9
Symbiosis as an Adaptive Process and Source of Phenotypic Complexity
NANCY A. MORAN
Genomics has revealed that inheritance systems of separate species are often not well segregated: genes and capabilities that evolve in one lineage are often stably acquired by another lineage. Although direct gene transfer between species has occurred at some level in all major groups, it appears to be far more frequent in prokaryotes than in multicellular eukaryotes. An alternative to incorporating novel genes into a recipient genome is acquiring a stable, possibly heritable, symbiotic association and thus enjoying benefits of complementary metabolic capabilities. These kinds of symbioses have arisen frequently in animals; for example, many insect groups have diversified on the basis of symbiotic associations acquired early in their evolutionary histories. The resulting associations are highly complex, often involving specialized cell types and organs, developmental mechanisms that ensure transfer of symbionts between generations, and mechanisms for controlling symbiont proliferation and location. The genomes of long-term obligate symbionts often undergo irreversible gene loss and deterioration even as hosts evolve dependence on them. In some cases, animal genomes may have acquired genes from symbionts, mirroring the gene uptake from mitochondrial and plastid genomes. Multiple symbionts often coexist in the same
Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ 85721.
host, resulting in coadaptation among several phylogenetically distant genomes.
Genomic sequence data, coupled with evolutionary analyses, have brought major new insights to our understanding of biological evolution. One of the biggest revelations is the extent to which biological adaptation and phenotypic innovation within a particular genetic lineage have depended on adopting already highly honed functional systems from other lineages, often only distantly related to the recipient. Traditional views of the evolutionary process, forged during the neo-Darwinian synthesis, focused on adaptation occurring as the result of natural selection acting on existing genes within a species. Most such adaptation occurs in small steps, although mutations in existing genes can sometimes cause major phenotypic changes. But the ability to reconstruct evolution at the molecular level, and especially the analysis of full genome sequences, has revealed that integration of genes originating from disparate sources has occurred on a very large scale.
Gene uptake confers novel adaptive capabilities, thereby enabling ecological expansion into new niches. But it also confers phenotypic complexity that is manifested at the genomic, the physiological, and the morphological levels. In many cases, and specifically in multicellular eukaryotes, the route to recruiting foreign genes and novel metabolic capabilities involves symbiotic association, that is, a persistent close interaction with another species. Comparative genomic studies now allow us to reconstruct the history of symbioses and episodes of genome amalgamation and to elucidate their contribution to the complexity evident in the dominant forms of life on earth.
Below, I briefly describe the routes by which organisms stably acquire capabilities evolved in other lineages, with emphasis on insights that have come from recent genome sequencing. I end with examples of the complex phenotypes generated by hereditary symbiosis in insects and with the consequences of this genome integration through symbiosis for animal evolution.
DISPARATE GENE SETS CONFER DISTINCT CAPABILITIES
The evolutionary motivation for assimilating foreign genes stems from the obvious fact that species differ in gene sets and corresponding capabilities. Thus, intimate association between two lineages can readily arise through natural selection acting within each species to fix alleles that promote close association with the other species. Although differences in metabolic capacities among species have long been evident, genomics is
providing a far more detailed view of how these differences have arisen. First, many genes and corresponding capabilities arose after lineage diversification had begun, so that some species descend from ancestors that never possessed the corresponding genes. Examples include fundamental metabolic innovations such as phototrophy (Mulkidjanian et al., 2006) and methanogenesis as well as specialties such as production of particular toxins or pathogenicity mechanisms, including the type III secretion system used by many bacteria for infecting host cells (Saier, 2004).
A second reason that species differ in gene sets and capabilities is that ancestral genes are often lost. Comparisons of genomic content among closely related species are now revealing that gene loss has been an important and ongoing process in evolution in all lineages. For example, tryptophan, required in proteins of all organisms, is produced by a single pathway requiring several enzymatic steps, and reconstructions of the evolution of this pathway point to a single origin before the divergence of the three domains of life (Xie et al., 2003). Many descendant lineages, including all animals and a variety of prokaryotes and parasitic protists, have lost the pathway and are dependent on acquiring tryptophan from ecologically associated species.
The loss of such useful capabilities may seem counterintuitive. But another major insight from comparative genomics is that genes are constantly being eliminated by mutation combined with insufficient purifying selection. The pathways lost most frequently are those with more enzymatic steps and higher energy requirements, suggesting that selection may favor pathway inactivation when the end products can be environmentally acquired. Outstanding examples of gene loss include many host-dependent microbial lineages; obligate pathogens, both bacterial and eukaryotic, lose genes for using substrates not encountered in their restricted environment and lose genes for synthesis of metabolic products that are dependably provided by the host cells (Shigenobu et al., 2000; Gardner et al., 2002; Tamas et al., 2002; Liu et al., 2006; Payne and Loomis, 2006). As a group, animals are unusual in lacking the ability to produce numerous universally required metabolic compounds such as vitamins and amino acids and also in lacking capabilities for producing bioactive secondary compounds that can act as toxins and defenses.
ACQUISITION OF FOREIGN GENES AND CAPABILITIES
For many genes, the distribution among species reflects not only the lineage of origin and subsequent losses in some descendants but also transfer to new lineages. Thus, a species can acquire, more or less instantaneously, traits that originated in unrelated lineages or that were lost ancestrally. Gene transfer is clearly important in prokaryotes, for which
hundreds of genomes have been sequenced. As an illustration of the extent of foreign-gene uptake, a study reconstructing the sources of genes in the genomes of numerous species in the Gammaproteobacteria showed that the overwhelming majority of genes in most genomes were acquired from external sources after these lineages diverged from a common ancestor (Lerat et al., 2005). The impact of gene uptake can be massive even on short time scales. For example, in comparisons of gene sets of distinct Escherichia coli strains, for which orthologous DNA sequences are 99% identical, indicating recent shared ancestry, 25% or more of the genes in one genome are absent from other strains, having arrived recently from more distant (often unidentified) sources (Welch et al., 2002). In Bacteria, such incorporation of foreign genes is the major route to the origination of novel capacities (Ochman et al., 2000), as illustrated in E. coli, in which recently acquired genes are the basis for strain-specific pathogenicity (e.g., Welch et al., 2002).
Firm estimates are not yet possible for rates of gene acquisition by eukaryotic genomes. For Bacteria, the evidence for rampant gene acquisition is primarily based on comparing related genomes by using complete gene inventories, sequence features, and gene arrangements. To date, the numbers of sequenced genomes for clusters of related eukaryotic species are relatively small for estimating total gene uptake by using comparisons of gene inventories and gene arrangements. Currently, the extent of foreign gene uptake, and specifically genes arriving from Bacteria, does appear to be substantial in certain groups of unicellular eukaryotes, including Dictyostelium (Eichinger et al., 2005) and other lineages of amoebae (Andersson et al., 2006). But even in unicellular eukaryotes, duplication and divergence of existing genes appear to be more prominent than gene uptake as a process generating change in genome contents (e.g., Kellis et al., 2004; Aury et al., 2006). Similarly, plant nuclear and plastid genomes have not been found to contain substantial numbers of acquired genes, although acquisition of genes by plant mitochondrial genomes does occur relatively frequently and is currently the major category of gene incorporation by multicellular eukaryotes (Richardson and Palmer, 2007). Despite increasing findings of horizontal transfer even in eukaryotes, the capacity to incorporate new genes underlying enzymatic pathways and processes has severe limits (Kurland et al., 2003). Some groups of organisms rarely incorporate foreign genes, and, even in those that do, such as most free-living Bacteria, many genes underlying important informational and metabolic processes seem to resist horizontal transfer, as illustrated by the case of the tryptophan biosynthetic pathway (Xie et al., 2003).
SYMBIOSIS AS A MECHANISM OF ADAPTATION AND AS A SOURCE OF PHENOTYPIC COMPLEXITY
An alternative to incorporating foreign genes directly into a recipient genome is to develop a close relationship with a species able to provide some beneficial product or process. Symbiotic associations that are mutually beneficial raise immediate issues involving evolutionary stability—issues that Darwin noted and also addressed in The Origin of Species: “Natural Selection cannot possibly produce any modification in a species exclusively for the good of another species; although throughout nature one species incessantly takes advantage of, and profits by, the structures of others” (Darwin, 1859a, p. 201). Because different species possess different capabilities, such as different abilities to use substrates or to produce required metabolic compounds, it is now evident that one species can profit through associations with another species and that this benefit can be mutual; that is, providing a benefit to another species need not entail a cost. These differences in capabilities have become more defined through genomic data, which allow us to use genome sequences to map many metabolic capabilities onto the branches of the tree of life. In many circumstances, one symbiotic partner immediately profits from providing some benefit to the other partner; for example, a compound available in excess to one species might act as a limiting substrate to its partner, which in turn can generate from this substrate additional compounds needed by the first species. Because of the different capabilities of different species, mutually beneficial associations can arise de novo from organisms that are not coevolved, and these associations can then become stabilized through natural selection acting within each species. Mutual advantage often can be enhanced by natural selection when the two lineages are associated across generations, although heritability of the symbiosis is not a requirement for mutual benefit (Sachs et al., 2004). Genomic data inform us that many symbioses are founded on the differences in metabolic capabilities that are enforced by differences in gene content of genomes.
Symbiosis binds organisms from all domains of life and has produced extreme modifications in genomes and structure (e.g., von Dohlen et al., 2001; Waters et al., 2003; Gilson et al., 2006; Nakabachi et al., 2006). In addition, symbiosis affects genome evolution by facilitating gene transfer from one genome to another and by facilitating the loss from one genome of genes that are present in both symbiotic partners. Both of these events can cause a facultative symbiosis to become an obligate one because one partner becomes dependent on products of genes that are restricted to the genome of the other partner. The result is a complex, fused metaorganism, with different compartments for different portions of its required genes, mechanisms for transporting compounds and gene products between compartments, complex development maintaining the different cell types
in proper proportions and arrangements, and different replication systems and population genetic processes applicable to different parts of the metagenome. In the following, I consider some of the most prominent symbioses in microorganisms and then focus on the role of symbiosis in generating phenotypic complexity in animals.
BACTERIOPHAGE AS GENE VECTORS AND SYMBIONTS OF BACTERIA
Although conventional views of bacteriophage have emphasized their role in killing bacterial hosts, it is now apparent that they often affect host ecology in other, more beneficial ways, including acting as vectors of genes that can enhance bacterial fitness in a particular environment (Comeau and Krisch, 2005; Hendrix, 2002). This realization comes in part from genome sequences for bacteria, which reveal that bacteriophage have made large ongoing contributions to bacterial genome contents and physiological capabilities, often sometimes becoming lasting parts of the bacterial genome even when genes required for the bacteriophage life cycle have been eliminated (Comeau and Krisch, 2005). The richest reservoir of gene diversity lies in the bacteriophage (e.g., Breitbart et al., 2002; Kwan et al., 2005), suggesting that the innovations that they are able to contribute are correspondingly diverse. For example, a very large proportion of pathogenic bacteria studied in humans and other mammals use pathogenicity mechanisms encoded by phage-borne genes (Wagner and Waldor, 2002), some of the competitive mechanisms used among bacterial strains are derived from phage-derived structures (e.g., Nakayama et al., 2000), and the central enzyme components underlying photosynthesis in ubiquitous marine cyanobacteria are transferred among bacterial hosts by bacteriophage (Lindell et al., 2004). Even phage-induced lysis of the bacterial host cell can be a mechanism favoring the growth and fitness of the bacterial host clone, as other cells containing the same phage genes persist and benefit from products released during the death of their sister cells (Wagner and Waldor, 2002).
SYMBIOTIC MICROBIAL CONSORTIA
One consequence of the fact that gene transfer is not without limits even in prokaryotes is the frequent evolution of microbial consortia or microbial syntrophy, that is, close, often obligate, associations of two (or a few) unrelated organisms that depend on one another for metabolic products or maintenance of chemically permissive conditions. Examples of close, coevolved associations include those between methanogenic archaea and bacteria or protists capable of fermentation (e.g., Schink,
2002), phototrophic aquatic consortia consisting of a flagellated bacterium coated with phototrophic bacteria (Overmann and Schubert, 2002; Glaeser and Overmann, 2004), and an archaean–bacterium partnership that links methane oxidation and denitrification (Raghoebarsing et al., 2006). More broadly, metabolic interdependencies among lineages are a major reason that most microbes in soil and other natural habitats cannot be established in pure laboratory culture (Schmidt, 2006). In some systems, detailed knowledge of the interactions of the different bacterial types shows that there has been extensive coadaptation, with recognition mechanisms for promoting the associations and with communication systems for coordinating the behavior of cells from phylogenetically distant groups (Schink, 2002).
SYMBIOSIS AS A ROUTE TO ADAPTATION AND COMPLEXITY IN EUKARYOTES
Molecular phylogenetics, based on sequence data from only a few genes, verified the hypothesis that mitochondria and plastids are derived from bacterial symbionts; these results also identified the bacterial lineages that gave rise to these symbionts and indicated a single primary origin in each case (Woese, 1987; Keeling, 2004; Embley and Martin, 2006; Kurland et al., 2006). Genomic data indicate that both plastids and mitochondria have transferred genes from the bacterial to the host genome, resulting in a genomic mélange (e.g., Martin et al., 2002; Keeling, 2004). Genome sequences have also helped to elucidate further complexities of the mitochondrial and plastid symbioses in some lineages: for example, secondary and tertiary symbioses in which a plastid-containing eukaryote itself becomes a symbiont in a new eukaryotic host, sometimes resulting in bizarre remnant genomes (e.g., Gilson et al., 2006) and complicated histories of gene transfers among several genomes (Keeling, 2004). Beyond cellular organelles, symbioses have arisen innumerable times in eukaryotic hosts. Protists often carry prokaryotic symbionts, and their nuclear genomes may have taken up genes from past symbionts (e.g., Eichinger et al., 2005; Andersson et al., 2005). Much of the complexity of modern eukaryotic cells arises from this divided ancestry involving gene movement between genomes and the evolution of mechanisms for targeting gene products to the correct cellular compartment. Even individual enzymatic pathways or functional systems can be encoded by complex combinations of genes with different histories of direct horizontal transfer, transfer through symbiosis, or vertical inheritance (e.g., Chen et al., 2006; Richards et al., 2006).
Symbioses originating in multicellular eukaryotes are rampant, with highly specialized obligate associations found in fungi, plants, sponges,
and most animal phyla (McFall-Ngai, 2002; Brundrett, 2003; Schardl et al., 2004). Many of these symbioses are vertically transmitted, resulting in continuous association of individual genetic lineages across generations and facilitating the evolution of mutually beneficial features. Others are reestablished each host generation from a dispersing symbiont population.
FORCES FAVORING SYMBIOSIS IN ANIMALS
Animals stand out as a group having lost many ancestral capabilities, making them unusually dependent on other organisms. In the Metazoa generally, gene loss has resulted in the inability to synthesize essential metabolic compounds, yielding a long list of required dietary components, including numerous cofactors (vitamins) as well as the 10 essential amino acids (Payne and Loomis, 2006). The losses of these pathways reflect the evolution of a digestive cavity by animals, which acquire diverse nutrients by eating tissues and cells of other organisms. If nutrients are readily available in the diet, selection to maintain pathways for production of these compounds will be relaxed, resulting in the inactivation of the underlying genes. Comparisons of recently sequenced animal genomes reveal that particular animal lineages have continued to eliminate particular sets of genes. For example, the Drosophila genome lacks many genes that are present in both honeybee and mammals, reflecting gene loss in the dipteran lineage (Honeybee Genome Sequencing Consortium, 2006).
Animals also appear to be limited in the ability to incorporate foreign genes directly into their nuclear genomes. To date, the complete genomes of several mammals, nematodes, and insects have not revealed large numbers of foreign genes. [In fact, the initial report that the human genome contains numerous genes acquired from Bacteria (Lander et al., 2001) was later shown to be unwarranted, reflecting artifacts of analysis and limited data from eukaryotic genomes (e.g., Stanhope et al., 2001).]. So, although uptake of nonfunctional DNA does occur (e.g., Sunnucks and Hales, 1996; Kondo et al., 2002) and sometimes may result in adaptive incorporation of functional genes from exogenous sources (e.g., Mallet et al., 2003; Daimon et al., 2003), current evidence indicates that this process is limited in animals. Duplication of existing genes and regulatory changes are far more important. Among potential barriers to gene acquisition in animals are the need for regulating expression in the context of the more complex development and also the separation of germ line. In contrast to organelles (mitochondria and plastids) that arose in single-celled hosts and are present in most or all cells in modern multicellular hosts, symbionts acquired by animals are typically restricted to specialized organs and often live primarily in somatic tissues, where they may be intracellular or
extracellular. This compartmentalization may act as an obstacle to gene transfer from symbiont to host because persistent gene transfer can only occur in germ line cells.
HEREDITARY SYMBIOSIS IN ANIMALS
Before molecular methods were available, Paul Buchner and his students conducted extensive surveys of specialized symbiosis in animals; this work was summarized in a book translated into English in 1965 (Buchner, 1965). Because symbionts are mostly noncultivable under typical laboratory conditions, the approaches of Buchner and his coworkers relied primarily on microscopy to trace the diversity of associations with microbes found in different invertebrate groups, with particular attention to insects. Buchner’s central theses included the idea that symbiotic microorganisms shared long evolutionary histories with their host clades and also the premise that the main role of animal symbionts was to provide nutrients to hosts that used deficient diets. The bulk of his work was devoted to describing the complex developmental adaptations that have allowed hosts to maintain stable associations.
Of all of the groups that Buchner studied, he devoted most attention to the sap-feeding insects, some of which possess unusually elaborate symbiotic systems involving multiple microbes. This group of insects serves as an exemplar of the remarkable complexity and variety that can arise in the context of evolving symbioses. One basis of the abundance of symbiotic interactions in this group is the poor diet of most species: plant phloem sap and xylem sap are both particularly unbalanced nutritionally, lacking essential amino acids, and, in the case of xylem sap, vitamins and carbohydrates. Thus, a phloem sap- or xylem sap-feeding animal, while enjoying the advantage of a constant food supply, must collaborate with a microbial symbiont able to synthesize missing nutrients from precursors that are available.
The group of insects that includes cicadas, treehoppers, planthoppers, leafhoppers, and spittlebugs, corresponding to the suborder Auchenorrhyncha in the order Hemiptera, shows a remarkable diversity of symbiotic associations. Buchner referred to this group as “the fairy land of symbiosis,” and his student H. J. Müller studied hundreds of auchenorrhynchan species in attempting to reconstruct the evolution of this bewildering diversity of associations (summarized in Buchner, 1965). Individual insects can possess up to six symbiont types, with each symbiont transferred from mother to progeny and packaged during development by means of specialized mechanisms.
Molecular phylogenetic studies have greatly extended our understanding of the origins and evolution of animal symbioses, validating and
extending Buchner’s thesis that many of these associations have long evolutionary histories. Such studies have shown repeatedly that nutritional symbionts have evolved in parallel with their hosts, starting with studies of aphids and Buchnera and extending to whiteflies, scale insects, psyllids (Baumann, 2005), tsetse flies (Chen et al., 1999), stinkbugs (Hosokawa et al., 2006), carpenter ants (Schröder et al., 1996), and cockroaches (Lo et al., 2003).
The oldest such example of bacterial symbiosis underlying nutrition in an insect is that of Sulcia muelleri, a symbiotic clade in the bacterial phylum Bacteroidetes found in most groups of Auchenorrhyncha (Moran et al., 2005b). The Sulcia phylogeny matches that of hosts, based on current understanding, although this symbiont has been lost from numerous subclades of Auchenorrhyncha. The most plausible explanation for its occurrence is that an ancestral member of the Bacteroidetes, possibly a gut-dwelling associate, became an obligate symbiont of an insect host that was beginning to feed by sucking liquid from primitive vascular plants. This transition would have occurred by the time of the common ancestor of modern Auchenorrhyncha, implying an origin by the late Permian, at least 270 million years ago, as based on the insect fossil record (Moran et al., 2005b). Thus, a symbiotic event was critical to the emergence of one of the earliest major groups of herbivores on vascular plants and has been retained by many thousands of descendant species (Fig. 9.1).
Sulcia is typical of insect nutritional symbionts in that it inhabits the cytosol of specialized cells, grouped into a specialized host organ called the bacteriome (Fig. 9.2). In this case, this structure is a paired laterally positioned organ in the abdomen of adult insects; this structure appears to be homologous across the Auchenorrhyncha (Moran et al., 2005b). Symbionts are packaged into these specialized cell types during development, requiring specialized mechanisms of part of host and/or symbiont for limiting the location and growth of the bacteria. In aphids, cells destined to become bacteriocytes exhibit distinctive patterns of gene expression very early in development, before colonization by the symbiont population acquired from the maternal bacteriocytes (Braendle et al., 2003).
MULTIPARTITE SYMBIOSES WITHIN INSECT LINEAGES
The elaborate symbiotic systems noted by Buchner for species of Auchenorrhyncha arise from the recruitment of additional symbionts in particular sublineages. In many cases, these later additions become obligate symbionts that coexist with Sulcia. The best-studied case to date is that of the sharpshooters, a subfamily of leafhoppers containing several thousand species. Sharpshooters are distinguished from related insects in that they have adopted a xylem sap diet, imposing distinct nutritional
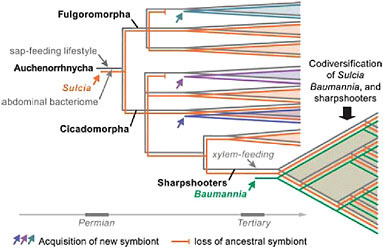
FIGURE 9.1 Schematic diagram of the evolutionary steps in the acquisition of bacterial symbionts and sap-feeding lifestyles in the insect group Auchenorrhyncha (Hemiptera), with emphasis on the sharpshooters (Cicadellinae). The bacterial symbionts S. muelleri (Bacteroidetes) and B. cicadellinicola (Gammaproteobacteria) colonized at different stages in sharpshooter evolution; they provide nutrients needed to supplement the xylem sap diet (Moran et al., 2005b; Takiya et al., 2006; Wu et al., 2006).

FIGURE 9.2 An individual sharpshooter (Cuerna sayi) dissected to reveal the bacteriomes on each side of the abdomen. These structures contain the intracellular symbionts, Sulcia and Baumannia. Photo by R. Rakitov and D. Takiya, University of Illinois.
needs, especially in the arena of vitamins and energy metabolism. They are also distinguished by the presence of a relatively restricted symbiont, Baumannia cicadellinicola, belonging to the Gammaproteobacteria and related to Buchnera (Moran et al., 2003; Takiya et al., 2006). Sharpshooters form a much younger group than the Auchenorrhyncha, with fossils not appearing until the Eocene. Phylogenetic analyses based on genes from both symbionts and insect hosts support the following evolutionary reconstruction: Sulcia was ancestrally present in a host lineage that acquired Baumannia at the same approximate time as the switch to xylem-feeding, consistent with the view that its nutrient-provisioning capabilities were a requirement for this lifestyle. After the acquisition of Baumannia, both Sulcia and Baumannia diversified in parallel with their sharpshooter hosts, through strict maternal transmission, based on the congruence of phylogenetic trees from the three clades (Takiya et al., 2006) (Fig. 9.1).
Other Auchenorrhynchan groups have recruited other bacterial and fungal symbionts, few of which have been studied beyond microscopy studies describing their morphology and transmission. Other cases of successive acquisition of symbionts are numerous, with cases documented in both aphids (Perez-Brocal et al., 2006) and weevils (Lefevre et al., 2004).
GENOME SEQUENCES ELUCIDATE COMPLEMENTARY FUNCTIONAL ROLES OF SYMBIONTS AND HOST
The nutritional role of symbionts associated with specialized organs (bacteriomes) (another of Buchner’s central theses) has been elucidated by cloning and sequencing of specific genes, complete genome sequencing, and studies of gene expression (Moran and Degnan, 2006). Thus, of four Buchnera genomes now fully sequenced, all are extremely small but nonetheless contain all or most pathways for synthesis of the amino acids that are required by animals (Shigenobu et al., 2000; van Ham et al., 2003; Perez-Brocal et al., 2006).
One genome of Sulcia has been partially sequenced, from the host species, Homalodisca coagulata (the “glassy-winged sharpshooter”) (Wu et al., 2006). As for Buchnera, Sulcia possesses a very small genome but retains pathways for synthesis of most essential amino acids, nutrients that are in short supply in both phloem and xylem sap, in both of which amino acid profiles are dominated by nonessential amino acids. A complete genome sequence for Baumannia of H. coagulata confirms that this symbiont plays a critical role in the dependence of sharpshooters on a xylem sap diet. Whereas Sulcia retains pathways for amino acid provisioning, Baumannia contains a large number of pathways for biosynthesis of vitamins (Wu et al., 2006). The complementarity between capabilities evident from the genomic sequences of the two symbionts is striking. For example,
the single essential amino acid biosynthetic pathway that is retained by Baumannia, that for histidine, appears to be the only such pathway missing from the Sulcia genome (Wu et al., 2006). The two symbionts live in close proximity within the host bacteriome and sometimes with a single Sulcia cell surrounded by closely adjacent Baumannia cells (Fig. 9.3).
GENOMIC DECAY IN OBLIGATE SYMBIONTS AND HOST DEPENDENCE
Obligate nutritional symbionts of insects provide prime examples of genome degradation in obligately host-associated bacterial lineages. These symbionts possess the smallest known genomes of cellular life forms with only 182–650 genes for fully sequenced cases (Wernegreen, 2002; Nakabachi et al., 2006; Perez-Brocal et al., 2006). In two symbiont clades, genomes of multiple representatives are available, with two for symbionts of carpenter ants [“Blochmannia” species (Degnan et al., 2005)] and four for Buchnera symbionts of aphids (Shigenobu et al., 2000; Tamas et al., 2002; van Ham et al., 2003; Perez-Brocal et al., 2006). Divergences in these two cases represent changes 30–200 million years of evolution, yet symbiont genomes show few changes. These cases are the extremes in genome
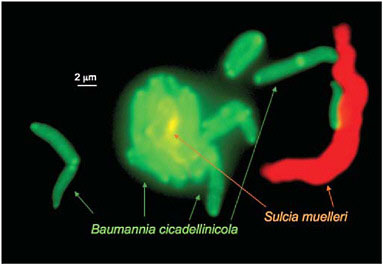
FIGURE 9.3 The two symbionts, Sulcia and Baumannia from the sharpshooter Graphocephala atropunctata. The cells are visualized by using fluorescent in situ hybridization with probes for taxon-specific 16S rRNA sequences (Wu et al., 2006). The large Sulcia cells are sometimes closely surrounded with Baumannia cells. (Photo by P. Tran and N. Moran, University of Arizona.)
stability among known genomes, lacking chromosomal rearrangements and, most significantly from the point of view of host biology, lacking any genes newly acquired from exogenous sources. The only substantial source of divergence is rapid sequence evolution affecting ancestral genes and elimination or inactivation of genes in individual lineages. The latter is of particular interest because such losses are irreversible given the lack of gene uptake; they represent loss of functions that cannot be reinstated. In several cases, eliminated genes are ones that affect host nutrition, such as those underlying pathways for sulfur fixation, arginine biosynthesis, and tryptophan biosynthesis (Tamas et al., 2002; van Ham et al., 2003; Perez-Brocal et al., 2006). The hosts must obtain these compounds from enriched diets available from certain plant species, from manipulation of plant phloem chemistry, or from additional symbionts that have been acquired subsequent to the acquisition of Buchnera >100 million years ago, as hypothesized for the smallest Buchnera genome sequenced to date (Perez-Brocal et al., 2006).
This genome degradation is not dependent on being intracellular, but rather it reflects long history of obligate host dependence and lack of recombination among strains, enforced by strict maternal transmission. The importance of population genetic structure rather than cellular location is confirmed by the observation that the symbiont, Ishikawaella capsulata, of plataspid stinkbugs (Hemiptera) shows reduced genome size and rapid protein evolution despite its location in the gut lumen rather than within cytoplasm of specialized cells (Hosokawa et al., 2006). Ishikawaella transmission, which occurs when progeny ingest an inoculum deposited on eggs by the mother, is strictly maternal, resulting in single infections and consequent lack of recombination among lineages. As for intracellular symbionts of other insect groups, Ishikawaella shows long-term parallel evolution with hosts, indicating an ancient origin (Hosokawa et al., 2006). These features of transmission enforce asexuality and small population size, as for intracellular symbionts such as Buchnera of aphids, and Ishikawaella shows similar patterns of gene and genome evolution.
The most extreme known case of degradation of a symbiont genome (other than those of organelles) occurs in Carsonella ruddii, the obligate symbiont of psyllids (a sap-feeding insect group related to aphids and whiteflies) (Baumann, 2005). This 160-kb genome contains only 182 protein-coding genes, a number considerably smaller than the proposed minimum gene number for cellular life, based on those required for essential metabolic and informational processes (Nakabachi et al., 2006). One of the most plausible explanations of how this symbiont functions with so few genes is that some genes have been stably transferred to the host genome, with their products reimported to the symbiont cellular compartment. The extent of gene transfer from symbionts to animal hosts will become appar-
ent as more genomes are sequenced for host species with long histories of symbiosis, such as aphids (Brisson and Stern, 2006). The Carsonella genome is also extreme in its base composition (16.5% GC content) and in the rate of sequence evolution of proteins; it is remarkable that the insect hosts are dependent on an organism that appears so degenerate.
Long-term coadaptation of hosts with symbionts can enforce dependence beyond the original basis for the symbiosis. For example, aphids require Buchnera for normal embryonic development and are unable to reproduce in the absence of Buchnera even when diets are supplemented with the nutrients that Buchnera normally provides.
ANIMAL SYMBIONTS RETAINING GENOME PLASTICITY
For an animal host, one potential consequence of acquiring a bacterial symbiont might be that it would serve as a portal for ongoing acquisition of novel genes, which is far more common in bacterial than in animal genomes. Although the bacteriome-associated nutritional symbionts provide the most extreme cases of genome stasis known in Bacteria and do not acquire novel genes (Tamas et al., 2002; Degnan et al., 2005), some heritable bacteria continue to undergo recombination, to harbor phage, and to incorporate foreign genes into their chromosomes. In many cases, these symbionts confer benefits such as protection against natural enemies (parasitoids and pathogens) (Oliver et al., 2005; Scarborough et al., 2005) or against variable abiotic conditions such as thermal stress (Russell and Moran, 2006). Although nutritional symbionts usually live in a specialized organ and are strictly required for normal host development, these bacteria are facultative for hosts and more varied in their locations within host bodies. Although they are maternally transmitted with high fidelity, they can also be transferred horizontally, sometimes through paternal transmission (Moran and Dunbar, 2006). As a result, different strains sometimes coinfect the same host individual, resulting in opportunity for recombination and transfer of phage and genes among strains (Moran et al., 2005a). In the case of the symbiont Hamiltonella defensa, which provides aphid hosts with protection against parasitoid wasps, phage-borne genes appear to contribute to defensive strategies that are observed to vary among symbiont strains (Oliver et al., 2005; Moran et al., 2005b). These symbionts use some of the same mechanisms for interacting with hosts as do mammalian pathogens, and many of these mechanisms are linked to capacity for gene uptake (Dale and Moran, 2006).
Mutualism is an obvious route for spread of heritable symbionts in a host population and has been the focus of this work. But heritable symbionts can spread among host lineages without conferring a benefit, by manipulating host reproduction to favor their own increase (Werren,
1997). The most well known symbionts in this category belong to the clade referred to as Wolbachia, an ancient group that contains members with a variety of kinds of interactions with hosts. In arthropods, Wolbachia is primarily exploitative, undergoes transfer among host lineages, and has a plastic genome with ongoing recombination and containing phage-derived elements (Masui et al., 2000; Wu et al., 2004). In contrast, Wolbachia in filarial nematodes appear to have been strictly vertically transmitted during host diversification, are required by hosts for normal development, and have a smaller and more static genome, lacking phage (Foster et al., 2005). Population studies indicate that exploitative symbionts can act as a force for reproductive isolation of populations with different infections (e.g., Jaenike et al., 2006). Thus, symbionts likely contribute to the species richness of hyperdiverse taxa such as the insects, not only by enabling expansion of lineages into novel ecological niches through augmentation of metabolic capabilities but also by affecting mating systems and reproductive compatibility of populations. As in the case of symbionts such as Buchnera that have evolved as beneficial symbionts, exploitative symbionts can become essential for host reproduction because of coadaptation of host genomes (e.g., Dedeine et al., 2005). Thus, complex development dependence on symbiotic partners is possible even when the original association was not beneficial for the host.
CONCLUSIONS
The literature on symbiosis is vast and growing quickly, largely because of the insights based in genomics. Although symbiosis was once discounted as an important evolutionary phenomenon (e.g., Sapp, 2004), the evidence is now overwhelming that obligate associations among microorganisms and between microorganisms and multicellular hosts have been crucial in many landmark events in evolution, in the generation of phenotypic diversity, and in the origin of complex phenotypes able to colonize new environments. Such evidence is abundant for the symbiotic systems found in insects, which are far better understood than in the recent past, largely because of molecular and genomic studies. Examples from insects show that symbioses can result in specialized organs with unique development, innovations in metabolic capabilities that allow new lifestyles, defenses against natural enemies and other environmental challenges, constraints on evolutionary range, and ongoing acquisition of novel genes and capabilities.
ACKNOWLEDGMENTS
I thank Francisco Ayala and John Avise for organizing this conference, Howard Ochman for critical comments on a draft of the manuscript, Daniela Takiya and Roman Rakitov for use of their photograph, Becky Nankivell for preparation of the manuscript and Fig. 9.1, and two reviewers for helpful comments. The work on symbionts of aphids and sharpshooters was supported by National Science Foundation Awards 0313737 and 0626716.


















