide or amino acid sequences of nucleic acids or proteins, respectively (which in both cases ultimately hold the information specifying the folding and thus the conformation of biologically active molecules), or for creating a diversity of small molecules with different shapes, sizes, and charge characteristics. In addition, some investigators are creating unnatural nucleic acids and amino acids in order to test and explore possible structural constraints on molecules with biological function. All of these approaches result in novel types of genetic or molecular diversity that then require assessment of functional potential. This assessment typically takes the form of a screening process (i.e., deliberate examination of all molecules for a desired feature or function) or a selective process (i.e., one that imposes a selective advantage on those molecules that have a property of interest). While the technological processes of assessing and selecting molecules of interest—high-throughput screening and selection—have some features in common with the next category of technologies (i.e., directed design), they are included in this first category because of their critical enabling role in the exploration of molecular and biological diversity.
DNA Synthesis
Description
DNA synthesis is a technology that enables the de novo generation of genetic sequences that specifically program cells for the expression of a given protein. It is not new, but technical enhancements continue to increase the speed, ease, and accuracy with which larger and larger sequences can be generated chemically. By the early 1970s, scientists had demonstrated that they could engineer synthetic genes.4 However, it was the automation of de novo DNA synthesis and the development of the polymerase chain reaction (PCR) in the early 1980s that spawned the development of a series of cascading methodologies for the analysis of gene expression, structure, and function. Our ability to synthesize short oligonucleotides (typically 10 to 80 base pairs in length) rapidly and accurately has been an essential enabling technology for a myriad of advances, not the least of which has been the sequencing of the human genome.
The past few years have seen remarkable technological advances in this field, particularly with respect to the de novo synthesis of increasingly longer DNA constructs. The chemical synthesis and ligation of large segments of a DNA template, followed by enzymatic transcription of RNA led to the de novo creation of the poliovirus genome in 2002 (about 7,500 nucleotides in length), from which the infectious, virulent virus was res-
cued following its transfection into permissive cells.5 The following year scientists announced the successful assembly of a bacterial virus genome.6 Parallel efforts in industry and academia led to the synthesis and assembly of large segments of the hepatitis C virus genome, from which replication competent RNA molecules were rescued. These studies raised concerns in the media that larger, more complex organisms, such as the smallpox virus (which is approximately 186,000 base pairs long), might be within reach.7
DNA synthesis technology is currently limited by the cost and time involved to create long DNA constructs of high fidelity as well as by its high error rate. Current estimates for generating even simple oligonucleotides are at least $0.10 per base (including synthesis of the oligonucleotides plus error correction).8 See Figure 3-1.
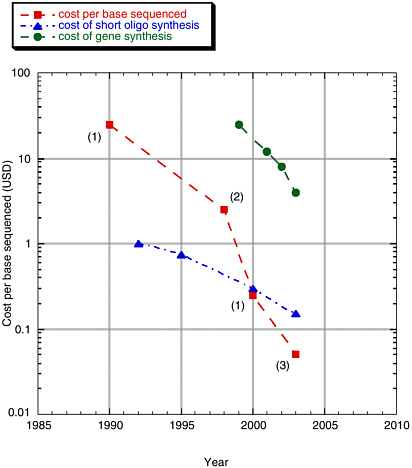
FIGURE 3-1 Cost per base of sequencing and synthesis.
SOURCE: Rob Carlson presentation to the committee, February 2004.
Current State of the Art
Several recent studies have demonstrated important steps toward making gene synthesis readily affordable and accessible to researchers with small budgets, by decreasing its cost and improving its error rate.9 For example, in December 2004, as this committee deliberated its charge, scientists described a new microchip-based technology for the semiautomated multiplex synthesis of long oligonucleotides.10 The researchers used the new technology to synthesize all 21 genes that encode proteins of the E. coli 30S ribosomal subunit. Almost simultaneously, another research group described a novel approach for reducing errors by more than 15-fold compared to conventional gene synthesis techniques, yielding DNA with one error per 10,000 base pairs.11
Future Applications
Developments in DNA synthetic capacity have generated strong interest in the fabrication of increasingly larger constructs, including genetic circuitry,12 the engineering of entire biochemical pathways,13 and, as mentioned above, the construction of small genomes.14 As a specific example of a potential future beneficial application of DNA synthesis, one research group has described a method for synthesizing terpenoid, a natural product used in commercial flavors, fragrances, and antimalarial and anticancer therapeutics, using recombinant DNA constructs.15 Terpenoids are normally isolated from plant tissue and can only be recovered in small amounts. DNA synthesis technology could be used as an alternative method for producing high-value compounds.
DNA synthesis technology could allow for the efficient, rapid synthesis of viral and other pathogen genomes—either for vaccine or therapeutic research and development, or for malevolent purposes or with unintentional consequences. Given the latter risks, in 2004, George Church (Harvard Medical School, Cambridge, MA) drafted a proposal for decreasing biohazard risks (i.e., creating nearly extinct human viruses, such as polio, or novel pathogens, like IL-4 poxvirus) while minimizing the impact on legitimate research. The proposal focuses on instrument and reagent licensing (e.g., restricting the sale and maintenance of oligonucleotide synthesis machines to licensed entities); regulation for the screening of select agents; establishing a method for testing these newly implemented licensing and, screening systems; criteria for exemption from the whole process; and, strategies for keeping the cost down.16 The proposal is mentioned here not to endorse it, but rather to highlight the need for a careful analysis and thoughtful discussion of the issues.
DNA Shuffling
Description
Classical genetic breeding has proven itself over and over again throughout human history as a powerful means to improve plant and animal stocks to meet changing societal needs. The late 20th century discovery of restriction endonucleases, enzymes that cut DNA molecules at sites comprising specific short nucleotide sequences, and the subsequent emergence of recombinant DNA technology provided scientists with high-precision tools to insert (or remove) single genes into the genomes of a variety of viruses and organisms, leading, for example, to the introduction of production-enhancing traits into crop plants.17 Most recently, a powerful mode of directed evolution known as “DNA shuffling”—also known as multigene shuffling, gene shuffling, and directed in vitro molecular evolution—has allowed scientists to greatly improve the efficiency with which a wide diversity of genetic sequences can be derived. A quantum leap in the ability to generate new DNA sequences, DNA shuffling can be used to produce large libraries of DNA that can then be subjected to screening or selection for a range of desired traits, such as improved protein function and/or greater protein production.
“Classical” single-gene breeding starts with a “parental” pool of related sequences (genes, etc.) and then breeds “offspring” molecules, which are subjected to screening and selection for the “best” offspring. The process is repeated for several generations. With DNA shuffling, sequence diversity is generated by fragmenting and then recombining related versions of the same sequence or gene from multiple sources (e.g., related species), resulting in “shuffling” of the DNA molecules. Basically, it allows for the simultaneous mating of many different species. The result is a collection of DNA mosaics. The reassortment that occurs during the shuffling process yields a higher diversity of functional progeny sequences than can be produced by a sequential single-gene approach.
In one of the earliest demonstrations of the technology, which involved shuffling four separately evolved genes (from four different microbial species), the shuffled “hybrids” encoded proteins with 270 to 540 times greater enzymatic activity than the best parental sequence.18 Even if that same recombined enzyme could have been evolved through single-gene screening, the process would have been dramatically slower. But chances are it never would have evolved. Evidence from at least one study shows that the best parent is not necessarily the one closest in sequence to the best chimeric offspring and thus would probably not represent the best starting point for single-gene evolution (i.e., some other better-look-
ing parental sequence would have been chosen for single-gene directed evolution).19
Current State of the Art
The technology has developed quickly, such that scientists are not just shuffling single genes, they are shuffling entire genomes. In 2002, biologists used whole-genome shuffling for the rapid improvement of tylosin production from the bacterium Streptomyces fradiae; after only two rounds of shuffling, a bacterial strain was generated that could produce tylosin (an antibiotic) at a rate comparable to strains that had gone through 20 generations of sequential selection.20 Also in 2002, a portion of the HIV genome was shuffled to create a new strain of HIV that was able to replicate in a monkey cell line that previously had been resistant to viral infection.21 By 2003 the technique had advanced to the point where many mammalian DNA sequences could be shuffled together in a single bacterial cell line. In one study, scientists shuffled one gene of a cytokine from seven genetically similar mammalian species (including human) to generate an “evolved” cytokine that demonstrated a 10-fold increase in activity compared to the human cytokine alone.22 It should be emphasized that the power of this technology (and any diversity generating procedure) is only fully realized if the molecules generated with the most enhanced, desired properties can be identified and isolated. Despite continual improvements in the throughput of current screening procedures, the use of conditions that impose strong selective pressures for emergence of molecules with the desired properties is far more efficient in finding the most potent molecule in the pool.
Future Applications
Ultimately, this rapid molecular method of directed evolution will allow biologists to generate novel proteins, viruses, bacteria, and other organisms with desired properties in a fraction of the time required with classical breeding and in a more cost-effective manner. For example, virologists are using DNA shuffling to optimize viruses for gene therapy and vaccine applications.23 Synthetic biologists are using the technology to speed up their discovery process (see “Synthetic Biology” later in this chapter).
Bioprospecting
Description24
Bioprospecting is the search for previously unrecognized, naturally occurring, biological diversity that may serve as a source of material for use in medicine, agriculture, and industry. These materials include genetic blueprints (DNA and RNA sequences), proteins and complex biological compounds, and intact organisms themselves. Humans have been exploiting naturally-derived products for thousands of years. Even as high-throughput technologies like combinatorial chemistry, described above, have practically revolutionized drug discovery, modern therapeutics is still largely dependent on compounds derived from natural products. Excluding biologics (products made from living organisms), 60 percent of drugs approved by the Food and Drug Administration and pre-new drug application candidates between 1989 and 1995 were of natural origin.25 Between 1983 and 1994, over 60 percent of all approved cancer drugs and cancer drugs at the pre-new drug application stage and 78 percent of all newly approved antibacterial agents were of natural origin.26 Taxol, the world’s first billion-dollar anticancer drug, is derived from the yew tree.27 Artemisinin, one of the most promising new drugs for the treatment of malaria, was discovered as a natural product of a fernlike weed in China called sweet wormwood. And aspirin—arguably one of the best known and most universally used medicines—is derived from salicin, a glycoside found in many species in the plant genera Salix and Populus.
Bioprospecting is not limited to plants, nor is drug discovery its only application. Most recently, with the use of molecular detection methods, scientists have uncovered a staggering number of previously unrecognized and uncharacterized microbial life forms.28 Indeed, microbial genomes represent the largest source of genetic diversity on the planet—diversity that could be exploited for medical, agricultural, and industrial uses. Natural products discovered through bioprospecting microbial endophytes—microorganisms that reside in the tissues of living plants—include antibiotics, antiviral compounds, anticancer agents, antioxidants, antidiabetic agents, immunosuppressive compounds, and insectides. With respect to the last, bioinsecticides are a small but growing component of the insecticide market. Bioprospected compounds exhibiting potent insecticidal properties include nodulisporic compounds for use against blowfly larvae (isolated from a Nodulisporium spp. that inhabits the plant Bontia daphnoides)29 and benzofuran compounds for use against spruce budworm (isolated from an unidentified endophytic fungus from winter-green, Gaultheria procumbens).30 Of note, naphthalene, the ingredient in
mothballs, is a major product of an endophytic fungus, Muscodor vitigenus, which inhabits a liana, Paullina paullinioides.31
Prospecting directly for DNA and RNA sequences that encode novel proteins with useful activities has become a potentially important scientific and business enterprise. This approach entails searches based on random expression of thousands or millions of sequences, followed by screening or selection for products with desired activities.32 Sometimes the search focuses on families of related sequences that are predicted to encode products of interest, which are recovered directly from environments using sequence amplification technology. This kind of approach can synergize with the DNA shuffling technology described above. Recent, early forays into “community genomics,” or large-scale random sequencing of the DNA from complex environmental microbial communities, reflect the immense future potential of this approach for the discovery and harnessing of previously unimagined biological activities.33
For example, Diversa Corporation (San Diego, CA) utilizes bioprospecting of microbial genomes to develop small molecules and enzymes for the pharmaceutical, agricultural, chemical, and industrial markets.34 After collecting environmental samples of uncultured microorganisms and extracting the genetic material, researchers search for novel genes and gene pathways for potentially useful products, like enzymes with increased efficiencies and stabilities (e.g., high and low temperature stability, high or low pH tolerance, high or low salt tolerance). The samples are collected from environments ranging from thermal vents to contaminated industrial sites to supercooled sea ice.
Bioprospecting has also been applied to the discovery of microbial agents in efforts to better understand the diversity of microbes in the environment that might serve as human pathogens if provided the opportunity. It has been argued that by deliberately scrutinizing the kinds of vectors and reservoirs that exist in a local environment for previously unrecognized microbes, novel agents might be identified long before they are discovered to be human, animal or plant pathogens, thus providing early warning of potential disease-causing agents.35 At the least, these surveys could expand our appreciation of microbial diversity and inferred microbial function.36 For example, in 2002, using a broad-range PCR approach (i.e., using conserved priming sites for a group of related sequence targets, as opposed to specific primers for single unique targets), scientists discovered four novel Bartonella DNA sequences in 98 arthropod specimens (fleas, lice, and ticks) from Peru; three of the sequences were significantly different from previously characterized Bartonella species.37Bartonella s are vectorborne bacteria associated with numerous human and animal infections.38 Rather than having any immediate known clinical
implications, this study illustrates the power of this generic approach as well as our incomplete understanding of Bartonella diversity.
Current State of the Art
Current methods include recovery of microbes using cultivation-based methods, serologic surveys of potential hosts, extraction/separation/purification of molecules with desired properties, amplification of families of related nucleic acid sequences using broad-range PCR (and similar techniques), shotgun cloning and sequencing of bulk DNA or cDNA from environments of interest, and the use of subtractive hybridization methods39 to enrich for novel nucleic acid sequences in hosts or environments.
Future Applications
One might consider both molecular and traditional cultivation-based approaches for examining hosts, such as fruit bats and small rodents, which are already known to serve as reservoirs for important human microbial pathogens (Hendra and Nipah viruses, Borrelia spp. and other genera, respectively). As described above, the potential benefits associated with the discovery of novel products and microbial genetic diversity are innumerable.
Combinatorial Chemistry: Generating Chemical Diversity
Description
Combinatorial chemistry refers to technologies and processes used for the rapid creation of large numbers of synthetic compounds (“libraries”), typically for the purposes of screening for activity against biological drug targets (see “High-throughput Screening”). Whereas DNA synthesis enables the acquisition of genetic sequence diversity, these techniques allow for the generation of libraries of chemical compounds having a diversity of shapes, sizes, and charge characteristics—all of which may be of interest for their varied abilities to interact with and bind to biologically active proteins or macromolecular complexes, thereby altering the biological properties of these proteins and complexes. Combinatorial chemistry techniques can be used to create a wide range of chemotypes or molecular motifs, ranging from large polycyclic compounds of a peptidic nature to smaller, presumably more druglike, compounds. Initially, it was believed that when used in combination with high-throughput screening technologies, combinatorial techniques would dramatically
accelerate the drug discovery process while reducing the associated up-front costs with the drug discovery effort. While this has not yet proven to be the case, most pharmaceutical companies are still heavily invested in combinatorial chemistry and are exploring the development and implementation of novel methods to create additional libraries of compounds. A recent trend noted in the pharmaceutical industry is the move from the development of large, unfocused, general screening libraries to smaller, less diverse libraries for screening against a particular target or family of related targets.
The origins of this new branch of chemistry can be traced back to the early 1960s, when methods were developed for the solid-phase synthesis of peptides.40 This involved attaching an amino acid to a solid support (i.e., beads of plastic resin) and then adding amino acid residues, one by one in a stepwise fashion through the creation of covalent peptide chemical bonds, until the desired peptide product is created. The final polypeptide is released by chemically breaking its bond with the solid support and washing it free.41 Subsequent modifications of the solid-phase synthesis process greatly enhanced the ability to generate a large number of peptides with specific amino acid sequences.42 Individual peptides were synthesized on the ends of “pins” that were spatially oriented in a two-dimensional array designed to match up with the wells of a 96-well microtiter plate. This reduced the scale of the process and greatly facilitated the parallel synthesis of large numbers of peptides. A further modification of the technique enhanced the ability to create a diversity of peptide sequences by incorporating a combinatorial approach.43 In this case, the solid-phase resin bearing the nascent synthetic peptide was enclosed in a mesh, or “tea bag.” Like the pin-based method, the tea-bag process facilitated the numerous washing and drying steps required for peptide synthesis and thus allowed for the parallel synthesis of many different peptides, each in its own tea bag. However, by mixing the resin from different tea bags after each individual stepwise addition of an amino acid residue, combinatorial peptide libraries involving a great diversity of amino acid sequences could be readily generated, in which each resin bead bears an individual peptide with a unique amino acid sequence.44
After the compounds are synthesized and a library is constructed, a selection or screening strategy is needed to identify unique compounds of interest to the biological sciences. The most obvious method involves affinity isolation of the peptide of interest on an immobilized target molecule, followed by release of the peptide and analysis utilizing combinations of gas-phase chromatography, high-performance liquid chromatography (HPLC), mass spectrometry, and nuclear magnetic resonance (NMR). It is also possible to determine the structure of compounds still
attached to the resin, using “on-bead” analytical techniques such as infrared analysis, gel-phase NMR, matrix-assisted laser desorption ionization time-of-flight mass spectrometry, electrospray mass spectrometry, and HPLC chemiluminescence nitrogen detection.45
While direct determination of structure, as described in the previous paragraph, works well for small libraries, these techniques are generally not applicable to large, mixture-based libraries. For libraries, various strategies have been developed that govern the reaction sequence by attaching a readable chemical “tag” to the bead while the molecule is being synthesized. One of the earliest tagging approaches employed the use of oligonucleotides.46 In this approach, for every amino acid added to the peptide chain, a specific set of oligonucleotides was added to a separate chain that was attached to the same bead. PCR and DNA sequencing techniques were then used to decode the structure of the peptide. Numerous additional tagging techniques and agents have since been developed.47
Current State of the Art
Solution-phase parallel synthesis is becoming the combinatorial chemistry technique of choice in the pharmaceutical industry, driven primarily by advances in laboratory automation, instrumentation, and informatics. Compounds can be synthesized either as single discrete compounds per reaction vessel or as mixtures of compounds in a single reaction vessel, so many of the same principles described above for solid-phase (resinbound) principles are applicable here as well. The primary advantage of solution-phase combinatorial chemistry lies in the increase in the number of chemical reactions/transformations that can be accessed, thereby greatly increasing the range of chemotypes (chemical scaffolds) that can be created.
The earliest reports of solution phase combinatorial chemistry techniques involved the use of a common multicomponent reaction, termed the Ugi reaction, in which an isocyanide, an aldehyde, an amine, and a carboxylic acid are combined in a single-reaction vessel to create a single major product. Using this synthetic approach coupled with advanced data analysis techniques, scientists were able to identify compounds with the desired biological effect after synthesizing only a 400-compound subset of the 160,000 possible products. This represents a 400-fold increase in discovery efficiency over conventional approaches.
The current trend in parallel solution-phase chemistry is leaning toward the development of smaller arrays (12 to 96 compounds) of simple to moderately complex chemical compositions. As the robotics and laboratory instrumentation required for parallel synthesis become more af-
fordable and readily accessible, the technology is being transferred into basic medicinal chemistry laboratories and becoming instrumental in the optimization of lead compounds (i.e., compounds that show potential to be developed into drugs). Such efforts are ideally carried out with knowledge of the structure of the target molecule, usually gained by application of either x-ray crystallography or NMR techniques. Structure-activity relationships are determined as lead compounds, identified initially through the screening of large libraries of compounds, are modified at specific sites, and the impact of the chemical modification on the desired biological properties of the compound is determined.
The purity and identity of combinatorially-produced compounds have been a source of recent great discussion and technological advance since, in order for any meaningful data to be produced from a biological assay, the purity of the compound of interest must be as high as possible.48 The activity of the compound must also be confirmed by resynthesis of the specific molecule and repeat assays for biological activity.
Future Applications
Combinatorial chemistry techniques are not only useful for drug discovery and development, they are being used in the search for better superconductors, better phosphors for use in video monitors (phosphors are substances that emit light), better materials for use in computer magnetic and other storage devices, and better biosensors for the detection of medically-important molecules and environmental toxins. Combinatorial approaches have been used to develop a “nose chip” sensor capable of detecting and distinguishing among seven common solvents (toluene, chloroform, tetrahydrofuran, acetone, ethyl acetate, ethanol, and methanol).49
Using combinatorial and high-throughput methods, the pharmaceutical industry synthesizes and screens several million new potential ligands annually. Although most companies have little use for the tens of thousands of these compounds identified each year as toxic, some might have potential as biochemical weapons (Chapter 1).50 Although most of the information derived from combinatorial and high-throughput technology is held in proprietary databases, a new public database recently proposed as part of the National Institutes of Health (NIH) Roadmap raises concerns about public access to dual-use information (Chapter 1, Box 1-1). The NIH Roadmap discovery effort is particularly worrisome in this regard, because of plans to optimize lead compounds shown to be capable of targeting specific cellular proteins. The goal is not to develop therapeutic agents but rather to provide a series of reagents, facilitating
further exploration of protein function and systems biology.51 Such compounds may be relatively potent poisons.
While the technologies applied in combinatorial chemistry are not exceedingly complex, a wide variety of laboratory automation and instrumentation is needed to stage an effective combinatorial chemistry campaign.
High-Throughput Screening52
Description
High-throughput screening (HTS) refers to the process of examining large numbers of diverse biomolecular or chemical compounds in a rapid and efficient manner for properties of interest. Such technologies are essential to achieving any benefit from the construction of large and diverse libraries of compounds, as they are used to select a particular compound having the desired properties. These properties might include biochemical or enzymatic activities desired of a potential therapeutic agent or toxicity in such an agent that under usual circumstances one would wish to avoid. Advances in miniaturized screening technologies, bioinformatics, robotics, and a variety of other technologies have all contributed to the improved biological assay efficiency that characterizes HTS. In contrast to this paradigm, in which a large library of compounds (i.e., samples) is tested for one specific activity or set of activities, a variation on the HTS theme involves the testing of a single biological sample for a wide variety of activities. The best example of this is the use of DNA or oligonucleotide microarrays—also known as DNA chips. These are routinely used in both basic and applied research to facilitate the large-scale screening and monitoring of gene expression levels, gene function, and genetic variation in biological samples, and to identify novel drug targets.
The process of screening large numbers of compounds against potential disease targets is characterized by a collection of technologies that strive to increase biological assay efficiency through the application of miniaturized screening formats and advanced liquid handling, signal detection, robotics, informatics, and a variety of other technologies. Over the past several years, the industry has witnessed an evolution in screening capabilities, resulting in the ability of a user to screen more than 100,000 compounds per day for potential biological activity. Evaluating upward of 1 million compounds for biological (or various other) properties in a screening campaign is now commonplace in the pharmaceutical industry.
Current State of the Art
Effective HTS relies on robust assays that can detect and then translate biological or other activities into a format that can be readily interpreted. A wide variety of assays are currently in use, including:
-
cell-free colorimetric or chemiluminescence assays;
-
cell-free fluorescence resonance energy transfer assays;
-
cell-based reporter gene assays, usually with an enzymatic read-out;
-
cell-based fluorescence imaging assays;
-
NMR assays, which involve identifying small molecule ligands for macromolecular receptor targets;
-
affinity chromatography assays;
-
DNA microarrays (high density arrangements of double-stranded DNA clones (cDNA) or oligonucleotides that serve as identical or complementary probes, respectively, for specific genes, transcripts, or genome sequences); and
-
Other types of microarrays, including high-density arrangements of antibodies, nucleic acid or peptide aptamers, antigens (protein or lipid), MHC53-peptide antigen complexes, and intact cells.
Future Directions
Future advances in HTS—such as the development of one-step assays and increased miniaturization—will continue to increase the throughput and reduce the cost of HTS assays and may eventually allow the simultaneous monitoring of multiple endpoints (e.g., biological, toxicological) across a wide variety of targets. An analysis of the current HTS technology landscape reveals the following as potential opportunities and future directions:
-
further development of one-step (homogeneous) assays;
-
development of improved primary screening hardware;
-
miniaturization as a means to increase throughput and decrease cost;
-
improvements in the capabilities and efficiency of robotic systems in the life sciences;
-
application of HTS to lead compound optimization; and,
-
novel approaches for identification of biologically-relevant targets.
In short, HTS assays and technologies will permeate new sectors in the life sciences, affecting the productivity and speed of advances and discoveries in these varied sectors. The cost effectiveness of HTS assays and technologies will improve, such that tasks previously believed to be impractical will become quite tractable. Coupled with methods to generate enhanced sequence and structural diversity beyond that seen in nature, these assays and technologies will permit the identification and selection of novel molecules with important biological functions, with ramifications for all of the life sciences.
2. DIRECTED DESIGN
There are other technologies, besides those described in the previous category of technologies, that seek to generate new kinds of genetic or molecular diversity. However, in contrast to the technologies in the first category, these “directed design” approaches are more deliberate, and rely on preexisting knowledge with regard to what needs to be created.
Rational Drug Design
Description
The methods described above, wherein a large library of diverse chemical compounds are screened using HTS methods to identify a smaller number of potential lead compounds with desired activities, are gradually being enhanced by less empirical approaches that are based on a greater understanding of biological systems (i.e., target: ligand interactions), identification of specific target molecules, and determination of the structure of a target molecule whose activity has been shown to be critical for the production of a particular disease or for maintenance of health. Such structural knowledge has grown rapidly over the past decade due to advances in x-ray crystallography, NMR technologies, and associated computational techniques that now allow for rapid determination of the structure of even large proteins or nucleic acid molecules at atomic-level resolution. A quick survey of the Protein Data Bank (PDB),54 the global resource for all publicly available biological macromolecular structures, reveals that the number of structures deposited on an annual basis witnessed nearly a 10-fold increase between 1994 (3,091) and 2004 (28,992); see Figure 3-2. With such structural knowledge of targets in hand, chemists can rationally pursue the design of novel chemical compounds that either bind to selected sites on the surface of these target molecules or mimic the structure of the target molecule and thereby compete for the binding to a receptor molecule.
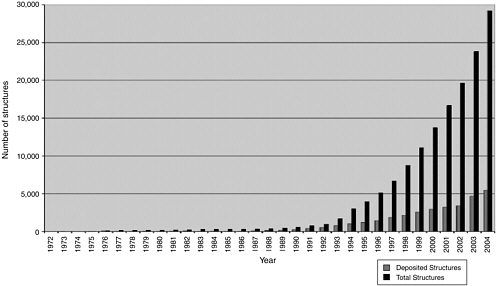
FIGURE 3-2 Growth in the number of structures deposited per year (gray) and total holdings of the PDB (black) from the time the bank was founded.
SOURCE: Reprinted from Dutta, S. and H.M. Berman. 2005. Large macromolecular complexes in the protein data bank: A status report. Structure 13(3):382, with permission from Elsevier.
An excellent example of technological convergence exists with the field of in silico, or virtual, screening. This methodology capitalizes on the advances described above with respect to the determination of structures for target molecules as well as advances in computer hardware and specialized chemical informatics algorithms, so-called docking and scoring programs. Many thousands of virtual compounds can be rapidly and effectively assessed for potential target molecule complementarity,55 as a prerequisite for biological activity, prior to any actual chemistry being carried out or biological assays being performed. The product of this computational effort is thus a rationally designed molecule that, once synthesized, can potentially serve as a lead compound in the drug discovery process.
Current State of the Art
Although rational drug design has received a great deal of attention from the pharmaceutical industry and is recognized as having great potential for the future, most efforts today by the drug discovery industry reflect a combination of structure-aided rational design of compounds and the HTS screening of libraries of diverse compounds. Thus, the use of structure, when known for a given molecular target, may come into play once a lead compound has been identified through an HTS process and efforts are made to optimize this lead and improve the biological activity or pharmacological properties of the compound. The field today is such that absence of knowledge of the structure of a targeted molecule is viewed as a critical impediment to the development of a new drug.
In contrast to the rational design of small-molecule therapeutics, the rational design of therapeutic nucleic-acid-based compounds is much easier in that such compounds are synthesized to be complementary to the targeted nucleic acid sequence. While nucleic acid therapeutics based on antisense oligonucleotides or ribozymes, enzymatically-active RNAs that cleave specific RNA target sequences, have been pursued for over a decade, their promise has not yet been realized due to difficulties in delivering stable compounds to desired sites. Significant advances are now occurring, however, in providing desired pharmacological properties to siRNA-based compounds and morpholino antisense oligonucleotides.
Future Applications
As the structure of greater numbers of potential target molecules are identified in the future and as both in silico screening and chemical synthesis methods continue to advance, it seems clear that a greater reliance
is likely to develop on these types of approaches. Greater application of rational, structure-based design approaches is likely to speed the discovery process significantly. While there are dual-use implications for such technologies, as there are for almost any advancing life sciences technology, the infrastructure required to pursue such structure-based design of novel biologically active compounds is likely to limit its use to the legitimate pharmaceutical industry for a number of years. It should be noted, however, that like the nucleotide sequence databases that are open to the public, rapidly growing numbers of protein structures are being placed in the public domain. This trend is likely to continue and even accelerate, and as the computer hardware and software requirements for viewing and interpreting such structures becomes increasingly simple, these approaches will become increasingly accessible to scientists outside the pharmaceutical industry.
Synthetic Biology
Description
The fledgling 5-year-old-field of synthetic biology—which is attracting engineers and biologists in equal measure—means different things to different researchers. Engineers view it primarily as a way to fabricate useful microbes to do what no current technology can do (i.e., they view it as an engineering discipline). Biologists see it as a powerful new way to learn about underlying principles of cellular function.
Unlike systems biologists (see description later in this chapter), who adopt a big-picture approach to biology by analyzing troves of data on the simultaneous activity of thousands of genes and proteins, synthetic biologists reduce the very same systems to their simplest components. They create models of genetic circuits, build the circuits, see if they work, and adjust them if they do not, learning underlying principles of biology in the process. By examining simple patterns of gene expression and treating pieces of DNA as modules, which, like Legos™, can be spliced together, synthetic biologists construct what are effectively biochemical logic boards that control both intra- and extracellular activity.
Because the molecular nature of many cellular reactions is only partially understood, most synthetic genetic circuits require considerable further empirical refinement after the initial computational work. Some scientists use DNA shuffling to streamline the empirical process. After inserting mutated DNA circuits into cells and selecting for those cells (and the circuits therein) that performed the best, researchers can evolve an effective product in just a couple of generations.56
Current State of the Art
One of the goals of the field is to transform bacteria into tiny programmable computers. Like electronic computers, the live bacterial circuits would use both analog and digital logic circuits to perform simple computations. For example, researchers are working to develop modular units, such as sensors and actuators, input and output devices, genetic circuits to control cells, and a microbial chassis in which to assemble these pieces. If they are successful, a “registry of biological parts” will allow researchers to go to the freezer, get a part, and hook it up.57 The computing power of programmable cells will likely never rival that of their electronic counterparts. Rather, the beauty of synthetic biology lies in what living cells can do.
In 2000, a genetic “circuit” was created in E.coli that caused the cells to blink like a lighthouse.58 The circuit, which was called “the repressilator,” was comprised of three repressor genes, one of which turned on a gene for green fluorescent protein (GFP), which, when activated, emits a green glow. Three years later another research group created a genetic circuit by crafting a “toggle switch” that could oscillate the circuit and alter its pattern depending on growth conditions.59 Using this technique, investigators subsequently developed a procedure to re-engineer a bacterial protein that binds to TNT (an explosive) and that, when bound, activates a gene circuit that produces GFP.60 This demonstrates an initial effort to engineer organisms that operate as biological sentinels, pinpointing explosives or detecting the presence of biological weapons.
In 2004, researchers in Israel designed a prototype “DNA computer” with the capacity to logically analyze mRNA disease indicators in vitro (i.e., in this case, early signs of prostate and lung cancer) and control the administration of biologically active ssDNA molecules, including drugs.61 The procedure is relatively innocuous, requiring the injection of a very small amount of fluid containing billions of nanoparticles, each of which operates as a tiny computer by effectively interrogating the cell and detecting the presence of diagnostic DNA markers (e.g., mutated mRNA sequences or underexpressed or overexpressed mRNA). If the markers are present, the nanoparticle sends out a therapeutic short nucleic acid that can affect the level of gene expression.
Future Applications
Synthetic biology technology has many potential applications, including designing bacteria that can detect chemical or biological agent signatures, engineering bacteria that can clean up environmental pollutants, and engineering organisms or compounds that can diagnose disease or fix faulty genes. Although initial efforts are focused on microbial cells, some synthetic biologists imagine a day when they will be able to pro-
gram adult stem cells for therapeutic purposes (e.g., to patch up a damaged heart).
Engineering ethicist Aarne Vesilind (Bucknell University) is one of many scientists promoting the idea that synthetic biologists and ethicists hold an Asilomar-like conference on synthetic biology—much like that held at the dawn of genetic engineering research in the mid-1970s—to define bioengineers’ “responsibilities to society” should these engineered organisms survive outside the laboratory to cause harm to human health or the environment.62 Several efforts have now been planned to examine the implications of this kind of work, including one foundation-funded study involving three institutions, two of which play a major role in synthetic genomics research.63 In addition, the National Science Advisory Board for Biosecurity has identified synthetic genomics as a major area of interest. Many of the same issues are raised by the genetic engineering of viruses.
Genetic Engineering of Viruses
Description
As described above, the development of recombinant DNA technology and the ability to manipulate DNA sequences in bacterial species such as E. coli has resulted over time in the capacity to insert almost any desired gene into almost any kind of prokaryotic or eukaryotic cell. Placing the DNA inserted under appropriate transcriptional controls, and the protein encoded by it under appropriate translational control, allows that gene to direct the expression of almost any kind of protein: a fluorescent marker (as in the GloFish described in Chapter 1), an enzyme that might function as a reporter, an antibiotic resistance marker, or even a toxin. Using very similar techniques, genes of interest (subject to size constraints) can be introduced into the genomes of many different types of DNA viruses, ranging from adenoviruses to herpesviruses. Such capabilities raise obvious and compelling dual-use concerns.
The introduction of heterologous gene sequences into the genomes of RNA viruses, or other types of modifications to the RNA genomes of these viruses, presents a special set of technical difficulties due to the fact that the genetic material is RNA, which is less stable than DNA and not as amenable to the genetic splicing techniques that have made recombinant DNA technology as versatile. However, this has been accomplished for a growing number of different types of RNA viruses. Moreover, given the small size of these RNA genomes, it has proven possible to synthesize completely de novo all the genetic material needed to recover fully infectious virus particles with near wild-type infectivity, virulence and replication potential.
RNA viruses come in several types, depending on the number of strands of RNA in each molecule of their genome (i.e., single-stranded or double-stranded RNA molecules) and the number of genomic segments (one or more). Genetic engineering of single-stranded RNA viruses in which the RNA is of positive polarity (i.e., the same sense as the messenger RNA that encodes the viral proteins) has proven most straightforward. It has been known for many years that genomic RNA isolated from positive-strand RNA viruses, such as poliovirus, is intrinsically infectious. When transfected (i.e., introduced) into a permissive cell in the absence of any accompanying proteins, such RNA will lead directly to the synthesis of the viral proteins, which will then begin to assemble the necessary replicative machinery to make additional copies of the RNA as well as more viral protein, leading ultimately to the assembly and “rescue” of fully infectious virus, which is then generally released from the cell.
To manipulate the viral RNA genome, scientists in the age of molecular biology have developed efficient enzymatic methods for creating complementary DNA (cDNA) copies of the viral genomic RNA using reverse transcriptase enzymes encoded by retroviruses. This cDNA can be engineered to have “sticky” ends, allowing it then to be molecularly cloned into E. coli, in which it can be manipulated by all the modern methods available. This can include the deletion of protein coding sequences, the creation of deletion or point mutations, or even the introduction of completely novel protein-coding sequences. The modified cDNA can then be placed downstream of an appropriate promoter sequence for a DNA-dependent RNA polymerase and a novel, molecularly engineered viral RNA genome efficiently transcribed in an in vitro transcription reaction. The transcribed RNA can then be transfected back into a permissive cell and, if the introduced mutations are compatible with continued viability of the virus, will give rise to novel infectious viruses.
The process by which virologists use this method, involving the conversion of the genetic sequence of the virus from RNA to DNA and back to RNA, generally in order to assess the impact of mutations on the viral life cycle or pathogenic properties, is known as “reverse genetic engineering.” This approach is widely used by positive-strand molecular virologists. First carried out in 1980 with poliovirus,64 infectious cDNA clones have now been constructed for members of many positive-stranded RNA virus families, including brome mosaic virus,65 yellow fever virus,66 Sindbis virus,67 citrus tristeza virus,68 and equine arteritis virus.69 In the case of hepatitis C virus, a positive-strand virus in the Flaviviridae, virus rescue has generally required injection of the synthetic RNA directly into the liver of a chimpanzee. On the other hand, fully infectious poliovirus, a member of the family Picornaviridae, has been recovered in a cell-free reaction carried out in vitro in an optimized cell extract system.
In the past, coronaviruses, which have the largest genomes of all positive-strand RNA viruses (around 30 kilobases long), were difficult to reverse engineer because of the sheer size and instability of their full-length cDNA clones in bacterial vectors.70 However, recent technological advances have made it possible to reverse engineer even these largest of all known RNA viruses,71 including the causative agent of severe acute respiratory syndrome (SARS), a previously undescribed coronavirus.72
Similarly, the reverse genetic engineering of negative-strand RNA viruses73 has proven much more difficult, given the fact that the RNA genomes of these viruses do not function directly as messenger RNAs and thus do not give rise to infectious virus progeny following their introduction into permissive cells. These RNAs require the expression of certain viral proteins, in order to make positive-strand copies of the negative-stranded RNA genome and to initiate the replicative cycle. The technology to accomplish this was first developed for influenza A virus in the late 1980s to early 1990s. Like the earlier efforts with positive-strand RNA viruses, these efforts not only have dramatically improved our understanding of how these viruses replicate, but have also created the means for genetically manipulating viral genomes in order to generate new viruses for use as live, attenuated vaccines or vectors.74
Initially, reverse engineering of the influenza virus required the use of helper viruses, which provided proteins and RNA segments that the reconstituted in vitro RNPs (i.e., reconstituted ribonucleoprotein complexes containing RNA transcribed from the molecularly cloned cDNA) needed in order to be infectious following transfection into cells. Later, alternative methods for introducing influenza RNPs into cells were developed, including entirely plasmid-driven rescue that did not require the involvement of a helper virus.75 The latter plasmid-based system allowed for easy engineering of viral genomes with multiple specific mutations. By 2001 at least one laboratory had generated a pathogenic H5N1 virus using reverse engineering.76
In addition to influenza A virus, and as summarized in a paper that appeared in the Journal of Virology in 1999,77 in its first decade the technology was used to reverse engineer, or “recover” many other negative-stranded RNA viruses including rabies virus,78 vesicular stomatitis virus,79 respiratory syncytial virus,80 measles virus,81 Sendai virus,82 human parainfluenza type 3,83 rinderpest virus,84 simian virus,85 bovine respiratory syncytial virus,86 Newcastle disease virus,87 and bunyavirus.88
Current State of the Art
Most recently, as mentioned in Chapter 1, reverse engineering has been used to produce infectious influenza A viruses containing the viral
haemagglutinin (HA) and neuraminidase (NA) genes of the strain that caused the devastating 1918-1919 “Spanish” influenza pandemic. Scientists demonstrated that the HA of the 1918 virus confers enhanced pathogenicity in mice to recent human viruses that are otherwise nonpathogenic in their murine host. HA is a major surface protein that stimulates the production of neutralizing antibodies in the host, and changes in the genome segment that encodes it may render the virus resistant to preexisting neutralizing antibodies, thus increasing the potential for epidemics or pandemics of disease. Moreover, the reverse engineered viruses expressing 1918 viral HA elicited hallmark symptoms of the illness produced during the original pandemic.89
With the complete genetic sequencing of the H1N1 influenza A virus, referred to in Chapter 1, some have questioned whether these studies should have been published90 in the open literature given concerns that terrorists could, in theory, use the information to reconstruct the 1918 flu virus.91 It should be noted that in addition to the “normal” scientific peer review, the editors of Science required the authors to demonstrate that they had obtained approval to publish their research from the director of the Centers for Disease Control and Prevention, and the director of the National Institute of Allergy and Infectious Diseases.92 Furthermore, the National Science Advisory Board for Biosecurity (NSABB) was asked to consider these papers prior to publication and determined that the scientific benefit of the future use of this research far outweighed the potential risk of misuse.93
Future Applications
Reverse engineering of the causative agent of SARS illustrates the many potential beneficial applications of the technology. In addition to opening up new opportunities for exploring the complexity of the SARS-coronavirus genome, the availability of a full-length cDNA provides a genetic template for manipulating the genome in ways that will allow for rapid and rational development and testing of candidate vaccines and therapeutics.94 By mutating the many small proteins seemingly expressed by this unique coronavirus, scientists will learn their function in viral replication and/or pathogenesis and potentially identify useful targets for drug discovery efforts.
The influenza A reverse genetic engineering system serves as an excellent example of the potential for this technology to be used with the intent to do harm. As summarized in a 2003 article on the potential use of influenza virus as an agent for bioterrorism, with respect to advances that allowed for helper virus free production of a pathogenic H5N1 virus, virologist Robert M. Krug (University of Texas, Austin) has written:
There is every reason to believe that the same recombinant DNA techniques can be used to render this H5N1 virus transmissible from humans to humans. Furthermore, it should be possible to introduce mutations into such a recombinant virus so that it is resistant to currently available influenza virus antivirals (M2 inhibitors: amantadine and rimantadine; and NA inhibitors: zanamivir and oseltamivir), and so that it possesses an HA antigenic site that is unlike those in recently circulating human viruses. In fact, several viruses with different HA antigenic sites could be generated. The human population would lack immunological protection against such viruses, existing antiviral drugs would not afford any protection, and these viruses could be spread simply by release of an aerosol spray in several crowded areas.95
3. UNDERSTANDING AND MANIPULATION OF BIOLOGICAL SYSTEMS
A more holistic understanding of complex biological systems (e.g., the workings of an intact cell, multicellular organism, or complex microbial community) is emerging through a set of technologies that allow for the collection of vast, comprehensive (highly parallel) sets of data for multiple kinds of biological processes, the integration of these data sets, and the identification of critical components or pathways. Critical components can then serve as targets for therapeutic and preventive intervention or manipulation; they can also serve as targets for malevolent manipulation and as the basis for novel kinds of biological attack. Concurrently, technologies that facilitate a better understanding of intracellular, organ, and whole-animal control “circuitry” will enhance the ability of scientists to manipulate these complex systems.
Examples of some technologies that are leading to this type of holistic overview include the emerging field/discipline of “systems biology”96 and genomic medicine. Examples of the tools that could be used to manipulate complex biological systems include gene silencing, novel binding reagents (e.g., nucleic acid and peptide aptamers, engineered antibodies), and small-molecule modulators of physiological systems. In many ways this category of technologies opens up entirely novel aspects of the future biodefense and biothreat agent landscapes and changes the fundamental paradigm for future discussions on this topic.
RNA Interference
RNA interference—also known as RNAi and RNA silencing—was first observed in plants when it was noted that endogenous and “foreign” genes appeared to be turning each other off by a process initially termed
“co-suppression.”97 What was initially thought to be peculiar to petunias was later found in other plants and also animals. The phenomenon is now known as RNA interference, and is recognized to be a common antiviral defense mechanism in plants and a common phenomenon in many other organisms, including mammals. It is also increasingly apparent that RNAi is intimately related to widespread regulation of gene expression by very small endogenously expressed RNA molecules, so-called micro-RNAs (miRNA). This field is exploding with new discoveries almost daily concerning the role of miRNAs in regulating gene expression during development and after. The interaction of endogenous miRNAs with cellular mRNAs encoding specific proteins leads to suppression of protein expression, either by impairing the stability of the mRNA or by suppressing its translation into protein. The fact that small, largely double-stranded RNAs of this type, about 21 nucleotides in length, could play such an apparently broad and fundamental role in development and in the control of cellular homeostasis was not at all appreciated just a few years ago and highlights the sudden, unpredictable paradigm shifts and sharp turns in the way scientists think that are possible in the advance of the life sciences (Figure 3-3).
The basic molecular mechanism of RNAi is as follows. Long, double-stranded RNAs (dsRNAs; typically >200 nucleotides long) silence the expression of target genes upon entering a cellular pathway commonly referred to as the RNAi pathway. First, in the so-called initiation step, the dsRNAs are processed into 20 to 25 nucleotide small interfering RNAs (siRNAs) by an Rnase III-like enzyme called Dicer. The siRNAs then assemble into endoribonuclease-containing complexes known as RNA-induced silencing complexes (RISCs), unwinding in the process. The siRNA strands subsequently guide the RISCs to complementary RNA molecules, where RISC complex cleaves and destroys the cognate RNA (i.e., this is the effector step). miRNAs are generated in a similar fashion from endogenously expressed RNAs containing short hairpin structures, using a related Dicer-like protein. They are capable of similarly silencing gene expression but can also direct post-transcriptional silencing by blocking translation of a targeted host mRNA. This later effect typically depends on binding to a partially complementary target sequence near the 3’ end of the mRNA.
RNAi is highly specific and remarkably potent (only a few dsRNA molecules per cell are required for effective interference), and the interfering activity can occur in cells and tissues far removed from the site of introduction.
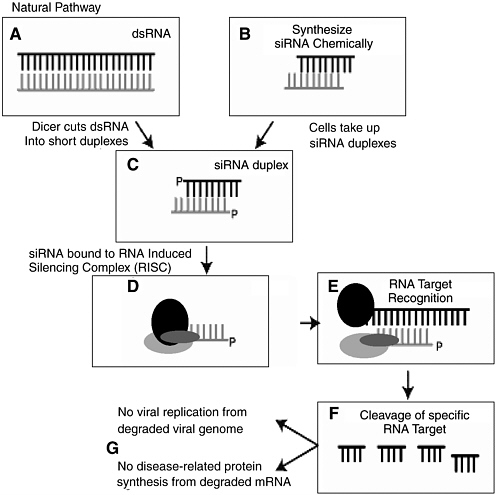
FIGURE 3-3 The process of RNA interference.
SOURCE: Steven Block, presentation to the committee, April 2004.
Current State of the Art
The technology is expected to prove particularly valuable in cases where the targeted RNA encodes genes and protein products inaccessible to conventional drugs (i.e., protein, small-molecule, and monoclonal antibody therapeutics). However, clinical delivery poses a significant challenge, as does the likelihood of undesirable silencing of nontargeted genes.98 Yet several recent experiments indicate that investigators are well on their way to overcoming these challenges and creating an emerging dual-use risk in the form of bioengineered RNAi-based pathogens. In 2003, a German research team announced the successful lentivirus vector
delivery of in vivo gene silencing with RNAi.99 Also in 2003, researchers announced the successful use of high-pressured, high-volume intravenous injection of synthetic siRNA.100 Other studies have demonstrated the potential to deliver RNAi to specific organs, such as the eyes,101 lungs,102 and central nervous system.103 Although human trials of RNAi have begun for the treatment of age-related macular degeneration,104 a systemic mode of delivery would arguably have greater clinical utility. Substantial progress is being made toward this aim, however, using liposome and lipid nanoparticle formulations of chemically modified, and hence stabilized, siRNAs. Scientists at Sirna, a small biotech company working for well over a decade on nucleic-acid-based therapies, have recently described a 1,000-fold reduction in the amount of hepatitis B virus present in the blood of mice replicating this virus in the liver, following a series of three separate intravenous inoculations of a lipid nanoparticle formulated, chemically modified, siRNA.
In November 2004, researchers from Alnylam Pharmaceuticals used chemically modified siRNAs to silence genes encoding Apolipoprotein B (ApoB) in mice, resulting in decreased plasma levels of ApoB protein and reduced total cholesterol.105 The study thus demonstrated systemic activity following a conventional clinical mode of delivery. Importantly, the delivery did not inadvertently impact nontargeted genes. Still, there are questions about the specificity of the siRNA, given that the investigators did not evaluate all proteins and given that they collected measurements over a relatively short period of time.106 A longer, more comprehensive study would be necessary to evaluate more fully the specificity of the technique. However, while “off- target” effects of siRNAs are certainly of concern to regulators and industry proponents as well, it is likely they can be managed in much the same way that “off target” effects (i.e., unexpected toxic effects) of small-molecule therapeutics have been in the past.
Potential Applications
Observations that RNAi works in vivo in mammals has not only created opportunities for the development of new therapeutic tools but also spawned a new generation of genetic research in mammals.107 For example, the vast majority of mammalian RNAi systems are driven by a polymerase III promoter, which can be manipulated such that the experimenter has the ability to turn the expression of a gene on and off at will, allowing for novel experimental designs. One could temporarily switch off a tumor suppressor gene suspected of providing genome protection (e.g., a checkpoint gene) and then turn it on again, allowing the experimenter to determine whether the gene is necessary for the initiation or
maintenance of tumorigenesis and whether it might be a good target for late-stage cancer treatments.
It is reasonable to expect significant additional advances in the formulation of siRNAs for use as pharmacological agents, particularly with contributions from the field of nanotechnology. As with so many of the technologies outlined in this chapter, just as RNAi promises new therapeutic options for cancer and other diseases, it could also be used to manipulate gene expression with the intent to do harm.
High-Affinity Binding Reagents (Aptamers and Tadpoles)
Description
Aptamers are short, single-stranded nucleic acid or peptidic ligands that fold into well-defined three-dimensional shapes, allowing them to inhibit or modulate their protein targets with high affinity and specificity. Since their discovery in the early 1990s,108 aptamers have been used in target validation, detection reagents, and functional proteomic tools.109 Over the past decade, several studies have explored the potential of aptamers for therapeutic intervention, including the inhibition of targets associated with inflammatory processes, cancer, and other disorders.110 Aptamers have been compared to monoclonal antibodies but with the added advantage that they are neither toxic nor immunogenic.
Current State of the Art
One of the first aptamers tested in an animal model was an antithrombin agent that blocks the proteolytic activity of thrombin, a protein involved in thrombosis (blood clot formation in a blood vessel).111 In June 2004, Archemix Corp. (Cambridge, MA) and Nuvelo, Inc. (San Carlos, CA) announced that an Investigational New Drug application had been submitted to the FDA to begin a Phase I clinical trial with an antithrombin aptamer, ARC183, for potential use in coronary artery bypass graft surgery.112 In another clinical trial, Eyetech Pharmaceuticals, Inc. (New York, NY) is testing Macugen, an aptamer that targets VEGF (vascular endothelial growth factor) as a treatment for age-related macular degeneration and diabetic macular edema.113
In January 2005, scientists reported that they had created a new type of high-affinity binding reagent—“tadpoles”—that bind to specific targets, such as Bacillus anthracis protective antigen and the enzyme cofactor biotin, as examples.114 Tadpoles are protein-DNA chimeras that contain a protein head coupled to an oligonucleotide tail. The head has an affinity for a specific target molecule; the tail, which contains a region for PCR
amplification, mediates detection. Tadpoles represent another type of high-affinity binding reagent with the power to not only detect but, with its DNA tail, “count” small numbers of proteins and other molecules in a precise fashion.
Future Applications
Their sensitivity, dynamic range, and, in the case of tadpoles, precise quantification make these high-affinity binding molecules potentially very useful tools for disease diagnosis and environmental detection, including pathogen and other biological agent detection in the event of a naturally occurring or deliberate biological attack.
Despite their promise as therapeutic agents, aptamers are very expensive to synthesize and are still a largely unknown entity (with respect to administration, formulation, adverse effects, etc.). So although several compounds have entered clinical trial, their future as biopharmaceuticals is unclear.115 More certain is their role as valuable lead structures in small-molecule drug discovery (because they can be so readily modified and adapted to almost any kind of high-throughput readout format) and as molecular detection reagents (because of their high specificity).
Computational Biology and Bioinformatics116
Description
Life scientists have exploited computing for many years in some form or another. But what is different today—and will be increasingly so in the future—is that the knowledge of computing and mathematical theory needed to address many of the most challenging biological problems can no longer be easily acquired but requires instead a fusion of the disciplines of biology, computation, and informatics. A National Research Council (NRC) report entitled Catalyzing Inquiry at the Interface of Computing and Biology (December 2005) has pointed out that the kinds and levels of expertise needed to address the most challenging problems of contemporary biology stretch the current state of knowledge of the field. The report identifies four distinct but interrelated roles of computing for biology:
-
Computational tools are artifacts—usually implemented as software but sometimes hardware—that enable biologists to solve very specific and precisely defined problems. Such biologically oriented tools acquire, store, manage, query, and analyze biological data in a myriad of forms and in enormous volume for its complexity. These tools allow bi-
-
ologists to move from the study of individual phenomena to the study of phenomena in biological context, to move across vast scales of time, space, and organizational complexity and to utilize properties such as evolutionary conservation to ascertain functional details.
-
Computational models are abstractions of biological phenomena implemented as artifacts that can be used to test insight, to make quantitative predictions, and to help interpret experimental data. These models enable biological scientists to understand many types of biological data in context, and even at very large volumes, and to make model-based predictions that can then be tested empirically. Such models allow biological scientists to tackle harder problems that could not readily be posed without visualization, rich databases, and new methods for making quantitative predictions. Biological modeling itself has become possible because data are available in unprecedented richness and because computing itself has matured enough to support the analysis of such complexity.
-
A computational perspective or metaphor on biology applies the intellectual constructs of computer science and information technology as ways of coming to grips with the complexity of biological phenomena that can be regarded as performing information processing in different ways. This perspective is a source for information and computing abstractions that can be used to interpret and understand biological mechanisms and function. Because both computing and biology are concerned with function, information and computing abstractions can provide well-understood constructs that can be used to characterize the biological function of interest. Further, they may well provide an alternative and more appropriate language and set of abstractions for representing biological interactions, describing biological phenomena, or conceptualizing some characteristics of biological systems.
-
Cyberinfrastructure and data acquisition are enabling support technologies for 21st century biology. Cyberinfrastructure—high-end general-purpose computing centers that provide supercomputing capabilities to the community at large; well-curated data repositories that store and make available to all researchers large volumes and many types of biological data; digital libraries that contain the intellectual legacy of biological researchers and provide mechanisms for sharing, annotating, reviewing, and disseminating knowledge in a collaborative context; and high-speed networks that connect geographically distributed computing resources—will become an enabling mechanism for large-scale, data-intensive biological research that is distributed over multiple laboratories and investigators around the world. New data acquisition technologies such as genomic sequencers will enable researchers to obtain larger amounts of data of different types and at different scales, and advances in informa-
-
tion technology and computing will play key roles in the development of these technologies.
Current State of the Art
A new level of sophistication in computing and informatics is required for interpretation of much of the data generated today in the life sciences. These data are highly heterogenous in content and format, multimodal in collection, multidimensional, multidisciplinary in creation and analysis, multiscale in organization, and international in collaborations, sharing, and relevance.117 Such data may consist of sequences, graphs, geometric information, scalar and vector fields, patterns of organization, constraints, images, scientific prose, and even biological hypotheses and evidence. These data may well be of very high dimension, since data points that might be associated with the behavior of an individual unit must be collected for thousands or tens of thousands of comparable units. The size and complexity of the data sets being generated require novel methods of analysis, which are being provided by computational biologists. For example, scientists at the U.S. Department of Energy’s Pacific Northwest National Laboratory have developed a new computational tool—called ScalaBLAST—that is a sophisticated “sequence alignment tool” and can divide the work of analyzing biological data into manageable fragments, so that large data sets can run on many processors simultaneously. The application of this technology means that large-scale problems—such as the analysis of an organism—can be solved in minutes rather than weeks.118
The NRC report notes that these data are windows into structures of immense complexity. Biological entities (and systems consisting of multiple entities) are sufficiently complex that it may well be impossible for any human being to keep all of the essential elements in his or her head at once. Thus, advances in computational biology will be driven by the need to understand how complex biological systems operate and are controlled and will contribute fundamentally to the development of a systems view in biology.
Future Applications
The NRC report emphasizes that the life sciences of the future will be an information science and will “use computing and information technology as a language and a medium in which to manage the discrete, nonsymmetric, largely nonreducible, unique nature of biological systems and observations. In some ways, computing and information will have a relationship to the language of 21st century biology that is similar to the
relationship of calculus to the language of the physical sciences. Computing itself can provide biologists with an alternative and possibly more appropriate language and sets of intellectual abstractions for creating models and data representations of higher-order interactions, describing biological phenomena, and conceptualizing some characteristics of biological systems.” This potential is nowhere more evident than in the nascent field of systems biology.
Systems Biology
Description
Systems biology—also known as integrative biology—uses high-throughput, genome-wide tools (e.g., microarrays) for the simultaneous study of complex interactions involving networks of molecules, including DNA, RNA, and proteins. It is, in a sense, classical physiology taken to a new level of complexity and detail. The term “systems” comes from systems theory or dynamic systems theory: Systems biology involves the application of systems- and signal-oriented approaches to understanding inter- and intracellular dynamic processes.119 Systems-level problem solving in biology is based on the premise that cellular behavior is a complex system of dynamically interacting biomolecular entities. A systems biologist seeks to quantify all of the molecular elements that make up a biological system and then integrate that information into graphical network models that can serve as predictive hypotheses.
A growing number of researchers in the life sciences community are recognizing the usefulness of systems biology tools for analyzing complex regulatory networks (both inside the cell, and the regulatory networks that integrate and control the functions of distinctly different cell types in multicellular organisms like humans) and for making sense of the vast genomic and proteomic data sets that are so rapidly accumulating.120 These efforts draw heavily on computational methods to model the biological systems, as described earlier. Systems biology is being seen as a valuable addition to the drug discovery toolbox.121 In medicine, where disease is being viewed as a perturbation of the normal network structure of a system (i.e., disease-perturbed proteins and gene regulatory networks differ from their healthy counterparts, because of genetic or environmental influences), a systems biology approach can provide insights into how disease-related processes interact and are controlled, guide new diagnostic and therapeutic approaches, and enable a more predictive, preventive, personalized medicine.122
Current State of the Art
This field is rapidly evolving, with the computational tools still in an immature state and inadequate for handling the reams of data derived from microarray assays and their functional correlates. Unconventional means of recording experimental results and conveying them rapidly to others in the field using an Internet-based approach are being pursued in an effort to manage the scale of data collection and analysis required for this effort.123 Whereas scientists previously may have examined only a single facet of a signal transduction pathway involved, for example, in control of a cellular response to infection, they are now looking more broadly at the effect of a particular stimulus on multiple different pathways, including what happens at common nodes and the counter-regulatory pathways that are activated in response to a particular signal. They are coming to realize that many novel molecular mechanisms are involved in controlling these signaling pathways, not only phosphorylation and kinase activation as classically recognized in signal transduction but also specific protein conformational changes, the translocation of proteins to different cellular compartments, proteolytic cleavage of signaling partners and latent transcription factors, and the binding and release of modulatory proteins from key signaling intermediates. A similar multiplicity of mechanisms exists within the extracellular regulatory networks, that must ultimately take their cues from intracellular events. In all of these signaling networks, tremendous specificity of responses stems from the timing, duration, amplitude, and type of signal generated and the pathways from which it emanates. At present, perhaps it could be said that while the magnitude and nature of the challenge posed by systems biology are increasingly well recognized, it remains unclear exactly how these challenges will be met, or how successful such attempts to do so will be.
Future Applications
The rise of systems biology is expected to have profound implications for research, clinical practice, education, intellectual property, and industrial competitiveness. As computational technologies advance, simulation of complex biological systems will have more predictive accuracy, aspects of laboratory experimentation will replaced by more cost-effective computational approaches, and physicians will have new decision support tools to help them identify the best preventative and therapeutic approaches for individual genotypes and phenotypes.
Just as systems biology will profoundly alter the way scientists and physicians conduct their analyses, the same global problem-solving ap-
proach could serve as a tool for the identification of ways to deliberately manipulate biological systems with the intent to do harm.
Genomic Medicine
Description
Genomic, or personalized, medicine refers to potential patient-tailored therapies made possible by improved molecular characterization of disease, technologies that allow for rapid genomic and proteomic analyses of individual patients, and advances in information technology that allow practitioners to access this information in meaningful ways. Scientists have known for a long time that human genetic variation is associated with many diseases and questions. With recent advances in technology that allow for quick, affordable genotypic assessments (i.e., from PCR to high-throughput sequencing), researchers have begun to understand the implications of human genetic variation for the treatment of disease.124 Patient-tailored therapies hold forth great promise as a new way of treating, or preventing, disease and are an active area of research and investment.
Current State of the Art
Recent accomplishments in the field include the use of an epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, gefitinib (i.e., Iressa), for use in the treatment of non-small cell lung cancer. Scientists have found that certain EGFR mutations may predict a patient’s sensitivity to the drug, meaning that some patients are more likely to benefit than others. Moreover, in one study the mutations (and benefits of treatment) were more prevalent among Japanese patients than U.S. patients, raising general questions about the ethnic or geographic specificity of this and other cancer drugs.125
Herceptin provides another more publicized example of the potential for genomic medicine. In 1997, this drug became the first gene-based therapeutic licensed and marketed for use against breast cancer. Women with metastatic breast cancer whose cells produce the proteins HER2 and HER2/neu were given new hope in the form of this monoclonal antibody drug developed and manufactured by San Francisco-based Genentech.126 Herceptin, an erbB2 monoclonal antibody, is now licensed for use in the 20 to 30 percent of breast cancer patients who overexpress this tyrosine kinase receptor.127 Although mechanism-based cardiotoxicity has been observed, response rates of up to 60 to 70 percent have been reported for Herceptin in combination with paclitaxel or doxorubicin.128 Similar
“proof of principle” is now emerging for the clinical activity of small-molecule inhibitors of oncogenic tyrosine kinases such as Glivec (imatinib) against chronic myeloid leukemia129 and preclinical activity in tumor models driven by the tyrosine kinase activity of the platelet-derived growth factor and c-kit receptors.130
Future Applications
Understanding and harnessing genomic variation are expected to contribute significantly to improving the health of people worldwide, including the developing world.131 In recognition of this, Mexico is in the process of delivering one of the first genomic medicine platforms in Latin America, one that is expected to serve as a regional model for other countries in their efforts to ease health and financial burdens. The Mexican government and medical and biomedical research communities view the present time as a window of opportunity for investing in this emerging technological trend, so as to minimize the likelihood of needing to depend on foreign aid and sources in the future.132 Likewise, genomic medicine activities in Singapore represent another national effort to gain leverage in this field. Already, high-tech manufacturing and financial services serve as the fulcrum of the Singaporean economy. Strengthening biotechnological capacity, including genomic medicine capacity, is viewed as the next high-tech step forward to accelerated economic growth.133
Integrating personalized, or genomic, medicine into regular health care (in any country) will require overcoming two major challenges. First, it will be necessary to make the “$1,000 genome” a reality. The $1,000 genome refers to the cost of sequencing an individual’s entire genomic sequence and, although a somewhat arbitrary threshold, has come to represent the point at which the technology is finally affordable enough for widespread use. It is not clear how the $1,000 genome hurdle will be jumped, although biotech companies are trying. Some experts believe it will require a new technology. The second and arguably more significant challenge will be making the philosophical jump from the highly interventional, British-style school of medicine to a preventative, predictive health care paradigm. Genomic medicine is expected to revolutionize human medicine by altering the nature of diagnosis, treatment, and prevention. In traditional medicine, diagnosis is based on clinical criteria, treatment is population-based, and prevention is based on late-stage identification of disease. In genomic medicine, diagnosis is based on molecular criteria (e.g., the use of microarrays in cancer diagnosis), treatment is highly individualized (i.e., genomic based), and prevention is based on early-stage identification of who is at risk.
The same genomic sequences that will one day allow health care pro-
viders to identify and provide genotype-specific (and phenotype-specific) treatment may some day be exploited as targets for novel biological agents. Knowledge generated from genomic medicine could potentially be used to target specific ethnic, racial, or other population characteristics. Such weapons need not be hugely effective or even completely selective; proportional selectivity would be sufficient since, in addition to the direct effect of the weapon itself, the social tension, erosion, and (potential) fragmentation resulting from headlines of the “Mystery Virus Strikes Blacks, Spares Whites” variety would be likely to trigger effects far in excess of those from the disease itself.
While knowledge spreading from the various genome projects has fueled speculation in this area, two points should be kept in mind when considering this topic. First, the hugely large number of point mutations and other polymorphisms within the genome are not likely to lead to any selective targeting in the near future. Although techniques such as RNAi, as discussed previously, certainly have the capability to inhibit the expression of key genes with relevant single nucleotide polymorphisms (SNPs) within them, the proportion of such mutations lying in functionally important areas of the genome is small and the technical difficulties associated with exploiting them are real. Second, the idea is not new; South Africa’s Project Coast reportedly conducted experiments on vaccines designed to target fertility.134
The technology to construct such weapons exists. For almost two decades, researchers have been using adenoviruses to target tumor cells in individuals and steadily refining their techniques for directing viral entry into cells. For example, it is now possible to modify through genetic approaches the fibers used by the virus for cellular attachment so that the virus attaches to particular cell types.135,136 Studies have also shown that preferential attachment and infection of target cells can be markedly elevated.137,138
Interestingly, while the availability of the complete human genome sequence has revealed numerous SNPs and other polymorphic elements—and has consequently raised greater concern about the possibility of using biological weapons to target specific racial or ethnic populations—the ability to identify139 and exploit genetic differences among such populations does not require this new information. Adenoviruses could be used to deliver antibodies that target distinct ethnic groups with characteristic cell surface molecules, without needing to identify population-specific SNPs.140 For example, human leukocyte antigens have distinctive distributions that vary with geographic origin (e.g., the common haplotype B8-DR3 is distributed almost uniquely among Northern European Caucasians; in the Mediterranean basin, this is replaced by B18-DR3).
Modulators of Homeostatic Systems
Description
The stability and integrity (homeostasis) of the molecular circuits, pathways and networks responsible for diverse body functions are altered by disease and by exposure to noxious environmental pollutants and toxins (xenobiotics). The quest to identify the molecular circuits and control systems in each specialized cell type in the body, and to understand the perturbations that give rise to disease, is a dominant research theme in contemporary biology. Mapping the “molecular signatures” of the body’s biocircuits in health and disease is the primary technological catalyst in the development of new molecular diagnostic tests for detecting disease and the emergence of new disease classification schemes based on causal molecular pathologies rather than clinical symptoms. Analysis of the disease-induced perturbations in biocircuits also provides the intellectual foundation of modern drug discovery, which is based increasingly on rational design therapeutic agents directed against the specific molecular lesions responsible in disease etiology.
Burgeoning knowledge about the composition and regulation of homeostatic molecular circuits in the body’s cells, tissues, and organs, and their dysregulation in disease, epitomizes the dual-use dilemma created by rapid advances in systems biology. The life sciences are undergoing a profound transformation from their historical reliance on descriptive and phenomenological observations to now focus on the detailed underlying mechanisms of disease and identification of the “rule sets” that govern the assembly and function of biological systems in both health and disease. These insights hold great promise for future advances in medicine, agriculture, ecology, and the environmental sciences. But the very same knowledge about the homeostatic control of body biocircuits can be usurped for less beneficent intentions.
The rapid pace of research progress in revealing the detailed molecular circuit diagrams and control processes for every body function, dictates that the risk of evolution of new threats will escalate in parallel. In this context, the concept of a “biothreat agent” will expand beyond the current limited perspective of biothreats as being only “bugs” (i.e., pathogenic organisms) to include an entirely new category of threats—the biological circuit disruptors.141
Current State of the Art
The commercial availability of large libraries of bioactive chemical compounds, together with automated high-throughput screening meth-
ods, allows the biological activities of thousands of chemical compounds to be assayed rapidly. Combinatorial chemistry and the directed evolution methods described earlier in this chapter are now used to routinely generate chemical libraries containing 104 to 107 compounds at relatively low cost (tens of thousands of dollars). In addition to “random” screening to identify compounds with the desired biological properties, robust knowledge about how the structure of a chemical compound correlates with causation of specific biological perturbations will permit increasingly accurate predictions about how the tertiary structure of bioactive molecules correlates with their affinity for, and reactivity with, specific molecular targets in the various cell lineages of the body.
The emerging field of toxicogenomics involves profiling the changes in gene and protein expression induced by chemicals found in the industrial workplace to assess potential risk from exposure to occupational and environmental hazards. The pharmaceutical industry and drug regulatory agencies such as the FDA (and their international counterparts) also have recognized the value of toxicogenomic profiling as a new tool to detect how investigational drugs might adversely affect genes important in drug metabolism or affect homeostatic genes that may lead to acute or chronic side effects. The current heightened public and legislative concern over drug safety will likely intensify pressures for the adoption of toxicogenomics as a routine part of the drug approval process. The benefits of toxicogenomics are self-evident. Once again, however, research that reveals structure-activity relationship (SAR) correlations between chemical structure(s) and specific toxicity events provide useful grist for the design of biological circuit disruptors in malevolent hands.
More robust correlations between chemical structure and therapeutic activity and absorption, distribution, metabolism, excretion, and toxicology (ADMET) properties will also come from research in the new field of chemical genomics (also referred to as chemogenomics or chemical biology). This emerging area of research seeks to establish the SAR rule of how chemical structure defines the selective interaction of different structural classes of molecules with various families of cellular proteins.
The Chemical Genomics Center, established in June 2004, by the Molecular Libraries and Imaging Implementation Group, as part of the NIH New Pathways to Discovery theme is but one example of an initiative that may eventually lead to potent new dual-use information. The center will be part of a consortium of chemical genomics screening centers to be located across the country whose purpose will be to identify small-molecule inhibitors of every important human cellular protein or signaling pathway. Part of the rationale for the chemical genomics initiative(s) is that, in contrast to researchers in the pharmaceutical industry, many academic and government scientists do not have easy access to large libraries
of small molecules (i.e., organic chemical compounds that are smaller than proteins and that can be used as tools to modulate gene function).
The database will give academic and government researchers an opportunity to identify useful biological targets and thereby contribute more vigorously to the early stages of drug development. With plans to screen more than 100,000 small-molecule compounds in its first year of operation, one of the goals of the Chemical Genomics Center network is to explore the areas of the human genome for which small-molecule chemical probes have yet to be identified. Data generated by the network will be deposited in a comprehensive database of chemical structures (and their biological activities). The database, known as PubChem, will be freely available to the entire scientific community. In addition to screening and probe data, it will list compound information from the scientific literature.
Should this come to pass, it will offer enormous opportunities for industry and academic scientists alike to pursue novel “drugable” targets in a search for small-molecule inhibitors of certain pathways that could offer substantial clinical benefit. However, the availability of information and reagents that enable one to disrupt critical human physiological systems has profound implications for the nature of the future biological and chemical threat spectrum. The difference between the NIH and industrial efforts resides in the fate of the information produced from these large-scale screening programs. Companies view their screening data and the accompanying SARs to be proprietary assets. Their data are viewed as a source of corporate competitive advantage and are not typically placed in the public domain. In contrast, the NIH data will be placed in the public domain, with the unavoidable accompanying complication of creating a rich source of SAR information that could potentially be exploited for malevolent use.
Future Applications
In the past, the dual-use risk of bioregulators was considered minimal because of their lack of suitability for aerosolization unless microencapsulated, their limited shelf life after atmospheric release, the fact that proteins denature at very high temperatures and lose activity at low temperatures, and high purchase costs. However, new knowledge and advancing technologies, particularly encapsulation technologies (as discussed elsewhere in this chapter), have raised concerns about the dual-use risk of bioregulators.142
A greater understanding of how small molecules and naturally occurring bioregulatory peptides function in higher organisms will open up novel opportunities to design agents—for good or bad—that target par-
ticular physiological systems and processes, such as the brain and the immune system, in very precise ways. Scientists’ understanding of neuropeptides and their role in diverse physiological processes has advanced considerably over the last several decades.143 This new knowledge, combined with the almost limitless size of the consumer market for pharmaceutical compounds that alleviate pain, depression, sleep disorders, and a wide range of other mental disorders, suggests that many new potentially dual-use psychoactive compounds will be discovered in the near future, including novel compounds that affect perception, sensation, cognition, emotion, mood, volition, bodily control, and alertness.144 Several so-called “smart drugs”—brain-boosting medications that enhance memory or cognition—are already being sold or are in development.145
4. PRODUCTION, DELIVERY, AND “PACKAGING”
The ability to manipulate “biological systems” in a defined, deliberate manner—for either beneficial or malevolent purposes—depends on the ability to produce and deliver such interventions. Technologies that allow for such production and delivery are evolving very quickly, driven by the goals and needs of the pharmaceutical, agricultural, and healthcare sectors. Some of these technologies, which clearly have immense potential future impact on biology, have not been traditionally viewed as biotechnologies or as having relevance to future biological threats. A prime example is the potential now offered by developments in nanoparticle science for the creation of novel and highly efficient delivery systems for previously difficult-to-deliver biologically-active compounds.
These technologies can be subdivided into those concerned with production, packaging, and delivery. Examples of production technologies with relevance to biology include microreactor technology (as used in the chemical engineering industrial sector), microfluidics and microfabrication technologies (e.g., currently being employed for next-generation detection tools), and transgenic plants. Examples of packaging technologies with relevance to biology include microencapsulation and nanotechnology. Examples of delivery technologies with relevance to biology include aerosol technology and gene therapy and gene vector technology.
Plants as Production Platforms—“Biopharming”
Description
“Biopharming,” also called “molecular pharming,” is the harvest of bioactive molecules from mass-cultured organisms and crops for use as ingredients in industrial products and pharmaceuticals. Transgenic crop
plants, into which genes for bioactive compounds from other species have been inserted, serve as the basis of biopharming. (Biopharming differs from bioprospecting in that the latter is sourced in wild populations.) A novel advantage of biopharming is the crop-based production of vaccines and antibodies otherwise not possible or too expensive to produce using conventional methods.146
Current State of the Art
As described in Chapter 1, many different genetically engineered crop varieties with genes for therapeutic products have been developed: transgenic rice (beta carotene, human milk proteins, higher iron content, higher zinc content, low phytic acid, high phytase); transgenic potato (gene from grain amaranth for high protein content, antigens of cholera and diarrheal pathogens, and hepatitis B vaccine); transgenic maize (AIDS antigens, higher content of lysine and tryptophan, nutritive value equivalent to that of milk); transgenic fruits and vegetables (bananas, melons, brinjals [Solanum melongena], and tomatoes with subunit vaccines against rabies; AIDS antigens in tomatoes; and human glycoprotein in tomatoes to inhibit Helicobacter pylori against ulcers and stomach cancer); transgenic tobacco (human hemoglobin, human antibody against hepatitis B virus, and 50 percent lower nicotine), and genetically engineered coffee (decaffeinated by gene splicing). However, despite the existence of functional prototypes and evidence that the technology works, there are some technical, delivery, and regulatory challenges that are slowing progress in the field.147
Future Applications
Plant manufacturing platforms may provide a cost-effective means to produce vaccines, offering the ability to address some of the problems associated with global vaccine manufacture and delivery.148 They are also being used to experiment with plant-derived microbicides, with the goal of finding a cost-effective way to block HIV transmission, and they are being explored as a possible cost-effective way to produce antibodies for use against potential biowarfare agents.149
However, transgenic plants could also be engineered to produce large quantities of bioregulatory or otherwise toxic proteins, which could either be purified from plant cells or used directly as biological agents. As with legitimate production, using transgenic plants as bioreactors would eliminate the need for mechanical equipment normally associated with the process. The technology would be limited to producing protein-based agents
(transgenic plants would be largely indistinguishable from nontransgenic crops), but it could potentially provide a covert means for producing large amounts of product.150
Microfluidics and Microfabrication
Description
Microfluidics and microfabrication are rapidly growing technologies in which a wide variety of processes and manipulations are carried out at miniaturized scales (e.g., nanoliter volumes) and in automated fashion. Microfluidic, or “lab-on-a-chip,” technology underlies many recent advances in point-of-care diagnostics, including DNA analysis, immunoassays, cell analysis, and enzyme-activity measurements.151 Microfabrication involves building functional devices at the molecular size.
Fabricated on glass or plastic chips ranging in size from a microscope slide to a compact disk, microfluidic arrays require only very small (on the order of picoliters, 10−12 liters) sample and reagent volumes. The most sophisticated systems are completely integrated, with sample introduction, preprocessing (e.g., cell lysis, dilution), reagent addition, and detection all conducted on the same chip. But most systems are bulkier and rely on external detector and other devices. Limitations of the current technology include reagent stability (or instability) and the need for liquid reagent reservoirs.
Microelectromechanical systems (MEMS) are a similar miniaturized technology. Unlike microfluidic systems, MEMS devices are self-contained and do not require reagents. Swallowed-capsule technology is a popular example of a MEMS: patients swallow a capsule containing all of the miniaturized equipment necessary for taking images in the gastrointestinal tract.
Current State of the Art
Nanotechnological advances are decreasing the size of microfluidic and other miniature diagnostic systems even further. For example, Biotrove, Inc. (Waltham, MA) has developed a nanoliter sample size real-time PCR machine that, when commercially available, will allow users to analyze thousands of samples simultaneously and for a much lower per-sample cost than with currently available high-throughput microarray systems. Other sampling problems come into play at smaller volumes (e.g., the small volume may not be representative of the whole sample or population).152
Future Applications
As stated in a recent Science review on miniaturized diagnostic systems: “Farther down the road may be personalized health care with diagnosis and disease-monitoring occurring in the home with easy-to-use miniature devices.” Although this possibility may be farther into the future than the scope of this report covers, for regulatory as much as technical reasons, steps are being taken in this direction. For example, there have been several recent advances in convenient sampling methods, including breath and saliva sampling, that would be necessary before personalized diagnostic devices become a widely accepted component of personal health care.
Nanotechnology
Description
Nanotechnology, which was defined in Chapter 2, started off as little more than a clever means of making incredibly small things. In 1990, IBM scientists made headlines by painstakingly arranging 35 xenon atoms to spell out the company’s three-letter name, creating the world’s smallest corporate logo. Other scientists followed with an invisibly small “nanoguitar.” Its strings, each just a few atoms across, could be plucked by laser beams to play notes 17 octaves higher than those produced by a conventional guitar—well above the human hearing range. Novelties though they were, these feats proved that,with new tools in hand, scientists could arrange atoms as methodically as masons arrange bricks—and in doing so build materials never made in nature.
Last year alone, hundreds of tons of nanomaterials were made in U.S. labs and factories. Microscopically thin sheets of tightly woven carbon atoms are being wrapped around the cores of tennis balls to keep air from escaping; new fabrics have been endowed with nanofibers that keep stains from setting; some sunscreens have ultraviolet-absorbing nanoparticles so small they cannot reflect light, making them invisible; and tennis rackets and airplane bodies are being made with nanomaterials whose atoms have been carefully arranged to make them especially strong.
Current State of the Art153
An intriguing feature of the nanoscale is that it is the scale on which biological systems build their structural components, like microtubules, microfilaments, and chromatin.154 In other words, biochemistry is a nanoscale phenomenon. Even more intriguingly, a key property of these
biological structural components—including, of course, the DNA double helix—is self-assembly. In their quest to emulate these biological phenomena, scientists have created the field of DNA nanotechnology155 and the closely related field of DNA-based computation by algorithm self-assembly.156
Some of the most interesting nanotech research being conducted today falls within the realm of so-called DNA nanotechnology. DNA nanotechnology is the design and development of objects, lattices, and devices made of synthetic DNA. Since the DNA helix is naturally linear (i.e., unbranched), the assembly of structures or devices built with synthetic DNA requires constructing branched molecules that can then be connected to form structural networks, or motifs. The DNA motifs are combined by sticky-end cohesion, a high-specificity DNA reaction.
The DNA can be used as either “brick” and “mortar” in the construction of various kinds of nano-objects (so-called “high structural resolution DNA nanotech”) or as just mortar to join non-DNA particles (“compositional DNA nanotech”). The latter, which laboratories worldwide are involved with, can be used in many ways to organize large complexes. There are only about a dozen labs worldwide involved in high structural resolution DNA nanotech, the potential applications—which are many and varied—include architectural control and scaffolding (e.g., DNA-based computation), nanomechanical devices (e.g., nanorobotics and nanofabrication), and self-replicating nano-systems.
Self-assembling systems are completely autonomous devices that do not require the input of a person (or a robot) in order to function (i.e., as nanomechanical devices do). Last year an investigator at Purdue University made one of the first self-assembling nano-devices, in this case a DNAzyme, which can bind and cleave RNA molecules one by one.157 Unimaginable just a couple of years ago, the creation of this device epitomizes the progress that the field of DNA nanotech has achieved in just a few years.
Future Applications
The future trajectory of the field, particularly the convergence of nanotechnology and molecular biology, is unclear, although it will almost certainly have multiple medical applications, including therapeutic delivery by nanoparticles.158 In October 2004, scientists from the Institute of Bioengineering and Nanotechnology (Singapore) reported having invented a contact lens capable of releasing precise amounts of medication to treat glaucoma and other eye diseases.159 Nanobiotechnology also promises multiple new approaches to molecular detection and diagnostics.160
Just as nanotubes and other nanodevices promise novel advantageous means of drug delivery, there is considerable concern that the very same devices and particles could have inadvertent dangerous (i.e., toxic) consequences. Several recent studies have examined the possible toxicity of nanotechnology-derived products.161 Likewise, the field opens up an entirely new means of potential deliberate misuse.
Aerosol Technology
Description
Very broadly, aerosol science is an interdisciplinary field focused on the study of the presence and movement of biological particles in the earth’s atmosphere, including the impact of such particles of human populations, agriculture, and animals (including insect control).162 The widespread aerial spraying of the Bacillus thuringiensis var. kurstaki (Btk), to protect forests from damage and defoliation caused by the spruce budworm, is a good example of how aerosol technology is being used and optimized.163 Other examples of recent research in this field include a study on the use of animal models for understanding the threat to human health caused by inhalation of toxic airborne particulate matter;164 a study on wind as a potential aerosolization mechanism for dispersing microorganisms at flooded wastewater irrigation sites (reuse of partially treated domestic wastewater is increasingly being done worldwide for agricultural irrigation purposes);165 studies of plume characteristics of bioaerosols generated during the application of liquid biosolids to farmland, and the microbial risk to human health associated with this practice;166 and studies on the aerial spraying of insecticides.167
In biomedical research, aerosol science revolves around the study of the use of inhaled particulate matter as a means to treat human disease. Although its current widespread use is for local treatment of asthma and chronic obstructive pulmonary disease, direct administration of drugs to the respiratory tract has been effectively used or is being tested to treat bacterial lung infections, cystic fibrosis, and lung carcinoma. The effectiveness of aerosol delivery for systemic action is also being explored, as a novel, injection-free way to control pain and deliver various therapeutics for the treatment of diabetes, human growth hormone deficiency (in children), prostate cancer, and endometriosis.168 Compared to oral delivery, advantages of aerosolized delivery169 include its rapid speed of onset and even biodistribution.170
Current State of the Art
In the drug delivery industry the three most common types of aerosol delivery devices currently in medical use are propellant metered-dose inhalers (pMDIs), dry powder inhalers (DPIs), and nebulizers.171 Propellant MDIs are the most popular, since they are small, convenient, and self-powered (i.e., by the high-pressured contents of the “metering chamber”). The aerosol is drawn into a metering chamber, followed by propulsion of the solution as droplets into the lung. In the past, most pMDIs utilized suspensions or solutions of drugs in chlorofluorocarbons (CFCs). But CFC propellants are being phased out in favor of non-ozone-depleting hydrofluoroalkane inhalers. But the latter require device components and formulations that are different than those of CFC pMDIs, which has necessarily led to the creation of novel delivery means.172
Many new DPI devices and technologies have been developed and patented since the first one was introduced in the 1960s.173 DPIs deliver powdered dry particles into the lungs, relying on the energy produced by the forces of the inhaled airflow. Most powder products are mixtures of drug particles and large lactose carrier particles. The smaller particles are delivered to the lungs while the larger particles, which help with dispersion, are deposited on the mouthpiece. A variety of different technologies have been used in the development of DPIs, and performance varies widely among different types of inhalers. Attempts to improve the delivery of respirable dry products to the lower airways and lungs remains an active area of research.174
Although air-jet nebulizers are inconvenient devices, due to their utilization of compressed gas (and thus requiring an air compressor) and their comparatively long aersolization time, their capability to deliver a high dose over an extended time period is widely considered an advantage over pMDIs and DPIs.175 Nebulizers work by passing an air-jet stream (which is created by using compressed gas or piezoelectric ceramics) through a capillary tube that runs through a reservoir containing the drug; the drug solution is drawn out of the reservoir and deposited into the lungs in droplet form. In addition to their inconvenience, other limitations of the technology include the partial loss of drug dose during exhalation (since nebulizers generate aerosol continuously) and the large size of some of the devices. A variety of newer, “next-generation” nebulizers, which overcome some of these limitations, are being developed and produced.
In addition to the quality and features of the delivery device, critical to the delivery of the drugs to the lungs is the preparation of particles of correct size and shape for incorporation into aerosol products. Advances in powder technology and particle engineering play a significant role in
improving powder production and aerosol drug formulation (e.g., by improving particle dispersibility, control of particle morphology, physical and chemical stability). For example, supercritical fluid (SCF) processing has recently emerged as an alternative technology for designing particles to use in metered-dose and dry powder inhalers.176 SCFs are substances that exist as a single phase but have the properties of both liquids and gases (at certain temperatures and pressures), and they can extract compounds from complex substrates much more quickly than liquid organic solvents can.
In addition to its pharmaceutical applications, SCF technology is being used in the food industry (for decaffeination of coffee and tea and extraction of edible oils); the flavor and fragrance industry (for extraction of aromas and flavors); the nutraceutical industry (for the extraction of active ingredients for nutraceuticals and purification of antioxidants for nutraceuticals), the paint/coating industry (for the production of small particles for paint coating applications); and for a variety of other industrial purposes (e.g., purification of natural and synthetic materials and polymers and production of small particles for explosives).
Future Applications
Biomedical advances in aerosol delivery technology are expected to improve drug delivery and patient adherence. Several companies are pursuing aerosolized insulin delivery as a non-invasive alternative to injectable insulin. It is widely believed that, once proven safe for prolonged use, aerosolized insulin delivery will stimulate further activity in this already very active field. Aerosol delivery is also being explored as a means of gene therapy.
Advances in drug delivery technology, including aerosol delivery, have raised concerns about the use of bioregulators for nefarious purposes. In the past, bioregulators have not generally been viewed as potential dual-use agents, largely because of the lack of effective delivery technology.177
The dual-use risk of bioregulators was considered to be minimal due to their lack of suitability for aerosolization unless microencapsulated, their limited shelf life after atmospheric release, the fact that proteins denature at very high temperatures and lose activity at low temperatures, and high purchase costs. However, new knowledge and advancing technologies, particularly delivery technologies, have raised concerns about the dual-use risk of bioregulators. Potential delivery platforms include the use of bacterial plasmids or viral vectors for cloning the genes that encode bioregulators; use of transgenic insects (i.e., to secrete and inoculate the bioregulators); nanoscale delivery systems (e.g., engineered pro-
teins either within or bound to nanotubes); and microencapsulated delivery systems (i.e., incorporating vectors or the proteins themselves into biodegradable microspheres or liposomes for controlled release).178 Given that anything less than 3 microns in diameter is respirable across what amounts to a 75-square-meter absorptive surface, miniaturization of respiratory delivery systems comes with considerable dual-use risk. Moreover, transgenic plants could be put to dual use as bioregulator-production factories.
Microencapsulation Technology
Description
Microencapsulation is the envelopment of small solid particles, liquid droplets, or gas bubbles with a protective coating derived from any of a number of compounds (organic polymer, hydrocolloid, sugar, wax, fat, metal, or inorganic oxide). The capsules, which are basically miniature containers that protect their contents from evaporation, oxidation, and contamination and can be engineered with any of a variety of unique release mechanisms (e.g., from controlled, delayed, targeted release to biodegradable or salt-induced release), have countless applications.
Microencapsulation is not a new technology. Between the late 1940s and early 1960s, the concept of chemical microencapsulation generated interest in the pharmaceutical industry as an alternative mode of drug delivery that could offer sustained controlled release. Researchers and entrepreneurs continue to utilize and investigate advances in microencapsulation technology in efforts to make dosages more palatable, make active ingredients more stable and/or soluble, and otherwise improve drug delivery.179 In the decades since the technology first emerged, many other life sciences industrial sectors have benefited tremendously from non-pharmaceutical applications of microencapsulation. In fact, it was partly in response to potential agrochemical applications of encapsulation technology that the Controlled Release Society, an international organization with 3,000 members from more than 50 countries, was formed in the mid-1970s (microencapsulation is the most common but not the only form of controlled release). As defined on its Web site, controlled release is “the field of scientific activity concerned with the control in time and space of the biological effects of therapeutic agents in human and animal health, and of other active agents in environmental, consumer and industrial applications.”180
According to data provided by the Southwest Research Institute, the number of U.S. patents for encapsulation processes increased from about 1,250 during 1976-1980 to about 8,500 during 1996-2001.181 U.S. patents
for nanoencapsulation grew from near zero to about 1,000 over the same time period.
There are two general categories of microencapsulation processes: physical (e.g., spray drying, fluid bed coating, co-extrusion, rotary disk atomization) and chemical (e.g., polymerization, phase separation, solvent evaporation, coacervation). Between 1996 and 2002, polymerization was the most commonly used process, based on U.S. patent data, followed by spray drying and rotary disk atomization.
Current State of the Art
Today, microencapsulation technology is being used in water treatment (to remove emulsified oils, heavy metals, phosphates, and suspended solids from wastewater), food and agriculture (to improve taste and mask odor; stabilize thermal, oxidative, and shelf-life properties of ingredients; and allow for more effective absorption of nutrients and vitamins), and in the cosmetics industry (to create “eye appeal” or a specific or special feel in a wide range of personal care products). It has also been used as a way to manage mercury-contaminated and other hazardous wastes.182
Examples of recent use and exploration of this technology include an investigation by University of Saskatchewan researchers into the use of microencapsulated engineered cells as an alternative approach to cancer treatment.183 The cells had been engineered to release a compound that kills tumor cells (i.e., functional necrosis factor-alpha). Implantation of encapsulated cells (into a mouse model system) led to tumor regression and slower tumor growth. In another study, researchers from the Netherlands tested the release, upon chewing, of flavored microencapsulates in Gouda cheese (the microencapsulates contained sunflower oil, lemon, and orange oil flavors).184 Japanese researchers recently demonstrated the use of a novel nanoencapsulation drug delivery method for the external treatment of photo-damaged skin.185 Advanced BioNutrition Corporation (Columbia, Maryland) was recently awarded a National Science Foundation grant to further develop its proprietary microencapsulation technology for the incorporation of functional ingredients—such as enzymes, fatty acids, probiotics, even vaccines—into its animal and human food products.186 The company will use the money to scale up its microencapsulation technology production process.
Future Applications
An exciting future application is the transplantation of encapsulated live cells for therapeutic purposes.187 Other future applications range from
teddy bears that release a scent to help children sleep to novel military applications. In January 2005, for example, Northrop Grumman (San Diego, CA) announced the development of new encapsulation technology that allows non-marinized188 weapons and vehicles to be released by submarines.
Gene Therapy Technologies
Description
Gene therapy is an experimental technique that uses “healthy” genes to treat or prevent disease. In most gene therapy studies, a “normal” gene is inserted into the genome to replace an “abnormal,” disease-causing gene. A carrier molecule—called a vector—must be used to deliver the “healthy” gene to a recipient’s target cells. Currently, the most commonly used vectors are viruses (including retroviruses, adenoviruses, adeno-associated viruses, and herpes simplex viruses) that have been genetically altered to carry normal human DNA. Nonviral options for gene delivery include the direct introduction of therapeutic DNA into target cells, although direct administration can only be used with certain tissues and requires large amounts of DNA (see Figure 3-4).
FIGURE 3-4 Viral vectors.
SOURCE: James Benjamin Petro, presentation to the committee, February 2004.
State of the Art
Gene therapy is still experimental, and most of the research performed to date has been conducted in animal trials (from rodents to primates). For example, in a study that appeared in Nature Medicine in March 2005, using a guinea pig model system, researchers from the University of Michigan and Kansai Medical University, Japan, reported that they had used gene therapy to restore hearing in mature deaf animals.189 The evidence suggests that gene therapy can be used to regenerate functional hair cells, which are necessary to restore hearing, by using (in this case) an adenovector to deliver the “healthy” gene into nonsensory cells that reside in the deaf cochlea. The introduced gene, Atoh1 (also known as Math1), encodes a basic helix-loop-helix transcription factor and key regulator of hair cell development. Upon delivery, hearing is substantially improved.
The few human clinical trials that have been conducted have not been as successful as originally hoped.190 Although substantial progress has been made, and some clinical successes seem to be on the horizon, further vector refinement and/or development is required before gene therapy will become standard care for any individual disorder.
Future Applications
When gene therapy does become a clinical reality, it will be used to correct faulty or defective disease-causing genes. But just as it will be used to delivery “healthy” genes into cells and tissues, gene therapy could potentially be used to deliver harmful genes.
Targeting Biologically-Active Materials to Specific Locations in the Body
The efficacy and safety of medical drugs, imaging agents, and vaccines depend on the ability to deliver these agents to the right location in the body and, ideally, with precision targeting only to the cells of interest. Selectivity in drug delivery reduces the exposure of nontarget tissues to the drug, thereby reducing the risk of unwanted drug actions and adverse events. However, this obvious therapeutic need is far from easy to achieve in practice. Selective targeting of bioactive molecules remains a largely unfulfilled objective in clinical therapeutics. The pharmaceutical and biotechnology industries, including companies that specialize only in the design of ways to optimize drug delivery, are investing substantial sums in research and development to achieve this attractive, yet elusive, goal.
Considerable ingenuity has been exhibited in designing “targeting”
vehicles and “homing” systems for precision delivery of drugs and imaging agents. These range from efforts to deliver materials to specific zones in the body (e.g., aerosol delivery to the lungs, selective drug delivery to different regions of the gastrointestinal tract) to the more challenging objectives of targeted delivery to specific cell types (e.g., cancer cells versus their normal counterparts) or delivery of a drug or other bioactive agents to a specific compartment inside the cell (e.g., nuclear uptake of genes into chromosomal DNA for gene therapy or targeted therapeutic ablation of deleterious genes).
A broad repertoire of targeting vehicles have been examined in this research effort. These include carrier particles containing encapsulated drugs (e.g., liposomes, nanoparticles, dendrimers); exploitation of the “homing” ability of microorganisms to bind selectively to specific cells (e.g., viruses or bacteria as vectors for targeted delivery of genes and proteins); and the coupling of drugs to cognate carrier molecules designed to recognize only the desired cell type and then release their therapeutic payload. A unifying theme linking these different approaches lies in engineering suitable “molecular recognition” systems whereby cognate molecules in/on the carrier system recognize and attach to molecules expressed exclusively on the desired target cell, tissue, or organ. Additional cognate molecular interaction systems can be designed to enhance the efficiency of drug uptake by cells once selective targeting has occurred and for directing the delivered drug or gene to the correct location inside the cell.
Two different technical approaches underpin technical strategies for targeted drug delivery. The first incorporates the targeting (homing) property into the drug itself so that it will interact only with target cells that bear a “receptor” molecule that recognizes a structural region (domain) on the drug molecule. In the second approach the cognate properties required for recognition and binding to target cells are engineered into a drug carrier rather than the drug itself. Drugs are associated with the carrier either via passive encapsulation (e.g., particulate carriers) or by chemical coupling to the carrier. Both approaches exploit cognate molecular interactions as a common design principle. Targeted delivery is achieved as a consequence of molecular recognition events that limit the interaction of the drug and/or the drug carrier to only those cells that express a specific molecular determinant that interacts with the drug or drug-carrier complex.
As emphasized in the earlier section on how knowledge of the body’s biocircuits can be used for both constructive and abusive purposes, the technical platforms for precision drug targeting pose a similar dual-use problem. Knowledge of how to target bioactive materials to specific cells can be usurped to disrupt or destroy vital functions in humans, animals,
or plants. However, the technical ease with which such assaults could be mounted will be influenced by the location of the target in the targeted host and the anatomic barriers that a targeting system must breach in order to reach its molecular locus of action.
One of the more concerning assaults, yet attainable even with today’s delivery technology, could arise from the use of targeted delivery systems to insert genes into chromosomal DNA. For example, viral delivery vectors developed for human gene therapy exploit the ability of viruses to bind selectively to specific cell types as a way to deliver genes encapsulated inside the viral particle into the target cells. The question of whether these viral delivery systems are applied to beneficent or malevolent goals is defined solely by the nature of the genetic payload incorporated into the vector. Although therapeutic gene therapy has yet to attain routine clinical utility, the extensive research literature on gene transfection technologies using viruses and various particulate carriers has demonstrated the feasibility of inserting exogenous genes in multiple cell types in the body. Future improvements in the efficiency of these delivery technologies can be confidently expected, with accompanying expansion in the horizons of both therapeutic and nefarious utility.
The delivery and expression of genes that code for the uncontrolled production of highly potent hormones and other natural bioactive mediators involved in homeostasis offer the simplest example of how this knowledge could be abused and used to expand the emerging threat spectrum. Alternatively, rather than using a transfected gene directly to produce a bioactive product to perturb body function, the transfected gene could act as a trigger for the abnormal expression or destruction of other genes vital to body homeostasis. In either of these settings the introduced gene is designed to integrate into the chromosomal DNA of the host. The disruptive effects could be manifest immediately as an acute event or the gene could lie silent in the genome for activation at a later time by a second external trigger.
An aphorism frequently cited in the design of drug delivery systems is that “the opportunities are limited only by the imagination of the inventor.” Theoretically, the ability to design drugs and carrier vehicles endowed with cognate molecular properties that enable them to home selectively to the desired target in the body is limited only by the availability of suitable molecular recognition molecules that can be incorporated into the delivery system to confer recognition and binding by molecules unique to the desired target cell. The availability of relevant molecular cognate pairs for the delivery system and for the target is an obligate prerequisite for targeting. However, this is but one component in the engineering of targeted delivery systems. For therapeutic applications, the tar-
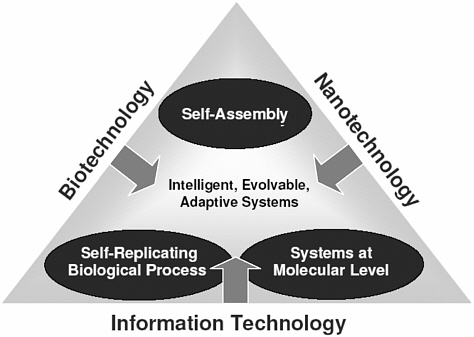
FIGURE 3-5 Converging technologies. Biotechnology, nanotechnology, and information technology are converging in ways that will enable humans to do things never dreamt of until now.
SOURCE: Michael Morgan, presentation during the Cuernavaca workshop, September 2004.
geting system must also exhibit suitable absorption, distribution, metabolism, excretion, and toxicology (ADMET) properties.
Complementarity and Synergy of Technologies
Some futurists consider the convergence of bio-, nano-, and information technologies, along with the neuro- and cognitive sciences, a transformation that will prove as powerful as the Industrial Revolution (Figure 3-5). However, the details and impact of possible convergent events are unclear at this time.
Enabling technologies are those that interact with each other to create novel products that would otherwise be impossible to achieve. Nanotechnology enables other technologies by providing a common hardware for molecular engineering and allowing for the realization of desirable architectures. Nanotechnology enables biotechnology by developing new imaging techniques, probes, and sensors; it also contributes to the miniatur-
ization demands of information technologies. Biotechnology enables other technologies by identifying chemical and physical processes and algorithmic structures in living systems that have a genetically based material organization. It enables nanotechnology by providing a paradigm that nanotechnologists use in developing systems; much of the work in nanotechnology involves mimicking biotechnological processes while simultaneously redesigning them to fit particular purposes. Biotechnology enables information technology by providing new systems of computing, some of which may be based on DNA. Information technology enables other technologies through its ability to represent physical states as information and model processes. It provides the computing power that is essential to all research; it enables nanotechnology through precision control of patterning and intervention; and it enables biotechnology by providing the means to model complex processes and thereby solve difficult research problems.
In addition to convergence, which leads to the emergence of entirely new disciplines such as DNA nanotechnology and bioinformatics, technologies combine and converge on a smaller, less dramatic scale all the time. In terms of future potential threats, one should note the importance of combinations or interactions involving technologies in any of the first three categories—the acquisition of biological or molecular diversity, directed design, and understanding and manipulation of biological systems—and technologies in the fourth category: production, delivery, and packaging. In other words, the impact, both beneficial and detrimental, of a small-molecule agent, synthetic agent, or an agent bred through “DNA shuffling” is enhanced by appropriate packaging and delivery. Indeed, growing concerns about the dual-use risk of bioregulators are partly in response to advances in microencapsulated delivery systems, which make the use of bioregulators for either beneficial or nefarious purposes more feasible.
Based on extensive deliberations on a wide range of advancing technologies with relevance to the life sciences, including many technologies and fields of knowledge not traditionally viewed within the rubric of biotechnology, the committee was particularly struck by the extent to which various tools and technologies are interacting and converging191—both additively and synergistically—and creating unanticipated opportunities for these technologies to be used for either beneficial or malicious intent (or with beneficial intent but unintended consequences). As already mentioned, the convergence of nanotechnology and molecular biology serves as a prime example of how an entirely new discipline, DNA nanotechnology, can emerge unexpectedly and with profound consequences. Nanotechnology is also merging with encapsulation and micro-
fluidic technologies, providing the means for further miniaturization of already very low-volume biological sampling, detection, delivery, and other processes.
As one example, synthetic biologists are using their new tools in conjunction with nanotechniques to program cells with decision-making therapeutic power. For example, researchers have designed a prototype “DNA computer” with the capacity to logically analyze mRNA disease indicators in vitro (i.e., in this case, early signs of prostate and lung cancer) and control the administration of biologically active ssDNA molecules, including drugs.192 The procedure is relatively innocuous, requiring the injection of a very small amount of fluid containing billions of nanoparticles, each of which operates as a tiny computer by effectively interrogating the cell and detecting the presence of diagnostic DNA markers (e.g., mutated mRNA sequences or underexpressed or overexpressed mRNA). If the markers are present, the nanoparticle sends out a therapeutic short nucleic acid that can affect the levels of gene expression.
The field of bioinformatics represents another key example of converging technologies—in this case biology, computer science, and information technologies—all of which have merged to form what is now a single discipline. Over the past 10 years, major advances in the field of molecular biology, coupled with advances in genomic technologies, have led to an explosive growth in biological information generated by the life sciences community. This deluge of genomic information, in turn, has led to an absolute requirement for computerized databases to store, organize, and index the data and for specialized tools to view and analyze the data. These databases and tools comprise the field of bioinformatics. Increasingly, biological studies begin with a scientist surveying databases to formulate specific hypotheses or design large-scale experiments, representing a dramatic shift in biology from a purely lab-based to an information-based science. Moreover, the growing availability of vast amounts of biological and other relevant information (e.g., small-molecule libraries) will also allow nonspecialists to tinker with or design constructs that, in the past, would have required years of education or training.
CONCLUSION
“During the century just begun, as our ability to modify fundamental life processes continues its rapid advance, we will be able not only to devise additional ways to destroy life but will also be able to manipulate it—including the processes of cognition, development, reproduction, and inheritance.”—Matthew Meselson193
It is difficult to predict what the global technology landscape will look like in 20, 10, or even 5 years into the future. But it is not difficult to anticipate that as advances are made, so too will opportunities for misuse. This chapter summarizes information on emerging technologies that are expected to have significant economic, societal, and dual-use risk impacts in the near future. As highlighted during the committee’s international workshop in Cuernavaca, prominent among these are advances in knowledge and delivery technology that have increased the dual-use potential and risk of nonlethal bioregulators and the convergence of nano- and biotechnology in the form of DNA nanotechnology.
A major theme that emerged from the committee’s deliberations in Mexico was the notion that pathogens are not the only problematic agents of biological origin. Some argue that bioregulators,194 which are nonpathogenic organic compounds, may pose a more serious dual-use risk than had previously been appreciated, particularly as improved targeted delivery technologies have made the potential dissemination of these compounds much more feasible than in the past. This shift in focus highlights the reality that the materials, equipment, and technology necessary for disseminating and delivering the agents to their intended recipient(s) are equally, if not more, important than the agents themselves in terms of their dual-use risk.
The immune and neuroendocrine systems195 are particularly vulnerable to bioregulator modification. In fact, the capacity to develop bioweapons that can be aimed at the interaction of the immune and neuroendocrine systems again points to a shift in focus from the agents to, in this case, how a range of agents can be exploited (or created) to affect the human body in targeted, covert, and insidious ways.
A controversial issue that arose from these discussions is how all research on immune system evasion could be considered potentially dangerous, thus highlighting the very important need to uphold the norms of the Biological and Toxin Weapons Convention. Another important theme that emerged from discussions of the material presented here is the notion of time and how the advancing technology landscape has an uncertain future and unpredictable dual-use risk implications. This unpredictability poses a significant challenge for developing and implementing a strategy to manage these risks. These challenges—and potential solution sets—are discussed in the following chapter.
ENDNOTES
|
|
minute fission time, only about 1045 have existed over the history of the earth, which is tiny compared to the number of possible 108 base pair DNA sequences. |
|
3 |
See discussion of virulence and evolution of pathogens in Chapter 1. |
|
4 |
Agarwal, K.L., et al. 1974. Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature 227(5253):27-34. |
|
5 |
Cello, J., et al. 2002. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297(5583):1016-1018. |
|
6 |
Smith, H.O., C.A. Hutchison, III, C. Pfannkoch, and J.C. Venter. 2003. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proceedings of the National Academy of Sciences 100(26): 15440–15445. |
|
7 |
Wade, N. 2005. “A DNA success raises bioterror concern. New York Times (January 12). Many experts in the field consider this view alarmist, since not only is the smallpox virus longer, but it cannot self-generate from its nucleotide sequence alone. |
|
8 |
Carlson, R. 2003. The pace and proliferation of biological technologies. Biosecurity and Bioterrorism: Biodefense Strategy, Practice, and Science 1(3):203-214. |
|
9 |
Carr, P.A., et al. 2004. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Research 32(20); Richmond, K.E., et al. 2004. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Research 32(17):5011–5018; Tian, J. et al. 2004. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432(7020):1050-1054. |
|
10 |
Tian, J. et al. 2004. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432(7020):1050-1054. |
|
11 |
Carr, P.A. et al. 2004. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Research 32(20):e162. |
|
12 |
Elowitz, M.B. and S. Leibler. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403(6767):335-338. |
|
13 |
Martin, V.J., et al. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nature Biotechnology 21(7):796-802. |
|
14 |
Hutchinson, C.A., et al. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286(5447):2165-2169. |
|
15 |
Martin, V.J. et al. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nature Biotechnology 21(7):796-802. |
|
16 |
Church, G. 2004. A synthetic biohazard non-proliferation proposal. Updated May 21, 2005. Available online at arep.med.harvard.edu/SBP/Church_Biohazard04c.htm [accessed January 5, 2006]. The Alfred P. Sloan Foundation recently funded an joint activity by the Massachusetts Institute of Technology, the Venter Institute, and the Center for Strategic and International Studies to examine the benefits and risks of synthetic genomics and develop and analyze policy options for governance of the relevant technologies. A press release issued by the three institutions describing this study may be found online at www.csis.org/press/pr05_23.pdf. |
|
17 |
Mann, C.C. 1999. Crop scientists seek a new revolution. Science 283(5400): 310-314. |
|
18 |
Crameri, A. et al. 1998. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391(6664):288-291. |
|
19 |
Ness, J.E. 1999. DNA shuffling of subgenomic sequences of subtilisin. Nature Biotechnology 17(9):893-896. |
|
20 |
Zhang, Y.Z. et al. 2002. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415(6872):644-646. |
|
21 |
Pekrun, K. et al. 2002. Evolution of a human immunodeficiency virus type 1 variant with enhanced replication in pig-tailed macaque cells by DNA shuffling. Journal of Virology 76(6):2924-2935. |
|
22 |
Leong, S.R. et al. 2003. Optimized expression and specific activity of IL-12 by directed molecular evolution. Proceedings of the National Academy of Sciences 100(3):1163-1168. |
|
23 |
Soong, N.W., et al. 2000. Molecular breeding of viruses. Nature Genetics 25 (4):436-439; Powell, S.K. et al. 2000. Breeding of retroviruses by DNA shuffling for improved stability and processing yields. Nature Biotechnology 18(12):1279-1282. |
|
24 |
Much of the information in this section is adapted from Strobel, G. and B. Daisy. 2003. Bioprospecting for microbial endophytes and their natural products. Microbiology and Molecular Biology Reviews 67(4):491-502. Available online at www.pubmedcentral.nih.govarticlerender.fcgi?tool=pubmed&pubmedid=14665674, [accessed March 24, 2005]. |
|
25 |
Grabley, S. and R. Thiericke, eds. 1999. Drug discovery from nature. Berlin: Springer-Verlag; 3-33. |
|
26 |
Concepcion, G.P. et al. 2001. Screening for bioactive novel compounds. In Pointing, S.B. and K.D. Hyde, eds. 2001. Bio-exploitation of filamentous fungi. Hong Kong: Fungal Diversity Press; 93-130 |
|
27 |
Wani, M.C. et al. 1971. Plant antitumor agents, VI. The isolation and structure of taxol, anovel antileukemic and antitumor agent from Taxus brevifolia. Journal of the American Chemical Society 93:2325-2327. |
|
28 |
Pace, N.R. 1997. A molecular view of microbial diversity and the biosphere. Science 276(5313):734-740; Venter, J.C. et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304(5667):66-74. |
|
29 |
Demain, A.L. 2000. Microbial natural products: a past with a future. In Wrigley, S.K., M.A. Hayes, R. Thomas, E.J.T. Chrystal, and N. Nicholson, eds. Biodiversity: new leads for pharmaceutical and agrochemical industries. The Royal Society of Chemistry, Cambridge, United Kingdom; 3-16. |
|
30 |
Findlay, J.A. et al. 1997. Insect toxins from an endophyte fungus from wintergreen. Journal of Natural Products 60:1214-1215. |
|
31 |
Strobel, G. and B. Daisy. 2003. Bioprospecting for microbial endophytes and their natural products. Microbiology and Molecular Biology Reviews 67(4):491-502. |
|
32 |
Lorenz, P. and J. Eck. 2005. Metagenomics and industrial applications. Nature Reviews. Microbiology 3(6):510-516. |
|
33 |
Tyson, G.W. et al. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428(6978):37-43; Venter, J.C. et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304(5667):66-74; Tringe, S.G. et al. 2005. Comparative Metagenomics of Microbial Communities. Science 308(5721):554-557. |
|
34 |
This search for novel microbial genomes to identify useful products is |
|
|
achieved through the use of laboratory methods and queries of bioinformatics “libraries.” |
|
35 |
Marshall, W.F. 3rd et al. 1994. Detection of Borrelia burgdorferi DNA in museum specimines of Peromyscus leucopus. Journal of Infectious Diseases 170:1027-1032; Mills, J.N. et al. 1999. Long-term studies of hantavirus reservoir populations in the southwestern United States: A synthesis. Emerging Infectious Diseases 5(1):135-142; Monroe, M.C. et al. 1999. Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerging Infectious Diseases 5(1):75-86. |
|
36 |
Relman, D.A. 2002. Mining the natural world for new pathogens. American Journal of Tropical Medicine and Hygiene 67(2):133-134. |
|
37 |
Parola, P. et al. 2002. First molecular evidence of new Bartonella s in fleas and a tick from Peru. American Journal of Tropical Medicine and Hygiene 67(2):135-136. |
|
38 |
Breitschwerdt, E.B. and Kordick, D.L. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clinical Microbiology Reviews 13(3):428-438. |
|
39 |
In higher eukaryotes, biological processes such as cellular growth and organogenesis are mediated by differential gene expression. To understand molecular regulation of these processes, differentially expressed genes of interest must be identified, cloned, and studied in detail. Subtractive cDNA hybridization has been a powerful tool in the identification and analysis of differentially expressed cDNAs. See www.evrogen.com/t6.shtml. |
|
40 |
Merrifield, R.B. 1963. Solid phase peptide synthesis: the synthesis of a tetrapeptide. Journal of the American Chemical Society 85:2149-2154. |
|
41 |
A more detailed understanding of how the technology works requires understanding the basic chemistry of polypeptide formation: the general chemical formula for amino acids is H2NCH(R)CO2H. Amino acids can be linked together to form peptides by reacting the −NH2 group of one amino acid with the −CO2H group of another, thus forming an amide bond. Solid-phase synthesis involves reacting the −CO2H group with a CH2Cl group on the resin, thereby leaving the −NH2 group free to form an amide bond with the second amino acid. The second amino acid is structurally modified, prior to mixing with first amino acid, in order to render its −NH2 group incapable of participating in an amide-forming reaction. The now protected second amino acid is added to the reaction mixture and a dipeptide, attached to the solid support, is created. The protecting group of the new dipeptide is removed, and a third protected amino acid is added to the mixture, resulting in a tripeptide. The process is continued until the desired product is created. |
|
42 |
Geyson, M.H. et al. 1984. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proceedings of the National Academy of Sciences 81(13):3998-4002. |
|
43 |
Houghton, R.A. 1985. General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proceedings of the National Academy of Sciences 82(15): 5131-5135. |
|
44 |
The general approach devised by Geyson and Houghten was modified further in the early 1990s, when Kit Lam developed a rapid method for producing and evaluating random libraries of millions of peptides. Initially applied to pep- |
|
|
tides, solid phase synthesis was gradually extended to produce libraries of druglike small molecules, which were of greater interest to the drug discovery industry. In the early 1990s, Jonathan A. Ellman, University of California, Berkeley, used Geyson’s multi-pin approach to create a library of 192 structurally diverse benzodiazepines. Concurrently, Sheila H. DeWitt, then at Parke-Davis Pharmaceutical Research, Michigan, reported a technique and apparatus for the multiple, simultaneous synthesis of so-called “diversomers” (collections of organic compounds, including dipeptides, hydantoins, and benzodiazepines). These studies represented some of the earliest techniques for generating small molecule libraries. |
|
45 |
Sanchez-Martin, R.M. et al. 2004. The impact of combinatorial methodologies on medicinal chemistry. Current Topics in Medicinal Chemistry 4(7): 653-669. |
|
46 |
Needles, M.C. et al. 1993. Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proceedings of the National Academy of Sciences 90(22):10700-10704. |
|
47 |
Ohlmeyer, M.H.J. et al. 1993. Complex synthetic chemical libraries indexed with molecular tags. Proceedings of the National Academy of Sciences 90(23):10922-10926; Moran, E. J., et al. 1995. Radio frequency tag-encoded combinatorial library method for the discovery of tripeptide-substituted cinnamic acid inhibitors of the protein tyrosinase phosphatase PTP1B. Journal of the American Chemical Society 117(43):10787-10788; Nicolau, K.C. et al. 1995. Radiofrequency encoded combinatorial chemistry. Angewandte Chemie International Edition 34(20): 2289-2291. |
|
48 |
Reader, J. C. 2004. Automation in medicinal chemistry.Current Topics in Medicinal Chemistry 4(7):671-686. |
|
49 |
Matzger, A.V. et al. 2000. Combinatorial approaches to the synthesis of vapor detector arrays for use in an electronic nose. Journal of Combinatorial Chemistry 2(4):301-304. |
|
50 |
Wheelis, M. 2002. Biotechnology and biochemical weapons. The Nonproliferation Review Spring:48-53. |
|
51 |
Austin, C.P., L.S. Brady, T.R. Insel, and F.S. Collins. 2004. NIH Molecular Libraries Initiative. Science 306(5699):1138-1139. |
|
52 |
See also, discussion of this issue in Chapter 1 “The NIH Roadmap.” |
|
53 |
Major Histocompatibility Complex (protein complexes that present antigens to lymphocytes). |
|
54 |
Berman, H.M., J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, and P.E. Bourne. 2004. The Protein Data Bank. Nucleic Acids Research 28(1):235-242. See pdbbeta.rcsb.org/pdb/static.do?p=general_information/pdb_statistics/content_growth_graph.html [accessed January 5, 2006]. |
|
55 |
The Smallpox Research Grid project distributed a screensaver to thousands of home computer owners to perform these calculations to identify drugs that might interfere with the enzyme that unwinds variola DNA to permit replication. The project is described at www.chem.ox.ac.uk/smallpox/news.html. (Altogether over 39,000 years of computer time were devoted to the project in less than six months, screening 35 million molecules against eight models of the target protein.) |
|
56 |
Yokobayashi, Y. et al. 2002. Directed evolution of a genetic circuit. Proceedings of the National Academy of Sciences 99(26):16587-16591. |
|
57 |
Registry of Standard Biological Parts. The Endy Lab, Massachusetts Institute of Technology. See parts.mit.edu/ [accessed January 5, 2006]. |
|
58 |
Elowitz, M.B. and S. Liebler. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403(6767):335-338. |
|
59 |
Atkinson, M.R. et al. 2003. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113(5):597-607. |
|
60 |
Looger, L.L. et al. 2003. Computational design of receptor and sensor proteins with novel functions. Nature 423 (6936):185-90; DeGrado, W.F. 2003. Biosensor Design. Nature 423(6936):132-133. |
|
61 |
Benenson, Y. et al. 2004. An autonomous molecular computer for logical control of gene expression. Nature 429(6990):423-429. |
|
62 |
Ferber, D. 2004. Microbes made to order. Science 303(5655):158-161. |
|
63 |
The Center for Strategic & International Studies (CSIS), the J. Craig Venter Institute (Venter Institute), and the Massachusetts Institute of Technology (MIT) have initiated a project, funded by the Alfred P. Sloan Foundation, to examine the societal implications of synthetic genomics, exploring risks and benefits as well as possible safeguards to prevent abuse, including bioterrorism. See further description online at www.csis.org/press/pr05_23.pdf. |
|
64 |
Racaniello, V.R. and Baltimore, D. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214(4523):916-919. |
|
65 |
Ahlquist, P. et al. 1984. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proceedings of the National Academy of Sciences 81(22):7066–7070. |
|
66 |
Rice, C.M., et al. 1989. Transcription of infectious yellow fever RNA from full-length cDNA templates produced by in vitro ligation. New Biologist1(3):285-296. |
|
67 |
Rice, C.M. et al. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants.Journal of Virology 61(12): 3809–3819. |
|
68 |
Satyanarayana, T. et al. 1999. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proceedings of the National Academy of Sciences 96(13):7433-7438. |
|
69 |
Van Dinten, L.C. et al. 1997. An infectious arterivirus cDNA clone: Identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proceedings of the National Academy of Sciences 94(3):991–999. |
|
70 |
Masters, P.S. 1999. Reverse genetics of the largest RNA viruses. Advances in Virus Research 53:245-64. |
|
71 |
Almazán, F., et al. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proceedings of the National Academy of Sciences 97(10):5516–5521. |
|
72 |
Yount, B., et al. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proceedings of the National Academy of Sciences 100(22):12995–13000. |
|
73 |
Negative-stranded RNA viruses have a genome consisting of one or more molecules of single-stranded RNA that is of opposite polarity (i.e., complementary) to the positive-sense mRNA that encodes their proteins. |
|
104 |
See phx.corporate-ir.net/phoenix.zhtml?c=141787&p=irol-newsArticle&ID=610478&highlight= |
|
105 |
Soutschek, J. et al. 2004. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432(7014):173-178. |
|
106 |
Check, E. 2004. Hopes rise for RNA therapy as mouse study hits target. Nature 432(7014):136. |
|
107 |
Voorhoeve, P.M. and R. Agami. 2003. Knockdown stands up. Trends in Biotechnology 21(1):2-4. |
|
108 |
Ellington, A.D. and J. W. Szostak. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346(6287):818-822; Tuerk, C. and L. Gold. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249(4968):505-510. |
|
109 |
Block, C. et al. 2004. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics 4(3):609-618; Jayasena, S.D. 1999. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clinical Chemistry 45(9):1628-1650; Mayer, G. and A. Jenne. 2004. Aptamers in research and drug development. Biodrugs 18(6): 351-359. |
|
110 |
Mayer, G. and A. Jenne. 2004. Aptamers in research and drug development. Biodrugs 18(6):351-359. |
|
111 |
Wang, K.Y. et al. 1993. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry 32(8):1899-1904; Li, W.X., et al. 1994. A novel nucleotide-based thrombin inhibitor inhibits clot-bound thrombin and reduces arterial platelet thrombus formation. Blood 83(3):677-82. |
|
112 |
See www.archemix.com/press/pr_jun04.html [accessed March 27, 2005]. |
|
113 |
See www.eyetk.com/clinical/clinical_index.asp [accessed March 27. 2005]. |
|
114 |
Burbulis, I. et al. 2005. Using protein-DNA chimeras to detect and count small numbers of molecules. Nature Methods 2(1):31-37. |
|
115 |
Mayer, G. and A. Jenne. 2004. Aptamers in research and drug development. Biodrugs 18(6):351-359. |
|
116 |
The following section is taken from the Executive Summary of an NRC report entitled: Catalyzing Inquiry at the Interface of Computing and Biology (December 2005). |
|
117 |
National Research Council. 2005. Catalyzing Inquiry at the Interface of Computing and Biology. Washington, DC: The National Academies Press. |
|
118 |
Pacific Northwest National Laboratory. 2005. Genomic sequences processed in minutes, rather than weeks. The Daily Nonproliferator, June 21. Available online at www.pnl.gov/news/2005/05-45.stm. |
|
119 |
Wolkenhauer, O. et al. 2005. The dynamic systems approach to control and regulation of intracellular networks. FEBS Letters 579(8):1846-1853. |
|
120 |
Goldbeter, A. 2004. Computational biology: a propagating wave of interest. Current Biology 14(15):601-602; Uetz, P. and R.L. Finley, Jr. 2005. From protein networks to biological systems. FEBS Letters 579(8):1821-1827; Aloy, P. and R.B. Russell. 2005. Structure-based systems biology: a zoom lens for the cell. FEBS Letters 579(8):1854-58; Rousseau, F. and J. Schymkowitz. 2005. A systems biology perspective on protein structural dynamics and signal transduction. Current Opinion in Structural Biology 15(1):23-30. |
|
121 |
Apic, G. et al. 2005. Illuminating drug discovery with biological pathways. |
|
|
FEBS Letters 579(8):1872-1877; Young, J.A. and E.A. Winzeler. 2005. Using expression information to discover new drug and vaccine targets in the malaria parasite Plasmodium falciparum. Pharmacogenomics 6(1):17-26. |
|
122 |
Hood, L. et al. 2004. Systems biology and new technologies enable predictive and preventative medicine. Science 306(5696):640-643. |
|
123 |
AfCS Nature; The Signalling Gateway; See www.signaling-gateway.org/update/updates/200201/nrn714.html. |
|
124 |
Balakrishnan, V.S. et al. 2005. Genomic medicine, gene polymorphisms, and human biological diversity. Seminars in Dialysis 18(1):37-40; Carr, K.M. et al. 2004. Genomic and proteomic approaches for studying human cancer: prospects for true patient-tailored therapy. Human Genomics 1(2):134-140. |
|
125 |
Guillermo Paez, J. et al. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304(5676):1497-1500. |
|
126 |
Marietti, C. 1999. Body Language; Health Care Informatics. Healthcare Informatics [Online]. Available online at www.healthcare-informatics.com/issues/1999/01_99/body.htm [accessed January 5, 2006]. |
|
127 |
Workman, P. 2001. New drug targets for genomic cancer therapy: Successes, limitations, opportunities and future challenges. Current Cancer Drug Targets 1(1):33-47. |
|
128 |
Ibid. |
|
129 |
Schinder, T., W. Bornmann, P. Pellicena, W.T. Miller, B. Clarkson, J. Kuriyan. 2000. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science 289(5486):1938-1941. |
|
130 |
Workman, P. 2001. New drug targets for genomic cancer therapy: Successes, limitations, opportunities and future challenges. Current Cancer Drug Targets 1(1):33-47. |
|
131 |
Daar, A.S. and P.A. Singer. 2005. Pharmacogenetics and geographical ancestry: implications for drug development and global health. Nature Reviews. Genetics 6(3):241-246. |
|
132 |
National Research Council/Institute of Medicine. 2005. An International Perspective on Advancing Technologies and Strategies for Managing Dual-Use Risks. Washington, DC: The National Academies Press. |
|
133 |
Ibid. |
|
134 |
While the vaccine was not one that would specifically target black as opposed to white people, it was clearly intended to be used to limit fertility in black women. |
|
135 |
Glasgow, J.N. et al. 2004. An adenovirus vector with a chimeric fiber derived from canine adenovirus type 2 displays novel tropism. Virology 324(1): 103-16. |
|
136 |
Nettelbeck, D.M. et al. 2004. Retargeting of adenoviral infection to melanoma: combining genetic ablation of native tropism with a recombinant bispecific single-chain diabody (scDb) adapter that binds to fiber knob and HMWMAA. International Journal of Cancer 108(1):136-45. |
|
137 |
Suzuki, T. et al. 2000. Adenovirus-mediated ribozyme targeting of HER-2/neu inhibits in vivo growth of breast cancer cells. Gene Therapy 7(3):241-248. |
|
138 |
Rein, D.T. et al. 2004. Gene transfer to cervical cancer with fiber-modified adenoviruses. International Journal of Cancer 111(5):698-704. |
|
139 |
A company called DNAprint Genomics has identified a number of genetic markers that correlate highly with racial or ethnic designations, many of them having to do with metabolizing toxins found in foods that are indigenous to certain areas. The markers identified by this firm are used to provide quantitative measures of an individual’s ancestry, according to four different “anthropological groups”—Native American; East Asian; Sub-Saharan Africa; and European. “European” can be broken down into Northern European; Southeastern European, Middle Eastern, and South Asian. For additional information on this company’s “products” see www.dnaprint.com/welcome/, and in particular a related site, www.ancestrybydna.com/welcome/home/. |
|
140 |
The Sunshine Project. 2003. Emerging Technologies: Genetic Engineering and Biological Weapons. Background Paper #12. Available online at www.sunshine-project.org/publications/bk/bk12.html#sec6 [accessed January 5, 2006]. |
|
141 |
Kagan, E. 2001 Bioregulators as instruments of terror. Clinics in Laboratory Medicine 21(3): 607-618. See also, Wheelis, M. 2004. Will the new biology lead to new weapons? Arms Control Today 34(July/August):6-13; and, Dando, M. 1999. Biotechnology, Weapons, and Humanity. British Medical Association. Amsterdam: Harwood Academic Publishers, especially Chapter 4 on “Genetic weapons.” See also Dando, M. 1996. A New Form of Warfare: The Rise of Non-Lethal Weapons. Dulles, VA: Potomac Books, Inc., especially Chapters 5 and 8. |
|
142 |
Wang, D. et al. 1999. Encapsulation of plasmid DNA in biodegradable poly(D, L-lactic-co-glycolic acid) microspheres as a novel approach for immunogene therapy. Journal of Controlled Release 57(1): 9-18; National Research Council/Institute of Medicine. 2005. An International Perspective on Advancing Technologies and Strategies for Managing Dual-Use Risks. Washington, DC: The National Academies Press. |
|
143 |
Neuropeptides, a type of bioregulator found in nervous system tissue, have a powerful modulatory effect on the nervous and immune systems. |
|
144 |
Wheelis, M. 2002. Biotechnology and biochemical weapons. The Nonproliferation Review 9(Spring):48-53. Available online at cns.miis.edu/pubs/npr/vol09/91/91whee.pdf [accessed January 5, 2006]. |
|
145 |
Healy, M. 2004. Sharper minds. Los Angeles Times (December 20): F1; Tully, T. et al. 2003. Targeting the CREB pathway for memory enhancers. Nature Reviews. Drug Discovery 2(4):267-77. |
|
146 |
National Research Council/Institute of Medicine. 2005. An International Perspective on Advancing Technologies and Strategies for Managing Dual-Use Risks. Washington, DC: The National Academies Press. |
|
147 |
Ibid. |
|
148 |
Arntzen, C. et al. 2005. Plant-derived vaccines and antibodies: potential and limitations. Vaccine. 23(15):1753-1756; Huang, Z. et al. 2005. Virus-like particle expression and assembly in plants: hepatitis B and Norwalk viruses. Vaccine 23(15): 1851-1858; Thanavala, Y. et al. 2005. Immunogenicity in humans of an edible vaccine for hepatitis B. Proceedings of the National Academy of Sciences 102(9):3378-3382. |
|
149 |
National Research Council/Institute of Medicine. 2005. An International Perspective on Advancing Technologies and Strategies for Managing Dual-Use Risks. Washington, DC: The National Academies Press. |
|
150 |
Petro, J.B. et al. 2003. Biotechnology: Impact on biological warfare and |
|
|
biodefense. Biosecurity and Bioterrorism: Biodefense Strategy, Practice, and Science. 1(3):161-168. |
|
151 |
Walt, D.R. 2005. Miniature analytical methods for medical diagnostics. Science 308(5719):217-219. |
|
152 |
Ibid. |
|
153 |
This section is based on the workshop presentation of N. Seeman, in National Research Council/Institute of Medicine. 2005. An International Perspective on Advancing Technologies and Strategies for Managing Dual-Use Risks. Washington, DC: The National Academies Press. |
|
154 |
Seeman, N.C. and A.M. Belcher. 2002. Emulating biology: building nanostructures from the bottom up. Proceedings of the National Academy of Sciences 99(Suppl 2):6451-6455. |
|
155 |
Seeman, N.C. 1982. Nucleic acid junctions and lattices. Journal of Theoretical Biology 99(2): 237-247; Seeman, N.C. 1999. DNA engineering and its application to nanotech-nology. Trends in Biotechnology 17(11):437-443. |
|
156 |
Winfree, E. 1995. On the computational power of DNA annealing and ligation. In Lipton, R. and E. Baum, eds. 1995. DNA Based Computers Volume 27, Proceedings of a DIMACS Workshop. Am. Math. Society:199-215; and Adleman, L. 1994. Molecular computation of solutions to combinatorial problems. Science 266(5187): 1021-1024. |
|
157 |
Chen, Y. and C. Mao. 2004. Putting a brake on autonomous DNA nanomotor. Journal of the American Chemical Society 126(28):8626-8627; Emerich, D.F. 2005. Nanomedicine—prospective therapeutic and diagnostic applications. Expert Opinion in Biological Therapy 5(1):1-5. |
|
158 |
Kohli, P. and Martin, C.R. 2005. Smart nanotubes for biotechnology. Current Pharmaceutical Biotechnology 6(1):35-47; Kubik, T. et al. 2005. Nanotechnology on duty in medical applications. Current Pharmaceutical Biotechnology 6(1):17-33. |
|
159 |
www.ibn.a-star.edu.sg/news_interface_article.php?articleid=54. |
|
160 |
Fortina, P. et al. 2005. Nanobiotechnology: the promise and reality of new approaches to molecular recognition. Trends in Biotechnology 23(4):168-173; Patolsky, F. et al. 2004. Electrical detection of single viruses. Proceedings of the National Academy of Sciences 101(39):14017-14022. |
|
161 |
Warheit, D. et al. 2004. Comparative pulmonary toxicity assessment of single-wall carbon nanotubes in rats. Toxicological Sciences 77:117-125; Oberdorster, E. 2004. Manufactured nanomaterials (fullerenes, C60) induce oxidative stress in the brain of juvenile largemouth bass. Environmental Health Perspectives 112(10):1058-62. |
|
162 |
Main, C.E. 2003. Aerobiological, ecological, and health linkages. Environment International 29(2-3):347-349. |
|
163 |
Bauce, E. et al. 2004. Bacillus thuringiensis subsp. kurstaki aerial spray prescriptions for balsam fir stand protection against spruce budworm (Lepidoptera: Tortricidae). Journal of Economic Entomology 97(5):1624-1634. |
|
164 |
Martonen, T.B. and J.D. Schroeter. 2003. Risk assessment dosimetry model for inhaled particulate matter: I. human subjects. Toxicology Letters 138(1-2): 119-132. |
|
165 |
Paez-Rubio, T. et al. 2005. Source bioaerosol concentration and rRNA gene- |
|
|
based identification of microorganisms aerosolized at a flood irrigation wastewater reuse site. Applied and Environmental Microbiology 71(2):804-810. |
|
166 |
Tanner, B.D. et al. 2005. Bioaerosol emission rate and plume characteristics during land application of liquid class B biosolids. Environmental Science and Technology 39(6):1584-90; Brooks, J.B. et al. 2005. Estimation of bioaerosol risk of infection to residents adjacent to a land applied biosolids site using an empirically derived transport model. Journal of Applied Microbiology 98(2):397-405. |
|
167 |
Brown, J.R. et al. 2005. Aerial optimization and canopy penetration study of Dibrom 14 Concentrate. Journal of the American Mosquito Control Association 21(1): 106-113. |
|
168 |
Chan, H.K. 2003. Inhalation drug delivery devices and emerging technologies. Expert Opinion on Therapeutic Patents 13(9):1333-1343. |
|
169 |
Edwards, D. 2002. Delivery of biological agents by aerosols. American Institute of Chemical Engineers Journal 48(1):2-6. |
|
170 |
LiCalsi, C., M.l Maniaci, T. Christensen, E. Phillips, G.H. Ward, and C. Witham. 2001. A powder formulation of measles vaccine for aerosol delivery. Vaccine 19(17-19):2629-2636. The authors describe a method to deliver live, attenuated, measles vaccine via the lungs. “In this study, live attenuated measles vaccine is micronized by jet milling to generate particle sizes appropriate for pulmonary delivery (1-5 µm). Milling does not induce detectable physical change and significant viral potency is maintained…. The measles vaccine formulation is dispersible …” |
|
171 |
Clark, A.R. 1995. Medical aerosol inhalers: past, present, and future. Journal of Aerosol Science and Technology 22:374-381. |
|
172 |
Chan, H.K. 2003. Inhalation drug delivery devices and emerging technologies. Expert Opinion on Therapeutic Patents 13(9):1333-1343. |
|
173 |
Crowder, T.M. 2002. Fundamental effects of particle morphology on lung delivery: predictions of Stokes’ Law and the particular relevance to dry powder inhaler formulation and development. Pharmaceutical Research 19(3):239-245. |
|
174 |
Garcia-Contreras, L. and H.D.C. Smyth. 2005. Liquid-spray or dry-powder systems for inhaled delivery of peptide and proteins? American Journal of Drug Delivery 3(1):29-45. |
|
175 |
Chan, H.K. 2003. Inhalation drug delivery devices and emerging technologies. Expert Opinion on Therapeutic Patents 13(9):1333-1343. |
|
176 |
Tan, H.S. and S. Borsadia. 2001. Particle formation using supercritical fluids: pharmaceutical applications. Expert Opinion on Therapeutic Patents 11(5):861-872. |
|
177 |
Based on Elliott Kagan’s presentation at the Cuernavaca workshop. See National Research Council/Institute of Medicine. 2005. An International Perspective on Advancing Technologies and Strategies for Managing Dual-Use Risks. Washington, DC: The National Academies Press. |
|
178 |
Wang, D. et al. 1999. Encapsulation of plasmid DNA in biodegradable poly(D,L-lactic-co-glycolic acid) microspheres as a novel approach for immunogene delivery. Journal of Controlled Release 57(1):19-18. |
|
179 |
Dai, C. et al. 2005. Microencapsulation peptide and protein drugs delivery system. Colloids and Surfaces B: Biointerfaces 41(2-3):117-20. |
|
180 |
See www.controlledrelease.org/about/index.cgi [accessed May 12, 2005]. |
|
181 |
See www.swri.org/4org/d01/microenc/microen/default.htm [accessed May 12, 2005]. |
|
182 |
Randall, P. and S. Chattopadhyay. 2004. Advances in encapsulation technologies for the management of mercury-contaminated hazardous wastes. Journal of Hazardous Materials 114(1-3):211-223. |
|
183 |
Hao, S. et al. 2005. A novel approach to tumor suppression using microencapsulated engineered J558/TNF-alpha cells. Experimental Oncology 27(1):56-60. |
|
184 |
Weinbreck, F. et al. 2004. Microencapsulation of oils using whey protein/gum Arabic coacervates. Journal of Microencapsulation 21(6):667-679. |
|
185 |
Yamaguchi, Y. et al. 2005. Successful treatment of photo-damaged skin of nano-scale atRA particles using a novel transdermal delivery. Journal of Controlled Release 104(1):29-40. |
|
186 |
See www.advancedbionutrition.com/html/news_press.html#2005_5 [accessed May 12, 2005]. |
|
187 |
Chang, T.M. 2005. Therapeutic applications of polymeric artificial cells. Nature Reviews. Drug Discovery 4(3):221-235; and Orive, G. et al. 2004. History, challenges and perspectives of cell microencapsulation. Trends in Biotechnology 22(2):87-92. |
|
188 |
A new technology, which will allow weapons and vehicles to be released from submarines even if they were not originally designed for undersea use. |
|
189 |
Izumikawa, M. et al. 2005. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nature Medicine 11(3):271-276. |
|
190 |
Parsons, D. 2005. Airway gene therapy and cystic fibrosis. Journal of Paediatrics and Child Health 41(3):94-96. |
|
191 |
Although “convergent technology” is a common term often used to refer to the convergence of specific types of technologies, we use it here loosely to refer to the convergence of any technologies. |
|
192 |
Benenson, Y. et al. 2004. An autonomous molecular computer for logical control of gene expression. Nature 429(6990):423-429. |
|
193 |
Kagan, E. 2001 Bioregulators as instruments of terror. Clinics in Laboratory Medicine 21(3): 07-618. See also, Wheelis, M. 2004. Will the new biology lead to new weapons? Arms Control Today 34(July/August):6-13; and Dando, M. 1999. Biotechnology, Weapons, and Humanity British Medical Association. Amsterdam: Harwood Academic Publishers, especially Chapter 4 on “Genetic weapons.” See also Dando, M. 1996. A New Form of Warfare: The Rise of Non-Lethal Weapons. Dullas, VA: Potomac Books, Inc., especially Chapter 8: “An assault on the brain?” and Chapter 5: “Lethal and non-lethal chemical agents.” |
|
194 |
Ibid. |
|
195 |
Nixdorff, K. and W. Bender. 2002. Ethics of university research, biotechnology and potential military spin-off. Minerva 40:15-35. See also Nixdorff, K., N. Davison, P. Millett, and S. Whitby. 2004. Technology and biological weapons: Future threats. Science and Technology Report, Number 2, University of Bradford, Department of Peace Studies. Available online at www.brad.ac.uk/acad/sbtwc/ST_Reports/ST_Report_No_2.pdf [accessed January 5, 2006]. |










































































