2
Role of Genomics: GTL in Achieving the Department of Energy’s Mission Goals: Promise and Challenges
The Department of Energy (DOE) has the mission of protecting our energy and economic security and our environment by promoting a diverse, reliable, affordable and environmentally sound domestic energy system. In carrying out that mission, DOE has recognized that genomics and systems biology research will enable development of novel strategies to address the agency’s three strategic challenges (DOE, 2005b):
-
To develop biofuels as a major secure energy source.
-
To develop biological solutions for remediation of soil, sediment, and groundwater contaminated with metals, radionuclides, and organic hazardous wastes.
-
To understand relationships between climate change and Earth’s microbial systems and to generate options for carbon sequestration.
The Genomics: GTL program is expected to provide the scientific underpinning for predicting and manipulating the behavior of complex biological systems, particularly systems that may play a central role in developing biotechnology solutions to fulfill DOE’s energy and environmental mandates. The Genomics: GTL program therefore becomes critical for strengthening the nation’s scientific leadership in systems biology and supporting an evolving industrial biotechnology sector that is essential for the nation’s economic competitiveness in the global economy. The following discussion is offered to illustrate how the science of the Genomics: GTL program can be used to address the three strategic challenges.
BIOENERGY
The Genomics: GTL program is addressing the needs for new sources of energy that could
-
Reduce the risk of global climate change by dramatically lowering the emission of greenhouse gases.
-
Have a favorable energy balance.
-
Have the potential to compete effectively with fossil fuels in the marketplace.
-
Reduce the adverse environmental effects of today’s pattern of energy production and consumption.
-
Meet a substantial fraction of U.S. (and global) energy demand.
One source of energy that could eventually meet those criteria is bioenergy produced by a variety of plants and microorganisms. The Genomics: GTL program could play a key role in realizing the potential of bioenergy by generating the fundamental knowledge that would make it technologically and economically feasible. Although it is premature to pick a “winner,” the research community has identified a number of promising directions, including
-
Genetic modification of crops to increase yields of usable energy per unit of cultivated land by a factor of 3-5 while maintaining nutrient and water requirements.
-
Conversion of cellulosic biomass to fuels by depolymerizing cellulose and hemicellulose into their component sugars and then converting the sugars to fuel.
-
Design of algae or bacteria that cost-effectively produce hydrogen or hydrocarbons.
Energy from biomass is the largest source of renewable energy in this country; it has surpassed hydropower and makes up 3 percent of the total energy consumed in the United States (Perlack et al., 2005). A recent study conducted by the Natural Resources Defense Council (NRDC, 2004) concluded that scientific and technological advances and sound public policies could rapidly expand the use of plants and plant-derived materials for energy. By 2050, biofuels could displace more than 7 million barrels of oil per day, the equivalent of nearly half of the oil that the United States use in the transportation sector. In that scenario, the United States would be able to reduce emissions of greenhouse gases by nearly 1.7 billion tons per year (as measured in tons of carbon dioxide [CO2] equivalents)—more than 22 percent of U.S. greenhouse-gas emissions in 2002. A transition to biofuels could also lead to improvements in air quality in that biofuels have almost no sulfur and produce fewer particles and toxic air pollutants (NRDC, 2004).
Development of advanced biological conversion processes (enzymatic, microbial, and plant processes) is central to the DOE biomass program and to the expanding industrial biotechnology sector. Biological processes are the preferred path because they tend to have higher reaction specificity, require milder reaction conditions, and produce fewer toxic byproducts. Those characteristics are consistent with the goal of developing industrial processes and systems that are environmentally friendly. However, the challenges are to increase rates and extents of conversion in an array of microbial and biochemical processes, to accelerate commercial development of biofuels, and to expand the portfolio of industrial enzymes, microorganisms, and plants for an expanding bioeconomy.
The Genomics: GTL program can promote the development of more-effective bioconversion processes and plant-based feedstocks by enhancing our understanding of biological conversion processes from a systems perspective. Understanding of systems biology will lead to better methods and tools for manipulating and controlling metabolic pathways that are important for bioenergy and industrial chemical production, for prospecting for novel industrial enzymes and microorganisms, and for bioengineering to enhance plants’ usability as feedstocks for energy and industrial chemicals. For example, one goal of the Genomics: GTL program is to discover functions of genes that could contribute to cheaper biofuels. The development of more-efficient and cost-effective enzymes is a critical step in making the abundant and diverse array of plant-derived polysaccharides available for the production of energy and industrial chemicals. A study commissioned by the National Research Council (NRC, 2000) and the roadmap from the Biomass Technical Advisory Committee (2002) established by the Biomass R & D Act of 2003 have each identified enzyme engineering as having one of the top three priorities in biological research to support the development of microbial or plant-based biofuels and industrial chemicals. The high priority is based on the recognition that enzymatic conversion of biomass is the preferred path for processing microbial or plant-based resources into industrial products because enzymes exhibit specific catalytic activities and enzymatic processes are environmentally more benign. It is also recognized that the industrial development of enzymes itself occupies an important industrial biotechnology sector that holds the promise of expanded economic growth. The enzyme market was estimated to be $2 billion per year in 2004 and to have an annual growth rate of 4-5 percent (Business Communication Company, 2004).
Plant cell walls comprise a highly complex matrix of polysaccharides, including cellulose, lignins, pectins, and diverse hemicelluloses. Microorganisms found in soils, compost piles, and other environments have been shown to produce enzymes effective for degrading each one of those polysaccharides into fermentable sugars. The Genomics: GTL program can play an important role in increasing understanding of the structure and functions of genes associated with degradation of polysaccharides by those microorganisms. It can also contribute to the discovery of new polysaccharide-degrading enzymes by prospecting for novel
microorganisms in exotic environments. For example, termites degrade polysaccharides. Bacteria that live in a termite’s hindgut break down plant matter and release hydrogen as a byproduct. The mechanism of hydrogen production in the termite hindgut is not yet known. The DOE Joint Genome Institute (JGI) is scheduled to sequence the community of microorganisms in the termite hindgut by 2006. That would enable the Genomics: GTL program to identify and characterize the enzymes associated with hydrogen production in the termite hindgut.
Another example involves the production of microbial polysaccharide-degrading enzymes by plants (Nuutila et al., 1999; Dai et al., 2000; Ziegler et al., 2000; Ziegelhoffer et al., 2001). Today, the National Renewable Energy Laboratory estimates that enzyme production costs account for $0.10 of the price of a gallon of ethanol. The challenge is to reduce that by half by increasing the activities of enzymes or reducing the production cost. One avenue toward that goal is the use of plants as biomolecular farms for the production of the enzymes. The concept has been demonstrated in a study in which a corn plant was used to produce an Acidothermus cellulolyticus endogluconase. In light of methods of bioconfinement of recombinant crops in the field (NRC, 2004), that technology could well become America’s standard technique for production of cellulases and other polysaccharide-degrading enzymes in biomass crops that are converted into fermentable sugars that can be fermented into ethanol biofuel. It is important to remember that the knowledge developed by producing polysaccharide-degrading enzymes in plant biomass should also be largely applicable to other proteins, including those which produce such valuable industrial products as 1,3-propanediol, a precursor of many important industrial polymers.
Cost-effective and efficient microbial conversion processes are necessary to convert low-cost sugars derived from plant-based resources to ethanol and other industrial chemicals. Although there are several key technological differences in how ethanol is produced from corn or cellulosic feedstock, both paths to ethanol production require a fermentation step that involves the conversion of glucose and other sugars to ethanol. Currently, baker’s yeast, Saccharomyces cerevisiae, provides the primary microbiological system used by the corn-based ethanol industry. As we seek to increase the amount of ethanol produced from biomass, we will have to increase our knowledge of the metabolism of important microbiological systems, particular those with the potential to enhance the production of useful biobased products. That need was clearly articulated in the National Research Council report on biobased industries (NRC, 2000) and in DOE’s Genomics: GTL roadmap (DOE, 2005b). Two of the key research activities identified by the Research Council are relevant to the Genomics: GTL program:
-
“Analysis of biochemical pathways that integrate basic intracellular measurements. Such analysis will provide fundamental understanding of the microbial metabolism and physiology necessary to focus metabolic engineering manipulations on enhancing organisms’ overall productivity.”
-
“Basic research on principles of intermediate microbial metabolism to gain a better understanding of how concentrations of substrate or product can inhibit rates of product formation. Such understanding will aid in engineering bioreactors control to enhance the rate and conversion of raw materials into useful products.”
Rapid advances in genomics have facilitated the manipulation of metabolic pathways to engineer organisms that can efficiently produce a desired metabolic product or reduce unwanted byproducts. Metabolic engineering allows a more directed and rational use of classical genetic or molecular biology tools to optimize the production of metabolites and proteins of interest. For example, the complete genome sequence of S. cerevisiae was published in 1996, and a complete collection of deletion mutants of yeast is commercially available (Goffeau et al., 1996). They create many opportunities to customize systems biology research and develop metabolic engineering tools to characterize the metabolic networks of wild-type S. cerevisiae and newly constructed mutant strains. For example, the Genomics: GTL program could provide new insight into the role of cellular myo-inositol in the physiological and metabolic behavior of S. cerevisiae that might reveal a clear link between high phosphatidylinositol concentration and ethanol tolerance.
Another important subject would be metabolic engineering of bacteria—such as Thermotoga neapolitana, Enterobacter aerogenes, and Clostridium butyricum—that are hydrogen producers that use fermentative pathways. Such microorganisms are known to ferment sugars to hydrogen at a relatively high rate by glycolytic breakdown of sugars through the anaerobic metabolism of pyruvate (Hallenbeck, 2005). The generation of hydrogen by fermentative bacteria is accompanied by the formation of organic acids as metabolic products that are not used by the microorganisms (Nath and Das, 2004). Thus, altering the metabolic pathway to shift more of the pyruvate to hydrogen is an important step for improving fermentative hydrogen production. The science, methods, and tools of the Genomics: GTL program would strengthen our understanding of the regulatory and metabolic pathways that influence hydrogen production and create opportunities for more-informed engineering of those pathways and others.
Although the focus of Genomics: GTL bioenergy research is on microbial processes, it should be clear from the preceding paragraphs that biomass for bioenergy is derived from plants. Better understanding of the mechanisms and regulation of polysaccharide and cell wall synthesis in plants is critical to meeting the goals of the nation’s bioenergy research agenda. For example, it may be possible to engineer plants for novel cell wall structures that enhance the efficiency of biomass conversion. The committee believes that bioenergy research through Genomics: GTL should include a parallel focus on polysaccharide and cell wall synthesis in plants. To that end, Arabidopsis thaliana provides an outstanding experimental platform for developing a systems-level analysis of plant
function. The resulting knowledge could greatly enable bioengineering applications involving biomass species, such as corn and poplar. The committee’s suggestion is to focus a portion of the Genomics: GTL program on specific aspects of plant biology (in this case, aspects relevant to biomass conversion), and not to develop a broad-based effort in plant biology.
CARBON SEQUESTRATION
About 6 billion tons of CO2, a greenhouse gas, is released into the atmosphere by anthropogenic activities each year. Atmospheric CO2 concentrations have increased because of human use of fossil fuels and changes in land use, such as deforestation. It is also known that microorganisms can be used to mitigate global change due to human activities, such as agriculture, mining, and waste treatment (ASM, 2004). It is estimated that atmospheric CO2 and methane concentrations are now increasing at about 0.4 percent and 1 percent each year, respectively. The growing scientific evidence that CO2 and other greenhouse gases are altering our climate has stimulated interest in CO2 sequestration as a means to counteract global climate change. The DOE mission with respect to carbon cycling and sequestration is to “understand the microbial mechanisms of carbon cycling in the earth’s ocean and terrestrial ecosystems, the roles they play in carbon sequestration, and how these processes respond to and impact climate change.” Photosynthetic terrestrial and aquatic organisms naturally perform biosequestration, and understanding how this is achieved at the whole-organism and microbial-community levels is one of the important roles of the Genomics: GTL program.
Microorganisms have a much greater role in mediating biogeochemical activities than previously thought, given that they outnumber all other forms of life on land and in rivers, lakes, and oceans. Therefore, it is of great importance to understand the genetic regulation behind these biogeochemical activities and the role of microorganisms in carbon sequestration. Many of the critical questions surrounding the role of microorganisms in biosequestration were addressed by the American Academy of Microbiology (ASM, 2001):
-
Which microorganisms are responsible for producing and consuming specific environmentally important compounds, and how does the diversity of microorganisms affect soil, water, and atmospheric concentrations of various chemicals?
-
How and to what extent do microorganisms and their recycling processes respond to climate change and other disturbances?
-
How can information about activities occurring on the scale of microorganisms (micrometers to millimeters) be integrated across scales of communities, landscapes, and ecosystems to help to explain phenomena observed on a global scale?
-
What new technologies and computational systems are needed to facilitate integration and understanding across scales?
Those are complex questions and can be addressed with a systems biology approach for generating realistic strategies for biosequestration. We provide some examples on how each of the above questions can be addressed by the Genomics: GTL program. First, it is known that phytoplankton photosynthesis in the oceans is an important subsystem in the recycling of CO2 in the biosphere. Variation in the species composition or population sizes of the ocean’s phytoplankton could theoretically have a great effect on the oceans’ ability to take up atmospheric carbon. The focus of the Genomics: GTL program is to understand how those microorganisms affect ocean ecosystems by cycling carbon and other important elements, such as nitrogen. For example, the program is supporting studies on and the sequencing of Emiliania huxleyi. DOE has also sequenced several species of ocean carbon-sequestering phytoplankton, such as the diatom Thalassiosira pseudonana and the cyanobacteria Prochlorococcus and Synechococcus. Diatoms account for about 20 percent of global carbon fixation, and the genome sequence of T. pseudonana sheds light on the silicic acid metabolism and energy storage and use strategies that allow diatoms to flourish in marine systems (Armbrust et al., 2004; DeLong and Karl, 2005). Many scientists believe that Prochlorococcus constitutes the most abundant photosynthetic organisms on Earth. Understanding those organisms’ roles in global carbon cycles is central to the issue of carbon sequestration.
As to the second question, this is being addressed by DOE studies on soil microorganisms. Soil respiration accounts for 75 percent of the carbon in the terrestrial ecosystem, and according to Rosenberg, Metting, and Izaurralde (2004), it returns nearly 10 times as much CO2 to the atmosphere as emissions from fossil-fuel combustion. Agriculture and fire also contribute substantially to the carbon being released to the atmosphere from soils. Metagenomic studies of soil samples indicate that microbial communities have a wide range of mechanisms and biochemical pathways for carbon metabolism, some of which may emerge as targets for the application of carbon management strategies. However, the mechanisms by which microbial populations adjust to climate change in an ecosystem are not well understood. Genomics, proteomics, and metabolomics could elucidate such mechanisms and thereby increase our understanding of the important role of microorganisms in carbon cycling. Moreover, the resulting knowledge could enable predictive models of system function that might presage changes in the global carbon economy.
Although currently missing from the Genomics: GTL research plan, plants contribute substantially to nutrient cycles in the soil through both photosynthesis and nitrogen fixation. As a consequence of these autotrophic processes, the soil zone around plant roots (the “rhizosphere”) is among the most important and diverse, yet least understood, of ecosystems. To fully address soil microbiology
and its relevance to carbon sequestration, nitrogen cycling, and response to climate change, it will be essential to include substantive efforts on rhizosphere biology in Genomics: GTL. By definition, such efforts will need to include studies of both plant and microbial systems, extending the systems biology analogy to multiorganism and cross-kingdom interactions.
Working across scales from whole organisms to the biosphere is daunting. The Biological and Environmental Research Division in DOE’s Office of Science established CSiTE (Carbon Sequestration in Terrestrial Ecosystems)—a research consortium—to perform fundamental research that will lead to acceptable methods of enhancing carbon sequestration in terrestrial ecosystems as one component of a carbon-management strategy. Three national laboratories are members of CSiTE—the Argonne, Pacific Northwest, and Oak Ridge National Laboratories. The goal of CSiTE (DOE-ORNL, 2002) is “to discover and characterize links between critical pathways and mechanisms for creating larger, longer-lasting carbon pools in terrestrial ecosystems. Research is designed to establish the scientific basis for enhancing carbon capture and long-term sequestration in terrestrial ecosystems by developing:
-
“Scientific understanding of carbon capture and sequestration mechanisms in terrestrial ecosystems across multiple scales from the molecular to the landscape,
-
“Conceptual and simulation models for extrapolation of process understanding across spatial and temporal scales,
-
“Estimates of carbon sequestration potential,
-
“Assessments of environmental impacts and economic implications of carbon sequestration.”
The fourth question, on needed new technologies and computational systems, highlights the need to develop infrastructure for advancing the Genomics: GTL program. For example, metagenomic methods can document the makeup and activity of ocean communities involved in CO2 recycling. The sequencing of microbial communities in the Sargasso Sea appears to have revealed 1.2 million previously unknown genes, including almost 800 genes coding for rhodopsins that are presumed to be involved in phototrophy (Venter et al., 2004). Those data serve as a starting point for further Genomics: GTL studies on the mechanism of this potential energy-yielding process. It is believed that the dynamics of carbon-assimilation and anabolic pathways that sequester carbon or return it to the atmosphere, respectively, will be elucidated and that biological models of carbon-sequestration activity can be developed to assess the effects of carbon-cycle perturbations on climate change. DOE can collaborate with and build on the joint National Science Foundation and U.S. Department of Agriculture program on microbial-genome sequencing that supports research on the diversity of microorganisms and their roles in complex ecosystems and in global geochemical cycles
(NSF, 2005). DOE is also exploring the use of biosensors based on genomic information that would detect changes in the levels of DNA, RNA, proteins, and metabolites in response to stress or population shifts.
Obviously, answers to those questions require a broad interdisciplinary approach and will benefit from genomic studies to identify key genes and pathways. Understanding the complexity of ecosystems in which many functions are being carried out simultaneously by millions of microorganisms of diverse species is no small task; it will take years of study, including the development of novel experimental and computational approaches. Effective implementation of the Genomics: GTL program would facilitate the broad interdisciplinary and multidisciplinary research collaborations needed to address the questions on a variety of biological and physical scales.
BIOREMEDIATION
During the 50 years of the nuclear era, the United States invested in facilities to do research on, develop, manufacture, and test nuclear weapons and materials. The environmental-remediation legacy left from those manufacturing and testing activities is staggering: DOE has the responsibility for monitoring and cleaning up more than 7,000 sites at 100 facilities. The groundwater and soil at those sites are contaminated with radionuclides, which are often mixed with other wastes, such as chlorinated hydrocarbons. Collectively, 2 trillion gallons of contaminated groundwater and 75 million cubic meters of soil and subsurface sediment must be remediated at these sites. For comparison, the groundwater volume equals 4 times the U.S. daily water consumption, and the sediment volumes would fill 17 professional sports stadiums.
With current technology, cleanup costs would run to $300 billion over a 70-year period. Hence, alternative strategies, methods, and technologies are being investigated throughout the many DOE remediation programs. The one with the most promise is bioremediation. Bioremediation is the use of microorganisms to contain or eliminate hazardous and radioactive wastes or decrease them to environmentally safe levels. Enhancing it with the modern tools of biotechnology could save 30-50 percent of the cost. The committee notes that, although they are not covered in the Genomics: GTL plan, plants have also been shown to have utility in bioremediation and thus should be considered among the targets for Genomics: GTL research. Enhancing the capacity and quality of bioremediation by means of the modern tools of biotechnology could lead to savings in the range of 30-50 percent. For example, at DOE’s Savannah River site in Aiken, South Carolina, bioremediation of subsurface solvent contamination cost two-thirds as much as a pump-and-treat method and was 40 percent more efficient. An Environmental Protection Agency study of 150 sites that use bioventing (a form of in situ bioremediation) showed cost savings of 50-90 percent.
Bioremediation of organic contaminants involves transforming them to be-
nign products, such as CO2. Metals and radionuclides must be immobilized to prevent subsurface travel to rivers or groundwater. All bioremediation systems have a common goal of stimulating and maintaining microbial metabolism (Hughes et al., 2002). Stimulation might involve optimizing the metabolic pathways of whole organisms or of a community of microorganisms to achieve the desired transformation. The optimization of metabolic activities of whole organisms and microbial communities is the key to converting hazardous materials to nonhazardous materials or nonbioavailable forms and is consistent with the research and development activities of the Genomics: GTL program.
The Genomics: GTL roadmap listed several research needs:
-
Assessment of benefits and effects.
-
Establishing links between biology and geochemistry.
-
Using genome sequences as a launching point for understanding communities.
-
Modeling microbial metabolic activities.
-
Merging metabolic and field-scale models.
Assessment of benefits and effects is going on through four DOE programs managed by the Environmental Remediation Sciences Division: the Natural and Accelerated Bioremediation Research (NABIR) program, the Environmental Management Science Program, the Environmental Molecular Sciences Laboratory, and the Savannah River Ecology Laboratory. Of all the programs reviewed, NABIR has had the most promising results for field applications and NABIR programs are linked to the Genomics: GTL program now (COV, 2004). The goal of the NABIR program is “to provide the fundamental science to serve as the basis for the development of cost-effective bioremediation of radionuclides and metals in the subsurface at DOE sites.” The program focuses on intrinsic bioremediation and accelerated bioremediation through the use of biostimulation (the addition of inorganic or organic nutrients).
Links between biology and geochemistry are being established through research focused on the survival of environmental microorganisms under stressful conditions, such as those at bioremediation sites. Researchers are integrating fields of biology—for example, genomics, ecology, molecular biology, proteomics, bioinformatics, and metagenomics. By understanding processes that allow specific bacteria to exploit different environments—such as water, air, soil, and the subsurface—scientists are identifying critical mechanisms for survival. For example, Gary Andersen and his group focus on understanding mechanisms of bacterial diversity by using 16S rRNA gene sequences to measure the relative abundance of individual members of microbial communities. In partnership with the DOE-JGI in Walnut Creek, California, they have developed novel microarray systems to measure dynamic changes and rapid systems for classifying the thousands of individual sequences from clone libraries that are being constructed.
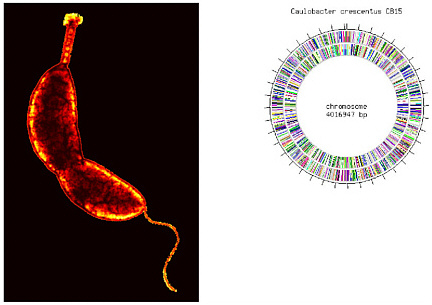
Another example of establishing links between biology and geochemistry is the research under way on the ubiquitous aquatic bacterium Caulobacter crescentus (Box 2-1). That organism was selected for extensive study by DOE because of its ability to survive in low nutrient habitats where contamination may be present. The completed genome sequence of strain CB15 has provided information needed to study genomewide response to heavy-metal stress. A customized 500,000-probe Affymetrix array was designed by Harley McAdams’s group at Stanford University to measure transcription levels of all 3,763 putative open reading frames (DOE, 2005c), both strands of genes for hypothetical proteins, and the intervening intergenic regions. The microarray was used to study transcriptional response to heavy-metal stress.
The work of Derek Lovley, of the University of Massachusetts, Amherst, on optimizing in situ bioremediation of uranium and harvesting electrical energy from waste organic matter by Geobacter species is an example of how genome sequences can be used as a launching point of understanding. His project addresses not only the identification and validation of the microbial community involved in the bioremediation of uranium in contaminated subsurface environments but also the use of this microbial community to harvest electricity from waste organic matter and renewable biomass. He is engaged in subsurface environmental studies in Colorado at the Old Rifle Uranium Mill Tailings Remedial Action site. His studies are supported primarily by the Genomics: GTL program, which focuses on detailed geochemical and microbiological characterization of the site.
Lovley’s research and the research of countless other environmental researchers across the country would benefit from a new generation of molecular ecology tools that might evolve from the Genomics: GTL program. For example, current methods for profiling 16S rDNA can provide information about the structure of a microbial community, but they tend to sample the most abundant species. Similarly, transcriptional profiles may provide information about community composition and their corresponding transcription and metabolic activities, but the methods are limited by the difficulty in reliably sampling environmental RNA. Moreover, RNA from only the most abundant organisms can be sampled, and potentially important details (such as spatial information) are lost. Thus, many of the current molecular ecology methods have reached their experimental limits. The Genomics: GTL program provides opportunities to address those and other technological limitations.
Modeling microbial metabolic activities is an important and challenging goal of the Genomics: GTL program. As articulated in the Genomics: GTL roadmap, the Genomics: GTL program enables three key modeling scenarios: microorganism-mineral interactions and resulting molecular structure and charge transfer, microbial-community responses (for example signaling, motility, and biofilm formation), and ensuing community functionality. There is a growing awareness in the systems biology research community that mathematical modeling is an essential tool for exploring those elements because it provides a framework for
structuring the understanding of complex biological systems; it can be used to extract insights and mechanisms from a rich set of empirical studies that have been sponsored by DOE, and it provides mathematical and computational tools that can be used in the design, analysis, and optimization of bioremediation strategies.
Thus far, the mathematical modeling and computer simulation tools being used to study bioremediation systems lack substantial biological detail. Bioremediation is an inherently spatial problem that operates under nonequilibrium and highly nonlinear conditions. It involves species that are potentially diverse genetically, and this results in complex and multi-dimensional models. Strong selection is likely to act on populations that are far from equilibrium and can result in considerable changes in the genetic composition of the population and thus potentially unpredictable changes in responses. Including evolutionary factors into standard ecological population models can yield behavior that differs from that of models in which evolutionary factors are absent (Neuhauser et al., 2003), but considerable experimental effort would be needed to develop such models and make them accurate and predictive. Beyond simulations, there is little available mathematical theory that could be applied to the transient behavior of such systems. Thus, there is a tremendous opportunity for the Genomics: GTL program to lead the effort to bring systems mathematics to the challenges of bioremediation.
Merging metabolic and field-scale models is a daunting task that requires a strong multidisciplinary approach. The Genomics: GTL program can address several essential elements, including the following problems:
-
Identify and characterize the multiprotein complexes—“protein machines”—that perform most cell functions in microorganisms.
-
Determine how the operations of the machines are orchestrated to allow organisms to thrive in diverse environments.
-
Describe the metabolic capabilities of complex microbial communities in their natural environments.
-
Develop new computational methods and tools to increase the understanding of complex biological systems and predict their behavior.
The Genomics: GTL program will also provide valuable data for improving field site treatments, for example, by developing computer models that would test and elucidate the activities and interactions occurring in microbial communities. Observations made with respect to syntrophic relationships, anaerobic degradation consortia, and shifts in the dominant terminal electron-accepting process observed in sediments could be integrated into models that describe the dynamic flow and transport regimes found at most DOE contaminated sites.
As stated earlier, bioremediation is considered to be the least expensive and
most versatile means of dealing with soil and groundwater contamination. Genomic studies being carried out by the Genomics: GTL program on microorganisms that have remediation potential will enable an assessment of the capability of individual species or strains and inform scientists and engineers how the bioremediation process might be better managed or improved. In addition, Genomics: GTL provides synergy with the NABIR program goals and supports research into the capabilities of microbial communities to promote metal and radionuclide precipitation. Finally, Genomics: GTL data may provide insight to guide the development of biosensors to monitor bioremediation over large areas and long durations and thus to help to sustain bioremediation activities in the field.
CHALLENGES TO THE ACHIEVEMENT OF THE DEPARTMENT OF ENERGY’S MISSION GOALS THROUGH SYSTEMS BIOLOGY
The primary cross-cutting theme of Genomics: GTL is systems biology—with the goal of developing predictive models of system function. The committee strongly endorses the notion that being able to predict the properties of DOE’s target systems would revolutionize energy-related and environment-related biotechnology. However the challenges to the achievement of the Genomics: GTL mission are immense; and, although they are likely to be solvable, the precise route to be taken is uncertain and subject to debate.
Where to begin depends largely on how one defines and sets priorities among the specific factors that limit progress. In its current form, Genomics: GTL has two parallel tracks: use of a traditional research funding process to identify and fund relatively large-scale, often multi-investigator projects focused on specific biological problems and a multidecade plan to construct and operate facilities that target high-throughput production and analysis of proteins, protein complexes, and microbial systems within which the proteins express their potential. The current facilities model assumes that progress in microbial systems biology is limited by lack of knowledge about proteins and their derived attributes within biological systems of interest to DOE, that acquiring such knowledge will require high-throughput facilities that can solve the problem by applying appropriate technology, and that knowledge of and access to all proteins in a range of target systems will revolutionize microbial systems biology in a way analogous to how genome sequencing has transformed biology in general.
Whether one agrees with that model depends largely on how one defines the primary barriers to progress. This section provides an overview of important gaps in knowledge and technology that must be filled to facilitate and expedite the achievement of the long-term goals of Genomics: GTL.
The Challenges
Multiple Scales of Systems Biology
In the context of DOE’s bioenergy mission, the relevant properties of a given system or molecular machine may span essentially all scales of biological organization, from simple binary interactions that may function at the subcellular level as regulatory switches to superdimensional interactions in microbial consortia, where the emergence of a particular property may depend on organism-organism or organism-environment interactions.
Diversity of Microorganisms
Many of the applications of bioremediation, carbon sequestration, and biofuel production will occur in situ, using endemic taxa. Biodiversity in terrestrial and marine environments is poorly understood. At the taxonomic level, recent estimates suggest that 1g of pristine soil may harbor as many as 106 distinct prokaryotic taxa and that most of the taxa are rare (Gans et al., 2005). Current sampling strategies, such as those based on metagenomic phenotype analyses (Williamson et al., 2005) or sequence analyses (Venter et al., 2004; Tringe et al., 2005), are adequate for sampling only the most abundant of these organisms with any certainty. Rare but stable components of ecosystems may contribute important properties to system function, but we lack routine methods for characterization of ultra-rare genomes, let alone for understanding the majority of such species.
Diversity within species is also relevant to DOE’s bioenergy mission. In recent years, high-quality assemblies of many microbial genomes have revolutionized microbiology. However, the sequences themselves are blueprints of reference genotypes, and in most cases the fraction of natural diversity that such reference genomes encompass is uncertain. Metagenomic sequencing is a means of surveying DNA from complex consortia. The results of several metagenomic projects highlight the wealth of information likely to accrue from ecosystem-level genome sequencing, both within and among species (Venter et al., 2004; Tringe et al., 2005). Nevertheless, current strategies for shotgun sequencing limit analysis to the most abundant genomes and typically yield only fragmentary assemblies. Recognition of that has sparked renewed interest in developing methods for enriching and culturing recalcitrant and rare species. Similar gains are likely to be realized by implementing nucleic acid normalization methods, such as Cot enrichment or suppressive-subtractive hybridization, which are well established in other genome investigations (for example, Yuan et al., 2003; Galbraith et al., 2004). The combination of new culture-independent technologies—such as single-cell sequencing, for example, the work of DOE-funded investigator George Church (DOE, 2005c)—with efforts to enrich and set priorities among specific genomes for analysis may have potential to expand our view of ecosystem biocomplexity.
Inferring Function from Sequence and Structure
Implicit in many of the arguments about the usefulness of data-gathering exercises, such as genome-sequencing projects and structural genomics initiatives, is the assumption that such data will illuminate the functions of many gene products. Several problems lead us to question the wisdom of accepting that assumption uncritically. First, the term function is imprecise (Ning et al., 2003; Fraser and Marcotte, 2004). Depending on who is the consumer of the information, function may refer to the biochemical activity of the isolated gene product, its role in metabolic or signal-transduction pathways in the cell, the phenotype of its knockout in a cell or model organism, or any combination of these. Even the seemingly simplest to ascertain, biochemical function, turns out to be loosely coupled to simple “determinants,” such as sequence and structure (see below). We are sympathetic with the need to obtain such information, given that 30-50 percent of the genes in most newly sequenced genomes have no established biochemical or cellular functions, but the goals of the Genomics: GTL program seem too sophisticated to adopt a single, restrictive view of function as a guiding principle.
The second problem is that much functional annotation depends on relating the sequence of a gene product to other sequences of known biochemical function, and the database of annotated functions is simply not as reliable as it needs to be (Gerlt and Babbitt, 2000). It is estimated that as many as 50 percent of the functional annotations based on sequence comparison may be wrong, at least in part; as more sequences are determined and annotations increase, the problem is likely to be compounded and should be rectified as soon as possible.
The third problem is that sequence and structure information rarely, if ever, increases understanding of whether a gene product has important interactions with others in the cell and, if it does, how those interactions affect its biochemical and cellular roles. Relying too heavily on such data-gathering for functional annotation risks taking a step backward, away from the more complex pictures demanded by systems biology.
Fourth is the increasing recognition that many or most of the gene products in higher organisms—and many in bacteria—have more than one function, no matter how one defines the term. Sequence and structural analyses hardly ever provide information on more than one prospective function and are usually silent about the conditions under which that function is biologically relevant. Focusing on a single function misses the point that systems biology is meant to address.
The most serious problem in using sequence and structure information to deduce function is that function changes much more rapidly than the other properties. In many instances in the database, two gene products that have more than 80 percent sequence identity—and correspondingly high structural identity—have biochemical functions that are completely different because of one or two changes in critical amino acid residues. Even when both sequence and tertiary structure are very similar, biochemical function may change. The ThiJ/DJ-1 su-
perfamily of proteins—which has members that are bacteria, archaeans, and eukaryotes—is a striking example. Members of that superfamily can share 40 percent sequence identity and monomer protein folds that superimpose to less than 1 angstrom of root-mean-square deviation and yet they have completely different functions as a result of different oligomerization states that bring completely different residues from one subunit into contact with those from the other (Wilson et al., 2005). Natural selection, which guides genome evolution, guarantees that examples like that will be common. The sequence of a protein can change easily. Overall structure is more robust but can still be affected considerably by a few mutational events. And neither is under the control of natural selection, which acts only on function.
The attempt to infer function from sequence and structure alone is an exercise in futility, particularly where complex phenomena are concerned. However, the committee does not wish to leave the impression that such data are of no value. On the contrary, they are a valuable part of the panoply of information that must be obtained to understand function. But they are data at the lowest level of complexity, involve the most routine and readily available technologies, and should not be a cornerstone of a program designed to advance the cutting-edge field of systems biology.
Computational Challenges
The Genomics: GTL roadmap states that “the goal is to create increasingly accurate mathematical models of life processes that enable predictions of cell and community behavior and create new and modified systems tailored for mission applications.” The systems biology approach of Genomics: GTL will integrate experiments, data acquisition and processing, modeling, and simulations in an iterative process in which model predictions inform experiments and experiments inform model development. The development and analysis of increasingly accurate models at all levels of biological organization pose mathematical and computational challenges. Vastly different time scales of the different biological processes can pose additional numerical challenges in simulation. The appropriate level of model complexity needs to be found because of tradeoffs between level of detail and computational complexity. The more detailed a model, the larger the number of variables and parameters, which further increases the difficulty of model validation and inference. Stochastic noise inherent in many of the processes makes accurate parameter estimation difficult. Many of the processes depend heavily on environmental conditions, so experiments with a wide array of environmental conditions will need to be integrated. We outline some of the modeling and simulation challenges on the different scales (see also Mathematics and 21st Century Biology, NRC, 2005).
At the cell level, the ultimate goal is to predict the cell phenotype from its
genotype and environmental conditions. A diverse set of mathematical and statistical approaches have been developed to unravel the topologies of networks (for example, “wiring diagrams”) that link cellular components, including gene networks, regulatory networks, and metabolic networks. Those networks provide a static view of cellular interactions. To model these accurately, spatiotemporally resolved data on numerous cellular processes must be integrated into dynamic models. That involves a large number of diverse components that range from rare to abundant. Many of the mathematical models that describe cellular processes are based on systems of ordinary differential equations. It is a mathematical framework that assumes that components are spatially homogeneous and can be approximated by continuously varying densities. That works well as long as the components under consideration are abundant in the entire cell with little spatial variation. Many components, however, are produced only in small quantities that are spatially localized within the cell and that exhibit considerable stochastic variability. Thus, accurate models of cellular processes will be mixtures of deterministic and stochastic models of discrete and continuous variables that vary both spatially and temporally. Little mathematical theory is available to deal with such models.
Microbial populations consist of genetically diverse cells, and individual cell responses even to the same environmental stimulus may vary. Ecological models of populations consider average responses and do not take genetic variation into account. Adding genetic heterogeneity to mathematical population models can considerably increase the complexity and dimensionality of the models. A considerable body of work deals with quantitative characters (see, for instance, Turelli and Barton, 1990 or Nuismer and Kirkpatrick, 2003). Including sequence variation in ecological models, however, would require a model framework that has not been established.
Modeling of ecological communities has a long history in mathematical ecology. Classical models use the simple framework of ordinary differential equations and consider few interacting species that are often highly unstable, exhibiting oscillations and chaotic behavior. Simulations of systems of larger numbers of interacting species have shown that they exhibit rich behavior but can be stabilized through interactions (see, for instance, Williams and Martinez, 2004). Attempts to gain a better understanding of the behavior of communities that consist of many interacting species have begun only recently, and there is no general theory that would allow prediction of the behavior of communities of thousands of interacting species. In addition, spatial heterogeneities and stochastic effects are rarely taken into account. Both are probably important, especially in soil microbial communities. There has been no attempt to integrate across all scales from molecules to ecosystems. Such multiscale models would span spatial and temporal scales of many orders of magnitude and would need to incorporate the genetic variation that is present in a community of interacting species, which
would require considerable computing power. Given the importance of this type of modeling activity to DOE’s missions, it is essential that the Genomics: GTL program seek to rectify the situation.
In addition to modeling and simulation challenges, some of which have been described above, there will be a need to develop bioinformatics tools further. The Genomics: GTL program will produce data at unprecedented rates and diversity, and they will need to be captured, archived, and annotated, preferably in an automated way. The types of data will go far beyond simple sequence data. They will include structural data from which three-dimensional models of molecules will be built, imaging data that track individual molecules in cells, and data that will track physiological responses of communities that consist potentially of thousands of different microorganisms in a wide array of environmental conditions. New methods in mathematics and computational biology to analyze such complex data will need to be developed, as will software and hardware to allow researchers to use these diverse datasets.
Issues to Be Addressed by Genomics: GTL Program and Facilities
A variety of issues will need to be addressed in the course of achieving the long-term goals of Genomics: GTL. Among these is the need to improve and implement genomics-enabled, high-throughput studies of genetic diversity in Genomics: GTL environments. The resulting information would contribute greatly to understanding aspects of ecosystem-level population biology, evolution, and function that are currently lacking and are critical to the mission of the Genomics: GTL program. The following are insights:
-
Description and then development of predictive models for how complex microbial consortia respond to natural and imposed selection.
-
Identification of the genomic diversity best suited to manipulation of Genomics: GTL target processes, for example, remediation of specific contaminants in unique environments.
-
Characterization of genotypes and the genes and proteins that most strongly influence system function.
-
Understanding how human intervention may alter community structure and function, and identifying and quantifying related risk factors, if any. In particular, if genetically modified organisms are to be released into open field settings for bioremediation, DOE should make strong efforts to gain public acceptance for such release.
Central to the Genomics: GTL mission is the need to identify the molecular machines that underlie target processes. The challenge is not simple, in that what we conceive of as distinct molecular entities may exist on any of a number of scales, from coherent protein complexes, to physically unrelated complexes in a
single cell, to proteins present in unrelated taxa but where complementary activities yield a desired outcome. Major challenges include the following:
-
Identification and functional characterization of the proteins and the complexes that underlie Genomics: GTL target processes.
-
Formulation of models that predict the function of these key cellular or organismal components in situ.
-
Development of strategies to improve the efficiency of these “molecular machines.”
-
Improving methods for analysis and interpretation of gene and protein function in heterologous systems, including both computational and experimental approaches.
Much of the progress envisioned under Genomics: GTL will require derivation and application of novel technologies, principles, and computational approaches that permit biologists and engineers to understand and manipulate the Genomics: GTL ecosystems. Key milestones toward this broad goal are
-
Improved technologies for surveying taxonomic and genetic diversity in target environments, including the development of tools for both culture-dependent and culture-independent methods and strategies to deal with ultrarare genomes.
-
Development of experimental tools, concepts, and mathematical methods that can model transient and stable states and identify the control points for particular system parameters.
-
Establishment of predictive models of microbial behavior during discrete phases of development and in response to external biotic and abiotic stimuli.
-
Development of new methods and instrumentation to measure key biological parameters that may be relevant to system function, including metabolite flow and protein function in vivo and in situ.
-
Establishment of methods to reproduce native ecologies in the laboratory or to analyze them in situ.
-
Understanding of the consequences and frequency of events that may alter population function, such as horizontal gene transfer, alterations of physical-chemical environments, and introduction of nonnative species.
Broadly stated, the goal of systems biology is to uncover properties of organisms and communities that would not be made apparent by analysis of their components in isolation. Few would argue that our current understanding and methods are adequate to develop a quantitative model of even one bacterium, much less a collection of genotypes in a single species, and even less an entire ecosystem. Systems biology suffers from a dearth of general principles that can guide further study. Nevertheless, although the challenges are substantial, the
situation is not impossible, and the general goal of understanding system function is worthy. Evolution solved the problem by making systems work. A question that is relevant to the proposed Genomics: GTL facilities is, What is the best route to an understanding of these systems?
Despite the temptation to draw analogies, biological systems are not similar to electrical circuits. Electrical circuits are composed of a rather small diversity of entities, whereas biological systems are composed of a multitude of dissimilar parts, even to the point of adaptive variation in apparently common components. Given the scale and complexity of the challenge, it is not obvious that a complete catalog and partial analysis of all proteins in a few target genomes would be a major advance toward understanding and predicting the function of complex microbial systems.
In going forward, some questions need to be considered:
-
Do we need a complete catalog of all parts, or do we want only to describe some parts?
-
If we want to focus only on some parts, what are they, and how do we identify them?
-
Given a complete catalog of parts, do we care equally about all interactions between parts?
-
Do we care equally about where every part resides, in every genotype, and under every condition, or do we care more about some parts under specific conditions?
It stands to reason that for any given situation only a small portion of the “parts” need to be understood in great detail to model system function. Such control points might be genes, alleles, proteins, metabolites, genotypes, or even taxa and their relative spatial distribution. Moreover, the specific control points may vary between different systems and situations. Developing methods of identifying such control points would be a major step toward predicting system function. Moreover, it would allow research to focus on relevant aspects of systems rather than all aspects irrespective of their relative importance.
It is not clear that describing the protein components of individual cells or multiorganism consortia is a necessary first step toward systems biology. Forefront science requires taking a step beyond that, into a detailed characterization of target systems by highly interdisciplinary teams of scientists. In the course of such an endeavor, enumeration of individual components and their interrelationships emerges naturally because it is driven by the complexity of the specific systems under study, as revealed by an integrated approach to their analysis. The technologies (both new and existing) are more effectively limited to those which are appropriate, and this is both time-efficient and cost-effective.