4
Cascades of Convergent Evolution: The Corresponding Evolutionary Histories of Euglenozoans and Dinoflagellates
JULIUS LUKEŠ,* BRIAN S. LEANDER,† and PATRICK J. KEELING†
The majority of eukaryotic diversity is hidden in protists, yet our current knowledge of processes and structures in the eukaryotic cell is almost exclusively derived from multicellular organisms. The increasing sensitivity of molecular methods and growing interest in microeukaryotes has only recently demonstrated that many features so far considered to be universal for eukaryotes actually exist in strikingly different versions. In other words, during their long evolutionary histories, protists have solved general biological problems in many more ways than previously appreciated. Interestingly, some groups have broken more rules than others, and the Euglenozoa and the Alveolata stand out in this respect. A review of the numerous odd features in these 2 groups allows us to draw attention to the high level of convergent evolution in protists, which perhaps reflects the limits that certain features can be altered. Moreover, the appearance of one deviation in an ancestor can constrain the set of possible downstream deviations in its descendents, so features that might be independent functionally can still be evolutionarily linked. What functional advantage
may be conferred by the excessive complexity of euglenozoan and alveolate gene expression, organellar genome structure, and RNA editing and processing has been thoroughly debated, but we suggest these are more likely the products of constructive neutral evolution, and as such do not necessarily confer any selective advantage at all.
The vast majority of eukaryotes on the planet, in terms of both abundance and diversity, are microbial. Generalities about fundamental biological processes are based on knowledge of a few model organisms, yet many microeukaryotes have deviated well beyond these generalities over the course of evolutionary history, which is a reflection of the deep phylogenetic distances between eukaryotic lineages that are neither plants nor animals nor fungi. It is also clear that some groups of protists have broken more rules than others, and 2 diverse lineages that particularly stand out in this regard are the Euglenozoa and the Alveolata (Fig. 4.1). In members of both of these groups, fundamental structures and processes have substantially deviated from those of other eukaryotes; however, perhaps even more interestingly, both groups have frequently departed in the same general fashion, resulting in surprising levels of convergence that suggest limits to the ways these features can be altered.
The Euglenozoa is a monophyletic group within the Excavata consisting of single-celled flagellates composed of 2 major subgroups (kinetoplastids and euglenids), and 1 smaller subgroup (diplonemids) (Fig. 4.1). Members of Euglenozoa have diverse modes of nutrition, including predation, parasitism, and photoautotrophy. Predatory euglenozoans are phylogenetically widespread within the group and tend to have diverse feeding apparatuses, feeding strategies, and prey preferences (Leander et al., 2007). For instance, some predatory species are limited to small prey such as bacteria, whereas other species frequently consume larger prey, such as other eukaryotic cells. Photoautotrophy is restricted to a specific subclade of euglenids and originated via secondary endosymbiosis between a predatory euglenid and a green algal prey (Leander et al., 2007). Parasitic and commensalic euglenozoans appear to have evolved independently several times within kinetoplastids (Simpson et al., 2006), and some species (e.g., Trypanosoma and Leishmania) cause important human illnesses such as African sleeping sickness, Chagas’s disease, and leishmaniases.
The Alveolata, another monophyletic group of primarily single-celled eukaryotes that have adopted similarly diverse modes of life, is composed of 3 major subgroups: ciliates, apicomplexans, and dinoflagellates (Fig. 4.1). All 3 subgroups contain predatory and parasitic species, and only dinoflagellates and an unusual lineage called Chromera are known
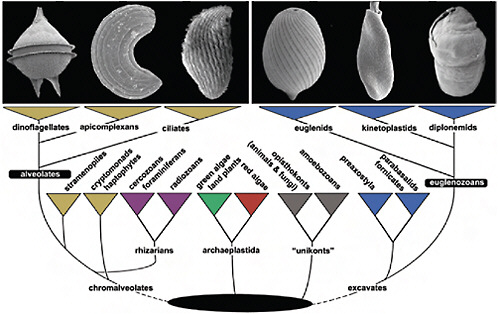
FIGURE 4.1 The tree of eukaryotes showing the relative positions of Alveolata and Euglenozoa. This tree is a reflection of many kinds of evidence that are summarized in Keeling et al. (2005), Hampl et al. (2009), and Keeling (2009). The Alveolata consists of 3 main groups, dinoflagellates, apicomplexans, and ciliates (illustrated in Upper Left; scanning electron micrographs of Protoperidinium, Selenidium, and an unidentified ciliate), whereas the Euglenozoa consists of euglenids, kinetoplastids, and diplonemids (illustrated in Upper Right; scanning electron micrographs of Lepocinclis, Leptomonas, and Diplonema). Alveolates are members of the supergroup Chromalveolata, whereas eugenozoans are members of a different supergroup, the Excavata. Neither supergroup is without controversy (e.g., the rhizarian supergroup is shown within chromalveolates to reflect recent phylogenetic analyses), but the relationships of alveolates and euglenozoans to some other eukaryotic groups to the exclusion of one another, and the relationships within alveolates and euglenozoans, are both consistently well supported by molecular analyses.
to contain fully integrated and photosynthetic plastids (Oborník et al., 2009). Photosynthetic dinoflagellates play important roles as planktonic primary producers in oceanic ecosystems, and some of the lineages form symbiotic relationships with corals (e.g., Symbiodinium) and are critical for maintaining the health of reef systems around the world. Nonphotosynthetic plastids have independently evolved in some dinoflagellates and in apicomplexans, which are all obligate parasites of animals and a few are exceedingly important disease organisms of vertebrates (e.g., Cryptosporidium, Toxoplasma, and Plasmodium). Although plastids have not been definitively demonstrated in ciliates, several independent lineages
in this group harbor photosynthetic symbionts that are intermittently replenished by feeding.
Both euglenozoans and alveolates have a reputation for “doing things their own way,” which is to say that they have developed seemingly unique ways to build important cellular structures or carry out molecular tasks critical for their survival. Why such hotspots for the evolution of novel solutions to problems should exist in the tree of life is not entirely clear. However, the deeper we look into these groups, the more often it is found that they are also evolving strikingly similar mechanisms for achieving these essential biological functions. Significantly, however, there is a great weight of phylogenetic data that show these lineages are not closely related: of the 5 eukaryotic supergroups hypothesized to explain all eukaryotic diversity, alveolates and euglenozoans fall into 2 different supergroups, chromalveolates and excavates, respectively (Fig. 4.1). The support for these supergroups as a whole remains contentious (Keeling et al., 2005; Leander, 2008; Hampl et al., 2009; Keeling, 2009), but there is strong support from phylogenomics and many individual phylogenies and rare genomic characters for a specific relationship between alveolates and stramenopiles on one hand, and euglenozoans and heteroloboseans on the other hand (Hampl et al., 2009). Moreover, no analysis of eukaryotic phylogeny has ever suggested they are closely related to one another. Still more significantly, the majority of the characteristics we discuss below are not universal to all members of either alveolates or euglenozoans, but rather appear to have evolved within a subgroup of each lineage. Altogether, the distribution of these characteristics can really only adequately be explained by convergent evolution. Below, we will examine some of these examples of convergence and what the cooccurrence of convergent traits may tell us about how they evolved.
CONVERGENT EVOLUTION
Recognizing the independent origins of similar traits in distantly related lineages—convergent evolution—allows us to better understand how different environmental and intrinsic conditions have shaped the characteristics of organisms over time; each specific example of convergence reflects a fundamental biological problem and its possible solutions. The causes of convergent evolution are varied and can involve camouflage, mimicry, biomechanical optimization, molecular constraints, developmental canalization, and character-state reversals. Examples of convergent evolution range from the biochemical level to the behavioral level and are best characterized within animals and land plants (Conway Morris and Gould, 1998; Zakon, 2002; Emery and Clayton, 2004; Arndt and Reznick, 2008), which collectively represent only a small portion of the full
tree of eukaryotes (Keeling et al., 2005; Leander, 2008). The occurrence and adaptive significance of convergent evolution in microbial eukaryotes, by contrast, is poorly understood, but it is clear from several examples that convergent traits can evolve over vast phylogenetic distances (Leander, 2008). Convergence in very distantly related lineages is particularly compelling because the influence of homologous developmental programs (i.e., intrinsic conditions) in constraining subsequent evolution should be minimal if not absent altogether (Leander, 2008). Therefore, improved understanding of convergent evolution in distantly related microbes will provide a much broader framework for evaluating the forces of natural selection and the potential role of constructive neutrality during the evolution of ultrastructural systems and complex molecular processes.
Eukaryotic cells are built from a few core systems that have become tremendously diverse over the course of evolutionary history. Some systems are remarkably conserved, in particular fundamental molecular processes such as information flow or core metabolism, but even in these systems substantial modifications accumulated in some lineages. In other cases, conserved ancestral building blocks (such as the proteinaceous cytoskeleton involved in locomotion and feeding) are widely shared, but have been used in different ways with diverse outcomes. The origins of other components are less clear and likely more recent, but also show a great deal of morphological variation (examples include photoreception systems or surface armor). Taken together, the diversity of cellular and molecular systems in microbial eukaryotes is simply staggering, and some emerging patterns indicate that convergent evolution played a major role in shaping the overall organization of eukaryotic cells at all levels (Arndt and Reznick, 2008; Leander, 2008).
Below, several features will be described, for which an excessive complexity is a common denominator. This is counterintuitive in single-celled organisms, especially when selective advantages for these complex structures and/or mechanisms remain elusive. We argue that the theory of constructive neutral evolution (Stoltzfus, 1999), which invokes nonselective factors such as excess capacities, can best account for their emergence.
CELLULAR ORGANIZATION OF EUGLENIDS AND DINOFLAGELLATES
The comparable combinations of ultrastructural features in euglenozoans and alveolates have been appreciated for decades (Taylor, 1987; Bouck and Ngo, 1996). For instance, the cells of benthic predatory species of euglenids and dinoflagellates are streamlined and dorsoventrally flattened and possess batteries of extrusive organelles, or extrusomes, that are similar in morphology and behavior (Fig. 4.2). The mucocysts of euglenids
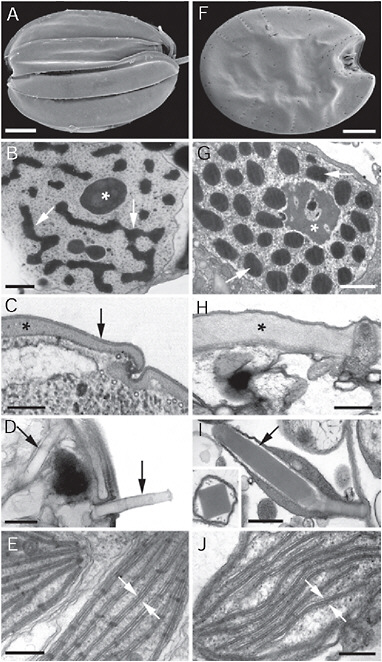
FIGURE 4.2 Electron micrographs showing convergent ultrastructural features in some euglenozoans (A–E) and dinoflagellates (F–J). (A) Scanning electron micrograph (SEM) of a benthic euglenid (Ploeotia) showing a dorsoventrally compressed cell with broad longitudinallyarranged pellicle strips. (B) Transmission electron micrograph (TEM) through the nucleus of a euglenid cell (Phacus) showing a large nucleolus (asterisk) and permanently condensed chromosomes (arrows). (C) TEM through the cell surface or pellicle of a euglenid cell showing the proteinaceous strips (asterisk) beneath the plasma membrane (arrow). (D) TEM through the cell surface of a euglenid cell (Peranema) showing mucocysts (arrows). (E) TEM through a euglenophyte cell (Euglena) showing chloroplasts with thylacoids in stacks of 3 (stack between the arrows). (F) SEM of a benthic dinoflagellate (Prorocentrum) showing a dorsoventrally compressed cell with broad thecal plates. (G) TEM through the nucleus of a dinoflagellate cell (Apicoporus) showing a large nucleolus (asterisk) and permanently condensed chromosomes (arrows). (H) TEM through the cell surface of a dinoflagellate cell (Apicoporus) showing the proteinaceous “dinoflagellate pellicle” (asterisk) that sits beneath the alveoli and plasma membrane (both missing in this preparation). (I) TEM through the cell surface of a dinoflagellate cell (Polykrikos) showing trichocysts (arrows). (Inset) TEM showing a cross section through a trichocyst of Polykrikos. (J) TEM through a dinoflagellate cell (Apicoporus) showing chloroplasts with thylacoids in stacks of 3 (stack between the arrows). (Scale bars: A and F, 10 μm; B and G, 1 μm; C, H, and J, 0.3 μm; D, E, and I, and 0.5 μm.)
and the trichocysts of dinoflagellates are compact, linear bodies containing a highly organized latticed framework of carbohydrates. When these bodies are released through discrete pores through the surface of the cell, the extrusomes become hydrated and rapidly extend in length as spear-like threads (Fig. 4.2D and I) (Hausmann, 1978). Although the origin and function of extrusomes in both groups are not clear, they probably play a role in escape responses, defense, and capturing prey cells.
Benthic euglenids and dinoflagellates, in particular, adhere to substrates and are capable of gliding motility using 2 heterodynamic flagella equipped with flagellar hairs (or mastigonemes). In both groups, the recurrent flagellum sits within a groove on the ventral surface of the cell and is oriented backward. Euglenids and dinoflagellates also possess cytoskeletal elements (called “paraxial/paraflagellar rods,” which run in parallel to the 9 + 2 microtubular axonemes within each flagellum) that are not found in any other group of eukaryotes. A major difference between euglenids and dinoflagellates, however, is the structure, orientation, and motility of the anterior flagellum. The anterior paraxial rod in euglenids is oriented on the ventral side of the axoneme, is stiff and held straight in front of the cell; the paraxial rod functions with the flagellar hairs to produce gliding forces (Saito et al., 2003). By contrast, the anterior flagellum of dinoflagellates forms a transverse loop or spiral around the circumference of the cell and usually sits within a transverse groove called the cingulum (Fig. 4.1). The coiled transverse flagellum bears hairs and a flagellar membrane that connects it to the base of the cingulum, and this entire apparatus is capable of producing forces on the surrounding medium that tend to spin the cell around its longitudinal axis.
Many free-living euglenids and dinoflagellates engulf prey organisms using sophisticated feeding apparatuses positioned on the ventral side of the cell. Although the evolution of these apparatuses is a shared fea-
ture, the details of these ultrastructural systems are quite distinctive. For instance, there are a few chief components present in most of the predatory species of euglenids described so far, namely “rods” and “vanes.” Two feeding rods oriented longitudinally within the cell are composed of microtubules and amorphous proteinaceous material. These stiff elements provide structural support for gripping and internalizing prey cells and work in concert with 4–5 membranous vanes that are usually reinforced with additional microtubules (Leander et al., 2007). The vanes originate from the rods, form the inside core of the feeding apparatus, and create space within the apparatus by opening up in a pinwheel-like fashion; the same mechanism can cause the apparatus to protrude from the cell when feeding. By contrast, the diversity and complexity of feeding apparatuses in dinoflagellates probably reflect independent origins in different lineages within the group. The feeding apparatus in dinoflagellates can be simple pockets that unzip when prey is drawn into the cell, dynamic siphons that suck out the cytoplasm of prey cells in a straw-like fashion or expansive veils that completely envelop large filamentous prey and fold it methodically into manageable packets small enough to ingest. Different kinds of feeding apparatuses are often associated with different kinds of photoreceptive eyespots and ocelloids, suggesting that in some dinoflagellates, photoreceptors are adaptations for detecting and capturing photosynthetic prey. Some predatory euglenids with a rod-and-vane feeding apparatus also possess a photoreceptor system, as a putative stigma and photosensory swelling (Leander et al., 2001), and this combination of features may serve the same basic function as in dinoflagellates.
Another convergent similarity between benthic euglenids and dinoflagellates is the tendency to reinforce their cell surfaces with robust proteinaceous layers beneath the plasma membrane (Fig. 4.2C and H). Euglenids possess a distinctive (and synapomorphic) pellicle consisting of discontinuous strips that run longitudinally or helically over the entire cell surface (Leander et al., 2007). The strips articulate along their lateral margins, and in many euglenids these zones facilitate sliding between strips that produce rhythmic deformations in cell shape, called “euglenoid movement.” Benthic dinoflagellates can also change their shape, especially after engulfing large and oddly shaped prey cells. The proteinaceous surface layer in dinoflagellates, called the “dinoflagellate pellicle” forms a continuous and flexible sheath beneath alveolar vesicles, which may in turn be filled with cellulosic material. Both the euglenid and the dinoflagellate pellicles comprise novel classes of proteins: articulins and epiplasmins (Bouck and Ngo, 1996). Although it is unclear whether these proteins represent an example of molecular convergence or distant homology, their presence in both euglenids and dinoflagellates underscores the striking similarities between these 2 very distantly related groups of eukaryotes.
At different points in their evolutionary history, both euglenids and dinoflagellates independently acquired photosynthesis via secondary endosymbiosis. Accordingly, some representatives of both groups contain at least 3 different genomes within 3 different cellular compartments: the nucleus, the plastid, and the mitochondrion. The general organization of the nucleus is a particularly notable feature that is shared by euglenids and dinoflagellates; both groups possess a conspicuous nucleus with a relatively large nucleolus and permanently condensed chromosomes (Fig. 4.2B and G). The plastids in both groups also share the unusual features of 3 envelope membranes and a tendency to have thylacoids in stacks of 3 (Fig. 4.2E and J) (Taylor, 1987). However, the analogous similarities between euglenozoans and dinoflagellates do not end at the ultrastructural level. As described in the next 3 sections, the molecular processes associated with the nucleus, plastid, and mitochondrion also reflect high levels of convergent evolution.
THE NUCLEUS: SPLICED LEADERS AND POLYCISTRONIC mRNA PROCESSING
The nuclear genomes of kinetoplastids and dinoflagellates have both acquired a long list of unusual characteristics. Some of these are unique to one lineage and very different in the other. For example, dinoflagellates have among the largest nuclear genomes known, and these genomes have a very low gene density and permanently condensed chromosomes that lack nucleosomes (McEwan et al., 2008). Kinetoplastid genomes, however, are relatively small, are gene-dense, and remain uncondensed during the cell cycle (Berriman et al., 2005). Both genomes are notorious for their rich representation of modified nucleotides, but the nucleotides themselves are not the same: the hypermodified base J (β-D-glucopyranosyloxymethyluracil) is common in kinetoplastid telomeric regions, whereas dinoflagellates have a high proportion of 5-hydroxymethyluracil and 5-methylcytosine.
However, other dramatic alterations to these genomes have taken place convergently, and interestingly, several characteristics have been altered in the same way in both lineages, in particular relating to how genes are arranged and transcribed, and how transcripts are processed. The canonical, simplified view of eukaryotic gene expression involves a single gene transcribed, capped, polyadenylated, spliced (if introns are present), and exported to the cytosol. Both kinetoplastids and dinoflagellates deviate from this canonical view in 2 significant ways that impact the way expression may be controlled.
The first of these is trans-splicing. The spliceosome is a large multisubunit complex that normally recognizes GT-AG bounded spliceosomal
introns within eukaryotic genes, and catalyzes their removal and the ligation of the flanking exons. Spliceosomal introns are very rare in trypanosomes (Berriman et al., 2005), and available evidence suggests they are relatively so in dinoflagellates as well (Bachvaroff and Place, 2008). In contrast, every mRNA in both groups has a 5′ spliced leader (SL) sequence that is added by trans-splicing. The SL, also called a miniexon, is a short conserved sequence that is encoded by a high-copy-number family of genes throughout the genome. In dinoflagellates, the same 22-bp fragment is added to all transcripts, and the sequence is also conserved across the entire group (Zhang et al., 2007; Slamovits and Keeling, 2008). In kinetoplastids, the SLs are conserved within a given genome, but vary in size and sequence between species (Campbell et al., 2003). The SL is expressed as a short RNA consisting of the leader sequence followed by a GT dinucleotide and a short stretch of sequence. Complimenting this, mRNAs for protein-coding genes begin with a short stretch of sequence ending with an AG dinucleotide, followed by the 5′ untranslated region and the coding region. The spliceosome brings these 2 elements together and mediates the removal of the 2 intronic fragments and ligation of the SL to the 5′ end of the mRNA (Fig. 4.3) (Campbell et al., 2003).
The second major oddity shared by kinetoplastid and dinoflagellate nuclear gene expression is the presence of polycistronic messages. Once again, the canonical view of nuclear gene expression in eukaryotes centers around the transcription of a single gene at a time; this stands in contrast to prokaryotes, where multiple genes can be expressed on a single, multifunctional mRNA and many genes can be coregulated in operons. Complete genomic sequences from trypanosomatids demonstrate an organization where genes are distributed in contiguous clusters, ranging in size from a handful of genes to several hundreds. In these clusters, stretching up to >1 Mb, genes are oriented on the same strand, usually toward the telomeres, with adjacent clusters located on opposite strands (Berriman et al., 2005). All of the genes within a contiguous cluster are transcribed on a single, sometimes very long, polycistronic mRNA. Relatively short AT-rich regions separate the clusters and are considered to contain the sites for transcription initiation and termination. Comparison of trypanosomatid genomes shows a high degree of conservation in gene order, even within clusters between flagellates that diverged 200–500 Mya (Ghedin et al., 2004).
It is important to point out that, in contrast to prokaryotes, these clusters do not contain genes of related function (Berriman et al., 2005) and they are not coordinately regulated like bacterial operons, so they should not be considered operons. These polycistronic messages are not even translated intact, but are processed to monomeric mRNAs before translation; these monomeric mRNAs are the substrate for trans-splicing by the
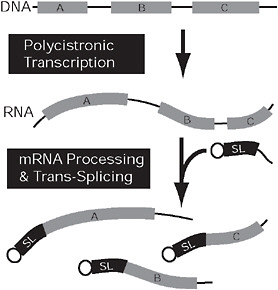
FIGURE 4.3 Convergent nuclear gene expression in kinetoplastids and dinoflagellates. In both kinetoplastids and dinoflagellates, genes (gray) may be arranged in tandem arrays (Top) that are expressed on a single polycistronic mRNA (Middle). This mRNA is broken into multiple monocistronic mRNAs concomitantly with trans-splicing, which adds a short, capped spliced leader sequence (black) to the 5′ end of all monocistronic mRNAs. In kinetoplastids, the genes (A–C) are arranged randomly and are functionally unrelated, but, in dinoflagellates, the coding sequences on a single polycistronic mRNA are tandem repeats of 1 gene.
addition, at the 5′ end, of an SL already equipped with a methylated cap, followed by the polyadenylation at the 3′ end (Campbell et al., 2003).
Far less is known about the organization of dinoflagellate genomes. Due to the enormous size of their nuclear DNA, nearly all sequencing of dinoflagellate genes performed to date has focused on expressed sequence tags, which do not provide information on the context of the gene. Nevertheless, what little is known about dinoflagellate genomes suggests a fascinating parallel with kinetoplastids. It now appears that some genes are isolated in the genome, but others are organized as tandem repeats (Bachvaroff and Place, 2008). These gene repeats are cotranscribed, resulting in polycistronic messages, and different from those of kinetoplastids because mRNAs have so far only been found to carry multiple copies of a single gene (Bachvaroff and Place, 2008). These transcripts are apparently processed into monocistronic mRNAs, which are presumably the substrates for trans-splicing.
In kinetoplastids, the presence of polycistronic mRNAs, together with the absence of introns, is frequently argued to be an ancient holdover,
frozen since their early divergence from other eukaryotes (Gunzl et al., 2007). However, this interpretation is flawed for several reasons, particularly because there is no evidence whatsoever for an ancient divergence of kinetoplastids (Keeling et al., 2005). Nonetheless, the independent origin of the same features in dinoflagellates raises an intriguing alternative explanation, namely that the evolutionary origins of polycistronic mRNAs and trans-splicing are linked. This is all the more compelling when one considers that both features are also found together in the nematode Caenorhabditis elegans (Graber et al., 2007). It is unlikely that this is either functionally advantageous or an evolutionary relict, but rather that the evolution of one feature preconditions the genome by removing deleterious effects of the second feature. For example, the establishment of widespread SL addition in a nuclear genome could precondition that genome for the subsequent establishment of polycistronic transcription. Polycistronic mRNAs that would otherwise be deleterious could flourish simply because the processing pathway eliminates their deleterious effect (the inability to translate all but the first cistron). SL addition appears to be universal in both dinoflagellates and kinetoplastids [in C. elegans 70% of mature mRNAs are produced through trans-splicing: Graber et al. (2007)]. Polycistronic messages, however, are also near universal in kinetoplastids, whereas in dinoflagellates (and C. elegans) only a subset of genes are expressed on polycistonic mRNAs (Bachvaroff and Place, 2008). Since so far only tandem duplications of closely related copies of the same gene are known in dinoflagellates, it would appear they may arise and dissolve continuously.
The functional impacts of SL addition and polycistronic transcription are also different in the 2 lineages. Posttranslational control may be somewhat restricted by the absence of sequence diversity at the 5′ end of mRNAs, but more importantly a heavy use of polycistronic messages eliminates the possibility of transcription-level differentiation of expression of any genes within the same cluster. In kinetoplastids, there is only a handful of promoters and a marked paucity of transcription factors (Gunzl et al., 2007), unavoidably leading to the general lack of control over transcription initiation. Indeed, in the well-studied T. brucei, virtually all nuclear DNA seems to be permanently transcribed. Consequently, control levels in kinetoplastids are confined to RNA processing, export, and half-life, as well as translation and protein stability (Clayton, 2002). This is a good illustration of how convergent processes differ in the details in different lineages. In this case, the kinetoplastids cotranscribe many different genes whereas dinoflagellates cotranscribe many copies of the same gene, and as a result, transcription-level control is likely not so severely affected in the latter group.
THE PLASTID: THREE MEMBRANE PLASTIDS AND UNIQUE TARGETING SYSTEM
Plastids are known in both alveolates and euglenozoans to have been derived from secondary endosymbiosis: the uptake of a eukaryotic alga by another eukaryote. In the Euglenozoa, plastids are derived from a green alga and are relatively restricted, being found in a subset of euglenids and nowhere else, namely the “euglenophytes” (Leander, 2004). In the Alveolata, plastids are derived from a red alga and are more widespread and ancient, being known in dinoflagellates and apicomplexans, and suspected of originating before the divergence of alveolates (Keeling, 2009). As with nuclear genomes, plastids have evolved a number of unusual characteristics, some unique and some arising convergently. Euglenophyte plastid genomes are home to some unique self-splicing introns (Copertino and Hallick, 1993), whereas the dinoflagellate plastid genome has been massively reduced in coding content and broken into single gene minicircles with polyuridylylated transcripts (Wang and Morse, 2006). Curiously, both features are also found in kinetoplastid mitochondria (Lukeš et al., 2005).
Once again, however, 2 probably interconnected features have arisen in both groups. The vast majority of secondary plastids are bounded by 4 membranes. Most proteins in these plastids are encoded in the nucleus and are posttranslationally targeted to the organelle by way of a 2-part pathway beginning with the endomembrane system and followed by the original primary plastid targeting system. In dinoflagellates and euglenophytes, however, the plastid is novel in that it is bounded by 3 membranes rather than 4. It was argued that this may reflect a different mechanism of plastid uptake, specifically that in these lineages plastids arose through myzocytosis whereas other secondary plastids arose through endocytosis. Myzocytosis is a mode of predation where a cell pierces its prey and sucks the prey cytoplasm directly into a digestive vacuole, leaving the prey wall and membrane behind. Although not as common as endocytosis of whole prey cells, myzocytosis is known in both dinoflagellates and euglenozoans, leading to the suggestion that their plastids originated from a myzocytosed alga, and therefore lacked its plasma membrane (Schnepf and Deichgraber, 1984). However, plastids in the closest relatives of dinoflagellates, apicomplexans and Chromera, are bounded by 4 membranes and have now been shown to be orthologous to the dinoflagellate plastid (Oborník et al., 2009). Accordingly, in at least dinoflagellates, plastids must have originated in the same fashion as some 4-membrane counterparts and at one time been bounded by 4 membranes, which means that the origins of 3 membranes around the plastids in dinoflagellates and euglenophytes cannot be attributed to a shared, unusual mechanism such as myzocytosis.
Interestingly, the system used to target proteins to 3 membrane plastids is also different in subtle but important ways to that of canonical secondary plastids with 4 enveloping membranes, and the same variations have been adopted in dinoflagellates and euglenophytes. The N-terminal leaders that direct proteins to canonical secondary plastids include a signal peptide (to enter the endomembrane system) and a transit peptide (to cross the 2 plastid membranes), and are similar in secondarily derived red and green plastids. In dinoflagellates and euglenophytes, however, an additional hydrophobic domain is found following the transit peptide of some, but intriguingly not all, proteins (Patron et al., 2005; Durnford and Gray, 2006). This domain is thought to anchor the proteins in the endomembrane, so as the protein moves through the Golgi apparatus the leader lays in the lumen but the mature protein remains in the cytosol (Sulli et al., 1999; Nassoury et al., 2003). The number of membranes and these unusual characteristics of targeting have both evolved convergently in dinoflagellates and euglenophytes, which suggests some link in how these 2 features evolved. Unfortunately, the mechanism by which proteins cross the membrane that is missing in both dinoflagellates and euglenophytes (the plasma membrane of the engulfed alga) is the most poorly understood step in the targeting pathway to canonical secondary plastids, so any specific model for preconditioning would be highly speculative.
THE MITOCHONDRION: RNA EDITING AND GENOME BREAKDOWN
The mitochondrial genomes of dinoflagellates and kinetoplastids are both highly unorthodox, and once again have evolved some unique features and several common complex characteristics. The kinetoplastid mitochondrion contains uniquely structured, protein-rich mitochondrial ribosomes with a reduced RNA component, unusual fatty acid synthesis and respiratory complexes such as the prokaryotic-like complex I, alternative terminal oxidase, massive tRNA import, and incomplete Krebs cycle. The complex genome of the kinetoplastid mitochondrion is known as kinetoplast DNA or kDNA, its genes being subjected to unprecedented levels of RNA editing (Fig. 4.4) (Lukeš et al., 2005). Dinoflagellate mitochondria have received far less attention, but it is now emerging that their genomes have also evolved a number of highly unusual characteristics, including trans-splicing, tRNA import, fragmented rRNAs, the loss of start and stop codons, and an oligouridine tail (Slamovits et al., 2007; Nash et al., 2008). Most strikingly, however, the structure of dinoflagellate mitochondrial genomes has also broken down into many fragments, the transcripts of which have high levels of RNA editing; however, as we discuss below, the details of both systems differ between kinetoplastids and dinoflagellates (Fig. 4.4).
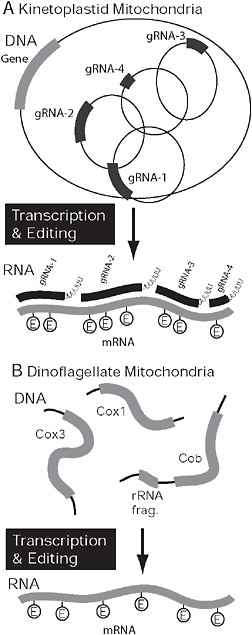
FIGURE 4.4 Covergent organization and processing in kinetoplastid and dinoflagellate mitochondria. (A) In kinetoplastids the genome is composed of maxi- and minicircles. mRNAs expressed from maxicircles (gray) require extensive RNA editing, specifically the insertion and deletion of uridine residues. The process is mediated by several mutiprotein complexes, but the information comes from small gRNAs (black), that are encoded on minicircles. In dinoflagellates, the genome is also fragmented but into multiple potentially linear pieces encoding 1 or more genes or gene fragments. Only 3 genes are encoded in the genome (cox1, cox3, and cob), and mRNAs are subject to extensive RNA editing. In this case, however, bases are substituted rather than inserted or deleted.
Within the Euglenozoa as a whole, mitochondrial genomes are generally odd. The euglenid mitochondrial genomes are experimentally refractive and remain poorly known (M. W. Gray, personal communication). The mitochondrion of related diplonemids was recently shown to harbor genomes of unprecedented organization, with fragments of genes residing on minicircles, which are assembled in the correct order posttranscriptionally by means as yet unknown (Marande et al., 2005). Virtually nothing is known about the form or content of the giant kDNAs in bodonid flagellates, which are estimated to comprise millions of base pairs (Lukeš et al., 1998), whereas the kDNA networks of trypanosomatids are among the best studied and most complex mitochondrial genomes known. They are composed of circular DNA molecules that are relaxed and catenated into a single 3-dimensional network. These networks are composed of dozens of maxicircles, which are equivalents of classical mitochondrial genome, and thousands of minicircles (Lukeš et al., 2005) involved in editing, discussed below. The gene content of the maxicircle genome is not unusual, except for the complete absence of tRNA genes. tRNAs have been demonstrated to be imported from the cytosol into the tRNA-lacking organelle of T. brucei, so that the prokaryotic translation system of the mitochondrion must cope with imported eukaryotic tRNAs (Crausaz-Esseiva et al., 2004). The only exception is tRNAMet-i, the import of which is blocked because it cannot function in the prokaryotic system. Instead, tRNAMet formyl-transferase is present, which formylates the translation initiator tRNAMet-e upon import (Tan et al., 2002).
Within alveolates, mitochondrial genome evolution has also taken more than its share of strange turns. Although the circular mitochondrial genome of ciliates is undistinguished in both form and content, the genomes in apicomplexans and dinoflagellates are both highly reduced and often scrambled (Feagin, 2000; Slamovits et al., 2007; Nash et al., 2008). These lineages have the smallest mitochondrial genomes known, with most species examined with just 3 protein-coding genes: cox1, cox3, and cob (strictly speaking, the dinoflagellate Oxyrrhis has only 2 genes since cob and cox3 are expressed as a fusion) (Slamovits et al., 2007). The only other coding regions are small fragments of rRNAs. These do not amount to an entire copy of either large or small subunit rRNAs, so fragments are all thought to be important and the functional RNAs assembled by base pairing interactions. As with kinetoplastids, no tRNAs are encoded in these genomes, and they have been shown to be imported into apicomplexan mitochondria. Moreover, apicomplexans also block the import of tRNAMet-i, and use tRNAMet formyl-transferase to formylate the translation initiator tRNAMet-e. Indeed, kinetoplastids and apicomplexans have independently evolved very similar tRNA import mechanisms to cope with this unique lack of tRNAs (Bouzaidi-Tiali et al., 2007). In apicomplexans,
the 3 protein-coding genes map to a linear, tandem repeat with rRNA fragments interspersed (Feagin, 2000). In dinoflagellates, the same coding regions are present, but the organization is much more complex. Here, multiple copies of each gene are found in various orientations on linear chromosomes of varying size. In some species, all possible permutations of 3 genes are adjacent, whereas in others chromosomes seem to contain copies of only 1 gene. Chromosomes also contain rRNA fragments, and substantial noncoding regions, and some have been shown to have structurally complex ends characterized by families of repeats (Jackson et al., 2007; Slamovits et al., 2007; Nash et al., 2008).
In kinetoplastids, the evolution of the complex genome organization is tightly linked to how genes are expressed, and specifically to RNA editing. The genes, such as they are, are encoded on the maxicircles and expressed as polycistronic mRNAs, but after processing into monocistrons these messages are then massively altered by the insertion and deletion of uridine residues (up to 553 insertions and 89 deletions in a single mRNA). Editing is mediated by hundreds of small guide (g) RNAs in an elaborate process involving numerous multisubunit protein complexes (Lukeš et al., 2005; Hashimi et al., 2008). The gRNAs that contain the information that directs editing are encoded on the minicircles, so the breakup of the genome into 2 chromosome types is likely linked to the evolution of editing.
In dinoflagellates, RNA editing has also been found to be widespread, but the process is mechanistically different and in no way related to the breakdown of the genome structure. Here, transcripts are edited at ≈2% of their positions via substitutional editing, as opposed to insertion/deletion editing (Lin et al., 2002; Nash et al., 2008; Zhang and Lin, 2008). Although A to G is the most common substitution, several others have been observed (U → C, G → C, G → A, A → C, and C → U), suggesting a highly flexible and sophisticated editing mechanism (Nash et al., 2008; Zhang and Lin, 2008). Fragments of edited gene sequences have been found in dinoflagellate mitochondrial genomes, prompting the suggestion that they employ gRNAs similar to that of kinetoplastids (Nash et al., 2007). However, the genomes are prone to recombination, so the significance of these fragments remains unclear; overall, there is no direct evidence for any particular editing mechanism at present. It is worth noting that mitochondrial transcripts in dinoflagellates have substantial polyadenylated tails, a feature linked to the editing process in kinetoplastids (Etheridge et al., 2008), and generally very rare in organelles.
The limited data further indicate that uridine insertion type of RNA editing might even coexist with trans-splicing in diplonemids (Marande and Burger, 2007). We predict that the extreme diversity of editing types documented in the dinoflagellate mitochondrion (Zhang and Lin, 2008) also requires poorly understood albeit complex protein machinery that is
the result of constructive neutrality similar to that described above for the kinetoplastid mitochondrion.
CONCLUSIONS
The deeper we look at protist biology, the greater the variety we discover in how cells can accomplish fundamental processes. Not only do protists represent the majority of the phylogenetic tree of eukaryotes, and therefore the greatest evolutionary diversity, but they also have pushed the limits of many biological systems and bending the “rules” of biology (such as the central dogma) far beyond what we see in the better studied multicellular eukaryotes. The alveolates and the euglenozoans may be “hotspots” for the generation of diverse solutions to fundamental processes, but it is also possible that they only appear this way because they are among the best studied protist groups. Other odd protists abound, but we know next to nothing about many of them, particularly at the molecular and genomics levels. All this is presently changing, and to interpret genomic diversity in eukaryotes we will have to set aside many of our preconceptions.
Comparing the alveolates and the euglenozoans is also appealing because they have broken many of the same rules in the same general way. Because they are so distant on the phylogenetic tree of eukaryotes (Keeling et al., 2005; Hampl et al., 2009), convergence between the 2 groups would ultimately be influenced only by intrinsic factors of a very basic nature (i.e., that are likely common to most or all eukaryotes) (Leander, 2008). In contrast, where multiple aspects of a system have all converged similarly, it is likely that the convergent appearance of one new characteristic can be a strong factor in the convergent evolution of others. Even if these characteristics are not obligatorily functionally linked, their evolution may be tightly linked. For example, polycistronic mRNAs can exist without an SL, but they are evolutionarily linked because adding the SL allows the polycistronic mRNA to function. Conversely, one can imagine other ways to get a polycistronic mRNA to function without SL processing (e.g., changes to translation initiation), but because no such system is known, these are evidently less likely than the advent of SL processing. In other words, within the limited universe of acceptable changes, one change closes some possibilities, but opens new ones as well.
So why have protists in general and alveolates and euglenozoans in particular engaged in so much evolutionary experimentation? Many characteristics discussed here have been considered individually and concluded to be ancient relicts, going back even so far as the RNA world, or to
have been favored by selection over the canonical way of accomplishing the same task (Speijer, 2007; Ochsenreiter et al., 2008). We find neither of these arguments to be particularly compelling given the narrow distribution of these characters in nature, and their often extreme complexity. For example, dozens of nuclear-encoded proteins are required for T. brucei to edit just 12 mRNAs (Lukeš et al., 2005; Etheridge et al., 2008; Hashimi et al., 2008). Despite considerable controversy, no obvious evolutionary advantage has ever been demonstrated for this type of editing, and such possible advantages that have been proposed (e.g., the generation of 2 proteins from 1 gene) (Ochsenreiter et al., 2008) are more than outweighed by the demonstrated cost (i.e., “save” 1 gene at the cost of dozens of genes). We argue that constructive neutral evolution offers a more compelling explanation (Covello and Gray, 1993; Stoltzfus, 1999). This is a very simple and intuitive way of explaining complexity in biological systems, but one that has not received much attention. Briefly, it is possible for a biological system to increase in complexity (i.e., to increase the number of components or interactions needed to sustain the system) by making a series of neutral changes that collectively do not affect fitness. Pan-editing is often thought of as an error correcting system, but as Stoltzfus (1999) pointed out, the duplicated information (e.g., gRNAs) must have been created before the mutations they are correcting, or they too would carry the mutations—so the error-then-solution model is backward. Instead, if a gratuitous duplication of information took place first (i.e., the origin of a gRNA), then a subsequent mutation could be neutralized by the presence of the duplicated information needed to change it. The fixation of such a mutation would render the gRNA essential, and would also allow for further mutations as long as the gRNAs could mediate their reversal. This last point is important because it would bias the system against the loss of the gRNA since mutations at many sites will further establish the gRNA as essential, whereas only complete reversion to the original sequence could render it unnecessary. Overall, the editing activity and the sites that are edited will coevolve, and the complexity of the system will inevitably grow while conferring no real selective advantage (for many other case studies and much greater detail) [see Covello and Gray (1993) and Stoltzfus (1999)].
Within this framework, together with the recognition that the evolution of an unusual character can be an intrinsic factor in the subsequent evolution of additional, specific characters, a complex cellular system may be explained simply by identifying the event(s) that preconditioned the cell for such a system. Convergence may offer a glimpse into these conditions by revealing how characters are linked when the same events are played out multiple times.
ACKNOWLEDGMENTS
We thank Mona Hoppenrath and Susan Breglia for providing the images in Figs. 4.1 and 4.2F, respectively. This work was supported by the Grant Agency of the Czech Republic Grant 204/09/1667 (to J.L.); Ministry of Education of the Czech Republic Grants LC07032, 2B06129, and 600766580 (to J.L.); a grant from the Tula Foundation to the Centre for Microbial Diversity and Evolution (to B.S.L. and P.J.K.); and the Canadian Institute for Advanced Research (J.L., B.S.L., and P.J.K.).




















