A5
IN VITRO AND IN VIVO CHARACTERIZATION OF NEW SWINE- ORIGIN H1N1 INFLUENZA VIRUSES23
Yasushi Itoh,24 Kyoko Shinya,25 Maki Kiso,26 Tokiko Watanabe,27 Yoshihiro Sakoda,28 Masato Hatta,27 Yukiko Muramoto,29 Daisuke Tamura,26 Yuko Sakai-Tagawa,26 Takeshi Noda,30 Saori Sakabe,26 Masaki Imai,27 Yasuko Hatta,27 Shinji Watanabe,27 Chengjun Li,27 Shinya Yamada,26 Ken Fujii,26 Shin Murakami,26 Hirotaka Imai,26 Satoshi Kakugawa,26 Mutsumi Ito,26 Ryo Takano,26 Kiyoko Iwatsuki-Horimoto,26 Masayuki Shimojima,26 Taisuke Horimoto,26 Hideo Goto,26 Kei Takahashi,26 Akiko Makino,25 Hirohito Ishigaki,24 Misako Nakayama,24 Masatoshi Okamatsu,28 Kazuo Takahashi,31 David Warshauer,32 Peter A. Shult,32 Reiko Saito,33 Hiroshi Suzuki,33 Yousuke Furuta,34 Makoto Yamashita,35 Keiko Mitamura,36 Kunio Nakano,36 Morio Nakamura,36 Rebecca Brockman-Schneider,37 Hiroshi Mitamura,38 Masahiko Yamazaki,39 Norio Sugaya,40 M. Suresh,27 Makoto Ozawa,27,30 Gabriele Neumann,27 James Gern,37 Hiroshi Kida,28 Kazumasa Ogasawara,24 and Yoshihiro Kawaoka25,26,27,29,30,41
Influenza A viruses cause recurrent outbreaks at local or global scale with potentially severe consequences for human health and the global economy. Recently, a new strain of influenza A virus was detected that causes disease in and transmits among humans, probably owing to little or no pre-existing immunity to the new strain. On 11 June 2009 the World Health Organization declared that the infections caused by the new strain had reached pandemic proportion. Characterized as an influenza A virus of the H1N1 subtype, the genomic segments of the new strain were most closely related to swine viruses (Novel Swine-Origin Influenza A [H1N1] Virus Investigation Team, 2009). Most human infections with swine origin H1N1 influenza viruses (S-OIVs) seem to be mild; however, a substantial number of hospitalized individuals do not have underlying health issues, attesting to the pathogenic potential of S-OIVs. To achieve a better assessment of the risk posed by the new virus, we characterized one of the first US S-OIV isolates, A/California/04/09 (H1N1; hereafter referred to as CA04), as well as several other S-OIV isolates, in vitro and in vivo. In mice and ferrets, CA04 and other S-OIV isolates tested replicate more efficiently than a currently circulating human H1N1 virus. In addition, CA04 replicates efficiently in non-human primates, causes more severe pathological lesions in the lungs of infected mice, ferrets and non-human primates than a currently circulating human H1N1 virus, and transmits among ferrets. In specific-pathogen-free miniature pigs, CA04 replicates without clinical symptoms. The assessment of human sera from different age groups suggests that infection with human H1N1 viruses antigenically closely related to viruses circulating in 1918 confers neutralizing antibody activity to CA04. Finally, we show that CA04 is sensitive to approved and experimental antiviral drugs, suggesting that these compounds could function as a first line of defence against the recently declared S-OIV pandemic.
Sequence analyses of recently emerged swine-origin H1N1 viruses (S-OIVs) revealed the absence of markers associated with high pathogenicity in avian and/or mammalian species, such as a multibasic haemagglutinin (HA) cleavage site (Kawaoka and Webster, 1988) or lysine at position 627 of the PB2 protein (Hatta et al., 2001). To characterize the new viruses in vitro and in vivo, we amplified the following S-OIVs in Madin–Darby canine kidney (MDCK) cells: A/California/04/09 (CA04), A/Wisconsin/WSLH049/09 (WSLH049), A/Wisconsin/WSLH34939/09 (WSLH34939), A/Netherlands/603/09 (Net603) and A/Osaka/164/09 (Osaka164). WSLH34939 was isolated from a patient who required hospitalization, whereas the remaining viruses were isolated from mild cases. These viruses represent the currently recognized neuraminidase (NA) variants among S-OIVs: CA04, NA-106V, NA-248N; Osaka164, NA-106I, NA-248N; WSLH049, NA-106I, NA-248D; WSLH34939, NA-106I, NA-248D; and Net603, NA-106V, NA-248N.
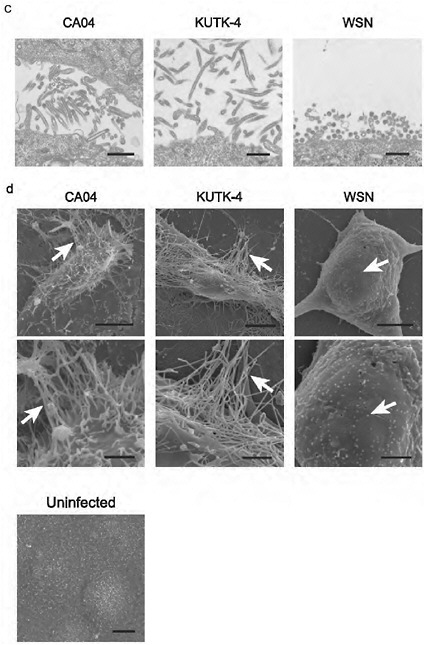
In MDCK cells and primary human airway epithelial cells, CA04 grew to titres comparable to those typically obtained for contemporary human H1N1 influenza viruses (Supplementary Figure A5-4). Confocal, transmission electron and scanning electron microscopy revealed virions of remarkably filamentous shape (Supplementary Figure A5-5), in marked contrast to the spherical shape observed with negatively stained virions (http://www.cdc.gov/h1n1flu/images.htm). The biological significance of the morphology of CA04 remains unknown.
To evaluate the pathogenicity of S-OIV in mammalian models, we conducted studies in mice, ferrets, non-human primates and pigs. BALB/c mice intranasally infected with a high dose (>104 plaque forming units (p.f.u.)) of CA04 (Supplementary Figure A5-6) experienced weight loss and those infected with the highest dose of this virus were humanely killed, in contrast to animals infected with a recent human H1N1 virus (A/Kawasaki/UTK-4/09, KUTK-4). The 50% mouse lethal dose (MLD50) was 105.8 p.f.u. for CA04 and .106.6 p.f.u. for KUTK-4. For the additional S-OIV isolates tested, the MLD50 values were >106.4 p.f.u. for Osaka164, >106.6 p.f.u. for WSLH049, 104.5 p.f.u. for WSLH34939 and >105.8 p.f.u. for Net603.
On day 3 after infection of mice, similar titres were detected in nasal turbinates of mice infected with 105 p.f.u. of S-OIVs or KUTK-4 (Supplementary Table A5-2); however, S-OIVs replicated more efficiently in the lungs of infected animals, which may account for the prominent bronchitis and alveolitis with viral antigen on day 3 after infection with CA04 (Supplementary Figure A5-7a, b). On day 6 after infection, virus titres followed a similar trend and the lungs of CA04-infected mice showed bronchitis and alveolitis with viral antigen, although signs of regeneration were apparent (Supplementary Figure A5-7c). We detected viral-antigen-positive bronchial epithelial cells, but not alveolar cells, on day 3 after infection of mice infected with KUTK-4 (Supplementary Figure A5-7e). By day 6, infection in KUTK-4-inoculated mice had progressed to bronchitis and peribronchitis; however, viral antigen was rarely detected in these lesions (Supplementary Figure A5-7f).
There were marked differences in the induction of pro-inflammatory cytokines in the lungs of mice infected with CA04 compared with KUTK-4 (Supplementary Figure A5-8a–c). Infection with KUTK-4 resultedin limited induction of pro-inflammatory cytokines/chemokines in the lungs, inmarked contrast to infection withCA04. Increased production of interleukin-10 (IL-10; Supplementary Figure A5-7a) in lungs of CA04-infected mice at day 6 after infection probably reflects a host response to dampen over-exuberant pulmonary inflammation and promote tissue repair. Infection with CA04 led to strong induction of both interferon-γ (IFN-γ) and IL-4 in the lungs. The selective induction of the TH2 cytokine IL-5 in CA04-infected, but not in KUTK-4-infected, mice on day 6 after infection is noteworthy (Supplementary Figure A5-7b), but further studies are needed to understand the relevance of this finding to viral control. IL-17 has been reported to have a role in protection against lethal influenza and also in eliciting
inflammatory responses (Iwakura et al., 2008; Hamada et al., 2009); however, the enhanced viral replication and lung pathology observed in CA04-infected mice was not linked to dysregulated IL-17 production.
Cynomolgus macaques (Macaca fascicularis) have been used to study highly pathogenic avian H5N1 viruses (Baskin et al., 2009; Rimmelzwaan et al., 2001) and the 1918 pandemic virus (Kobasa et al., 2007). Infection of cynomolgus macaques with CA04 (see Methods for detailed procedures) resulted in a more prominent increase in body temperature than infection with KUTK-4 (Supplementary Fig. A5-9). This difference might originate from the observed differences in virus titres (Table A5-1 and Supplementary Table A5-3). No remarkable difference in body weight loss was found between the two groups (data not shown). CA04 replicated efficiently in the lungs and other respiratory organs of infected animals, similar to highly pathogenic influenza viruses (Baskin et al., 2009; Kobasa et al., 2007) (Table A5-1). By contrast, conventional human influenza viruses are typically limited in their replicative ability in the lungs of infected primates (Baskin et al., 2009; Kobasa et al., 2007) (Table A5-1), although a seasonal H1N1 virus was isolated from one animal on day 7 after infection. Pathological examination revealed that CA04 caused more severe lung lesions than did KUTK-4 (Fig. A5-1 and Supplementary Fig. A5-10). On day 3 after infection with CA04, alveolar spaces were occupied by oedematous exudate and inflammatory infiltrates (Fig. A5-1a, b); severe thickening of alveolar walls was also observed (Fig. A5-1b). Viral-antigen-positive cells were distributed in the inflammatory lesions, and many of these cells were elongated with thin cytoplasm and hemming around the alveolar wall, indicating type I pneumocytes (Fig. A5-1c). In addition to type I pneumocytes, CA04 viral antigens were also detected in considerable numbers of cuboidal, cytokeratin-positive cells, hence identified as type II pneumocytes (Fig. A5-1d and Supplementary Fig. A5-11), as has been reported for highly pathogenic avian H5N1 influenza viruses6. Upon infection with KUTK-4, large sections of infected lungs showed thickening of the alveolar wall on day 3 after infection (Fig. A5-1e). Although the infiltration of inflammatory cells was prominent at the alveolar wall (Fig. A5-1f), viral antigens were sparse and detected in type I (but not type II) pneumocytes (Fig. A5-1g). By contrast, the lungs of non-infected animals show clear alveolar spaces (Fig. A5-1h).
On day 7 after infection, lung pathology remained more severe for CA04-than for KUTK-4-infected lungs (Supplementary Fig. A5-10), although regenerative changes were seen for CA04. Nonetheless, considerable numbers of antigen-positive cells were still detectable (Supplementary Fig. A5-10c). Collectively, these findings demonstrate that CA04 causes more severe lung lesions in non-human primates than does a contemporary human influenza virus.
Induction of pro-inflammatory cytokines/chemokines in the lungs of CA04-infected macaques was variable at day 3 after infection (Supplementary Fig. A5-12). However, consistent with persisting lung pathology and inflammation on day 7 after infection, the levels of MCP-1, MIP-1α, IL-6 and IL-18 were markedly higher in the lungs of two of three CA04-infected macaques.
TABLE A5-1 Virus Titres in Organs of Infected Cynomolgus Macaques
|
Organ |
A/California/04/09 (H1N1) |
A/Kawasaki/UTK-4/09 (H1N1) |
||||||||||
|
Day 3 after infection |
Day 7 after infection |
Day 3 after infection |
Day 7 after infection |
|||||||||
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
|
|
Nasal mucosa |
4.7 |
3.3 |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
|
Oro/nasopharynx |
6.3 |
4.4 |
4.7 |
— |
7.9 |
— |
— |
— |
4.3 |
— |
— |
4.8 |
|
Tonsil |
6.4 |
— |
— |
— |
7.1 |
— |
— |
— |
2.8 |
— |
— |
3.0 |
|
Trachea |
5.9 |
2.0 |
5.6 |
— |
— |
— |
2.0 |
4.1 |
— |
3.7 |
— |
5.4 |
|
Bronchus (right) |
5.7 |
2.9 |
4.3 |
— |
5.1 |
— |
— |
2.5 |
— |
3.5 |
— |
3.8 |
|
Bronchus (left) |
5.9 |
— |
6.1 |
— |
5.1 |
— |
— |
— |
— |
3.3 |
— |
5.1 |
|
Lung (upper right) |
5.7 |
5.6 |
4.5 |
— |
— |
— |
2.7 |
— |
— |
— |
— |
— |
|
Lung (middle right) |
5.6 |
6.4 |
6.9 |
— |
— |
— |
2.3 |
2.6 |
2.5 |
— |
— |
— |
|
Lung (lower right) |
6.1 |
4.5 |
6.0 |
— |
— |
— |
2.6 |
2.6 |
— |
— |
— |
3.4 |
|
Lung (Upper left) |
4.7 |
4.3 |
6.4 |
— |
— |
— |
— |
— |
— |
— |
— |
— |
|
Lung (middle left) |
5.8 |
4.3 |
6.3 |
— |
— |
— |
— |
— |
— |
— |
— |
— |
|
Lung (lower left) |
6.7 |
4.5 |
6.6 |
— |
— |
— |
— |
— |
— |
— |
— |
2.3 |
|
Conjunctiva |
3.6 |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
|
Cynomolgus macaques were inoculated with 107.4 p.f.u. of virus (6.7 ml) through multiple routes (see Methods). Three macaques per group were killed on days 3 and 7 after infection for virus titration. No virus was recovered from lymph nodes (chest), heart, spleen, kidneys or liver of any of the animals. A dash indicates that virus was not detected (detection limit: 2 log10 p.f.u.g–1). Numbers 1-12 indicate animal identification number. Values indicate virus titre (mean log10 p.f.u.g–1). |
||||||||||||
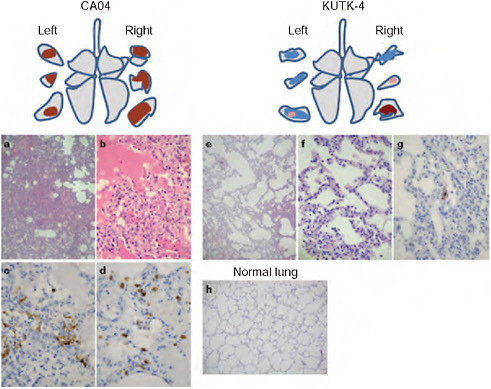
FIGURE A5-1 Pathological examination of the lungs of infected cynomolgus macaques. A-h Representative pathological images of CA04-infected (macaque no. 1, a-d), KUTK-4 infected (macaque no. 7, e-g) and mock-infected (h) lungs on day 3 after infection. One or two sections per lung lobe were examined. Representative findings are shown to depict the distribution of lesions in the sections (shown as cross-sections placed next to illustrations of ach lung lobe), with or without viral antigen, as follows: brown, severe lung lesion containing moderate to many viral-antigen-positive cells; pink, mild lung lesions containing a few viral-antigen-positive cells; blue, lung lesions with alveolar wall thickening, with remaining air spaces unaffected. Original magnification: a, e, h, ×40; b-d-f, g, ×400.
Ferrets are widely accepted as a suitable small-animal model for influenza virus pathogenicity and transmissibility studies. Infection of ferrets with S-OIVs or KUTK-4 did not cause marked changes in body temperature or weight in any group (data not shown). Although all test viruses were detected in nasal turbinates at similar titres on day 3 after infection (Supplementary Table A5-4), S-OIVs replicated to higher titres in trachea and lungs.
Pathological examination detected similar levels of viral antigen in the nasal mucosa of both CA04- and KUTK-4-infected ferrets (Supplementary Fig. A5-13a and e). However, the lungs of CA04-infected ferrets showed more severe broncho-
pneumonia with prominent viral antigen expression in the peribronchial glands and a few alveolar cells (Supplementary Fig. A5-13b–d) on day 3 after infection. By contrast, most of the lung appeared normal after infection with KUTK-4 (Supplementary Fig. A5-13f and g). Thus, in all three mammalian models tested, CA04 seemed to be more pathogenic than a contemporary human H1N1 virus, KUTK-4.
Efficient human-to-human transmission is a critical feature of pandemic influenza viruses. To assess the transmissibility of CA04, naive ferrets in perforated cages were placed next to ferrets inoculated with 106 p.f.u. of CA04 (see Methods for detailed procedures). This experimental setting allows for aerosol transmission (that is, the exchange of respiratory droplets between the inoculated and noninoculated ferrets) but prevents transmission by direct and indirect contact. All three contact ferrets were positive for CA04 virus on days 3 and 5 after infection (Supplementary Table A5-5). This transmission pattern is comparable to those of two human control influenza viruses that are known to transmit among ferrets: KUTK-4 and A/Victoria/3/75 (H3N2) (Maines et al., 2006). By contrast, an avian influenza virus (A/duck/Alberta/35/76; H1N1) did not transmit (Supplementary Table A5-5).
Genetic analysis suggests that S-OIV originated in pigs (Novel Swine- Origin Influenza A (H1N1) Virus Investigation Team, 2009). However, there were no confirmed influenza virus outbreaks in Central American pigs before the reported S-OIV infections in humans. To assess S-OIV replication in pigs, we inoculated specific-pathogen-free miniature pigs, which are easier to manage, with CA04 or a classical swine influenza virus (A/swine/Hokkaido/2/81, H1N1). No signs of disease were observed (data not shown), although both viruses replicated efficiently in the respiratory organs of these animals (Supplementary Tables A5-6 and A5-7). Slightly higher titres of CA04 were detected in lungs on day 3 after infection, which is supported by pathological findings that show more apparent bronchitis and bronchiolitis in pigs infected with CA04 (Supplementary Fig. A5-14). The asymptomatic infection of CA04, despite efficient virus replication, might explain the lack of reports of S-OIV outbreaks in pigs before virus transmission to humans.
Antiviral compounds are the first line of defence against pandemic influenza viruses. Sequence analysis suggests that S-OIVs are resistant to ion channel inhibitors such as amantadine and rimantadine (Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, 2009). We therefore tested the licensed neuraminidase inhibitors oseltamivir and zanamivir, the experimental neuraminidase inhibitor R-125489 (the active form of CS-8958 [Yamashita et al., 2009]) and the experimental compound T-705 (a broad-spectrum viral RNA polymerase inhibitor [Furuta et al., 2002]) for their efficacy against CA04. In cell culture, CA04 was highly susceptible to all compounds tested (Supplementary Table A5-8), as were the human H1N1 control viruses A/Kawasaki/UTK-23/08 and KUTK-4, with the exception of the known oseltamivir resistance of KUTK-4. Comparable sensitivities were also found in an enzymatic neuraminidase inhibition assay (Hayden et
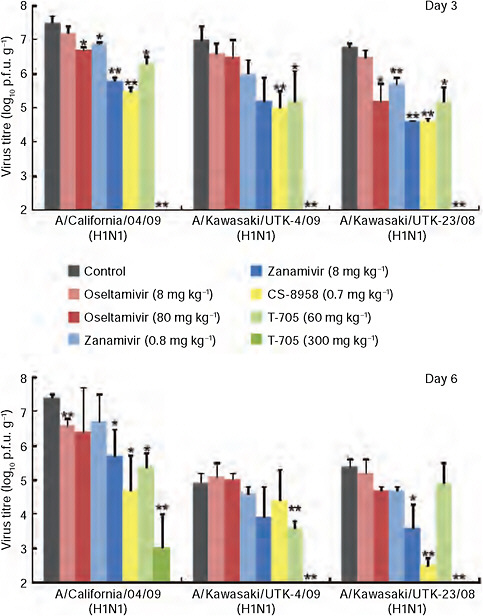
FIGURE A5-2 CA04 sensitivity to antiviral compounds in mice. Mice were intranasally inoculated with 104 p.f.u. (50 μl) of CA04, KUTK-4 or A/Kawasaki/UTK-23/08 (H1N1). At 1 h after infection, mice were administered oseltamivir phosphate, zanamivir, CS-8958, T-705, or distilled water and PBS (control). Three mice per group were killed on days 3 and 6 after infection and the virus titres in lungs were determined by plaque assays in MDCK cells; results are reported as means ± s.d. The statistical significance of differences in lung virus titres of control mice and those treated with antivirals were assessed by use of the Student’s t-test (asterisk, P < 0.05; double asterisk, P < 0.01).
al., 2000) (Supplementary Table A5-9) and in mice (Fig. A5-2), consistent with observations in clinical settings.
A recent report suggested that 33% of individuals over 60 years of age had neutralizing antibodies to CA04 (http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5819a1.htm; Morbidity and Mortality Weekly Report, Centers for Disease Control and Prevention), probably due to previous exposure to antigenically similar H1N1 viruses. In fact, both the human H1N1 viruses that circulated until 1957 and the classical swine virus HA gene of S-OIVs are descendants of the 1918 pandemic virus, possibly explaining their antigenic relatedness. In 1977, H1N1 viruses re-emerged that were genetically and antigenically very closely related to viruses circulating in the 1950s (Nakajima et al., 1978) and should thus have elicited neutralizing antibodies to CA04 among younger age groups; however, this does not seem to be the case, according to the above described report. To resolve this puzzling finding, we assessed the neutralizing activities of sera collected from a broad range of age groups against CA04 and KUTK-4. We used two sets of donor sera, collected in 1999 from residents and workers in a nursing home (donor set 1), and in April 2009 from workers and patients in a hospital (donor set 2). High neutralizing activity against KUTK-4 was detected for many sera in donor set 2 (Fig. A5-3), but not for sera in donor set 1, probably because these sera were collected before the emergence of the current human H1N1 viruses. Interestingly, with few exceptions, no appreciable neutralizing antibodies against CA04 were found for individuals born after 1920; however, many of those born before 1918 had high neutralizing antibody titres (individual neutralizing antibody titres are shown in Supplementary Table A5-9). These data indicate that infection with the 1918 pandemic virus or closely related human H1N1 viruses, but not infection with antigenically divergent human H1N1 viruses circulating in the 1920s to 1950s, and again since 1977, elicited neutralizing antibodies to S-OIVs.
Our findings indicate that S-OIVs are more pathogenic in mammalian models than seasonal H1N1 influenza viruses. In fact, the ability of CA04 to replicate in the lungs of mice, ferrets and non-human primates, and to cause appreciable pathology in this organ, is reminiscent of infections with highly pathogenic H5N1 influenza viruses (Peiris et al., 2004), as acknowledged in a recent report by the World Health Organization (http://www.who.int/wer/2009/wer8421/en/index.html). We therefore speculate that the high replicative ability of S-OIVs might contribute to a viral pneumonia characterized by diffuse alveolar damage that contributes to hospitalizations and fatal cases where no other underlying health issues exist (http://www.who.int/wer/2009/wer8421/en/index.html). In addition, sustained person-to-person transmission might result in the emergence of more pathogenic variants, as observed with the 1918 pandemic virus (reviewed in Wright et al., 2007). Furthermore, S-OIVs may acquire resistance to oseltamivir through mutations in their NA gene (as recently witnessed with human H1N1 viruses [Moscona, 2009]), or through reassortment with co- circulating,
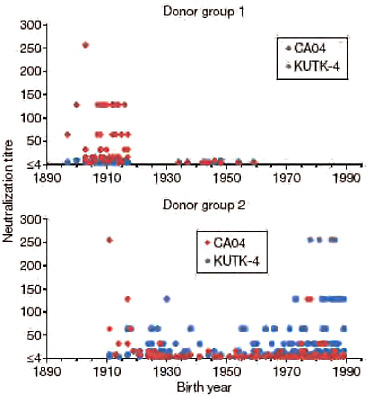
FIGURE A5-3 Neutralization activities in human sera against viruses. Human sera of donor groups 1 (collected in 1999) and 2 (collected in April and May of 2009) were subjected to neutralization assays with CA04 and KUTK-4. Because the sera of donor group 1 were collected in 1999, little neutralization activity was expected against KUTK-4, which was isolated in 2009.
oseltamivir- resistant seasonal human H1N1 viruses. Collectively, our findings are a reminder that S-OIVs have not yet garnered a place in history, but may still do so, as the pandemic caused by these viruses has the potential to produce a significant impact on human health and the global economy.
Methods Summary
Viruses and Cells
All swine-origin H1N1 viruses were isolated and passaged in MDCK cells to produce viral stocks. The viruses and their passage histories are described in Methods. All experiments with S-OIVs were performed in approved enhanced biosafety level 3 (BSL3) containment laboratories.
MDCK cells and MDCK cells overexpressing the β-galactoside α2,6-sialyltransferase I gene (Hatakeyama et al., 2005) were maintained in Eagle’s minimal essential medium (MEM) containing 5% newborn calf serum. Human airway epithelial (HAE) cells were obtained from residual surgical tissue trimmed from lungs during the process of transplantation. The bronchial specimens were dissected and enzymatically digested, and monolayers of HAE cells were isolated, cultured and differentiated as previously described (Jakiela et al., 2008).
Animals
Five- and six-week-old female BALB/c mice (Jackson Laboratory and Japan SLC Inc.), approximately three-to-four-year-old cynomolgus macaques (Ina Research Inc.), five-to-eight-month-old male ferrets (Marshall Farms and Triple F Farms) and two-month-old female specific-pathogen-free miniature pigs (Nippon Institute for Biological Science) were used according to approved protocols for the care and use of animals. Detailed procedures are provided in Methods.
Antiviral Sensitivity of Viruses in Mice
Five-week-old female BALB/c mice (Japan SLC Inc.; groups of six) were anaesthetized with sevoflurane and intranasally inoculated with 104 p.f.u. (volume, 50 μl) of CA04, KUTK-4, or A/Kawasaki/UTK-23/08 (H1N1). At 1 h after infection, mice were administered antiviral compounds as described in detail in Methods. Three mice per group were killed on days 3 or 6 after infection and the virus titres in lungs were determined by plaque assays in MDCK cells.
Methods
Viruses
A/California/04/09 (H1N1; CA04) was provided by the Centers for Disease Control (CDC). A/Wisconsin/WSLH049/09 (H1N1) was isolated from a patient with mild symptoms, whereas A/Wisconsin/WSLH34939/09 (H1N1) was isolated from a hospitalized patient. A/Netherlands/603/09 (H1N1) was isolated from a patient with mild symptoms and was provided by R. Fouchier. A/Osaka/164/09 (H1N1) was also isolated from a patient with mild symptoms.
The following influenza viruses served as controls: A/Kawasaki/UTK-4/09 (H1N1; KUTK-4; passaged twice in MDCK cells), an oseltamivir-resistant seasonal human virus; A/WSN/33 (H1N1; generated by reverse genetics and passaged twice in MDCK cells), a typical spherical influenza virus (Noda et al., 2006); A/Kawasaki/UTK-23/08 (H1N1; passaged twice in MDCK cells), an oseltamivir-sensitive seasonal human virus; A/Victoria/3/75 (H3N2; passaged several times in eggs after it was obtained from the CDC), a human virus; A/swine/Hokkaido/2/81
(H1N1; passaged several times in eggs), a classical swine virus; and A/duck/Alberta/35/76 (H1N1; passaged several times in eggs), an avian virus. All experiments with S-OIV viruses were performed in enhanced biosafety level 3 (BSL3) containment laboratories at the University of Wisconsin-Madison, which are approved for such use by the CDC and the US Department of Agriculture, or in BSL3 containment laboratories at the University of Tokyo, the Shiga University of Medical Science, or the Hokkaido University, all of which are approved for such use by the Ministry of Agriculture, Forestry and Fisheries, Japan.
Viral Pathogenesis in Mice
Six-week-old female BALB/c mice (Jackson Laboratory) were used in this study. Baseline body weights were measured before infection. Three mice per group were anaesthetized with isoflurane and intranasally inoculated with 102, 103, 104, or 105 p.f.u. (50 µl) of CA04 and KUTK-4, or undiluted virus from virus stocks (CA04, 106.5 p.f.u.; KUTK-4, 106.6 p.f.u.). Body weight and survival were monitored daily for 14 days and mice with body weight loss of more than 25% of pre-infection values were killed. For virological and pathological examinations, 6 mice per group were intranasally infected with 105 p.f.u. of S-OIVs and KUTK-4 and 3 mice per group were killed on days 3 and 6 after infection. The virus titres in various organs were determined by plaque assays in MDCK cells.
Growth Kinetics of Virus in Human Airway Epithelial (HAE) Cells
Cultures of differentiated HAE cells were washed extensively with PBS to remove accumulated mucus and infected with virus at a multiplicity of infection (MOI) of 0.001 from the apical surface. The inoculum was removed after 1 h of incubation at 35°C, and cells were further incubated at 35°C. Samples were collected at 12, 24, 48, 72 and 96 h after infection from the apical surface. Apical harvesting was performed by adding 500 µl of medium to the apical surface, followed by incubation for 30 min at 35°C, and removal of the medium from the apical surface. The titres of viruses released into the cell culture supernatant were determined by plaque assay in MDCK cells.
Experimental Infection of Cynomolgus Macaques
Approximately three-to-four-year-old cynomolgus macaques (Macaca fascicularis) from the Philippines (obtained from Ina Research Inc.), weighing 2.1–3.0 kg and serologically negative by AniGen AIV antibody ELISA, which detects all influenza A virus subtypes (Animal Genetics Inc.), were used in this study. Baseline body weights were established by two or three measurements before infection. Under anaesthesia, telemetry probes (TA10CTA-D70, Data Sciences International) were implanted in the peritoneal cavities of animals to monitor body temperature. Six macaques per group were intramuscularly
anaesthetized with ketamine (5 mg per kg) and xylazine (1 mg per kg) and inoculated with a suspension containing 106.5 p.f.u. ml–1 of CA04 or KUTK-4 virus through a combination of intratracheal (4.5 ml), intranasal (0.5 ml per nostril), ocular (0.1 ml per eye) and oral (1 ml) routes (resulting in a total infectious dose of 107.4 p.f.u.). Macaques were monitored every 15 min for changes in body temperature. On days, 1, 3, 5 and 7 after infection, nasal and tracheal swabs and bronchial brush samples were collected. On days 3 and 7 after infection, 3 macaques per group were killed for virological and pathological examinations. The virus titres in various organs and swabs were determined by plaque assays in MDCK cells. Experiments were carried out in accordance with the Guidelines for the Husbandry and Management of Laboratory Animals of the Research Center for Animal Life Science at Shiga University of Medical Science, Shiga, Japan, and approved by the Shiga University of Medical Science Animal Experiment Committee and Biosafety Committee.
Experimental Infection of Ferrets
We used five-to-eight-month-old male ferrets (Marshall Farms and Triple F Farms), which were serologically negative by haemagglutination inhibition (HI) assay for currently circulating human influenza viruses. Baseline body temperatures and body weights were established by one or two measurements before infection. Six ferrets per group were intramuscularly anaesthetized with ketamine and xylazine (5 mg and 0.5 mg per kg of body weight, respectively) and intranasally inoculated with 106 p.f.u. (500 μl) of S-OIVs or KUTK-4. On days 3 and 6 after infection, 3 ferrets per group were killed for virological and pathological examinations. The virus titres in nasal washes and various organs were determined by plaque assays in MDCK cells.
Experimental Infection of Miniature Pigs
Two-month-old female specific-pathogen-free miniature pigs (Nippon Institute for Biological Science), which were serologically negative by AniGen AIV antibody ELISA for currently circulating influenza viruses, were used in this study. Baseline body temperatures were measured once before infection. Four pigs per group were intranasally inoculated with 106.2 p.f.u. (1 ml) of viruses. Nasal swabs were collected daily. On day 3 after infection, two pigs per group were killed and their tissues collected for examination. On day 14 after infection, the remaining two pigs per group were killed for virological and pathological examinations. Virus titres in various organs and swabs were determined by plaque assays in MDCK cells. The miniature pigs used in this study were housed in self-contained isolator units (Tokiwa Kagaku) at a BSL3 facility and experiments were conducted in accordance with guidelines established by the Animal Experiment Committee of the Graduate School of Veterinary Medicine, Hokkaido University, Japan.
Pathological Examination
Excised tissues of the nasal turbinates, trachea and/ or lungs of killed mice, macaques, ferrets and pigs were preserved in 10% phosphate-buffered formalin. Tissues were then processed for paraffin embedding and cut into 5-μm-thick sections. One section from each tissue sample was stained using a standard haematoxylin-and-eosin procedure, whereas another one was processed for immunohistological staining with an anti-influenza virus rabbit antibody (R309; prepared in our laboratory) that reacts comparably with CA04 and KUTK-4. Specific antigen–antibody reactions were visualized by 3,3′-diaminobenzidine tetrahydrochloride staining using a Dako EnVision system (Dako Co. Ltd).
Ferret Transmission Study
For transmission studies in ferrets, animals were housed in adjacent transmission cages that prevent direct and indirect contact between animals but allow spread of influenza virus through the air. Three or two 5-to-8-month-old ferrets were intranasally inoculated with 106 p.f.u. (500 μl) of CA04, KUTK-4, A/Victoria/3/75 (H3N2) or A/duck/Alberta/35/76 (H1N1) (inoculated ferrets). One day after infection, three or two naive ferrets were each placed in a cage adjacent to an inoculated ferret (contact ferrets). All ferrets were monitored daily for changes in body temperature and weight, and the presence of clinical signs. To assess viral replication in the upper respiratory tract, viral titres were determined in nasal washes collected from virus-inoculated and contact ferrets on day 1 after inoculation or co-housing, respectively, and then every other day (up to 9 days).
Cytokine and Chemokine Measurement
For cytokine and chemokine measurement, homogenates of mouse lungs were processed with the Bio-Plex Mouse Cytokine 23-Plex and 9-Plex panels (Bio-Rad Laboratories), whereas macaque lung homogenates were measured with the MILLIPLEX MAP Non-human Primate Cytokine/Chemokine Panel–Premixed 23-Plex (Millipore). Array analysis was performed by Bio-Plex Protein Array system (Bio-Rad Laboratories).
Antiviral Sensitivity of Viruses in Mice
To test the antiviral sensitivity of viruses in mice, animals were infected as described in the Methods Summary section and 1 h later administered the follow ing antiviral compounds: (1) oseltamivir phosphate: 8 or 80 mg per kg per 400 μl (divided into two oral administrations per day) for 5 days; (2) zanamivir: 0.8 or 8 mg per kg per 50 μl in one daily intranasal administration for 5 days; (3) CS-8958: 0.7 mg per kg per 50 μl in one intranasal administration; (4) T-705:
60 or 300 mg per kg per 400 μl (divided into two oral administrations per day) for 5 days; (5) or distilled water orally (200 μl) and PBS intranasally (50 μl). Three mice per group were killed on days 3 or 6 after infection and the virus titres in lungs were determined by plaque assays in MDCK cells.
Sensitivity to Antiviral Compounds in Tissue Culture
MDCK cells overexpressing the β-galactoside α2,6-sialyltransferase I gene (or, for studies with T-705, regular MDCK cells) were infected with CA04, KUTK-4, or A/Kawasaki/UTK-23/08 (H1N1) at a multiplicity of infection of 0.001. After incubation for 1 h at 37°C, growth medium containing various concentrations of oseltamivir carboxylate (the active form of oseltamivir), zanamivir, R-125489 (the active form of CS-8958), or T-705 was added to the cells. Twenty-four hours later, the culture supernatants were harvested and the 50% tissue-culture infectious dose (TCID50) in MDCK cells determined. On the basis of the TCID50 value, the 90% inhibitory concentration (IC90) was calculated.
Neuraminidase Inhibition Assay
To assess the sensitivity of viruses to neuraminidase inhibitors (that is, oseltamivir, zanamivir and CS-8958), neuraminidase inhibition assays were performed as described previously (Kiso et al., 2004). Briefly, diluted viruses were mixed with various concentrations of oseltamivir carboxylate, zanamivir, or R-125489 in 2-(N-morpholino) ethanesulphonic acid containing calcium chloride, and incubated for 30 min at 37°C. Then, we added methylumbelliferyl-N-acetylneuraminic acid (Sigma) as a fluorescent substrate to this mixture. After incubation for 1 h at 37°C, sodium hydroxide in 80% ethanol was added to the mixture to stop the reaction. The fluorescence of the solution was measured at an excitation wavelength of 360 nm and an emission wavelength of 465 nm and the 50% inhibitory concentration (IC50) was calculated.
Neutralization Assay with Human Sera
Human sera were collected in 1999 or 2009 from donor group 1 (age range: 50–112 years as of 2009, mean = 92.7 ± 15.0 years) or 2 (age range: 20–68 years as of 2009, mean = 48.2 ± 23.7 years), respectively. These sera were treated with receptor destroying enzyme (DENKA SEIKEN CO.) to remove inhibitors of influenza virus replication. One hundred TCID50 (50% tissue culture infectious dose) of CA04 and KUTK-4 were pre-incubated with twofold serial dilutions of treated sera, incubated for 60 min on MDCK cells, which were then observed for cytopathic effects to determine the neutralizing activity of the test sera. Our research protocol was approved by the Research Ethics Review Committee of the Institute of Medical Science, the University of
Tokyo (approval numbers: 21-6-0428 for donor group 1; 21-7-0529 for donor group 2).
Immunofluorescence Microscopy
MDCK cells were infected with CA04, KUTK-4, or WSN and fixed with 4% paraformaldehyde 16–24 h later. Infected cells were incubated with the following primary antibodies: mouse anti-HA (7B1b), anti-HA (IVC102), or mouse anti-HA (WS3-54) antibody against CA04, KUTK-4 or WSN, respectively. Cells were then incubated with Alexa Fluor 488 goat anti-mouse immunoglobulin G (Invitrogen), and examined with a confocal laser-scanning microscope (LSM510META; Carl Zeiss).
Electron Microscopy
MDCKcells were infected with CA04, KUTK-4 orWSNat a multiplicity of infection of 10. At 16–24 h after infection, cells were processed for ultrathin section electron microscopy and scanning electron microscopy as described previously (Noda et al., 2006; Neumann et al., 2005).
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank the Centers for Disease Control (CDC) for A/California/04/09 virus and R. Fouchier for A/Netherlands/603/09 virus. We thank K. Wells for editing the manuscript, and M. McGregor, R. Moritz, A. Hanson, H. Ishida, H. Tsuchiya, R. Torii, N. Yamamoto, K. Soda, N. Nomura and H. Yoshida for technical assistance. We also thank T. Umemura, Y. Sunden and T. Tanaka for pathological analyses of virus-infected pigs. This work was supported by National Institute of Allergy and Infectious Diseases Public Health Service research grants, by an NIAID-funded Center for Research on Influenza Pathogenesis (CRIP, HHSN266200700010C), by Grant-in-Aid for Specially Promoted Research, by a contract research fund for the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology, and by grants-in-aid from the Ministry of Health and by ERATO (Japan Science and Technology Agency).
Author Contributions
Y. I., K. S., M. K., T. W., Y. S., M. H., Y. M., D. T., Y. S.-T., T. N., M. Imai, S. W., K. I.-H., T. H., N. S., H. K., K. O. and Y. K. designed the experiments;
Y. I., K. S., M. K., T. W., Y. S., M. H., D. T., Y. S.-T., T. N., S. S., M. Imai, Y. H., S. W., C. L., S. Y., K. F., S. M., H. Imai, S. K., M. Ito, R. T., K. I.-H., M. S., T. H., Kei Takahashi, A. M., H. Ishigaki, M. Nakayama, M. Okamatsu, Kazuo Takahashi, D. W., P. A. S., R. S., H. S., Y. F., M. Yamashita, K. M., K. N., M. Nakamura, R. B.-S., J. G., H. M. and M. Yamazaki performed the experiments; Y. I., K. S., M. K., T. W., Y. S., M. H., Y. M., Y. S.-T., T. N., M. Imai, S. W., C. L., S. Y., K. I.-H., T. H., H. G., M. S., M. Ozawa, G. N., H. K., K. O. and Y. K. analysed data; Y. I., K. S., M. K., T. W., Y. S., M. H., Y. M., Y. S.-T., T. N., M. Imai, K. I.-H., M. S., M. Ozawa, G. N., K. O. and Y. K. wrote the manuscript. Y. I., K. S., M. K., T. W., Y. S., M. H. and Y. M. contributed equally to this work.
Author Information
Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Correspondence and requests for materials should be addressed to Y. K. (kawaokay@svm.vetmed.wisc.edu).
Supplementary Information is linked to the online version of the paper at www.nature.com/nature and also following this paper.
References
Baskin, C. R. et al. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc. Natl Acad. Sci. USA 106, 3455–3460 (2009).
Furuta, Y. et al. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 46, 977–981 (2002).
Hamada, H. et al. Tc17, a unique subset of CD8 T cells that can protect against lethal influenza challenge. J. Immunol. 182, 3469–3481 (2009).
Hatakeyama, S. et al. Enhanced expression of an α2,6-linked sialic acid on MDCK cells improves isolation of human influenza viruses and evaluation of their sensitivity to a neuraminidase inhibitor. J. Clin. Microbiol. 43, 4139–4146 (2005).
Hatta, M., Gao, P., Halfmann, P. & Kawaoka, Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293, 1840–1842 (2001).
Hayden, F. G. et al. Inhaled zanamivir for the prevention of influenza in families. Zanamivir Family Study Group. N. Engl. J. Med. 343, 1282–1289 (2000).
Iwakura, Y., Nakae, S., Saijo, S. & Ishigame, H. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunol. Rev. 226, 57–79 (2008).
Jakiela, B., Brockman-Schneider, R., Amineva, S., Lee, W. M. & Gern, J. E. Basal cells of differentiated bronchial epithelium are more susceptible to rhinovirus infection. Am. J. Respir. Cell Mol. Biol. 38, 517–523 (2008).
Kawaoka, Y. & Webster, R. G. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc. Natl Acad. Sci. USA 85, 324–328 (1988).
Kiso, M. et al. Resistant influenza A viruses in children treated with oseltamivir: Descriptive study. Lancet 364, 759–765 (2004).
Kobasa, D. et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445, 319–323 (2007).
Maines, T. R. et al. Lack of transmission of H5N1avian-human reassortant influenza viruses in a ferret model. Proc. Natl Acad. Sci. USA 103, 12121–12126 (2006).
Moscona, A. Global transmission of oseltamivir-resistant influenza. N. Engl. J. Med. 360, 953–956 (2009).
Nakajima, K., Desselberger, U. & Palese, P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature 274, 334–339 (1978).
Neumann, G. et al. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J. Virol. 79, 10300–10307 (2005).
Noda, T. et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439, 490–492 (2006).
Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360, 2605–2615 (2009).
Peiris, J. S. et al. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363, 617–619 (2004).
Rimmelzwaan, G. F. et al. Pathogenesis of influenza A (H5N1) virus infection in a Primate model. J. Virol. 75, 6687–6691 (2001).
Wright, P. F., Neumann, G. & Kawaoka, Y. Fields Virology (eds Knipe, D. M. et al.) 1691–1740 (Wolters Kluwer/Lippincott Williams & Wilkins, 2007).
Yamashita, M. et al. CS-8958, a prodrug of the new neuraminidase inhibitor R-125489, shows long-acting anti-influenza virus activity. Antimicrob. Agents Chemother. 53, 186–192 (2009).
Supplementary Information
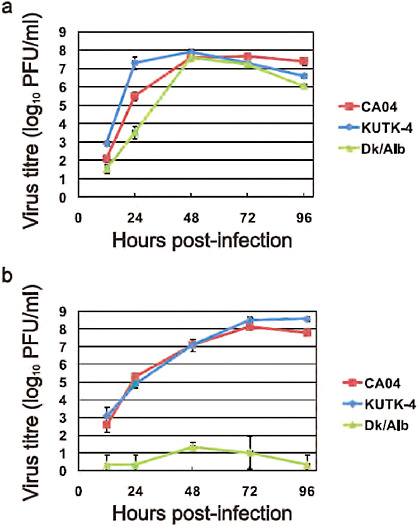
FIGURE A5-4 Growth properties of viruses in cells. MDCK cells were infected with CA04 (red), KUTK-4 (blue), or A/duck/Alberta/35/76 (H1N1; Dk/Alb, green) at an MOI of 0.001 (a). Differentiated human airway epithelial cells were infected with CA04 (red), KUTK-4 (blue), or A/duck/Alberta/35/76 (H1N1; Dk/Alb, green) at an MOI of 0.001 (b). The supernatants of infected cells were harvested at the indicated times and virus titres were determined by plaque assays in MDCK cells. Error bars indicate standard deviations from three independent experiments.
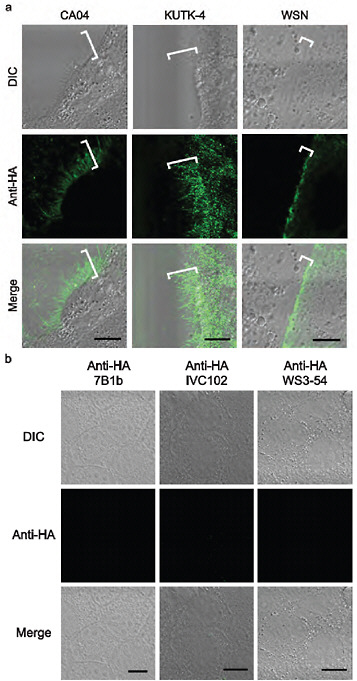
FIGURE A5-5 a-d Morphology of budding CA04 virions. MDCK cells infected with A/California/04/09 (H1N1) (CA04, left), A/Kawasaki/UTK-4/09 (H1N1) (KUTK-4, middle), or A/WSN/33 (H1N1) (WSN, right) were examined with confocal microscopy (a); mock-infected cells were processed similarly (b). Virus-infected cells were observed by transmission electron (TEM, c) and scanning electron microscope (SEM, d) (Kawaoka and Webster, 1988). Brackets in a and arrows in d indicate budding viruses. Scale bar: a and b, 10 µm; c, 500 nm; d, 5 (upper panels) and 2 µm (lower panels). DIC, differential interference contrast.
FIGURE A5-6 Body weight changes in infected mice. Three mice per group were intranaasally inoculated with 102, 103, 104, or 105 PFU (each in 50 μl) of CA04 (red) or KUTK-4 (blue), or undiluted virus (106.5 PFU for CA04 and 106.6 PFU for KUTK-4). Body weights were monitored daily. Mice with body weight loss of more than 25% of pre-infection values were euthanized. The values are means ± SD from three mice.
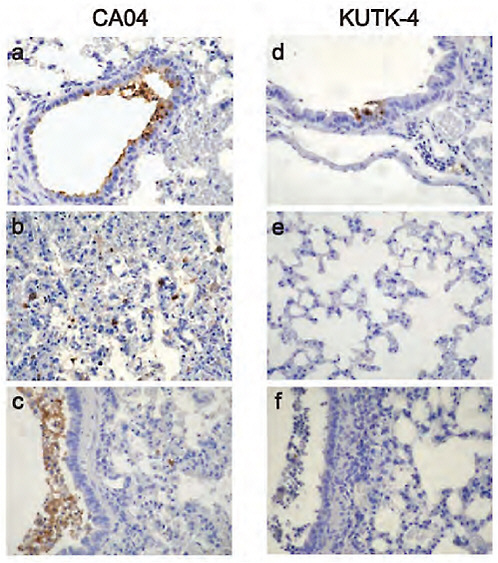
FIGURE A5-7 Pathological findings in infected mice. Representative pathological findings for the lungs of mice infected with CA04 (a-c), or KUTK-4 (d-f). infection with CA04 resulted in detectable viral antigen in bronchiolar epithelia and desquamated cells in the bronchial lumen on day 3 pi (a). Also, prominent alveolar thickening with scattered antigen-positive cells in the alveolus was observed (b). By day 6, epithelia were regenerative but accumulation of antigen-positive cell debris in the lumen was prominent (c). Upon infection with KUTK-4, a small number of viral antigen-positive cells was detected in the bronchial and bronchial epithelia on day 3 pi (d), but no viral antigen was detected in the alveolar area (e). On day 6 pi, accumulation of cell debris in the bronchiolar lumen with peribronchiolitis was observed, but viral antigens were rarely detected in these lesions (f).
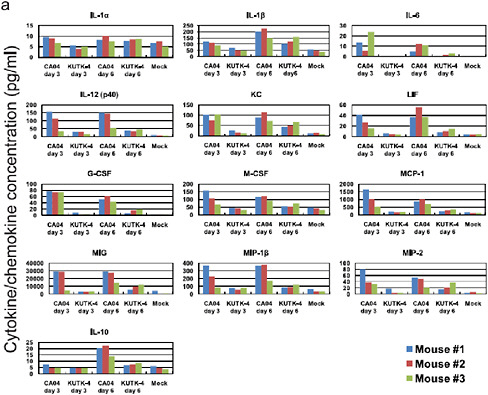
FIGURE A5-8 Pro-inflammatory cytokine/chemokine responses in the lungs of infected mice. The concentrations of various cytokines/chemokines were measured in the lungs of mice by use of a protein array analysis with the Bio-Plex Mouse Cytokine 23-Plex and 9-Plex panels (Bio-Rad laboratories). IL-12 (p70) was not detected. IL-18 data are not available due to technical problem of the manufacturer.
FIGURE A5-9 Body temperatures of infected cynomolgus macaques. Six macaques per group were inoculated with 107.4 PFU (total volume: 6.7 ml) of CA04 (red #1-6) or KUTK-4 (blue, #7-12) through multiple routes (see Supplementary materials and methods). Temperatures were monitored every 15 minutes by telemetry probes implanted in the peritoneal cavities. The periodic sharp reduction in body temperatures on days 0, 1, 3, and 7 was caused by anesthesia required for sampling. Monkeys #1-3 and #7-9 were euthanized on day 3.
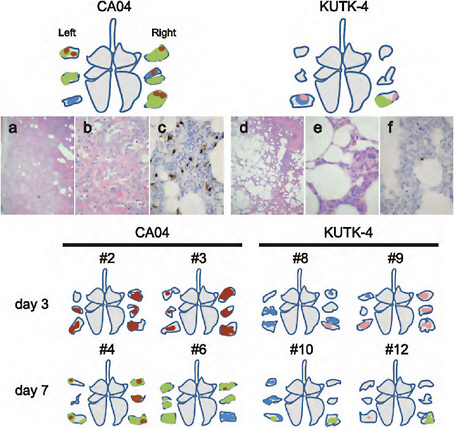
FIGURE A5-10 Pathological findings in infected cynomolgus macaques. Shown are representative pathological findings in the lungs of cynomolgus macaques on day 7 post infection with CA04 (macaque #5, a-c) or KUTK-4 (macaque #11, d-f) (upper portion). Schematic figures summarize the distribution of lesions, with or without viral antigen, in the lungs of the remaining virus-inoculated macaques. Colors: green, severe lung lesions where alveolar spaces were filled with edema fluid, inflammatory cells, or cell debris; brown, severe lung lesions containing moderate to many viral antigen-positive cells; pink, mild lung lesions containing a few viral antigen-positive cells; blue, lung lesions where severe alveolar wall thickening was prominent, but air spaces were preserved. (a) Alveolar spaces were not clear because of inflammatory exudate. (b) Large areas of affected lung contained accumulated cell debris, inflammatory infiltrates, fibrin, and edema fluid; alveolar walls were thickened by infiltration of inflammatory cells. (c) Viral antigen-positive cells were detected extensively in lung lesions in some areas. (d) In most areas, alveolar spaces were still clear, although thickening of the alveolar walls was apparent. (e) Most lung lesions consisted of thickening of alveolar walls by mononuclear cells. (f) A few antigen-positive cells were detected in lung lesions.
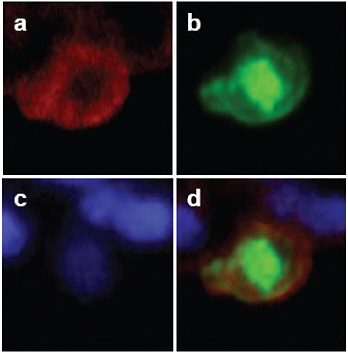
FIGURE A5-11 Detection of viral antigens in type II pneumocytes in the lungs of CA04-infected cynomolgus macaques. On day 3 post-infection, cells were stained with anti-cytokeratin (N1590, DAKO) antibody (a; red) and anti-influenza (H1N1) antibody (b; green). The nucleus was stained with DAPI (c). Considerable amounts of viral antigen were detected in type II pneumocytes (d).
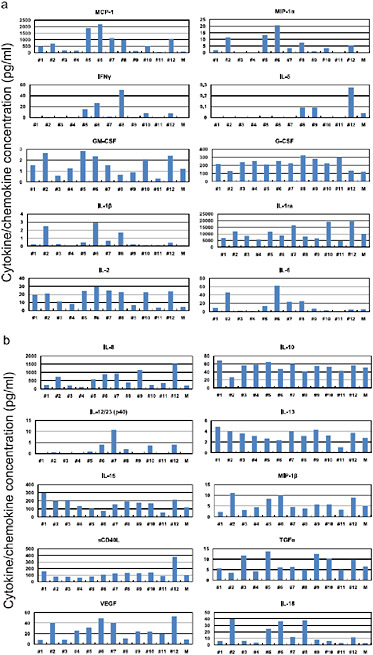
FIGURE A5-12 Pro-inflammatory cytokine/chemokine responses in the lungs of infected cynomolgus macaques. The concentrations of various cytokines/chemokines in the lungs of infected cynomolgus macaques on day 3 post-infection were measured by protein array analysis with the MILLIPLEX MAP Non-human Primate Cytokine/Chemokine Panel – Premixed 23-Plex (Millipore, Bedford, MA). We did not detect IL-4, IL-17, or TNFα in the lungs or G-CSF and IL-4 in the sera.
FIGURE A5-13 Pathological findings in infected ferrets. CA04-omfected ferret lungs showed severe and wide lung lesions with viral antigen on day 3 pi and without viral antigen on day 6 pi. KUTK-4-infected ferret lungs showed limited lung lesopns with viral antigen on days 3 and 6 pi. Representative pathological findings of nasal mucosa and lungs of CA04-(a-d), and KUTK-4-(e-g) infected ferrets on days 3 and 6 pi. (a) Extensive viral antigen present at the nasal epithelium on day 3 pi in CA04-infected ferret. (b) and (c) In the lungs, viral antigen was mainly detected in the peribroncial glands with severe peribronchitis and bronchopneumonia (d) Sparse viral antigen was detected within alveolar lesions. (e) Extensive viral antigen expression at the nasal mucosa on day 3 pi in KUTK-4-infected ferret. (f) Most of the lung was not affected by viral infection in KUTK-4-infected ferrets. (g) Most of the peribronchial gland appeared normal in KUTK-4-infected ferret lungs at day 6 pi. Colors: green, severe lung lesions where alveolar spaces were filled with edema fluid, inflammatory cells, or cell debris; brown, severe lung lesions containing moderate to many viral antigen-positive cells.
FIGURE A5-14 Pathological findings in infected miniature pigs. Shown are representative pathological findings for the lungs of miniature pigs on day 3 post infection with CA04 (miniature pig #1, a-c) or A/swine/Hokkaido/2/81 (H1N1) (miniature pig #5, d-f). The distribution of viral antigen (brown) is shown in the schematic figures. (a) The lumens of the bronchus and bronchioles were filled with inflammatory infiltrates. (b) Viral antigen was detected along bronchus and bronchiole. (c) Viral antigen was mainly detected in epithelial cells and desquamated cells. (d) The bronchial lumen remained clear with limited inflammatory reactions. (e) and (f) Few antigen-positive cells were detected at the epithelium, with minimum inflammatory reaction. The areas delineated by green boxes in the schematic diagrams correspond to the histopathological sections.
TABLE A5-2 Virus Titres in Organs of Infected Micea
TABLE A5-3 Virus Titres in Respiratory Swabs from Infected Cynomolgus Macaquesa
|
|
Virus titre (log10 PFU/ml) of animals infected with: |
||||||||||||
|
A/California/04/09 (H1N1) |
A/Kawasaki/UTK-4/09 (H1N1) |
||||||||||||
|
Animal ID |
#1 |
#2 |
#3 |
#4 |
#5 |
#6 |
#7 |
#8 |
#9 |
#10 |
#11 |
#12 |
|
|
Nasal swab |
Day 1 |
5.8 |
1.0 |
1.5 |
4.7 |
4.5 |
2.6 |
3.0 |
2.9 |
1.6 |
3.6 |
2.4 |
3.6 |
|
|
Day 3 |
5.2 |
2.1 |
2.5 |
3.7 |
2.6 |
3.3 |
2.8 |
3.2 |
2.4 |
4.1 |
1.5 |
3.3 |
|
|
Day 5 |
|
|
|
4.7 |
4.6 |
3.4 |
|
|
|
1.3 |
3.6 |
2.5 |
|
|
Day 7 |
|
|
|
—b |
3.5 |
— |
|
|
|
— |
— |
5.0 |
|
Tracheal swab |
Day 1 |
3.4 |
2.3 |
3.6 |
2.3 |
3.5 |
2.0 |
1.3 |
1.3 |
— |
2.0 |
2.3 |
2.1 |
|
|
Day 3 |
4.3 |
— |
2.6 |
2.6 |
2.4 |
2.0 |
1.0 |
1.8 |
— |
4.0 |
— |
— |
|
|
Day 5 |
|
|
|
3.5 |
2.5 |
3.7 |
|
|
|
5.6 |
— |
— |
|
|
Day 7 |
|
|
|
— |
2.0 |
— |
|
|
|
3.4 |
— |
2.6 |
|
Bronchial brush |
Day 1 |
2.9 |
2.4 |
3.7 |
2.2 |
3.3 |
— |
1.5 |
— |
— |
— |
— |
— |
|
|
Day 3 |
3.5 |
— |
— |
3.1 |
— |
— |
— |
1.5 |
— |
— |
— |
— |
|
|
Day 5 |
|
|
|
4.4 |
2.4 |
1.8 |
|
|
|
4.4 |
— |
1.3 |
|
|
Day 7 |
|
|
|
— |
— |
— |
|
|
|
1.5 |
— |
4.4 |
|
aCynomolgus macaques were inoculated with 107.4 PFU of virus (6.7 ml) through multiple routes. Nasal and tracheal swabs and bronchial brush samples were collected every other day for virus titration. b—. virus not detected (detection limit: 1.0 log10 PFU/ml). Blank: not applicable, animals euthanized on day 3 pi. |
|||||||||||||
TABLE A5-4 Virus Titres in Respiratory Organs of Infected Ferretsa
|
Virus |
|
Virus titres (mean log10 PFU ± SD/g) in: |
||
|
Nasal turbinates |
Trachea |
Lungs |
||
|
A/California/04/09 (H1N1) |
Day 3 |
6.7±0.7 |
5.9±0.4 |
3.53, 4.12 |
|
|
Day 6 |
2.40 |
3.1±0.1 |
2.95 |
|
A/Netherlands/603/09 (H1N1) |
Day 3 |
7.3±0.6 |
6.0±1.7 |
5.15 |
|
|
Day 6 |
3.26, 4.77 |
4.36, 5.14 |
—b |
|
A/Wisconsin/WSLH049/09 (H1N1) |
Day 3 |
7.8±0.6 |
6.0±0.9 |
6.49, 3.23 |
|
|
Day 6 |
4.3±1.1 |
4.57, 3.40 |
5.48 |
|
A/Wisconsin/WSLH34939/09 (H1N1) |
Day 3 |
8.3±0.1 |
4.6±0.3 |
4.5±1.8 |
|
|
Day 6 |
4.5±1.0 |
3.8±1.4 |
3.6±1.1 |
|
A/Osaka/164/09 (H1N1) |
Day 3 |
6.9±1.0 |
6.4±1.0 |
6.8±0.8 |
|
|
Day 6 |
— |
— |
— |
|
A/Kawasaki/UTK-4/09 (H1N1) |
Day 3 |
6.5±0.5 |
2.45, 3.81 |
— |
|
|
Day 6 |
— |
3.18 |
— |
|
aFerrets were intranasally infected with 106 PFU (500 μl) of virus. Three ferrets from each group were euthanized on days 3 and 6 pi for virus titration. When virus was recovered from all three animals, average titres are presented. When virus was not recovered from all three ferrets, individual titres were recorded. None of the viruses tested was recovered from the spleens, kidneys, brains, intestines, or livers of infected animals. b—. virus not detected (detection limit: 2.3 log10 PFU/g). |
||||
TABLE A5-5 Virus Titres in Nasal Swabs of Inoculated and Contact Ferretsa
|
Virus |
Virus titres (mean log10 PFU/ml) in nasal swabs |
||||||
|
Day 1 |
Day 3 |
Day 5 |
Day 7 |
Day 9 |
|||
|
A/California/04/09 (H1N1) |
Pair 1 |
i |
7.1 |
4.0 |
—b |
— |
— |
|
|
|
c |
— |
6.8 |
4.3 |
— |
— |
|
|
Pair 2 |
i |
7.1 |
5.3 |
3.4 |
— |
— |
|
|
|
c |
— |
6.2 |
4.0 |
— |
— |
|
|
Pair 3 |
i |
5.9 |
5.3 |
3.9 |
— |
— |
|
|
|
c |
— |
6.4 |
5.9 |
2.1 |
— |
|
A/Kawasaki/UTK-4/09 (H1N1) |
Pair 4 |
i |
6.6 |
5.3 |
4.3 |
— |
— |
|
|
|
c |
— |
5.0 |
4.5 |
2.5 |
— |
|
|
Pair 5 |
i |
6.1 |
5.9 |
1.3 |
— |
— |
|
|
|
c |
— |
— |
— |
— |
— |
|
A/Victoria/03/75 (H3N2) |
Pair 6 |
i |
6.3 |
3.9 |
2.0 |
— |
— |
|
|
|
c |
— |
— |
6.0 |
3.7 |
2.5 |
|
|
Pair 7 |
i |
5.8 |
2.3 |
— |
— |
— |
|
|
|
c |
— |
— |
— |
— |
2.3 |
|
A/duck/Alberta/35/76 (H1N1) |
Pair 8 |
i |
— |
2.8 |
2.6 |
— |
— |
|
|
|
c |
— |
— |
— |
— |
— |
|
|
Pair 9 |
i |
— |
2.3 |
— |
— |
— |
|
|
|
c |
— |
— |
— |
— |
— |
|
|
Pair 10 |
i |
— |
4.9 |
3.7 |
4.0 |
— |
|
|
|
c |
— |
— |
— |
— |
— |
|
aFor pairs of ferrets, one animal was intranasally inoculated with 106 PFU of virus (500 μl) (inoculated ferret, i) and one day later, a naïve ferret was placed in an adjacent cage (contact ferret, c). Nasal swabs were collected from inoculated and contact ferrets every other day for virus titration. b—. virus not detected (detection limit: 1.3 log10 PFU/ml). |
|||||||
TABLE A5-6 Virus Titres in Organs of Infected Miniature Pigsa
|
|
Virus titres (log10 PFU/g) of infected animals with: |
|||
|
A/California/04/09 (H1N1) |
A/swine/Hokkaido/2/81 (H1N1) |
|||
|
Animal ID |
#1 |
#2 |
#5 |
#6 |
|
Nasal mucosa |
6.7 |
5.0 |
5.1 |
4.8 |
|
Oro/nasopharynx |
3.1 |
3.3 |
6.8 |
5.0 |
|
Tonsil |
3.2 |
—b |
4.5 |
4.4 |
|
Trachea |
6.3 |
5.5 |
5.8 |
5.3 |
|
Bronchus (right) |
5.6 |
6.1 |
5.9 |
6.5 |
|
Bronchus (left) |
6.7 |
6.5 |
5.4 |
6.3 |
|
Lung (upper right) |
7.8 |
6.6 |
6.1 |
4.5 |
|
Lung (middle right) |
7.5 |
6.7 |
6.1 |
5.5 |
|
Lung (lower right) |
6.4 |
6.8 |
5.3 |
4.5 |
|
Lung (upper left) |
6.8 |
6.4 |
6.8 |
5.1 |
|
Lung (middle left) |
8.0 |
7.6 |
4.7 |
5.5 |
|
Lung (lower left) |
6.2 |
7.4 |
5.5 |
4.7 |
|
Ileum |
— |
— |
3.5 |
— |
|
Jejunum |
— |
— |
2.8 |
— |
|
aSpecific-pathogen free miniature pigs were intranasally infected with 106.2 PFU (1 ml) of virus. Two animals from each group were euthanized on day 3 pi for virus titration. No virus was recovered from heart, spleen, kidneys, liver, duodenum, rectum, bladder, cerebrum, cerebellum, or brain stem. b—. virus not detected (detection limit: 2.0 log10 PFU/g). |
||||
TABLE A5-7 Virus Titres in Nasal Swabs from Infected Miniature Pigsa
|
|
|
Virus titers (log10 PFU/ml) of infected animals with: |
|||||||
|
A/California/04/09 (H1N1) |
A/swine/Hokkaido/2/81 (H1N1) |
||||||||
|
Animal ID |
|
#1 |
#2 |
#3 |
#4 |
#5 |
#6 |
#7 |
#8 |
|
|
Day 1 |
5.6 |
6.5 |
6.4 |
6.1 |
6.3 |
5.3 |
3.6 |
4.1 |
|
|
Day 2 |
6.5 |
6.5 |
7.4 |
6.7 |
5.5 |
5.5 |
5.1 |
5.6 |
|
|
Day 3 |
5.7 |
5.3 |
7.2 |
5.3 |
5.0 |
4.4 |
4.7 |
5.5 |
|
|
Day 4 |
|
|
3.7 |
3.6 |
|
|
4.9 |
4.6 |
|
|
Day 5 |
|
|
4.5 |
5.4 |
|
|
2.7 |
3.2 |
|
|
Day 6 |
|
|
4.3 |
5.3 |
|
|
2.8 |
3.2 |
|
|
Day 7 |
|
|
3.3 |
3.4 |
|
|
1.3 |
1.6 |
|
|
Day 8 |
|
|
—b |
— |
|
|
— |
— |
|
|
Day 9 |
|
|
— |
— |
|
|
— |
— |
|
aMiniature pigs were intranasally infected with 106.2 PFU of virus (1 ml) of virus. b —. virus not detected (detection limit: 1.0 log10 PFU/ml). Blank: not applicable, animals euthanized on day 3 pi. |
|||||||||
TABLE A5-8 Virus Susceptibility to Antiviral Compounds in Cell Culture
|
|
IC90 |
||
|
A/California/04/09 (H1N1) |
A/Kawasaki/UTK-4/09 (H1N1) |
A/Kawasaki/UTK-23/08 (H1N1) |
|
|
Oseltamivir carboxylatea |
10.56c |
2971.30 |
5.58 |
|
Zanamivir |
17.67 |
42.33 |
21.93 |
|
R-125489b |
4.24 |
11.70 |
10.17 |
|
T-705 |
0.16 |
0.23 |
0.13 |
|
aOseltamivir carboxylate is the active form of oseltamivir. bR-125489 is the active form of CS-8958. cIC90 value: mean µg/ml or nM of triplicate reactions for T-705 and other compounds tested, respectively. |
|||
TABLE A5-9 Virus Sensitivity in Neuraminidase Assays
|
|
IC50 |
|||
|
A/California/04/09 (H1N1) |
A/Osaka/164/09 (H1N1) |
A/Kawasaki/UTK-4/09 (H1N1) |
A/Kawasaki/UTK-23/08 (H1N1) |
|
|
Oseltamivir carboxylatea |
0.96c |
1.6 |
1313 |
1.88 |
|
Zanamivir |
0.32 |
0.43 |
0.79 |
0.36 |
|
R-125489b |
0.41 |
0.44 |
0.34 |
0.20 |
|
aOseltamivir carboxylate is the active form of oseltamivir. bR-125489 is the active form of CS-8958. cIC50 value: mean nM of duplicate reactions. |
||||