6
Manipulating Host-Microbe Interactions: Probiotic Research and Regulations
OVERVIEW
As defined by an expert panel convened in 2002 by the Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO), probiotics are “live microorganisms administered in adequate amounts that confer a beneficial health effect on the host.” (FAO/WHO, 2002). Contributors to this chapter discuss the meaning of “beneficial health effects” in this context, and more importantly, how such overtly qualitative notions can be replaced by quantifiable variables. The first two papers explore genetic and molecular mechanisms specific to interactions between probiotic bacteria and their hosts; two additional papers describe how the health benefits of probiotic bacteria are currently understood and how their safety and efficacy could be characterized in the future.
Recently developed genomic technologies provide a promising route to evaluating the effects of probiotics on the host-microbe relationship, according to Michiel Kleerebezem of the Wageningen Centre for Food Sciences in the Netherlands. In the first contribution to this chapter, he notes that comparative genomic studies of probiotic bacteria may lead to insights on the nature of molecular mechanisms that confer probiotic effects—findings that could be complemented by DNA microarray technology that analyzes host responses to probiotic microbes. Further, Kleerebezem describes how techniques previously used to elucidate in vivo responses of pathogenic bacteria to environmental parameters in such complex niches as the gastrointestinal (GI) tract, and that are currently being applied to probiotic bacteria, may permit the construction of site-directed bacterial vehicles for delivering beneficial molecules to the human GI tract: the next generation of probiotics.
Research in Suzanne Cunningham-Rundles’ lab concerns the possible role of probiotic bacteria in the modulation of the host immune response. In their contributed paper, Cunningham-Rundles and coworkers describe recent studies characterizing the initial formation of mucosal immunity through interaction with the GI microflora—an interaction that may induce specific and persistent immune response patterns in the host, and which could potentially be manipulated with probiotics. In particular, they explore the potential of probiotic lactic acid bacteria to enhance systemic, as well as mucosal, immune response in infants and young children and the use of probiotic bacteria as antigen delivery vehicles or adjuvants in HIV-1-positive patients.
Based on the results of their efforts, Cunningham-Rundles noted in workshop discussion that she and her coworkers are preparing an investigational new drug (IND) application for probiotic-fortified formula intended for use in low birth-weight infants. The IND application process and its implications for probiotic development are described in the subsequent essay on U.S. probiotics regulatory issues by workshop participant Julienne Vaillancourt of the FDA’s Office of Vaccine Research and Review in the Center for Biologics Evaluation and Research, which has regulatory authority over the development and marketing of probiotics for clinical treatment indications.
To date, all probiotics on the U.S. market fit the FDA definition of a dietary supplement (“a product taken by mouth that contains a dietary ingredient intended to supplement the diet”). Although this situation is expected to change, there is little incentive for manufacturers of probiotics—currently marketed as dietary supplements—to develop them as biotherapeutics given the rigors and expense of the associated review and regulation process. A similar situation currently exists in Europe, where several countries are currently considering legislation to require proof for health claims by manufacturers of dietary supplements. In the United States, the dietary supplement/biotherapeutic dichotomy is likely to remain a part of probiotic regulations for some time. However, as Vaillancourt notes, the regulatory process for biotherapeutics is likely to expand and change to reflect new knowledge; she identifies several issues that need to be addressed through collaborative efforts between all involved parties.
It is not only clear that guidelines and regulations governing probiotics must be revised to reflect recent research findings, but also that this goal is a fast-moving target. In the final contribution to this chapter, workshop presenter Lorenzo Morelli and coworkers describe the process and considerations that produced the recent FAO/WHO guidelines for the evaluation of probiotics in food and raise a variety of cutting-edge issues that need to be addressed in subsequent guidelines, including:
-
new genomic techniques that allow enhanced characterization of the gut microbiota,
-
evaluation of emerging methods that allow assessment of bacteria-epithelial interactions,
-
potential for targeted probiotic or biotherapeutic methods, such as to enhance the production of a specific cytokine or to suppress a specific pathogen.
The authors emphasize the need for such guidelines to be flexible and to reflect ongoing communication between regulatory bodies and the scientific community.
MOLECULAR ANALYSIS OF PROBIOTIC-HOST INTERACTIONS IN THE GASTROINTESTINAL TRACT
Michiel Kleerebezem1
Abstract
Recent years have seen an explosion in the number of complete, and almost complete, genome sequences of lactic acid and other food-grade bacteria, including probiotic strains that are applied as functional food ingredients to increase the health of the consumer. This information is crucial for the development of functional, comparative, and other postgenomic approaches to unravel the in situ functionality of these bacteria in the human intestinal tract and how they affect consumer health at the molecular level. These advances can ultimately be exploited to develop novel and designer probiotics with a predestined impact on consumer gut health.
Introduction
The term probiotics was coined in the 1960s, although, the probiotic concept has existed for a much longer time. The definition of the term has changed through the years, but perhaps the most appropriate definition was published by an expert consultation at a meeting convened by the FAO and the WHO in October 2001, which states, “probiotics are live microorganisms which when administered in adequate amounts confer a health benefit on the host” (FAO/WHO, 2001). Although relatively simple, it defines six aspects that must be fulfilled by probiotic culture applications, while it encompasses probiotic applications outside the food market (Sanders, 2003):
-
Probiotics must be alive, which redirects reference to physiological effects in hosts of the administration of dead cells or cellular fractions to an alternative term.
-
Probiotics must deliver a measured physiological benefit that requires substantiation by studies performed in the target host organism.
|
1 |
Wageningen Centre for Food Sciences, NIZO Food Research. P.O. Box 20, 6710 BA Ede, The Netherlands, Phone: 31-318-659629, Fax: 31-318-650400, E-mail: Michiel.Kleerebezem@nizo.nl. |
-
Probiotics are not necessarily administered as food products via the oral route, but could encompass other applications.
-
Pharmaceutical and therapeutic applications of probiotics are not excluded.
-
Restrictions in terms of mode of action are not defined; therefore, aspects such as survival of the gastrointestinal (GI) tract passage or affect on the normal microflora are not required.
-
Because it is scientifically untenable to envision validated host-physiology effects convened by undefined microbial preparations, the definition also implies that probiotics are defined strains.
Despite this definition of the term probiotics, regulatory restrictions for the use of the term in specific applications are lagging. Moreover, probiotic health claims on products in general tend to be vaguely phrased. As a consequence, in most cases it is virtually impossible for consumers to perceive what to expect from a product carrying the probiotic designation.
Most probiotic preparations currently marketed aim at functional modulation of specific physiological aspects of the GI tract of the consumer. In this respect, it is important to note that these probiotic cultures are to exert their effects on host physiology within this highly complex ecosystem that is colonized by a myriad of endogenous microbes (microbiota, as described below). The species most commonly encountered in these products are specific strains of bifidobacteria and lactobacilli. The health benefit(s) attributed to these products is highly diverse, and the quality of the scientific evidence underlying these benefits is highly variable. The evidence is mostly descriptive and frequently lacks comparison of different strains of bacteria to validate the probiotic’s specificity. In addition, the molecular basis of the effects on host physiology of specific probiotics remains largely unexplored. Therefore, correlating specific physiological effects measured in the target host to a specific characteristic of a bacterial strain is not yet possible. Such correlations could provide avenues toward second-generation probiotics with predestined or improved health effects.
Probiotics and the Human GI Tract Microbiota
The human GI tract is colonized by a vast and complex consortium of mainly bacterial cells. This microbiota consists of at least 1013 microbes, dominated by anaerobic bacteria, comprising over 1,000 species, of which the majority cannot yet be cultured under laboratory conditions (Vaughan et al., 2000; Zoetendal et al., 2004). Culture-independent molecular approaches that utilize the 16S ribosomal RNA (rRNA) genes as a universal bacterial biomarker have been used to monitor the composition of the dominant GI tract microbiota in different individuals at different time points in their lives. These approaches revealed a relatively stable composition in individual adults, but appeared to vary considerably when differ-
ent individuals were compared (Zoetendal et al., 1998). Moreover, host development, host genotype, and environmental factors influence the composition of the microbiota, illustrating how microbiologically challenging this environmental niche is (Zoetendal et al., 2004). Many functions are associated with the bacterial GI tract communities and their interaction with the host system including roles in host nutrition, intestinal epithelial development and activity, education of the immune system, maintenance of the integrity of the mucosal barrier, and contribution to drug and xenobiotic metabolism. However, we are only beginning to understand the dimensions of these activities and interactions.
Next to the global microbiota-composition profiling efforts described above, dedicated 16S rRNA-based methods have been developed to track and trace specific species within the GI tract (Vaughan et al., 2005). These studies not only enable profiling the microbiota at the level of a specific species that is endogenously present in the GI tract, but also allow more detailed analyses of the fate of orally administered (probiotic) bacteria during their transit through this complex system. Nevertheless, these technologies do not allow the study of microbiota or probiotic in situ activity, and thus do not provide insight in their mode of action in relation to host functionality. In contrast, genomics-based approaches should allow the functional evaluation of host and microbe interactions at the molecular level. Such molecular studies should reveal the molecules involved in these interactions, which would allow the development of molecular models that underlie host-microbe interactions. Such mechanistic insight into the way probiotic bacteria affect health will not only contribute to novel, improved functional foods but also to the support of their health claims that will most likely be subject to an increased scientific scrutiny in the future. Therefore, these developments will not only provide benefits for the scientific or industrial community, but will also meet the interest of the consumer (de Vos et al., 2004) (Figure 6-1).
Bacterial Genomics
Over the past decade, the sequences of more than 200 bacterial genomes have become available in the public domain. Considerable attention has focused on pathogenic bacteria, including food-borne pathogens. However, over the last years, sequencing the genomes of GI tract commensals and symbionts as well as food-grade bacteria has received considerable attention. This progress includes elucidation of the (partial) genome sequences of more than 20 lactic acid bacteria and bifidobacteria (Table 6-1; Klaenhammer et al., 2005). Among these are the published complete genome sequences of species associated with probiotic health effects in humans, including Lactobacillus plantarum (Kleerebezem et al., 2003; van Kranenburg et al., 2005), Lactobacillus acidophilus (Altermann et al., 2005), Lactobacillus johnsonii (Pridmore et al., 2004), and Bifidobacterium longum (Schell et al., 2002). Additionally, the release of genomic sequences of commensal human GI bacteria such as Bacteroides thetaiotaomicron (Xu et al., 2003)
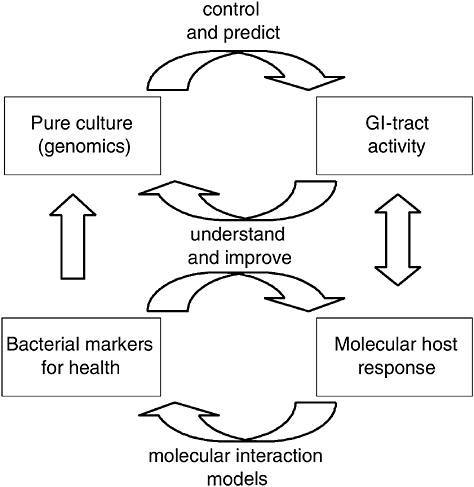
FIGURE 6-1 Schematic representation of pure culture genomics in relation to GI tract behavior, and corresponding molecular analyses of host responses. The molecular interaction models should allow the identification of bacterial and host markers involved in probiotic-mediated host health benefits. Such information can subsequently be employed to construct or select specific strains or mutants that have improved or enhanced health of the host, which would generate second-generation probiotic cultures with science-based health claims and validated modes of action.
SOURCE: Kleerebezem (2005).
will add valuable information to this field. Moreover, rapid expansion of this knowledge base is anticipated upon publication of the genomic sequences of several other species that can be categorized as (potential) probiotics (Table 6-1). The availability of these bacterial genome sequences and their annotated functions provides valuable clues towards the survival strategy of these bacteria during their residence in the human GI tract. Moreover, in silico comparative
TABLE 6-1 Overview of Genome Sequences of Food-Grade Bacteria. Adapted from Klaenhammer et al., 2005
|
Genus |
Species |
Strain(s) |
Genome size (Mbp) |
|
Bifidobacterium |
longum |
NCC2705, DJ010A |
2.3, 2.4 |
|
|
breve |
NCIMB 8807 |
2.4 |
|
Brevibacterium |
linens |
ATCC9174 |
4.4 |
|
Enterococcus |
faecalis |
V583 |
3.2 |
|
Lactobacillus |
acidophilus |
NCFM |
2.0 |
|
|
gasseri |
ATCC333323 |
1.8 |
|
johnsonii |
NCC533 |
2.0 |
|
|
plantarum |
WCFS1 |
3.3 |
|
|
casei |
ATCC334, BL23 |
2.5, 2.6 |
|
|
rhamnosus |
HN001 |
2.4 |
|
|
helveticus |
CM4, CNRZ32 |
2.0, 2.4 |
|
|
sakei |
23K |
1.9 |
|
|
delbrueckii |
ATCCBAA365, ATCC11842, DN-100107 |
2.3, 2.3, 2.1 |
|
|
reuteri |
|
2.0 |
|
|
salivarius |
UCC118 |
|
|
|
brevus |
ATCC367 |
2.0 |
|
|
Lactococcus |
lactis spp. lactis |
IL1403 |
2.3 |
|
|
lactis spp. cremoris |
SK11, MG1363 |
2.3, 2.6 |
|
Leuconostoc |
mesenteroides |
ATCC8293 |
2.0 |
|
Oenococcus |
oeni |
ATCCBAA331, IOEB84.13 |
1.8, 1.8 |
|
Pediococcus |
pentacosus |
ATCC25745 |
2.0 |
|
Propionibacterium |
freundereichii |
ATCC6207 |
2.6 |
|
Streptococcus |
thermophilus |
LMD9, LMG 18311, CNRZ1066 |
1.8, 1.9, 1.8 |
|
SOURCE: Kleerebezem, 2005. |
|||
genomics can provide important insight in diversity, evolutionary relationship, and functional variation between bacteria (Boekhorst et al., 2004, 2005), which might eventually generate a comprehensive prediction of microbe behavior during residence in the human GI tract. Based on the probiotic definition, it is postulated that probiotic features can be attributed to specific strains rather than to any particular species as a whole. This notion suggests that activities proposed to confer probiotic effects are expected to be variable among different strains of a species.
A clear example where this could experimentally be established is the mannose-specific adherence capacity observed in L. plantarum that is proposed to be involved in inhibition of pathogenic Escherichia coli infections via a mechanism of competitive exclusion (Adlerberth et al., 1996; Pretzer et al., 2005). It had been reported earlier that the capacity to adhere specifically to mannose moieties is a variable phenotype among L. plantarum strains. Nevertheless, thanks to the availability of the L. plantarum WCFS1 template genome and the corresponding DNA microarrays, this relevant phenotype could be correlated to a genotypic diversity
that corresponded to a single gene that could subsequently be shown to encode the mannose-specific adhesion (therefore designated: msa gene) in this species by mutation analysis (Molenaar et al., 2005; Pretzer et al., 2005). The functional proof of the E. coli inhibitory, probiotic character mediated by this msa gene can now be established using isogenic variants that either lack or overexpress a functional copy of this gene. In analogy, application of host and microbe postgenomics tools in the actual in vivo setting enables new avenues towards more exploratory research aiming to unravel the molecular interactions between host and microbe that underly the health benefits mediated by consumption of probiotic bacteria (Figure 6-1).
Monitoring Bacterial Responses to Intestinal Conditions: In Vitro and In Vivo Approaches
Severe barriers are met by bacteria that we consume as part of our diet. These include several physical and chemical conditions, such as gastric acidity, the presence of bile salts, and stress conditions associated with oxygen gradients that are steep at the mucosal surface while the intestinal lumen is virtually anoxic. In addition, bacterial competition is high throughout the intestinal tract, but reaches a climax in the colon where bacterial densities are highest. Because the GI tract environment is highly complex and differs in different regions of this niche, many studies describe the in vitro response of bacteria to a simplified model that mimics a single host GI tract parameter. Most of these studies have focussed on intestinal pathogens, including studies describing the response toward acid and bile stress in bacteria such as Salmonella species, Escherichia coli, Listeria monocytogenes, and Enterococcus faecalis. More recent studies describe the molecular responses of food-grade and probiotic bacteria to similar stress conditions. Only a few studies describe genomewide approaches aiming at the identification of genes and proteins important for acid- and bile-resistance in lactobacilli or bifidobacteria. With Lactobacillus plantarum, a genetic screen resulted in the identification of more than 30 genes that are induced by bile-stress in vitro, and for a selection of these genes it could be established that induction of these genes also occurs in vivo in the duodenum of mice (Bron et al., 2004a). Additionally, a recent DNA microarray-based study in the same species revealed a number of genes and gene clusters of which the expression is significantly affected by the presence of bile acids (Bron et al., 2006). Many of these genes are predicted to play a role in the cell membrane or cell wall, which is in good agreement with several physiological studies in GI tract bacteria, including Lactobacillus plantarum and Propionibacterium freudenreichii, demonstrating how bile salts induce severe changes in the morphology of the cell membranes and/or cell walls of these organisms (Bron et al., 2004a; Leverrier et al., 2003). Overall, these in vitro studies have generated valuable insight in the responses of food-grade and probiotic bacteria to the individual parameters that are encountered in the host GI
tract. However, the extrapolation of these results to the real-life situation in this highly complex niche is certainly not straightforward and requires a substantial amount of additional research to establish the importance of the in vitro responses in terms of the full response repertoire triggered in vivo (Bron et al., 2004a).
To experimentally approach the elucidation of in vivo responses of bacteria to environmental parameters in complex niches such as the GI tract, more sophisticated methods have been developed. Three main strategies have been employed for the identification of genes that are highly expressed in vivo as compared to laboratory conditions: in vivo expression technology (IVET) and recombination-based in vivo expression technology (R-IVET), signature-tagged mutagenesis (STM), and selective capture of transcribed sequences (SCOTS) (Bron et al., 2005). These in vivo gene-identification strategies were initially used exclusively in pathogenic bacteria, aiming at the identification of genes important to the virulence of these species. Recently, the first two reports appeared that describe the utilization of R-IVET strategies in food-grade or commensal microorganisms in order to determine the specific induction of gene expression in these bacteria after introduction in the GI tract of animal models. With L. reuteri an IVET strategy based on in vivo selection of an antibiotic-resistant phenotype led to the identification of three in vivo induced (ivi) genes during colonization of the GI tract of Lactobacillus-free mice (Walter et al., 2003). One of these genes encodes a peptide methionine sulfoxide reductase that has previously been identified using IVET in Streptococcus gordonii during endocarditis. Although not noticed by the authors at the time of publication, this finding already hinted at overlap in the genetic response triggered in the pathogenic and nonpathogenic bacteria upon contact with the host intestine. This notion was further exemplified by an R-IVET approach in L. plantarum (Bron et al., 2004b). This study revealed 72 L. plantarum genes that were ivi during passage of the GI tract of conventional mice. Functional classification of these genes indicated that prominent in vivo responses include functions associated with nutrient acquisition, intermediate and/or cofactor biosynthesis, stress response, and cell surface proteins. However, also a number of (conserved) hypothetical proteins were identified among the L. plantarum ivi genes (Bron et al., 2004b). Remarkably, one of the latter group of genes displayed significant homology (32 percent identity) to the conserved hypothetical protein that was identified with IVET in L. reuteri (Bron et al., 2004b; Walter et al., 2003), indicating conservation of GI tract responses in different lactobacilli. Moreover, a striking number of the ivi functions and pathways identified by R-IVET in L. plantarum were previously found among the in vivo response in pathogens, suggesting that survival rather than virulence is the explanation for the importance of these genes during host residence (Table 6-2; Bron et al., 2004b). Following the above-mentioned ivi gene-identification strategies, the actual in situ expression of a selection of the L. plantarum ivi genes could be obtained by quantitative reverse transcription polymerase chain reaction (qRT-PCR) using RNA preparations from mouse intestinal samples that had consumed
TABLE 6-2 Functional Classification of Predicted ivi Gene Functions in L. plantarum and Pathogenic Bacteria. Adapted from Bron et al., 2004b; de Vos et al, 2004
L. plantarum containing food. Time-range analysis in a series of these mice revealed geographical differentiation of the induction patterns displayed by these ivi genes in the various regions of the mouse intestine (Unpublished observations, M. Marco, R. Bongers, and M. Kleerebezem. Wageningen Centre for Food Sciences, The Netherlands, 2005). This information could be exploited to construct dedicated, site-directed bacterial delivery vehicles aiming to deliver health beneficial (therapeutic) molecules in the GI tract of humans. The potential of lactic acid bacteria in this type of application has recently been reviewed by Hanniffy et al. (2004). In addition to in situ transcript detection, the importance for bacterial functionality in the host GI tract of the identified ivi genes can be established by comparative persistence and survival analysis of wild-type and isogenic mutant analysis in vivo. Such approaches have been performed in both lactobacilli. In L. reuteri, mutation of the methionine sulfoxide reductase-encoding msrB gene (one of the ivi genes) resulted in reduced ecological performance in the mouse GI tract as compared to the wild-type strain. Analogously, mutation of the or—the gene encoding the Lsp surface protein of L. reuteri that is implicated in adherence of this bacterium to epithelial cells—also resulted in similar in vivo performance defects (Walter et al., 2005). In L. plantarum, nine isogenic ivi mutants
were constructed, mainly focusing on genes that encode proteins with a predicted role in cell envelope functionality, stress response, and regulation. Quantitative PCR experiments were performed to monitor the relative population abundance of the group of the L. plantarum ivi mutants in fecal samples after competitive passage through the GI tract of mice; the experiments revealed that the relative abundance of three of the ivi gene mutants was reduced by 100- to 1,000-fold compared to other mutant strains and the wild-type strain, suggesting a critical role for these ivi genes in GI tract survival and persistence (Bron et al., 2004c).
Global bacterial expression profiling of Bacteroides thetaiotaomicron residing in the cecum of monoassociated mice has recently been reported for the first time. By varying the mouse diet from a polysaccharide-rich diet to isocaloric diets that only contained simple sugars, this pioneering study revealed that B. thetaiotaomicron adapts its enzyme expression patterns to the sugars available; furthermore, upon paucity of the polysaccharides, the commensal microbe shifted its glycan-foraging behavior to the host mucous as a nutritive source, a property that aids its stability in the intestine (Sonnenburg et al., 2005). This work establishes that—although technically difficult—it is certainly feasible to monitor bacterial gene expression in the GI tract at a global level.
The experiments described above were all performed using mouse model systems, and an obvious question that should be raised is whether these bacterial responses to the residence in the mouse GI tract can be extrapolated to bacterial activities in the human GI tract. This question was addressed in a study aiming to measure L. plantarum gene expression in the human GI tract by analyzing RNA extracted from mucosa-associated cells and hybridising to L. plantarum WCFS1 microarrays or gene-specific qRT-PCR. Appropriate controls were performed and included the absence of specific hybridization in biopsies from a subject that had not consumed the L. plantarum cells, the absence of interference by human nucleic acids on the microarray, and confirmation of the specificity by sequence analysis of the gene-specific qRT-PCR (de Vries MC, Marco M, Kleerebezem M, Mangell P, Ahrne S, Molenaar D, de Vos Wm, Vaughan EE, Unpublished data). The ingested L. plantarum cells were found to be metabolically active in all subjects, and differences between gene expression between the individuals and intestinal location were apparent. Moreover, significant parallels were observed in the mouse intestine-induced ivi genes and those that were detected as being highly expressed in the human intestinal system. These findings support an at least partially conserved response of this bacterium to the GI tract conditions encountered in different host systems (de Vries et al., unpublished data).
The studies highlighted above give a first glimpse of what the near future could bring in terms of understanding probiotic activity in situ in the GI tract of host organisms. This information will be of critical importance to construct molecular host-microbe interaction models. However, the responses by the host to probiotic encounter should also be addressed to fill in both sides of the interaction model.
Monitoring Host Intestine Responses to Bacterial Encounters
There are not many studies available that detail the molecular response of the host intestinal system to the interaction with bacteria using a genomic-based approach. Studies in gnotobiotic mice have indicated that there is specific signalling between the commensal bacterium B. thetaiotaomicron and its host. Synthesis of host epithelial glycans is elicited by a B. thetaiotaomicron signal of which the expression is regulated by a fucose-binding bacterial transcription factor. This factor senses environmental levels of fucose and coordinates the decision to generate a signal for production of host fucosylated glycans when environmental fucose is limited or to induce expression of the bacteria’s fucose utilization operon when fucose is abundant (Hooper et al., 1999).
Additional studies have evaluated the global intestinal response to colonization of gnotobiotic mice with B. thetaiotaomicron. This colonization dramatically affected the host’s gene expression, including several important intestinal functions such as nutrient absorption, mucosal barrier fortification, and postnatal intestinal maturation (Hooper et al., 2001). From the in situ global transcription profiles mentioned above and follow-up experiments, it could be established that the production of a previously uncharacterised angiogenin is induced when gnotobiotic mice are colonized with B. thetaiotaomicron, revealing a mechanism whereby intestinal commensal bacteria influence GI tract bacterial ecology and shape innate immunity (Hooper et al., 2003). In addition, the cellular origin of the angiogenin response was investigated when different intestinal cell types were separated by laser-capture microdissection and analysed by qRT-PCR, revealing that angiogenin-3 mRNA is specifically induced only in crypt epithelial cells. Hence, these experiments strongly suggest an intestinal tissue-specific response of the host during colonization (Hooper et al., 2001). Interestingly, comparison of the changes in global host gene expression in mice after colonization with B. thetaiotaomicron, Bifidobacterium infantis, or E. coli led to the observation that part of this host response was only induced in mice by colonization with B. thetaiotaomicron (Hooper et al., 2001). However, analysis of a broader range of members of the intestinal microbiota will reveal what the level of bacterial response specificity within the host’s tissues actually is.
One such study is currently being performed with L. plantarum (Personal communication, Erik Peters, M. Marco, J.I. Gordon, and M. Kleerebezem, Wageningen Centre for Food Sciences, The Netherlands, 2005). Overall, the aforementioned studies on B. thetaiotaomicron colonization of gnotobiotic mice provided valuable information on the influence of one particular member of the microbiota on the host. However, the host response during colonization by more complex mixtures of microbes, and/or the host response in other animal systems, remained to be investigated at that time. Recently, it was found that conventionalization of adult gnotobiotic mice with normal microbiota harvested from the distal intestine of conventionally raised mice produced a 60 percent increase in body fat content and insulin resistance despite reduced food intake. Studies of
gnotobiotic and conventionalized mice revealed that the microbiota promotes absorption of monosaccharides from the gut lumen, resulting in induction of de novo hepatic lipogenesis. Fastin-induced adipocyte factor (Fiaf), a member of the angiopoietin-like family of proteins, is selectively suppressed in the intestinal epithelium of normal mice by conventionalization. Analysis of gnotobiotic and conventionalized, and normal and Fiaf knockout mice established that Fiaf is a circulating lipoprotein lipase inhibitor and that its suppression is essential for the microbiota-induced deposition of triglycerides in adipocytes. These results suggest that the gut microbiota have a major impact on food-derived energy harvest and storage in the host (Bäckhed et al., 2004). Another recent study investigated the host response during colonization of a different animal model. DNA microarray comparison of gene expression in the digestive tracts of six days postfertilization gnotobiotic, conventionalized and conventionally raised zebrafish (Danio rerio) revealed 212 genes regulated by the microbiota. Notably, 59 of these genes were also found to be regulated in the mouse intestine during colonization, including genes that encode functions involved in stimulation of epithelial proliferation, promotion of nutrient metabolism, and innate immune response, indicating a substantial overlap in the genetic response of mice and zebrafish towards intestinal colonization (Rawls et al., 2004). Despite these recent developments, an important future challenge lies within the translation of these animal host response analyses to the human system.
Concluding Remarks and Future Perspectives
At present, a large part of the consortium of bacteria residing in the GI tract has not been cultured in vitro. Because most genetic approaches require the cultivability of the microbe under investigation, the expansion of our knowledge of this group of bacteria is highly challenging and very limiting at this stage. The development of effective and robust methods to assess microbiota activity in situ in a culture-independent manner combined with metagenomic approaches will be critical for our understanding of the large number of uncultivable bacteria in the GI tract.
Historically, research on the bacterial flora of the GI tract has concentrated on pathogenic bacteria. More recently, research has expanded to nonpathogenic bacteria, including symbionts, commensals, and food bacteria. One obvious reason for this is the accumulating evidence that certain bacteria, especially strains from the genera Lactobacillus and Bifidobacterium, may have probiotic effects in humans and animals. At present, we start to appreciate the complexity of microbial distribution of specific bacteria along the human colon and the variations that can occur between different individuals. Moreover, knowledge on the activity and response of specific species to the conditions encountered when they transit through this complex niche is starting to accumulate. A promising prospect from the increasing availability of complete genome sequences is the construction of
DNA microarrays in several laboratories working on food-associated microbes. Genomics-based global investigations of gene expression in food-grade microbes under various conditions will further detail our understanding of their behavior. Besides the application of DNA microarray technology to reveal the bacterial side of host-microbe interactions, this technology also allows host response analyses. Combination of these knowledge datasets should enable scientists to unravel the mechanisms that are underlying the effects of intestinal and food-derived bacteria on host physiology, and will provide the probiotic arena with new and more scientifically solid consumer health benefits.
In conclusion, the genomewide transcript-profiling approaches that have been performed to date have provided us with clues of the possible role of individual host and bacterial genes during host-microbe interactions. Bacterium and host transcriptome information should allow the construction of molecular models that describe host-microbe interactions, allowing more pinpointed experiments in the future, designed on the basis of a molecular interaction hypothesis. Because GI tract bacteria such as L. plantarum and B. thetaiotaomicron are genetically accessible, gene deletion and overexpression mutants can be employed to study the impact of a single bacterial gene on host gene expression. Alternatively, knockout mice and/or antisense RNA approaches might allow gene silencing on the host side of the spectrum, thereby enabling the study of single host gene mutations on the in situ functionality of microbes.
Acknowledgments
My WCFS colleagues that shared unpublished information are cordially acknowledged.
ROLE OF PROBIOTICS IN MODULATION OF HOST IMMUNE RESPONSE
Susanna Cunningham-Rundles, Siv Ahrne, John Peoples, Francesca Tatad, Mohamed Mohamed, and Mirjana Nesin2
Abstract
Mucosal immune response is primed at birth, and responses generated at this time support specific immunity in later life. Recent studies show that mucosal
|
2 |
Host Defenses Program, Department of Pediatrics, Weill Cornell Medical College, New York, NY 10021. Sources of Support: NIH NCI 29502, NIH NCRR M01RR00047, NIH NCRR M01RR6020, Probi, and The Children’s Blood Foundation. Address for correspondence: Susanna Cunningham-Rundles, Professor of Immunology, Department of Pediatrics. Cornell University Weill Medical College, 1300 New York Avenue, New York, NY 10021. Telephone: (212) 746 3414; fax: (212) 746 8512; e-mail: scrundle@med.cornell.edu. |
immunity is initially formed through interaction with the GI microflora. The composition of the developing microbiota in the neonatal period may induce specific immune response patterns that persist in the host. Environmental antigens, dietary factors, and nutrition can alter this interaction. Dietary factors also stimulate the growth of flora and may have selective effects on the composition of the microbiota. Infections and expression of host modifier genes can block or disrupt the establishment of controlled inflammation in the GI tract. Several lines of investigation suggest that alteration of gut microflora in association with lack of appropriate initial colonization, overuse of antibiotics, chronic infection, or malnutrition has a negative effect on host immune response. Probiotic lactic acid bacteria offer a possible approach for stimulating the GI immune system thereby enhancing systemic as well as mucosal immune response. Rigorous studies are needed to assess whether the microbial microenvironment can be modulated by the introduction of defined probiotic bacteria and to analyze the long-term potential for host defense.
Introduction
Mucosal membrane surfaces provide the strategic interface between the internal and external world and contain a large and variable antigenic load (Didierlaurent et al., 2002). In addition to the indigenous mucosal microbiota, the mucosal immune system must respond to potential microbial pathogens, food antigens, and environmental allergens (Eberl, 2005). Most infectious diseases are acquired by, or affect, mucosal surfaces such that preventive and protective host defenses require mucosal response. The gut immune system contains the highest numbers of macrophages, plasma cells, and T lymphocytes in the body. This highly specialized immune system is based on anatomical separation of inductive and effector sites, compartmentalization of pattern recognition receptors, and distinctive cellular functions that bridge innate and adaptive immune response (Hall et al., 1994; Macdonald and Monteleone, 2005; Pabst, 1987). Mucosal immune response is characteristically skewed towards a T helper type2 (Th2) cytokine pattern dominated by production of interleukin 4 (IL-4) and IL-5, which supports B-cell differentiation and the development of antibodies. Response to infection is mediated by secretory IgA antibodies and mucosal cytotoxic T cells and a highly developed innate immune response (Kelly and Conway, 2005; Knight et al., 2004; Rakoff-Nahoum et al., 2004).
The presence of large populations of commensal bacteria has a major influence on the development of immune response that is only now beginning to be studied in detail. Commensal flora directly activate the mucosal immune system leading to the development of Peyer’s patches and to IgA plasma cells and CD4+T cells in the lamina propria. Maturation of the mucosal immune system and establishment of protective immunity varies between individuals but is usually fully developed in the first year of life, regardless of gestational age at birth. Experi-
mental studies have shown that mucosal immune response is primed in the neonate and that responses generated at this time are retained in later life (Harrod et al., 2001). Host response exerts a selective pressure on colonizing species, as shown by longitudinal study of secretory immune response in saliva to colonizing bacteria in infants during tooth eruption (Cole et al., 1998). The evolution of bacterial species in the gut microflora over a lifetime has not been studied extensively, but preliminary studies indicate a possible increase in the species diversity of the dominant microflora with aging (Blaut et al., 2002; Saunier and Dore, 2002). One study indicates that a general reduction in specific serum IgM antibody to commensals may occur in old age, which could lead to relaxation of selective pressure and outgrowth of less benign bacterial populations. This may have relevance for increasing infections and decreased immune response in older people (Percival et al., 1996). A relationship between altered microflora and cancer may also exist, for example, through effects on levels of isothiocyanates, which mediate the cancer-protective effects of cruciferous vegetables (Rouzaud et al., 2003). Generally, microflora influence maintenance of intestinal homeostasis through direct effects on the development of organized lymphoid tissue (Rhee et al., 2004) and prevention of inflammation.
Recent studies show that gut bacteria induce cross-talk between epithelial cells and dendritic cells that leads to the release of IL-10 and IL-6 and, therefore, drive the polarization of T cells toward a noninflammatory Th2 response that is maintained even after exposure to Th1-inducing pathogens (Rimoldi et al., 2005). Intestinal immune stability depends upon the function of toll-like receptors (TLRs) and the nucleotide oligomerization domain NOD1 and NOD2 pattern-recognition receptors to convert the recognition of pathogen-associated molecules expressed by enteric bacteria in the gut into signals for antimicrobial peptide expression, barrier fortification, and proliferation of epithelial cells. This process can be blocked or disrupted by infections, or by the expression of certain host polymorphisms in the TLR or NOD genes that lead to uncontrolled gut inflammation, as in inflammatory bowel disease (IBD) (Abreu et al., 2005). Other critical influences on gut microflora include environmental antigens, dietary factors, and nutrients (Cunningham-Rundles, 2001; Macdonald and Monteleone, 2005; Pacha, 2000). Nondigestible dietary factors stimulate the growth of flora and have selective effects on the composition of the microbiota (Roller et al., 2004). The gut microbiota as a whole is essential for production of short-chain fatty acids from polysaccharides and has been shown to regulate host metabolism through direct effects on fat storage (Bäckhed et al., 2004). The interaction of host immune response and the microbiota may, therefore, indirectly influence metabolism.
How the gut immune system distinguishes between pathogenic and commensal bacteria is unclear. A pathogenic immune response in the gut wall characterized by a highly polarized Th1 response is the hallmark of Crohn’s disease, and this response is probably directed against antigens of the commensal flora
(Macdonald et al., 2005). Commensal flora interact with the gut epithelium to stimulate the development of gut barrier function and this mutualism nonspecifically regulates inflammation (Macdonald and Monteleone, 2005). Development of immune tolerance towards food and environmental antigens, including commensal bacteria, is a central requirement for gut homeostasis. The primary mediators now appear to be regulatory T cells and dendritic cells that down regulate inflammatory response through production of IL-10 and transformation of growth factor beta-1 (Bilsborough and Viney, 2004; Caramalho et al., 2003). Tolerance to commensal antigens involves active suppression of T cell response has not been established.
Host-bacterial interaction has general implications for the development of adaptive immune response to environmental antigens and allergens in early childhood. A functional relationship between the composition of microflora and presence or absence of allergies and atopic disease in children has been shown by several well-constructed and thorough studies. These include a report of reduced colonization with lactobacilli and higher counts of aerobic bacteria in a large study of allergic children (Bjorksten et al., 1999) and the demonstration that characteristic differences in neonatal gut flora precede development of allergic responses (Kalliomaki et al., 2001a). In particular the microbiota of allergic children were less often colonized with lactobacilli and bifidobacteria. These findings led to a definitive study in which perinatal administration of probiotic lactobacilli was shown to decrease presentation of eczema in high-risk children (Kalliomaki et al., 2001b).
The concept that supplementation with some bacteria would lead to elective colonization and would prevent outgrowth of more pathogenic bacteria was originally proposed by Metchnicoff (1907). The term probiotic is generally accepted to include, “live microorganisms which when administered … confer a health benefit on the host” (Reid et al., 2003). Probiotic bacterial supplementation has shown consistent effects on resolution of antibiotic-associated diarrhea (Arvola et al., 1999) and recovery from acute diarrhea caused by rotavirus infection (Guandalini et al., 2000). In addition, combinations of lactobacilli, bifidobacteria, and Streptococcus salivarius prevent relapse of recurrent pouchitis, and may decrease the initial onset of pouch inflammation. Also, Escherichia coli strain Nissle 1917 is reported to maintain remission in ulcerative colitis (Sartor, 2005). These effects may be mediated by production of antimicrobial substances, local competition for adhesion receptors and nutrients, and stimulation of intestinal antigen-specific and nonspecific-immune responses. However, efforts to improve IBD and irritable bowel syndrome with probiotic bacterial supplementation have only had limited success, perhaps because the central mechanism of these disorders appears to involve altered host response to commensal flora (Schultz et al., 2004b). With the exception of prophylactic treatment of children at risk for atopic disease that clearly involves direct immunoregulatory effects, probiotic bacteria appear to stimulate enhancement of recovery through acute effects on the micro-
biota. It is possible that in adults the resident flora and interactive host response compose a formidable barrier that is not as readily affected by short-term colonization. Adult human flora appear to have a constrained diversity as only a limited number of bacterial divisions have been identified (Bäckhed et al., 2005). One study suggests that gut populations are fairly stable within individuals over time (Bäckhed et al., 2005; Zoetendal et al., 2004). However, another report, also on a few subjects, suggests marked variability over time (Barcenilla et al., 2000).
In summary, current studies suggest that host interaction with the microbiota shapes mucosal immune response. These studies also support the concept that probiotic bacteria could influence host response, as well as composition of the microbiota, at least transiently. Because host microbial cross-talk is initiated at birth and the immune system is primed by this exposure over the first year of life, studies in children may have unique potency. Settings of interest, including the low birth-weight infant and the immune compromised child, are discussed in the following sections.
Neonatal Response to Commensal Microbes
After birth, bacteria are always present in great abundance on skin and in the GI tract, and critical development of immune function occurs in early neonatal life in response to microbial colonization. Th2-skewed immune response prevails systemically in the neonate, and contact with microbial antigens acts to repolarize this orientation gradually during the first months of life (Prescott et al., 1998). Studies strongly suggest that absence of exposure to appropriate microbial signals and lack of a Th2 to Th1 switch is associated with allergic disease in high-risk children. Generally infants are initially colonized with Escherichia coli and streptococci and then by anaerobes belonging to Bacteroides, Bifidobacterium, and Clostridia. In contrast to formula-fed infants, bifidobacteria and lactobacilli predominate in the breast-fed infant. Breast-fed infants have higher levels of lactic acid bacteria and enhanced mucosal immune response (Martin et al., 2003). Our recent studies in healthy infants show that certain lactobacillus species, especially L. rhamnosus, thrive in the intestinal flora of breast-fed infants. After weaning, they are replaced by other lactobacillus species found in food (Ahrne et al., 2005).
Neonates usually acquire bacterial pathogens transplacentally. Most commonly these are Group B beta-hemolytic Streptococcus (GBBS), and gram-negative flora, E. coli, and Listeria monocytogenes. Premature neonates can acquire nosocomial infections and develop staphylococcal, enterococcal, and P. aeruginosa infections in association with hospitalization. Because the neonate is born with an immature immune system, there is a critical period when encounter with microbes may cause serious infection. Exposure to pathogens in association with prematurity adds an additional stress that can alter the development of normal immune response and is also likely to affect the composition of the
microbiota. Some microorganisms, like coagulase negative staphylococci, rarely enter an uninterrupted mucosal barrier, although they can colonize it. Others, like GBBS, are actively transferred into cytoplasm of respiratory epithelial cells. Because maternal IgG antibodies acquired passively drop in the first days of life, pathogen-specific antibodies are decreased or absent, and neonates must depend upon the innate immune system (Cunningham-Rundles and Nesin, 2000).
Probiotic bacteria have been given to infants primarily for prophylaxis and alleviation of diarrheal disease, reduction in atopic disease, reduction in necrotizing enterocolitis, and reduction in infection, with generally beneficial effects (Kliegman, 2005; Schrezenmeir et al., 2004). In addition, there is increasing interest in the possible use of probiotics to promote the development of immune response (Huang et al., 2003). Immaturity of the neonatal immune response contributes significantly to the severity and morbidity of response to infections and to bacterial antigens. Recent studies indicate that both term and preterm infants have an increased capacity to produce inflammatory cytokines (Schultz et al., 2004a) and a decreased compensatory anti-inflammatory response syndrome that may predispose preterm infants to harmful effects of proinflammatory cytokines (Schultz et al., 2004a). The consequences of enhanced proinflammatory responses are implicated in the pathogenesis of neonatal disease (Goepfert et al., 2004; Huang et al., 2004). Although probiotic bacteria are generally regarded as safe and benefits from treatment are substantial, translocation of indigenous lactobacillus from a catheter is possible, and two cases of lactobacillus bacteremia from probiotic treatment in short bowel syndrome have been reported (Kunz et al., 2004; Thompson et al., 2001).
In previous studies, we observed that premature infants had a heightened proliferative response in vitro to S. epidermidis compared to adults in the first weeks of life suggesting a strong modulatory effect postbirth (Veber et al., 1991). Although S. epidermidis is a normal colonizer of human skin, rarely causing disease in term infants, this is the most common cause of late-onset sepsis in preterm infants. To ascertain if the neonate has a stereotypical response to microbes, we are analyzing the relative response of memory and naive T cells from cord blood of healthy term infants to specific microbial activators, L. plantarum, S. epidermidis, GBBS, and E. coli to determine if lack of memory T cells might be a barrier to response. Both CD4+ and CD8+RA+ cord blood T cells responded to L. plantarum and S. epidermidis, but only CD8+RA+ cells showed significant response to GBBS. Data in Figure 6-2 show the response of CD8+T cells as detected by the up-regulation of CD69 expression. Although memory response was essentially absent, as there are very few memory T cells in cord blood, naive cells were strongly activated by all tested microbial antigens. In addition, we are evaluating the specificity of cytokine response at the level of the producer cell using intracellular cytokine detection by flow cytometry. As shown in Figure 6-3, E. coli induced higher levels of both IL-6 and IL-8 production in cord blood monocytes compared to levels induced by L. plantarum. Insignificant production
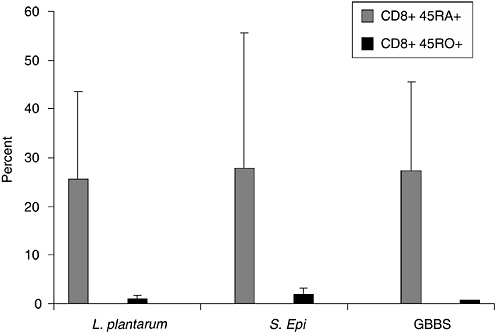
FIGURE 6-2 Response of CD8+ naive (CD45RA+) and CD8+ memory (CD45 RO+) cord blood T cells to bacterial antigens. Data are shown as percent of gated populations expressing CD69 after 18 hours of culture with whole inactivated bacterial cells. The three microbial activators on the x axis are L. plantarum, S. epidermidis, and Group B beta-hemolytic Streptococcus (GBBS).
SOURCE: Cunningham-Rundels (2005).
of either mediator was observed in T cells. These results indicate that the innate immune response in newborns to commensal bacteria is strong, and they also suggest that specific bacterial strains may have differential effects on the maturation of the immune system.
Response to Lactobacillus in HIV-1 Immune Deficiency
Primary immune deficiency is associated with sinopulmonary or GI infections, autoimmunity, and neoplasia (Kalha and Sellin, 2004, Cunningham-Rundles, 1994). Selective IgA deficiency carries an increased risk of celiac disease, IBD, and perhaps also GI malignancy. Recent studies suggest that IgA-deficient individuals carry an increased frequency of E. coli with potentially inflammatory properties in their microflora, which may contribute to the development of GI disorders such as IBD (Friman et al., 2002). Secondary immune deficiency caused by HIV-I infection is associated with increased risk of
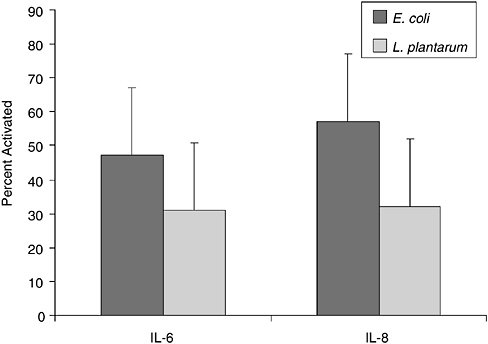
FIGURE 6-3 Intracellular cytokine response of cord blood monocytes from term infants to bacterial antigens. Data are shown as percent of monocytes producing the denoted cytokines after 18 hours of culture.
SOURCE: Cunningham-Rundels (2005).
GI malignancy (Crump et al., 1999). CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the GI tract (Brenchley et al., 2004). The introduction of highly active antiretroviral therapy has resulted in a significant decrease in the incidence of opportunistic enteric pathogens as a consequence of immune recovery. Nonetheless, patients with advanced HIV-1 disease who were recently diagnosed or have a poor response to HAART can still suffer from a wide range of enteric pathogens. Bacterial vaginosis is associated with human immunodeficiency virus (HIV) acquisition, and this condition is associated with reduction of lactobacilli. Interestingly, inhibition of HIV infectivity has been shown with a natural human isolate of lactobacilli engineered to express functional 2-domain CD4 9 (Chang et al., 2003).
HIV-1 may play a direct enteropathogenic role and is implicated in both diarrhea and intestinal dysfunction. HIV-1-associated failure to thrive was a relatively common presentation in congenital HIV infection before the introduction of HAART, yet less severe growth problems still persist (Nachman et al., 2005; Watson and Counts, 2004). We have studied the effect of L. plantarum 299v in
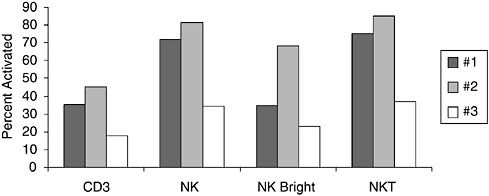
FIGURE 6-4 Differential response of T cells (CD3+) and natural killer (NK) subpopulations defined as NK (CD3–CD56+), NK bright (CD3–CD56+bright), and NKT (CD56+ CD3+) lymphocytes from HIV-positive children to bacterial antigens. Data are shown as percent of activated cells identified by up-regulation of CD69 after 18 hours of culture.
SOURCE: Cunningham-Rundles (2005).
the HIV-1-positive child for impact on growth and on specific systemic immune response following oral supplementation and found a growth benefit in advanced HIV-1 infection (Cunningham-Rundles et al., 2000). The level of immune response in vitro in HIV-1-positive children was essentially independent of percentage of CD4+ T cells unlike response to any other activator where level of response is strongly associated with CD4 T cell level (Cunningham-Rundles and Nesin, 2000). Comparison of T cell and natural killer (NK) cell subset responses to L. plantarum are shown in Figure 6-4. Although intersubject variation is evident, the NK and NKT responses are distinctly increased compared to the CD3+ T cell responses in HIV-1-positive children. These studies support current interest in commensal bacteria as antigen delivery vehicles as well as potential adjuvants in the immunocompromised host.
In summary, commensal bacteria have a modulatory effect on GI immune function that is critical for host interaction with neoantigens and also for the development and maintenance of systemic immune response. Probiotic bacteria have been shown to support gut defense through immune exclusion, immune elimination, and immune regulation in several settings and have potential future use as immune modulators. Further studies addressing critical mechanisms of action, specificity, and long-term consequences are warranted.
REGULATING PRE- AND PROBIOTICS: A U.S. FDA PERSPECTIVE
Julienne Vaillancourt3
Introduction
Probiotics is a term applied to microorganisms that are administered with the intention of providing a health benefit. Probiotic preparations are usually administered orally, and in some cases intravaginally or topically, as they are thought to act locally or regionally rather than systemically. Probiotic products typically contain bacteria that are considered nonpathogenic and commensal in humans, such as lactic-acid producing strains of Lactobacillus and Bifidobacterium. Many probiotic organisms have been traditionally used in food preparation, particularly as fermenting agents in dairy products. With the increased interest in alternative and complementary medicine, there has been a surge in the marketing and availability of these agents as dietary supplements. As dietary supplements, probiotics may be marketed in the United States based on “structure/function” claims or health claims, rather than on treatment claims. The latter would apply to market-approved drugs. However, recently there has been active interest across many medical disciplines in the use of probiotics for a myriad of clinical indications. Such proposed and investigated uses for these products exceed the definition of a dietary supplement and instead meet the definition of a drug per the Food, Drug, and Cosmetic Act of 1938. Probiotics used for clinical indications also meet the definition of a biological product per the Public Health Service Act of 1944 and are regulated overall as such. The Office of Vaccines Research and Review (OVRR) at the Center for Biologics Evaluation and Research (CBER) at the U.S. Food and Drug Administration (FDA) has regulatory authority over the development and marketing of probiotics for clinical treatment indications.
FDA/CBER’s Experience in Regulating Probiotics as Biological Drug Products
As interest has grown in the United States over the last few years in studying the effects of probiotics on specific diseases and conditions, CBER/OVRR has provided much regulatory advice to individuals including practitioners interested in conducting clinical research to evaluate probiotics, recipients of government funding for such research, as well as probiotic manufacturers. These regulatory interactions have focused on a variety of proposed uses for probiotics, most of
|
3 |
Division of Vaccines and Related Products Applications, Office of Vaccines Research and Review, Center for Biologics Evaluation and Research, U.S. Food and Drug Administration. 1401 Rockville Pike, Suite 370N, Rockville, MD 20852. Phone: 301-827-3070; Fax: 301-827-3532, e-mail: Julienne.Vaillancourt@fda.hhs.gov |
which have been gastrointestinal in nature. Intended study populations have varied from premature neonates to older adults, and from healthy individuals who may be at risk for specific diseases to individuals afflicted with particular diseases or conditions. Single strain and multistrain probiotic products have been proposed for investigation. Also, most probiotic products that have been the subject of regulatory interaction with CBER have been intended for oral administration, which is logical given the GI nature of most proposed diseases of study and the intended local effect of these products. However, some products have been intended for other routes of administration, such as intravaginal. Finally, proposed indications have varied with respect to prevention versus treatment, as well as with respect to stand-alone therapy versus use of a probiotic preparation as adjunct therapy to antimicrobial or other therapy. The discussion that follows will reflect CBER/OVRR’s experience and perspective on the regulation and clinical development of these products as biological drug products.4
Prebiotic substances, such as those that are available as food ingredients, are typically regulated by the FDA’s Center for Food Safety and Nutrition (CFSAN) unless a prebiotic substance is formulated in a biological drug product intended to treat or prevent disease. In this case, the prebiotic substance is subject to regulation by CBER as a component or ingredient of a biological drug product.
Nonregulatory Product Terms
It is important to clarify for regulatory purposes that currently probiotic, prebiotic, and live biotherapeutic are not regulatory terms, even though these terms are used to describe particular products that if introduced into interstate commerce within the United States would be regulated by the FDA. Specifically, these terms are not used or defined in any law or regulation that pertains to food and drugs. Nonetheless, a brief description of these terms is provided for the purpose of this discussion.
Probiotic. There is wide and varied use of the term probiotic in the medical literature, lay literature, and product advertising. Frequently it is used to describe an oral preparation containing live, lactic acid-producing bacteria for intended GI benefits. A commonly cited definition from a report of a joint working group of the FAO and the WHO on guidelines for the evaluation of probiotics in food refers to probiotics as live microorganisms, which when administered in adequate amounts confer a health benefit on the host (FAO/WHO, 2002). A more inclusive
definition that considers nonviable forms of probiotics (e.g., dead cells and bacterial cell fragments) has been proposed and defines probiotics as microbial cell preparations or components of microbial cells that have a beneficial effect on the health and well-being of the host (Salminen et al., 1999). The term probiotic has also been proposed to describe microorganisms that have general beneficial effects on the health of animals or humans, whereas the term biotherapeutic agent has been proposed to describe microorganisms having therapeutic effects in humans (Elmer et al., 1999).
Prebiotic. As with the term probiotic, different definitions have been used and proposed for the term prebiotic. A prebiotic may simply be defined as a substance that serves as nutrition or fermentation media for probiotic organisms. The International Scientific Association for Probiotics and Prebiotics (ISAPP) defines a prebiotic as a nondigestible substance that provides a beneficial physiological effect on the host by selectively stimulating the favorable growth or activity of a limited number of indigenous bacteria (Reid et al., 2003). A prebiotic may be administered on its own, usually as an oral preparation, as a food ingredient, or as a component of a product containing probiotic organisms to enhance their growth. Oligosaccharides are commonly used as prebiotics.
Live Biotherapeutic. Currently CBER/OVRR uses the term live biotherapeutic in a working manner to refer to those products under its regulatory jurisdiction that contain whole, live microorganisms (e.g., bacteria, yeast) with an intended therapeutic effect in humans. As noted above in this section, this term is not currently used or defined in any law or regulation pertaining to food and/or drugs. Live biotherapeutic products differ from traditional vaccine products because their dosing regimens more closely resemble those of drugs (i.e., they are typically administered on a daily basis for a given period of time or indefinitely), the postulated effects are frequently local rather than systemic, and the intended therapeutic effect is typically not induction of an adaptive immune response to a pathogenic organism but rather an effect that may be directly attributable to component organisms in the product (e.g., production of antibacterial substances). Probiotics intended for clinical or therapeutic use are categorized as live biotherapeutic products by CBER/OVRR.
Regulatory Definitions
The key to understanding how the FDA regulates probiotics (or any product under its jurisdiction) is summarized by the following statement: Intended use determines how a substance is regulated. If a probiotic product is intended to be used as a drug, it is regulated as a drug. If a probiotic product is intended to be used as a food or dietary supplement, it is regulated as such. The Food, Drug, and Cosmetic Act defines a drug and a dietary supplement, whereas the Public Health
Service Act defines a biological product. A set of corresponding regulations exists for each of these distinct areas, and these regulations are found in Part 21 of the Code of Federal Regulations (CFR), which is updated annually. For the purpose of this discussion, simplified regulatory definitions for these terms are provided below. A definition for the acronym GRAS (generally recognizable as safe), which CBER/OVRR has encountered in a number of regulatory submissions concerning probiotic products, is also provided. Of note, GRAS has been frequently misused as a regulatory term, concerning probiotic products that are considered biological drug products, because use of the term GRAS is limited to food regulation.
-
Drug. An article intended for use in the diagnosis, cure, mitigation, treatment, or prevention of disease.5
-
Dietary Supplement. A product taken by mouth that contains a “dietary ingredient” intended to supplement the diet.6
-
Biological product. Includes any product containing microorganisms applicable to the prevention, treatment, or cure of a disease or condition of human beings,7 and to which the Federal Food, Drug and Cosmetic Act applies.8
-
GRAS (generally recognizable as safe). A food ingredient classification that distinguishes a substance from a food additive on the basis of common knowledge about safety for its intended use.9
|
5 |
FD&C Act (Federal Food, Drug, and Cosmetic Act), June 25, 1938, as amended through December 31, 2004, codified at Title 21 United States Code (U.S.C.) available online at http://www.access.gpo.gov/uscode/title21/chapter9_.html. |
|
6 |
The current definition of a dietary supplement is a result of the Dietary Supplement Health Education Act of 1994 (DSHEA), P.L. 103-417, 21 U.S.C. 321 which amended the FD&C Act to provide standards for dietary supplements. The actual definition as written in the FD&C Act is more detailed than presented here and discusses claims and labeling. The full text of the DSHEA is available online at http://www.fda.gov/opacom/laws/dshea.html). |
|
7 |
The actual definition of a biological product per the Public Health Service Act, uses the term virus rather than microorganism. According to 21 CFR 600.3(h) a virus is interpreted to be a product containing the minute living cause of an infectious disease and includes, for example, bacteria and fungi. As use of the term virus might cause confusion, the term microorganism is used instead in an attempt to simplify the definition. According to 21 CFR 600.3(h)(5)(i) a product is considered “analogous to a virus,” if it is prepared from or with a virus or agent actually or potentially infectious, without regard to the degree of virulence or toxicogenicity of the specific strain used. |
|
8 |
PHS Act (Public Health Service Act), July 1, 1944, Chap. 373, Title III, Sec. 351, 58 Stat. 702, currently codified at 42 U.S.C., Sec. 262 indicates that a biological product is a drug by definition. Therefore, biological products are subject to both drug and biological product regulations. See http://www.fda.gov/opacom/laws/phsvcact/sec262.htm); 21 CFR (Code of Federal Regulations, Title 21) Parts 1–1299. Washington, DC, Office of the Federal Register, National Archives and Records Administration, 2004. |
|
9 |
Use of the term GRAS is appropriate in the context of food regulation but not in the context of drug and biologic regulation. GRAS pertains to the use of a food substance, rather than the substance itself. Information about GRAS, 21 CFR 170.3 and 170.30, is available from FDA/CFSAN at http://www.cfsan.fda.gov/~dms/grasguid.html. |
U.S. FDA Regulation of Probiotics: Dietary Supplements Versus Biological Products
Probiotic products for ingestion may be lawfully marketed in the United States as dietary supplements under the Dietary Supplement Health and Education Act of 1994 (DSHEA) based on limited claims such as those that pertain to affecting the structure and function of the human body or general well-being,10 but not based on drug or disease claims. Probiotic products intended for intravaginal or topical use are not dietary supplements by definition, because they are not intended for ingestion. If a probiotic product is intended to be used in a manner that meets the definition of a drug per the Food, Drug, and Cosmetic Act as indicated above, it cannot be introduced into interstate commerce unless it is either approved for such use by the U.S. FDA or an investigational new drug application (IND) is in effect with the U.S. FDA for that specific use. Most proposed uses of probiotics for clinical investigation meet the definition of a drug from a regulatory perspective.
The OVRR at the CBER has regulatory jurisdiction over most probiotic products intended for use as drugs. Because probiotic products intended for use as drugs are regulated as biological products under section 351 of the Public Health Service Act (PHS Act), they are subject to the biologics regulations (21 CFR 600).11 For example, a probiotic for an intended clinical use must be marketed under an approved biologics license application (BLA) (21 CFR 601)12 unless the product is excluded from the definition of a new drug under section 201(p) of the Food, Drug, and Cosmetic Act. Probiotic products intended for use as drugs are also subject to pertinent drug regulations. For example, when a probiotic product is proposed for evaluation in a clinical study, it may be viewed as an unapproved biological product and an investigational drug product, and thus would be subject to the regulations for an IND (21 CFR 312).13 FDA reviews INDs to assure the safety and rights of patients in all clinical studies and to assure that studies to evaluate both safety and effectiveness are adequately designed to do so [21 CFR 312.22(a)].14
Any clinical investigator or probiotic manufacturer interested in evaluating a probiotic product for clinical use is encouraged to request a pre-IND meeting
with CBER prior to submitting an IND.15 A pre-IND meeting is an opportunity for a potential IND sponsor to obtain useful regulatory advice from FDA that, if heeded, might prevent regulatory delays (e.g., clinical holds) in the conduct of a proposed clinical trial (Miller and Ross, 2005).
In light of the current trend whereby practitioners recommend probiotics to their patients as a therapeutic modality for particular diseases or clinical indications, it is important to keep in mind the following:
-
Biological products require premarket review and approval by the FDA. Dietary supplements do not.
-
The safety, purity, and potency, as well as efficacy, of a biological product must be demonstrated for approval. Dietary supplements need not demonstrate any of these to be marketed.
-
A probiotic product that is marketed or promoted as a treatment, prevention, or cure for a specific disease or condition without an approved indication for such is considered an unapproved and, thus, illegal drug.
-
A probiotic manufacturer who intends to market a probiotic product based on a drug claim must seek approval for that claim.
The Biological Drug Development Pathway for Live Biotherapeutics
Data to support approval of a biological product are provided in a BLA to CBER for review. Clinical data for inclusion in a BLA are generated during the investigational stage of drug development. Adequate preclinical and chemistry, manufacturing, and controls (CMC) data are needed to support the initial clinical evaluation of a live biotherapeutic in human subjects. Figure 6-5 presents the typical stages of product development and regulatory review for live biotherapeutic products including probiotics for therapeutic uses. As with traditional pharmaceutical development, a live biotherapeutic product is expected to be evaluated in phase 1, 2, and 3 studies, which progressively build upon each other to provide supportive data for a license application. Safety is evaluated in all phases of investigational biological drug development. Other parameters such as colonization, which has yet to be consistently defined, may be important in live biotherapeutic development and recommended for evaluation during all three phases as well. Typically, dose selection for phase 3 efficacy studies—and often the proposed dose for marketing—are based on the results of phase 2 dose ranging studies. Pivotal phase 3 studies should be adequately designed to provide safety and efficacy data to support a licensed indication.
|
15 |
The following FDA/CBER website provides many informational resources concerning how to submit an IND and how to conduct a clinical trial under IND. See http://www.fda.gov/cber/ind/ind.htm. Also, CBER’s Office of Communication, Training, and Manufacturer’s Assistance (OCTMA) may be contacted directly at (301) 827-1800 for assistance in the process. |
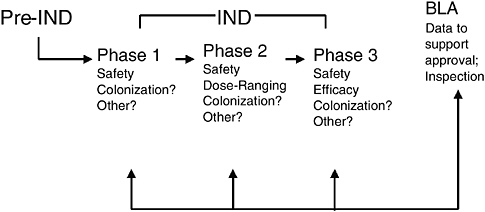
FIGURE 6-5 Stages of premarket review and regulation for live biotherapeutics.
SOURCE: Vaillancourt (2005).
Issues for Consideration in the Development of Live Biotherapeutic Products
The CBER/OVRR has identified the following eight issues that merit consideration in the development of live biotherapeutic products:
-
The term probiotic is inconsistently used and lacks a standard definition.
-
The regulatory food ingredient term GRAS is inappropriately used to describe component microorganisms of live biotherapeutic products.
-
Submitted INDs frequently lack adequate chemistry, manufacturing, and controls (CMC) data that are required to support the clinical evaluation of a live biotherapeutic product in human subjects.16 In this regard, probiotic manufacturers may not be familiar with biological product manufacturing requirements, which greatly exceed those for foods or dietary supplements.
-
There is a need for education among investigators and sponsors concerning how to prepare INDs and master files and submit them to CBER, and drug and biologics regulations in general.
|
16 |
When the IND sponsor is not the manufacturer of the product, the IND sponsor must rely on obtaining CMC data from the manufacturer for submission in the IND, or alternatively, the manufacturer may submit such proprietary data directly to CBER for review in a master file (21 CFR 314.420) to support a proposed clinical study. A guideline on drug master files is available from FDA’s Center for Drug Evaluation and Research at http://www.fda.gov/cder/guidance/dmf.htm. Prior to submitting a master file, a manufacturer is strongly advised to contact the Division of Vaccines and Related Products Applications at CBER/OVRR at (301) 827-3070. |
-
Live biotherapeutic product development for any given clinical indication should focus on demonstrating a measurable treatment effect that reflects a meaningful clinical benefit.
-
Colonization should be explored as a valid function of the intended treatment effect and/or a postulated mechanism of action. In this regard, discussion is necessary on how it should be defined and how and when it should be evaluated in the clinical development of live biotherapeutic products.
-
There is a need for scientifically sound approaches to defining and evaluating the potency of live biotherapeutic products.17
-
With regard to the safety of live biotherapeutic products, there is a need for effective evaluation and management of the potential pathogenicity of product strains, particularly in targeted patient populations. Some factors to consider include virulence, host translocation, environmental persistence and the concomitant risk of infection to others, alterations in normal flora that may increase disease risk or susceptibility, and gene transfer in the therapeutic niche.
If these issues can be addressed, perhaps progress can be made in the overall development and regulatory evaluation of live biotherapeutic products, including probiotics for therapeutic uses, as a promising new class of biological drug products. However, efforts in addressing these issues should be collaborative and include funding institutions, manufacturers of live biotherapeutic products, clinical investigators, and regulatory agencies such as the U.S. FDA.
Conclusions
Although many probiotic products are marketed and available in the United States as dietary supplements, interest in the therapeutic use of probiotic products for a variety of clinical diseases and conditions is growing. Despite extensive retail and research experience with probiotics, there is a need for medical and scientific validation of their value in disease management. This may well require rigorous clinical trials and regulatory evaluation (Shanahan, 2003; Tannock, 2003). The FDA’s CBER has regulatory jurisdiction over probiotic products intended for therapeutic use and refers to them as live biotherapeutic products. Specifically, CBER’s OVRR has gained tremendous experience in this area in recent years. In general, the development of a live biotherapeutic product as a biological drug product should proceed along the established product and clinical development pathway for drugs and biologics, with safety being continuously
evaluated. Many resources are available from FDA’s CBER for guidance on U.S. regulatory standards for biological products and clinical study design that can be applied to the development of live biotherapeutic products. Finally, in its recent experience in regulating these products, CBER’s OVRR has encountered numerous challenges and issues that impact valid evaluation of these products for therapeutic uses. Collaborative efforts in addressing these challenges and issues appear to be necessary.
FROM RESEARCH IN MICROBIOLOGY TO GUIDELINES
Lorenzo Morelli and Elena Bessi18
New Aspects of the War Metaphor: Good Bacteria as Potential Allies
From the point of view of a microbial ecologist the war metaphor describing the fight between humans and bacterial pathogens could still be useful if widened to include not only the human body and bacterial pathogens, but also potential bacterial allies already inhabiting, or parachuted into, the battleground to ensure support to the body.
A dramatic increase worldwide in research efforts (hundreds of papers are published every year with the key words probiotics or prebiotics), as well as a booming market of drugs, dietary supplements, and food claiming “contains good bacteria” or “good for feeding your good bacteria,” urged the FAO and the WHO to hold two expert consultations to both review the state of the field and prepare guidelines for assessing probiotics (FAO/WHO, 2001; FAO/WHO, 2002). This overview will try to establish a link between cutting-edge research and regulatory efforts, with special attention paid to FAO/WHO documents19 that are in progress.
In the following sections, some points of this potential new war strategy will be addressed by focusing on items that have been considered by the joint FAO/WHO consultations held in Argentina during 2001 and in Canada during 2002 (FAO/WHO, 2001; FAO/WHO, 2002). In addition to these documents, it could be worthwhile to recall the excellent documents prepared by the European Society for Paediatric Gastroenterology Hepatology and Nutrition (ESPGHAN) (Agostoni et al., 2004a,b), the French Food Safety Authority (AFSSA, 2005), and the Italian Ministry of Health (Ministero della Salute, 2006). These documents altogether provide a state-of-the-art assessment of this field as well as some guidelines for assessing safety and efficacy of pre- and probiotics.
|
18 |
L Morelli and E Bessi are from the Istituto di Microbiologia, UCSC; Via Emilia Parmense 84, 29100, Piacenza, Italy. Phone: +39-523-599248 Fax: +39-523-599246. E-mail: lorenzo.morelli@unicatt.it E Bessi is also with AAT, Srl, a spin-off company of UCSC; Via Emilia Parmense 84, 29100, Piacenza, Italy. E-mail: aat@libero.it. |
|
19 |
All paragraphs marked with this symbol are taken from documents (FAO/WHO, 2001) or (FAO/WHO, 2002). |
Why FAO/WHO Issued Guidelines on Probiotics
A joint FAO/WHO expert consultation on health and nutritional properties of powdered milk with live lactic acid bacteria was held in Córdoba, Argentina, October 1–4, 2001. The consultation focused on the evaluation of the scientific evidence available on the properties, functionality, benefits, safety, and nutritional features of probiotic foods. Following are the reasons to convene such a consultation:
The beneficial effects of food with added live microbes (probiotics) on human health, and in particular of milk products for children and other high-risk populations, are being increasingly promoted by health professionals. It has been reported that these probiotics can play an important role in immunological, digestive, and respiratory functions and could have a significant effect in alleviating infectious disease in children. As there is no current international consensus on the methodology to assess the efficacy and safety of these products it was considered necessary to convene an Expert Consultation to evaluate and suggest general guidelines for such assessments.
Bacterial Armies of the Gut
In order to assess the properties of probiotics, the joint FAO/WHO consultation suggested that (FAO/WHO, 2001):
Probiotic microorganisms should not only be capable of surviving passage through the digestive tract, but also have the capability to proliferate in the gut. This means they must be resistant to gastric juices and be able to grow in the presence of bile under conditions in the intestines, or be consumed in a food vehicle that allows them to survive passage through the stomach and exposure to bile. They are gram-positive bacteria and are included primarily in two genera, Lactobacillus and Bifidobacterium (Holzapfel et al., 1998; Klein et al., 1998).
A microbial ecologist has to add that probiotic microorganisms, to be successful, have to take their place among the bacterial population already inhabiting the gut. To evaluate the potential of good bacterium to proliferate into a gut already heavily inhabited is not an easy task. Technical difficulties abound in analysing the gut microbiota as a whole. If we do not have a clear picture of what we are going to impact, then it is difficult to prepare guidelines or to regulate.
Composition of the vast and complex ecosystem formed by bacteria inhabiting the various sites of the intestine is only beginning to be understood due to the availability of molecular tools that allow culture independent assessment. However, the complexity of this research effort must be considered. It is generally estimated that each of us has about 1011 viable bacteria per gram of large bowel content. This multitude comprises at least 400–500 different species. The physiological, and also the clinical, potential of this bacterial organ is underlined when we consider that the total number of genes contained in the microbiota is about
50–100 times greater than in the human genome. A further level of complexity is the spatial and temporal diversity of the microbiota. The bacterial distribution varies greatly at different levels of the GI tract, ranging from less than 103 colony-forming units/mL (CFU/mL) in the stomach, where the number of ingested bacteria is dramatically reduced by the hostile gastric environment, to 1011 CFU/mL within the colon, where anaerobes outnumber aerobes by a ratio of 1000:1. Environmental conditions of the colon seem to favour the growth of selected bacterial groups; however, a clear picture of this complex process, in which ingested bacteria are greatly reduced by extremely hostile conditions and then survivors are hosted in a kind of bacterial luxury resort is not yet available. This could be a first suggestion for developing a strategy of alliance between humans and “good enteric bugs” to unravel the mechanisms used by the human gut for sorting out some bacterial groups to be favoured for reproduction in this environment.
Selection of Probiotic Strains for Human Use
Probiotics must be able to exert their benefits on the host through growth and/or activity in the human body (Collins et al., 1998; Morelli, 2000). However, it is the specificity of the action, not the source of the microorganism, that is important. Indeed, it is very difficult to confirm the source of a microorganism. Infants are born with none of these bacteria in the intestine, and the origin of the intestinal microbiota has not been fully elucidated. It is the ability to remain viable at the target site and to be effective that should be verified for each potentially probiotic strain (FAO/WHO, 2001).
This statement is in full opposition with “conventional wisdom,” and even with many recent scientific reviews on the selection of probiotic bacteria stating that probiotic bacteria must be selected among “bacteria isolated from human beings.” Even acknowledging the difficulty with determining the natural source of a particular strain, species including Saccharomyces boulardii and Bifidobacterium animalis, which are not considered to have humans as their natural habitat, are documented as very effective probiotics (see also comments on the efficacy of Saccharomyces boulardii, in the section “Good Bugs: Data with Consensus”). The assertion that probiotics must be of human origin should be laid to rest.
There is a general agreement that the term dominant bacteria is used to indicate species that are present at a level not less than one percent of the total fecal microbiota. As such, bacteria at a minimum concentration of 108 could be considered the likely players in gut health (Ducluzeau, 1988). To what extent exogenous bacteria can influence the microbe-associated characteristics is key to the probiotic concept. Some authors suggest that each person has his or her own rather stable profile of dominant (therefore more than 108 CFU/g) species. These observations still need to be confirmed by studies involving larger numbers of
individuals. This is not a trivial task as the assessment of the ecological composition of the gut microbiota will depend on (1) the section of the GI tract sampled, (2) the “layer” of the GI pipeline, (3) age of the considered individual, (4) diet, and (5) the analytical methodology used.
Disproportionately, our knowledge of microbiota composition of human beings has been determined investigating the stool microbiota by means of microbiological plating techniques. However, this approach is laborious, time consuming, and limited in scope as a majority of the bacterial species present in feces are not culturable using standard microbiologic techniques (Blaut et al., 2002; Langendijk et al., 1995; Suau et al., 1999) or could become unviable during sample handling. Among those microorganisms that are culturable, species identification by traditional biochemical identification methods is often difficult. Assessment of gut microbiota using genetic techniques, applicable also to unviable bacterial cells, is a relatively new approach able to overcome at least some of these problems.
Molecular tools based on 16S rDNA sequence similarities such as fluorescent in situ hybridization (FISH), denaturing gradient gel electrophoresis (DGGE), quantitative dot blot hybridization, restriction fragment length polymorphism, and large scale 16S rDNA sequencing have helped to overcome limitations of conventional microbiological plating methods in studying the fecal microbiota composition (Suau, 2003). In addition, DNA microchips that allow for more efficient identification of bacterial species present in complex communities are currently under development in various laboratories. However, bias introduced through PCR or subcloning might affect such analyses (McCartney, 2002); the extent of such potential distortions has not been thoroughly investigated.
Both FAO/WHO documents strongly recommend the use of molecular biology techniques. In the 2001 document, section 11, you find “Potential probiotic strains must be identified by methods including internationally accepted molecular techniques.” This should be taken as a strong recommendation that assessment of the identity of bacteria potentially used to promote health must be done by means of the best techniques available, which may be a combination of classical and DNA-based techniques. But new identification techniques are providing a novel assessment of the composition of the intestinal microbiota, suggesting that bacterial populations of the gut are not a single body. At the moment, these observations have not evolved into a new regulatory framework.
The Dominant Bacterial Population
Studies based on plating and cultivating reveal a fecal microbiota dominated by Bacteroides, Eubacterium, Ruminococcus, Clostridium, and Bifidobacterium (Finegold and Rolfe, 1983; Moore and Holdeman, 1974), but recent investiga-
tions based on genetic tools suggest that 70–80 percent of fecal bacteria are unculturable or extremely fastidious in their cultivation requirements (Blaut et al., 2002; Langendijk et al., 1995; Suau et al., 1999; Suau, 2003). It is possible that a new understanding of the bacterial composition of the intestinal microbiota will emerge given time. For example, in past investigations, Fusobacterium prausnitzii was reported either as one of the most frequent and numerous species or was seldom retrieved. A specific rRNA-targeted oligonucleotide probe (Suau et al., 2001) showed that F. prausnitzii and phylogenetically related species represent a dominant group within the human fecal microbiota, ranging from 5 percent to 16 percent of the total microbiota.
Anaerobes are the dominant component of the stool microbiota and very possibly of the colon. These groups of bacteria seem to be quite stable over time in healthy adults allowing for a bacterial species profile typical of each individual. Differences among individuals seem to be at the species, but not at the genus, level. Compositional changes of the dominant microbiota are rarely reflected by changes in metabolic activities generally assigned to the microbiota. However, the particular species responsible for specific metabolic activities is largely unknown.
Another puzzling observation is that genetic analysis will probably move some bacterial groups, now allotted among the dominant population on the basis of cultivation techniques, to a lower ranking in the list of the bacterial armies of the gut. They may move from the threshold between the large, “heavy infantry,” and the highly specialized, parachuted corps. One example could be bifidobacteria, up to now considered a dominant population, but perhaps simply because they are less common among anaerobes.
The Subdominant Population
If human beings are looking for allies in the war against pathogens, they should not forget the so-called subdominant population bacteria that are detected at levels below 108 per gram of faeces. Among these groups we can encounter the most studied and used “healthy bacteria,” the lactic acid bacteria, mainly belonging to the genus Lactobacillus. This bacterial population is always a subdominant one, but it is credited with an impressive number of positive actions (FAO/WHO, 2001), but meta-analysis has revealed a puzzling similarity among the efficacy of products containing bacteria of different origin and species. (Further details will be provided in the “Probiotic Activities with Consensus” section). As microbiologists, there are opportunities for new selection criteria and more potent allies to be used as biotherapeutics pharmaceuticals. If it were possible to identify mechanisms used by the human gut to sort out some ingested bacteria, favour their reproduction, and adopt them as “indigenous” we could have in our hands better companions in our fight against pathogens (Vinderola et al., 2004).
Mucosa-Associated Bacteria
If we are engaged in combat, the location of potential allies is another relevant aspect. The subdominant bacterial population may play a specific role by adhering to mucosa. Few data are available, but genetic analysis and biopsy samples clearly suggest that mucosa-associated bacteria differ from those recovered from feces. Even more interesting is that, for each individual, the same bacterial strains are uniformly distributed along the colon (Morelli, 2005a; Nelsen et al., 2003; Zoetendal et al., 2002).
Zoetendal et al. (2002) investigated the distribution of mucosa-associated bacteria in the ascending, transverse, and descending colon of 10 individuals and found that the predominant bacterial community was host specific and uniformly distributed along the colon. Lactobacillus community was dominated by a single species, Lactobacillus gasseri, in all sampling sites. In 7 out of the 10 subjects, no variation in the Lactobacillus population was detected, whereas minor differences among sampling sites were observed in the other three individuals.
Nielsen et al. (2003) investigated 10 volunteers for the presence of mucosa-associated lactobacilli and bifidobacteria. They confirmed that mucosa-associated bacterial communities are clearly different in composition from those recovered from fecal samples of the same subjects. High similarity between bacterial communities from different locations of the colon was also confirmed.
In our laboratory, we have focused our attention on the Lactobacillus population adhering to biopsies obtained from six individuals from the same three sites of the large bowel and the ileum. In all cases, Lactobacillus fermentum was isolated. These data suggest that the bacterial profile of mucosa-associated bacteria is much less complex than the profile obtained by fecal samples. In addition, we have recently characterised the Lactobacillus biota of seven adults; biopsies were obtained from the terminal ileum and from the ascending, descending, and transverse colon. From all subjects stool samples were also obtained. Results are summarized in Table 6-3.
TABLE 6-3 Lactobacillus Species Recovered and Genetically Identified from Biopsies of Seven Adults
|
Subjects |
Lactobacillus species |
|
A |
L. rhamnosus; all sites |
|
B |
L. rhamnosus; L. paracasei; L. fermentum |
|
C |
L. rhamnosus; L. paracasei; L. fermentum |
|
D |
L. rhamnosus; L. fermentum; L. gasseri; L. paracasei; L. salivarius |
|
E |
L. rhamnosus; L. fermentum; L. gasseri; |
|
F |
L. fermentum; a new species of the genus Lactobacillus |
|
G |
L. fermentum all sites |
|
SOURCE: Morelli, 2005. |
|
Puzzling is that the species L. fermentum has been found in six out the seven subjects studied but only in biopsy samples and never in stools. This species of Lactobacillus species has been recovered from human stools, but it is not one of the most commonly isolated species.
Caution is to be exerted, as available data are still scarce (limited number of individuals sampled), but what is becoming evident is that strains stably entrapped into mucus or adhering to deeper layer intestinal tissues are different from those living as planktonic cells sometimes associated with food particles and found in stool.
It may be relevant to have a more clear picture of the biota established near the intestinal tissues, as these bacteria are probably in a better position to exert positive action for the human hosts such as the interaction with the gut associated lymphoid tissue (GALT), the competition for receptor sites of pathogens, or as players in cross-talk with the genes of enterocytes.
These bacteria could be considered the real potential allies in our war against pathogens. It could be argued that their total number is much lower than those found in the stool or as planktonic, free-flowing cells in the colon, but they are in better position to establish interactions with GALT and intestinal tissues or to block pathogens in their struggle for adhesion.
An interesting observation is that the large majority of scientific evidence reviewed by the FAO/WHO consultation was obtained by papers dealing with probiotics isolated from human stools. Furthermore, according to the latest results obtained by molecular techniques, we have to assume that all—or nearly all—of them belong to the subdominant population. This could open a world of opportunities for new research and new biotherapeutic bacteria.
Good Bugs Meet Us and We Become Friends
To end this section devoted to the considerations on microbial ecology of the gut as reviewed in the FAO/WHO documents, it is interesting to note that becoming friends may be easy but remaining friends for a lifetime is definitely a hard task and requires specific feelings and attitudes. Genetic techniques allow us to learn a little more about the human host-bacteria relationship. For example, consider the species distribution of lactobacilli among a range of higher animals including, quite surprisingly, fish and insects (Table 6-4).
Recently we identified an abundant Lactobacillus microbiota in honeybees, and, reinforcing the concept of host specificity, the four Lactobacillus species identified were all new species never before isolated (Bessi and Morelli, 2005). This data strongly suggests that a specific link between some species and specific hosts exists. However, more needs to be known about the nature of this host specificity. If unique species with a true affinity for humans are discovered, this could lead to a new generation of probiotics which, in turn, might necessitate new
TABLE 6-4 Host Specificity of the Lactobacillus Species
|
Animal Source |
Genetic Taxonomy |
References |
|
Pigs |
L. amylovorus |
Axelsson and Lingren, 1987 |
|
Pigs |
L. amylovorus |
Pryde et al., 1999 |
|
Calves |
L. johnsonii |
Sarra et al., 1980 |
|
Poultry |
L. crispatus |
Morelli et al., unpublished results; Sarra et al., 1986 |
|
Poultry |
L. johnsonii |
|
|
Humans |
L. gasseri, L. rhamnosus, |
Dunne et al., 1999; Morelli et al., 1998 |
|
|
L. paracasei, L. reuteri |
Song et al., 1999, 2000; Tannock et al., 2000 |
|
Honeybees |
L. plantarum, L. cellobiosum |
Bessi et al., 2005 submitted; Kvasnicov et al., 1971 |
|
Fishes |
L. fermentum |
Kvasnicov et al., 1977; Rawls et al., 2004 |
|
SOURCE: Morelli, 2005. |
||
safety considerations. For example, probiotic bacteria currently on the market persist for weeks, at a maximum, in the gut of fed volunteers. Probiotics able to persist long term in the human gut may need to be reassessed for safety (Fujiwara et al., 2001).
To summarize:
-
The gut microbiota compose a complex ecosystem with an enormous potential for influencing the whole health status of human beings.
-
The composition of this ecosystem requires further investigation, and available genetic techniques are promising, but further improvements are needed.
-
The analysis of fecal microbiota alone is not sufficient for comprehension of the complex situation occurring in this body site.
-
Stool bacteria do not reflect the composition of mucosa-associated bacteria, but the latter are probably the most interesting allies in the war against pathogens.
-
It is of interest to distinguish between dominant and subdominant bacteria among the total gut microbiota.
-
Guidelines available at the moment consider probiotic products based on a relatively “old” concept. New products and new guidelines are emerging.
Looking for Allies in the Gut Microbiota: From History to Guidelines
E. Metchnikoff, the Nobel Prize winner, is credited with being the first to suggest the value of ingestion of beneficial bacteria. He asserted, “the dependence of the intestinal microbes on the food makes it possible to adopt measures to modify the flora in our bodies and to replace the harmful microbes by useful microbes” (Metchnikoff, 1907). He put into practice his observations by commercializing fermented milk, as well as a pharmaceutical preparation containing the same Lactobacillus strain selected for the food product. This little piece of
history is useful to point out that good microorganisms have been used either as healthy food ingredients or as therapeutic products since the very beginning of the probiotic story. After one century of research and development, this duality is creating not only opportunities but also problems.
Metchnikoff’s observations were useful to the dairy food industry, and several fermented dairy products were marketed during the last century, claiming to favor a good microbial balance in the gut. The rationale, according to the Metchnikoff concept, was clearly an ecological one—to modify the ratio among some components of intestinal microbiota. For example, Metchnikoff favoured lowering the number of putrefactive bacteria by replacing them with acid-producing ones in order to protect the human body from noxious substances. Unfortunately, only carefully selected strains, and not those generally contained in yogurt, are able to colonize the gut. It is now established that some yogurt starter cultures can survive—albeit in low quantities—to the intestinal transit, but they are unable to actively reproduce themselves in the gut. The evolution from Metchnikoff’s work to probiotic is reflected in changes that, over time, occurred in the meaning of the same word.
At first probiotics was used to name “substances produced by microorganisms which promoted the growth of other microorganisms” (Lilly and Stillwell, 1965). A few years later, Parker redefined the word as “organisms and substances that contribute to intestinal microbial balance” (Parker, 1974). Then Fuller specified that probiotics are “live microbial feed supplements that beneficially affect the host animal by improving its intestinal balance” (1989). In more recent years, the meaning of this word has been refined several times and today a widely accepted definition of probiotics is “live microorganisms, which when consumed in adequate amounts, confer a health effect on the host” (Guarner and Schaafsma, 1998).
Some notes on different wording of the various definitions could be helpful:
-
In the first definition the subject of the definition was substances then viable bacteria appear, and nowadays live microorganisms remained alone.
-
The probiotic activity was originally quite similar to a feed supplement for bacteria, then it turned into an ecological action towards the intestinal microbiota. Nowadays, there is increasing evidence that the presence of specific bacterial strains in the human gut could have a favourable action per se, without any detectable change in the overall composition of the gut microbiota.
-
At the end, today the positive action is no more related to intestinal ecology but is simply a “health effect.”
-
In the more recent definitions, a dose effect is included.
If the above are definitions derived from the scientific community, here is the regulatory counterpart:
-
Expert consultation held jointly by the FAO and the WHO redefined the term probiotics as “live microorganisms which when consumed in adequate amounts as part of food (water is included as a food) confer a health benefit on the host.”
-
The consultation agreed that the scope of the meeting would include probiotics and prebiotics in food and exclude reference to the term biotherapeutic agents and beneficial microorganisms not used in food.
Probiotic Activities with Consensus
The expert consultation reviewed
-
disorders associated with the GI tract,
-
prevention of diarrhea caused by certain pathogenic bacteria and viruses,
-
Helicobacter pylori infection and complications,
-
inflammatory diseases and bowel syndromes,
-
cancer,
-
constipation,
-
mucosal immunity,
-
allergy,
-
cardiovascular disease,
-
urogenital tract disorders,
-
bacterial vaginosis,
-
yeast vaginitis,
-
urinary tract infections, and
-
use of probiotics in otherwise healthy people.
What seems relevant to a microbiologist in this section of the presentation is that the above evidence of probiotic activities have been obtained with bacterial probiotic populations detected in fecal samples at a level less than 1/100 of the total bacterial population.
The same levels have been detected when probiotics were found active in
-
significant reduction in atopic dermatitis in allergic children,
-
treatment of pouch infections,
-
gut-associated immune response modulation,
-
lactose intolerance reduction,
-
gastric inflammation in H. pylori disease reduction,
-
urogenital infections prevention and treatment,
-
intestinal transit modulation,
-
IBD and Crohn’s diseases symptom reduction,
-
an active role in arthritis prevention and treatment, and
-
an active role in autoimmune diseases treatment and incidence reduction.
All the above observations have led the FAO and the WHO to formulate these optimistic but prudent statements:
-
The experts agreed that adequate scientific evidence exists to indicate that there is potential for the derivation of health benefits from consuming food containing probiotics. However, it was felt that additional research data are needed to confirm a number of these health benefits in humans, applying a systematic approach and following the guidelines for the assessment of probiotics suggested in this report.
-
There is good evidence that specific strains of probiotics are safe for human use and able to confer some health benefits on the host, but such benefits cannot be extrapolated to other strains without experimentation.
If we look to the scientific literature produced after the two FAO/WHO documents were released, we can note that the attention is now focused on the bacteria actually present in the dominant population. Commensal dominant microbiota coevolved with their hosts and express a number of components able to activate innate and adaptive immunity (Vaarala, 2003); the mucosal immune system has developed specialized regulatory, anti-inflammatory mechanisms for eliminating or tolerating nondangerous food, antigens, and commensal microorganisms (Tlaskavola-Hogenova et al., 2004). The important role of commensal bacteria in mucosal immunity development was first demonstrated in a germ-free animal model; colonization of germ-free zebrafish with individual members of its microbiota revealed the bacterial species specificity of selected host responses. Rawls et al. 2004 studies revealed 212 genes regulated by the microbiota, and 59 responses that are conserved in the mouse intestine, including those involved in stimulation of epithelial proliferation, promotion of nutrient metabolism, and innate immune responses (2004).
These studies are elucidating the role of bacteria that form the vast majority of the gut microbiota. These studies are strategically different from all the scientific literature produced on subdominant probiotic bacteria. It could be speculated that when these studies are turned into applications, a new regulatory framework will be necessary.
Good Bugs: Data with Consensus
To reinforce the opinion that currently available guidelines have been generated by an initial, but still evolving, generation of probiotics, it is worthwhile to look at some more consensus analysis. At least five meta-analyses (Cremonini et al., 2002; D’Souza et al., 2002; Elmer et al., 1996; Szajewska and Mrukowicz, 2001; Van Niel et al., 2002) showed that some selected bacteria could be active in reducing the severity of antibiotic-associated diarrhea. Diarrhea associated with antibiotics is presumed to result from the antibiotics disrupting the normal biota
in the gut of a healthy person. Such disruptions cause dysfunction of the gut’s ecosystem, and they may allow pathogenic bacteria to colonize the gut and gain access to the mucosa. Whether probiotic supplements stop this process by reducing the disruption or by acting as substitutes for the healthy microbiota is unclear. It is also unclear if the mechanism of action is competition with pathogens for the nutrients, or adhesion sites, or pathogen inhibition by means of antimicrobial production, or something else. If we consider all meta-analyses supporting the efficacy of probiotics when applied to antibiotic-associated diarrhea, it is possible to point out some further observations.
All meta-analyses suggest that probiotics can be used to prevent antibiotic-associated diarrhea, while statistical comparison suggests that S. boulardii and lactobacilli have a similar potential to be used in this situation. On the contrary, the efficacy of probiotics in treating antibiotic-associated diarrhea remains to be proven. All meta-analyses include clinical trials in which a range of totally different antibiotics have been used, but it has been shown that the positive action of probiotics in preventing the onset of diarrhea is not influenced by the drug(s) used. Moreover, it is also interesting to note that the trial scored negative, in terms of odds ratio, in all meta-analyses performed with a daily dose of S. boulardii reduced to one-fifth of the recommended dosage. This could be a positive observation stressing the importance of the dosage. One recent study supporting this hypothesis is a controlled trial of three probiotic regimens (Lactobacillus GG, Saccharomyces boulardii, or a mixture of Lactobacillus acidophilus and Bifidobacterium) to prevent diarrhea associated with anti-Helicobacter pylori antibiotic treatments. Authors showed a 5 percent incidence of diarrhea with all probiotic used, compared to 30 percent with placebo. It is possible to speculate that all the above observations could be explained by some (still unknown) nonspecific mechanisms, possibly based on general ecological considerations more than on specific, strain-related activity.
This observation could also be reinforced by data presented on the efficacy of probiotics in prevention of recurrent Clostridium difficile disease. In this case the efficacy seems to be related to only some specific probiotic preparations; for at least one of them, there are also some suggestions about the mechanisms active against C. difficile. Here, we are probably crossing the boundaries between probiotics and biotherapeutics.
FAO/WHO constructed a decision tree (Figure 6-6) to obtain data with consensus for new probiotic products (FAO/WHO, 2002). A detailed characterization of the strain is required and at least one double blind, randomised efficacy trial is mandatory to use the term probiotic for a food product containing viable bacterial cells endowed with some specific properties beneficial for the consumers.
FIGURE 6-6 Guidelines for the evaluation of probiotics for food use.
SOURCE: FAO/WHO (2002).
Are These Allies Safe for Us?
When dealing with war, you need to be sure that your allies are true allies and not imposing undue risk. As far as the lactobacilli and bifidobacteria are concerned, there is a general consensus that they are “safe” but several new items have recently raised some worries.
-
There is concern over the use in foods of probiotic bacteria that contain specific drug resistance genes.
-
The consultation recognized the need for the development of standardized assays for the determination of drug insensitivity or resistance profiles in lactobacilli and bifidobacteria.
-
Due to the relevance of this problem, further research is suggested for antibiotic resistance of lactobacilli and bifidobacteria.
-
Research is required to relate the antibiotic resistance of lactobacilli and bifidobacteria and the potential for transmission of genetic elements to other intestinal and/or food-borne microorganisms.
The European Union has recently funded a research project, named ACE-ART (Assessment and Critical Evaluation of Antibiotic Resistance Transferability in Food Chain). The project (www.aceart.net) started one year ago and aims to provide a critical evaluation of the presence of antibiotic resistance genes in nonpathogenic bacteria belonging to Lactobacillus, Bifidobacterium, Lactococcus, and Streptococcus thermophilus. One of us (Morelli) is the coordinator of this project, involving 14 laboratories spread all over Europe. At the moment more than 1,300 strains belonging to 20 species have been studied; a little more that 100 strains with “atypical” profiles of antibiotic resistance have been detected.
Prebiotics or the Modification of the Host Nutritional Environment
Prebiotics were not assessed by FAO/WHO consultation and only the following remarks were made:
-
Prebiotics are generally defined as “nondigestible food ingredients that beneficially affect the host by selectively stimulating the growth and/or activity of one or a limited number of bacterial species already established in the colon, and thus improve host health” (Gibson and Roberfroid, 1995).
-
The concept of prebiotics essentially has the same aim as probiotics: to improve host health via modulation of the intestinal flora, although by a different mechanism. However, there are some cases in which prebiotics may be beneficial for the probiotic, especially with regard to bifidobacteria; that is the synbiotic concept. Synbiotics are defined as “mixtures of probiotics and prebiotics that beneficially affect the host by improving the survival and implantation of live microbial dietary supplements in the gastrointestinal tract of the host” (Andersson et al., 2001). If a synbiotic relationship is intended, then it should be verified scientifically, following the guidelines outlined in section 5 of this report.
Conclusion
More than a dozen years of intense microbiological, genetic, and clinical research have provided enough sound science to convince regulatory bodies that it is necessary to provide suggestions and guidelines for this evolving field; however, it is paramount to realize the difficulty in establishing guidelines for a field which is in fast progress. New techniques that allow enhanced characterization of the gut microbiota are now a long way from “culture and phenotype” and we are
approaching a no-culture, genotype-only approach. Moreover, emerging methods that allow assessment of bacteria-epithelial interactions are also to be evaluated. As it is more a possibility that lifelong cross-talk between the host and the gut microbiota determines whether health is maintained or disease intervenes, it is clear that understanding of these bacteria-bacteria and bacteria-host immune and epithelial cell interactions is likely to lead to a greater insight of disease pathogenesis. It is highly possible that a new generation of probiotic or biotherapeutic could be used for obtaining a more targeted use, such as enhancing the production of a specific cytokine or suppressing a specific pathogen.
If guidelines are prepared to provide suggestions and regulatory boundaries, it is important to take note that flexibility as well as constant dialogue between regulatory bodies and the scientific community is necessary. To manage this new war we must look for new allies, but we need also to write down new “alliance treaties” or guidelines for an optimal use of these alliances.
Acknowledgments
We would like to thank Mary Ellen Sanders, president of the International Scientific Association for Probiotics and Prebiotics (ISAPP), for critically reading and editing the manuscript.
REFERENCES
Abreu MT, Fukata M, Arditi M. 2005. TLR signaling in the gut in health and disease. Journal of Immunology 174(8):4453–4460.
Adlerberth I, Ahrne S, Johansson ML, Molin G, Hanson LA, Wold AE. 1996. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Applied Environmental Microbiology 62(7):2244–2251.
AFSSA (Agence Francaise de Sécurité Sanitaire des Aliments/French Food Safety Agency). 2005 (February). Effects of probiotics and prebiotics on flora and immunity in adults. [Online]. Available: http://www.usprobiotics.org/docs/AFFSA%20probiotic%20prebiotic%20flora%20immunity%2005.pdf [accessed January 12, 2006].
Agostoni C, Axelsson I, Braegger C, Goulet O, Koletzko B, Michaelsen KF, Rigo J, Shamir R, Szajewska H, Turck D, Weaver LT (ESPGHAN Committee on Nutrition). 2004a. Probiotic bacteria in dietetic products for infants: A commentary by the ESPGHAN Committee on Nutrition. Journal of Pediatric Gastroenterology and Nutrition 39(4):365–374.
Agostoni C, Axelsson I, Goulet O, Koletzko B, Michaelsen KF, Puntis JW, Rigo, J, Shamir R, Szajewska H, Turck D (ESPGHAN Committee on Nutrition). 2004b. Prebiotic oligosaccharides in dietetic products for infants: A commentary by the ESPGHAN Committee on Nutrition. Journal of Pediatric Gastroenterology and Nutrition 39(5):465–473.
Ahrne S, Lonnermark E, Wold AE, Aberg N, Hesselmar B, Saalman R, Strannegard IL, Molin G, Adlerberth I. 2005. Lactobacilli in the intestinal microbiota of Swedish infants. Microbes and Infection 7(11–12):1256–1262.
Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, Lick S, Hamrick A, Cano R, Klaenhammer TR. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proceedings of the National Academy of Sciences USA 102(11):3906–3912.
Andersson H, Asp N-G, Bruce A, Roos S, Wadstrom T, Wold AE. 2001. Health effect of probiotics and prebiotics: A literature review on human studies. Scandanavian Journal of Nutrition 45: 58–75.
Arvola T, Laiho K, Torkkeli S, Mykkanen H, Salminen S, Maunula L, Isolauri E. 1999. Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: A randomized study. Pediatrics 104(5):e64.
Axelsson L, Lindgren S. 1987. Characterization and DNA homology of Lactobacillus strains isolated from pig intestine. Journal of Applied Bacteriology 62(5):433–440.
Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich, CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences USA 101(44):15718–15723.
Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307(5717):1915–1920.
Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Applied Environmental Microbiology 66(4):1654–1661.
Bessi E, Morelli L. 2005. Lactobacillus apis, Lactobacillus alvei, Lactobacillus larvae and Lactobacillus insectis spp. nov., isolated from larvae, faeces and guts of healthy honeybees. Systematic and Applied Microbiology. In press.
Bilsborough J, Viney JL. 2004. In vivo enhancement of dendritic cell function. Annals of the New York Academy of Sciences 1029:83–87.
Bjorksten B, Naaber P, Sepp E, Mikelsaar M. 1999. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clinical and Experimental Allergy 29(3):342–346.
Blaut M, Collins MD, Welling GW, Dore J, van Loo J, de Vos W. 2002. Molecular biological methods for studying the gut microbiota: The EU human gut flora project. British Journal of Nutrition 87(Suppl 2):S203–S211.
Boekhorst J, Siezen RJ, Zwahlen MC, Vilanova D, Pridmore RD, Mercenier A, Kleerebezem M, de Vos WM, Brussow H, Desiere F. 2004. The complete genomes of Lactobacillus plantarum and Lactobacillus johnsonii reveal extensive differences in chromosome organization and gene content. Microbiology 150(pt 11):3601–3611.
Boekhorst J, de Been MW, Kleerebezem M, Siezen RJ. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. Journal of Bacteriology 187(14):4928–4934.
Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. Journal of Experimental Medicine 200(6): 749–759.
Bron PA, Marco M, Hoffer SM, Van Mullekom E, de Vos WM and Kleerebezem M. 2004a. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. Journal of Bacteriology 186(23): 7829–7835.
Bron PA, Grangette C, Mercenier A, de Vos WM, Kleerebezem M. 2004b. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. Journal of Bacteriology 186(17):5721–5729.
Bron PA, Meijer M, Bongers RS, De Vos WM, Kleerebezem M. 2004c. In: The Molecular Response of Lactobacillus plantarum to Intestinal Passage and Conditions [thesis]. (Ponsen en Looijen, Wageningen, Wageningen University). Pp. 129–153.
Bron PA, De Vos WM, Kleerebezem M. 2005. In: Vaughan EE, Ouwehand A, eds. Gastrointestinal Microbiology. New York: Marcel Dekker, Inc. In press.
Bron PA, Molenaar D, De Vos WM, Kleerebezem M. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. Journal of Applied Microbiology 100(4): 728–738.
Caramalho I, Lopes-Carvalho T, Ostler D, Zelenay S, Haury M, Demengeot J. 2003. Regulatory T cells selectively express toll-like receptors and are activated by lipopolysaccharide. Journal of Expimental Medicine 197(4):403–411.
Chang TL, Chang CH, Simpson DA, Xu Q, Martin PK, Lagenaur LA, Schoolnik GK, Ho DD, Hillier SL, Holodniy M, Lewicki JA, Lee PP. 2003. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proceedings of the National Academy of Sciences USA 100(20):11672–11677.
Cole MF, Bryan S, Evans MK, Pearce CL, Sheridan MJ, Sura PA, Wientzen R, Bowden GH. 1998. Humoral immunity to commensal oral bacteria in human infants: Salivary antibodies reactive with Actinomyces naeslundii genospecies 1 and 2 during colonization. Infection and Immunity 66(9):4283–4289.
Collins K, Salminen S, von Wright A, Morelli L, Marteau P, Brassart D, de Vos WM, Fonden R, Saxelin M, Mogensen G, Birkeland SE, Mattila-Sandholm T. 1998. Demonstration of safety of probiotics, a review. International Journal of Food Microbiology 44(1–2):93–106.
Cremonini F, Di Caro S, Santarelli L, Gabrielli M, Candelli M, Nista EC, Lupascu A, Gasbarrini G, Gasbarrini A. 2002. Probiotics in antibiotic-associated diarrhoea. Digest of Liver Diseases 34(suppl 2):S78–S80.
Crump M, Gospodarowicz M, Shepherd FA. 1999. Lymphoma of the gastrointestinal tract. Seminars in Oncology 26(3):324–337.
Cunningham-Rundles C. 1994. Clinical and immunologic studies of common variable immunodeficiency. Current Opinion in Pediatrics 6(6):676–681.
Cunningham-Rundles S. 2001. Nutrition and the mucosal immune system. Current Opinion in Gastroenterology 17(2):171–176.
Cunningham-Rundles S. 2005 (March 17). Session IV: Novel Approaches for Mitigating the Development of Resistance. Presentation at the Forum on Microbial Threats Workshop Ending the War Metaphor: The Changing for Unraveling the Host-Microbe Relationship, Washington, D.C., Institute of Medicine, Forum on Microbial Threats.
Cunningham-Rundles S, Nesin MC. 2000. Bacterial infections in the immunologically compromised host In: Nataro JP, Blaser MJ, Cunningham-Rundles S, eds. Persistent Bacterial Infections. Washington, DC: American Society of Microbiology Press. Pp.145–164.
Cunningham-Rundles S, Ahrne S, Bengmark S, Johann-Liang R, Marshall F, Metakis L, Califano C, Dunn AM, Grassey C, Hinds G, Cervia J. 2000. Probiotics and immune response. American Journal of Gastroenterology 95(1 Suppl):S22–S25.
de Vos WM, BronPA, Kleerebezem M. 2004. Post-genomics of lactic acid bacteria and other food-grade bacteria to discover gut functionality. Current Opinion in Biotechnology 15(2):86–93.
Didierlaurent A, Sirard JC, Kraehenbuhl JP, Neutra MR. 2002. How the gut senses its content. Cell Microbiology 4(2):61–72.
D’Souza AL, Rajkumar C, Cooke J, Bulpitt CJ. 2002. Probiotics in prevention of antibiotic associated diarrhoea: Meta-analysis. British Medical Journal 324(7350):1361.
Ducluzeau R. 1988. Role of experimental microbial ecology in gastroentrology. In: Bergogne-Berezin E, ed. Microbial Ecology and Intestinal Infection, Springer-Verlag, France. Pp. 7–26.
Dunne C, Murphy L, Flynn S, O’Mahony L, O’Halloran S, Feeney M, Morrissey D, Thornton G, Fitzgerald G, Daly C, Kiely B, Quigley EM, O’Sullivan GC, Shanahan F, Collins JK. 1999. Probiotics: From myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials [review]. Antonie Van Leeuwenhoek 76(1–4):279–292.
Eberl G. 2005. Inducible lymphoid tissues in the adult gut: Recapitulation of a fetal developmental pathway? Nature Reviews Immunology 5(5):413–420.
Elmer GW, Surawicz CM, McFarland LV. 1996. Biotherapeutic agents. A neglected modality for the treatment and prevention of selected intestinal and vaginal infections. Journal of the American Medical Association 275(11):870–876.
Elmer GW, McFarland LV, Surawicz CM. 1999. Biotherapeutic Agents and Infectious Diseases. Totowa, NJ: Humana Press.
FAO/WHO (Food and Agriculture Organization/World Health Organization). 2001 (October). Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Córdoba, Argentina. [Online]. Available: http://www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf [accessed January 3, 2005].
FAO/WHO. 2002 (April 30, May 1). Guidelines for the evaluation of probiotics in food. Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Ontario, Canada.
Finegold SM, Rolfe RD. 1983. Susceptibility testing of anaerobic bacteria. Diagnostic Microbiology and Infectious Disease 1(1):33–40.
Friman V, Nowrouzian F, Adlerberth I, Wold AE. 2002. Increased frequency of intestinal Escherichia coli carrying genes for S. fimbriae and haemolysin in IgA-deficient individuals. Microbial Pathogenesis 32(1):35–42.
Fujiwara S, Seto Y, Kimura A, Hashiba H. 2001. Establishment of orally administered Lactobacillus gasseri SBT2055SR in the gastrointestinal tract of humans and its influence on intestinal microflora and metabolism. Journal of Applied Microbiology 90(3):343–345.
Fuller R. 1989. Probiotics in man and animals. Journal of Applied Bacteriology 66:365–378.
Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. Journal of Nutrition 125(6):1401–1412.
Goepfert AR, Andrews WW, Carlo W, Ramsey PS, Cliver SP, Goldenberg RL, Hauth JC. 2004. Umbilical cord plasma interleukin-6 concentrations in preterm infants and risk of neonatal morbidity. American Journal of Obstetrics and Gynecology 191(4):1375–1381.
Guandalini S, Pensabene L, Zikri MA, Dias JA, Casali LG, Hoekstra H, Kolacek S, Massar K, Micetic-Turk D, Papadopoulou A, de Sousa JS, Sandhu B, Szajewska H, Weizman Z. 2000. Lactobacillus GG administered in oral rehydration solution to children with acute diarrhea: A multicenter European trial. Journal of Pediatric Gastroenterology and Nutrition 30(1):54–60.
Guarner F, Schaafsma GJ. 1998. Probiotics. International Journal of Food Microbiology 39(3): 237–238.
Hall PA, Coates PJ, Ansari B, Hopwood D. 1994. Regulation of cell number in the mammalian gastrointestinal tract: The importance of apoptosis. Journal of Cell Science 107(pt 12): 3569–3577.
Hanniffy S, Wiedermann U, Repa A, Mercenier A, Daniel C, Fioramonti J, Tlaskolova H, Kozakova H, Israelsen H, Madsen S, Vrang A, Hols P, Delcour J, Bron P, Kleerebezem M, Wells J. 2004. Potential and opportunities for use of recombinant lactic acid bacteria in human health. Advanced Applications in Microbiology 56:1–64.
Harrod T, Martin M, Russell MW. 2001. Long-term persistence and recall of immune responses in aged mice after mucosal immunization. Oral Microbiology and Immunology 16(3):170–177.
Holzapfel WH, Haberer P, Snel J, Schillinger U, Huis in’t Veld JH. 1998. Overview of gut flora and probiotics [review]. International Journal of Food Microbiology 41(2):85–101.
Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proceedings of the National Academy of Sciences USA 96(17):9833–9838.
Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291(5505):881–884.
Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. 2003. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nature Immunology 4(3):269–273.
Huang Y, Shao XM, Neu J. 2003. Immunonutrients and neonates. European Journal of Pediatrics 162(3):122–128.
Huang HC, Wang CL, Huang LT, Chuang H, Liu CA, Hsu TY, Ou CY, Yang KD. 2004. Association of cord blood cytokines with prematurity and cerebral palsy. Early Human Development 77(1–2):29–36.
Kalha I, Sellin JH. 2004. Common variable immunodeficiency and the gastrointestinal tract. Current Gastroenterology Reports 6(5):377–383.
Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. 2001a. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. Journal of Allergy and Clinical Immunology 107:129–134.
Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. 2001b. Probiotics in primary prevention of atopic disease: A randomised placebo-controlled trial. Lancet 357(9262): 1076–1079.
Kelly D, Conway S. 2005. Bacterial modulation of mucosal innate immunity. Molecular Immunology 42(8):895–901.
Klaenhammer TR, Barrangou R, Buck BL, Azcarate-Peril MA, Altermann E. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiology Reviews 29(3): 393–409.
Kleerebezem M. 2005 (March 17). Session IV: Novel Approaches for Mitigating the Development of Resistance. Presentation at the Forum on Microbial Threats Workshop Ending the War Metaphor: The Changing Agenda for Unraveling the Host-Microbe Relationship, Washington, D.C., Institute of Medicine, Forum on Microbial Threats.
Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos W M, Siezen RJ. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proceedings of the National Academy of Sciences USA 100(4):1990–1995.
Klein G, Pack A, Bonaparte C, Reuter G. 1998. Taxonomy and physiology of probiotic lactic acid bacteria. International Journal of Food Microbiology 41(2):103–125.
Kliegman RM. 2005. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Journal of Pediatrics 146(5):710.
Knight PA, Pemberton AD, Robertson KA, Roy DJ, Wright SH, Miller HR. 2004. Expression profiling reveals novel innate and inflammatory responses in the jejunal epithelial compartment during infection with Trichinella spiralis. Infection and Immunity 72(10):6076–6086.
Kunz AN, Noel JM, Fairchok MP. 2004. Two cases of Lactobacillus bacteremia during probiotic treatment of short gut syndrome. Journal of Pediatric Gastroenterology and Nutrition 38(4): 457–458.
Kvasnikov EI, Kovalenko NK, Nesterenko OA. 1971. Lactobacilli of bees. Veterinariia 8:38–39.
Kvasnikov EI, Kovalenko NK, Materinskaia LG. 1977. Lactobacilli of freshwater fishes. Mikrobiologiia 46(4):755–760.
Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MH, Welling GW. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Applied Environmental Microbiology 61(8):3069–3075.
Leverrier P, Dimova D, Pichereau V, Auffray Y, Boyaval P, Jan G. 2003. Susceptibility and adaptive response to bile salts in Propionibacterium freudenreichii: Physiological and proteomic analysis. Applied Environmental Microbiology 69(7):3809–3818.
Lilly DM, Stillwell RH. 1965. Probiotics: Growth-promoting factors produced by microorganisms. Science 147:747–748.
Macdonald TT, Monteleone G. 2005. Immunity, inflammation, and allergy in the gut. Science 307(5717):1920–1925.
Macdonald TT, Disabatino A, Gordon JN. 2005. Immunopathogenesis of Crohn’s disease. Journal of Parenteral and Enteral Nutrition 29(4 Suppl):S118–S125.
Martin R, Langa S, Reviriego C, Jiminez E, Marin ML, Xaus J, Fernandez L, Rodriguez JM. 2003. Human milk is a source of lactic acid bacteria for the infant gut. Journal of Pediatrics 143(6):754–758.
McCartney AL. 2002. Application of molecular biological methods for studying probiotics and the gut flora. British Journal of Nutrition 88(Suppl 1):S29–S37.
Metchnikoff E. 1907. The prolongation of life. In Optimistic Studies Heinemann W. ed., London: G. P. Putnam & Sons. Pp. 1–100.
Miller D, Ross J. 2005. Vaccine INDs: Review of clinical holds. Vaccine 23(9):1099–1101.
Ministero della Salute. 2006. Ministero della Salute homepage. [Online]. Available: http://www.ministerosalute.it/ [accessed January 12, 2006].
Molenaar D, Bringel F, Schuren FH, de Vos WM, Siezen RJ, Kleerebezem M. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. Journal of Bacteriology 187(17):6119–6127.
Moore WE, Holdeman LV. 1974. Human fecal flora: The normal flora of 20 Japanese-Hawaiians. Applied Microbiology 27(5):961–979.
Morelli L. 2000. In vitro selection of probiotic lactobacilli: A critical appraisal [review]. Current Issues in Intestinal Microbiology 1(2):59–67.
Morelli L. 2005 (March 17). Session IV: Novel Approaches for Mitigating the Development of Resistance. Presentation at the Forum on Microbial Threats Workshop Ending the War Metaphor: The Changing for Unraveling the Host-Microbe Relationship, Washington, D.C., Institute of Medicine, Forum on Microbial Threats.
Morelli L, Cesena C, de Haen C, Gozzini L. 1998. Taxonomic Lactobacillus composition of feces from human newborns during the first few days. Microbial Ecology 35(2):205–212.
Nachman SA, Lindsey JC, Moye J, Stanley KE, Johnson GM, Krogstad PA, Wiznia AA. 2005. Growth of human immunodeficiency virus-infected children receiving highly active antiretroviral therapy. Pediatric Infectious Disease Journal 24(4):352–357.
Nielsen DS, Moller PL, Rosenfeldt V, Paerregaard A, Michaelsen KF, Jakobsen M. 2003. Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon. Applied Environmental Microbiology 69(12):7545–7548.
Pabst R. 1987. The anatomical basis for the immune function of the gut. Anatomy and Embryology 176(2):135–144.
Pacha J. 2000. Development of intestinal transport function in mammals. Physiological Reviews 80(4):1633–1667.
Parker RB. 1974. Probiotics: The other half of the antibiotic story. Animal Nutrition and Health 29:4–8.
Percival RS, Marsh PD, Challacombe SJ. 1996. Serum antibodies to commensal oral and gut bacteria vary with age. FEMS Immunology and Medical Microbiology 15(1):35–42.
Prescott SL, Macaubas C, Holt BJ, Smallacombe TB, Loh R, Sly PD, Holt PG. 1998. Transplacental priming of the human immune system to environmental allergens: Universal skewing of initial T cell responses toward the Th2 cytokine profile. Journal of Immunology 160(10):4730–4737.
Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA, Lambert J, de Vos WM, van der Meer R, Smits MA, Kleerebezem M. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. Journal of Bacteriology 187(17): 6128–6136.
Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet AC, Zwahlen MC, Rouvet M, Altermann E, Barrangou R, Mollet B, Mercenier A, Klaenhammer T, Arigoni F, Schell MA. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proceedings of the National Academy of Sciences USA 101(8):2512–2517.
Pryde SE, Richardson AJ, Stewart CS, Flint HJ. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Applied Environmental Microbiology 65(12):5372–5327.
Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118(6): 229–241.
Rawls JF, Samuel BS, Gordon JI. 2004. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proceedings of the National Academy of Sciences USA 101(13): 4596–4601.
Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, Roberfroid M, Rowland I, Cherbut C, Klaenhammer TR. 2003. New scientific paradigms for probiotics and prebiotics. Journal of Clinical Gastroenterology 37(2):105–118.
Rhee KJ, Sethupathi P, Driks A, Lanning DK, Knight KL. 2004. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. Journal of Immunology 172(2):1118–1124.
Rimoldi M, Chieppa M, Salucci V, Avogadri F, Sonzogni A, Sampietro GM, Nespoli A, Viale G, Allavena P, Rescigno M. 2005. Intestinal immune homeostasis is regulated by the crosstalk between epithelial cells and dendritic cells. Nature Immunology 6(5):507–514.
Roller M, Rechkemmer G, Watzl B. 2004. Prebiotic inulin enriched with oligofructose in combination with the probiotics Lactobacillus rhamnosus and Bifidobacterium lactis modulates intestinal immune functions in rats. Journal of Nutrition 134(1):153–156.
Rouzaud G, Rabot S, Ratcliffe B, Duncan AJ. 2003. Influence of plant and bacterial myrosinase activity on the metabolic fate of glucosinolates in gnotobiotic rats. British Journal of Nutrition. 90(2):395–404.
Salminen S, Ouwehand A, Benno Y, Lee YK. 1999. Probiotics: How are they defined? Trends in Food Science and Technology 10:107–110.
Sanders ME. 2003. Probiotics: Considerations for human health. Nutrition Reviews 61(3):91–99.
Sarra PG, Magri M, Bottazzi V, Dellaglio F. 1980. Genetic heterogeneity among Lactobacillus acidophilus strains. Antonie Van Leeuwenhoek 46(2):169–176.
Sarra PG, Vescovo M, Fulgoni M. 1986. Study on crop adhesion genetic determinant in Lactobacillus reuteri. Microbiologica 9(3):279–285.
Sartor RB. 2005. Probiotic therapy of intestinal inflammation and infections. Current Opinion in Gastroenterology 21(1):44–50.
Saunier K, Dore J. 2002. Gastrointestinal tract and the elderly: Functional foods, gut microflora and healthy ageing. Digest of Liver Diseases 34(Suppl 2):S19–S24.
Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proceedings of the National Academy of Sciences USA 99(22):14422–14427.
Schrezenmeir J, Heller K, McCue M, Llamas C, Lam W, Burow H, Kindling-Rohracker M, Fischer W, Sengespeik HC, Comer GM, Alarcon P. 2004. Benefits of oral supplementation with and without synbiotics in young children with acute bacterial infections. Clinical Pediatrics 43(3):239–249.
Schultz C, Temming P, Bucsky P, Gopel W, Strunk T, Hartel C. 2004a. Immature anti-inflammatory response in neonates. Clinical and Experimental Immunology 135:130–136.
Schultz M, Timmer A, Herfarth HH, Sartor RB, Vanderhoof JA, Rath HC. 2004b. Lactobacillus GG in inducing and maintaining remission of Crohn’s disease. BMC Gastroenterology 4:5.
Shanahan F. 2003. Probiotics: A perspective on problems, pitfalls. Scandinavian Journal of Gastroenterology 237(Suppl):34–36.
Song YL, Kato N, Matsumiya Y, Liu CX, Kato H, Watanabe K. 1999. Identification of Lactobacillus species of human origin by a commercial kit, API50CHL. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi 10(2):77–82.
Song Y, Kato N, Liu C, Matsumiya Y, Kato H, Watanabe K. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23SrRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiology Letters 187(2):167–173.
Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307(5717):1955–1959.
Suau A. 2003. Molecular tools to investigate intestinal bacterial communities. Journal of Pediatric Gastroenterology and Nutrition 37(3):222–224.
Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Applied Environmental Microbiology 65(11):4799–4807.
Suau A, Rochet V, Sghir A, Gramet G, Brewaeys S, Sutren M, Rigottier-Gois L, Dore J. 2001. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Systematic and Applied Microbiology 24(1):139–145.
Szajewska H, Mrukowicz JZ. 2001. Probiotics in the treatment and prevention of acute infectious diarrhea in infants and children: A systematic review of published randomized, double-blind, placebo-controlled trials. Journal of Pediatric Gastroenterology and Nutrition 33(Suppl 2): S17–S25.
Tannock GW. 2003. Probiotics: Time for a dose of realism. Current Issues in Intestinal Microbiology 4(2):33–42.
Tannock GW, Munro K, Harmsen HJ, Welling GW, Smart J, Gopal PK. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Applied Environmental Microbiology 66(6):2578–2588.
Thompson C, McCarter YS, Krause PJ, Herson VC. 2001. Lactobacillus acidophilus sepsis in a neonate. Journal of Perinatology 21(4):258–260.
Tlaskalova-Hogenova H, Stepankova R, Hudcovic T, Tuckova L, Cukrowska B, Lodinova-Zadnikova R, Kozakova H, Rossmann P, Bartova J, Sokol D, Funda DP, Borovska D, Rehakova Z, Sinkora J, Hofman J, Drastich P, Kokesova A. 2004. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases [review]. Immunology Letters 93(2–3):97–108.
Vaarala O. 2003. Immunological effects of probiotics with special reference to lactobacilli [Review]. Clinical and Experimental Allergy 33(12):1634–1640.
Vaillancourt R. 2005 (March 17). Session IV: Novel Approaches for Mitigating the Development of Resistance. Presentation at the Forum on Microbial Threats Workshop Ending the War Metaphor: The Changing Agenda for Unraveling the Host-Microbe Relationship, Washington, D.C., Institute of Medicine, Forum on Microbial Threats.
van Kranenburg R, Golic N, Bongers R, Leer RJ, de Vos WM, Siezen RJ, Kleerebezem M. 2005. Functional analysis of three plasmids from Lactobacillus plantarum. Applied Environmental Microbiology 71(3):1223–1230.
Van Niel CW, Feudtner C, Garrison MM, Christakis DA. 2002. Lactobacillus therapy for acute infectious diarrhea in children: A meta-analysis. Pediatrics 109(4):678–684.
Vaughan EE, Schut F, Heilig HG, Zoetendal EG, de Vos WM, Akkermans AD. 2000. A molecular view of the intestinal ecosystem. Current Issues of Interest in Microbiology 1(1):1–12.
Vaughan EE, Heilig HG, Ben-Amor K, de Vos WM. 2005. Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiology Review 29(3):477–490.
Veber MB, Cunningham-Rundles S, Schulman M, Mandel F, Auld PA. 1991. Acute shift in immune response to microbial activators in very-low-birth-weight infants. Clinical and Experimental Immunology 83(3):391–395.
Vinderola CG, Medici M, Perdigon G. 2004. Relationship between interaction sites in the gut, hydrophobicity, mucosal immunomodulating capacities and cell wall protein profiles in indigenous and exogenous bacteria. Journal of Applied Microbiology 96(2):230–243.
Walter J, Heng NC, Hammes WP, Loach DM, Tannock, GW, Hertel C. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Applied Environmental Microbiology 69(4):2044–2051.
Walter J, Chagnaud P, Tannock GW, Loach DM, Dal Bello F, Jenkinson HF, Hammes WP, Hertel C. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Applied Environmental Microbiology 71(2):979–986.
Watson DC, Counts DR. 2004. Growth hormone deficiency in HIV-infected children following successful treatment with highly active antiretroviral therapy. Journal of Pediatrics 145(4): 549–551.
Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299(5615):2074–2076.
Zoetendal EG, Akkermans AD, de Vos WM. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Applied Environmental Microbiology 64(10):3854–3859.
Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Applied Environmental Microbiology 68(7):3401–3407.
Zoetendal EG, Collier CT, Koike S, Mackie RI, Gaskins HR. 2004. Molecular ecological analysis of the gastrointestinal microbiota: A review. Journal of Nutrition and Dietetics 134(2):465–472.






















































