A9
RUMORS OF PANDEMIC: MONITORING EMERGING DISEASE OUTBREAKS ON THE INTERNET
Lawrence C. Madoff, M.D.70
International Society for Infectious Diseases
University of Massachusetts Medical School
John Brownstein, Ph.D.71
HealthMap
Children’s Hospital and Harvard Medical School
Summary
Unofficial or informal sources (also called “rumors” or “unstructured data”) of emerging disease outbreaks such as media reports and firsthand accounts have become an important mechanism for detecting these outbreaks (Brownstein et al., 2009a). These sources are disseminated by a variety of human-based and automated biosurveillance networks that are now routinely monitored by public health authorities at all levels. The 2005 revisions to the International Health Regulations (IHR) recognize that these sources often appear in advance of official notification of disease threats and are important in allowing the timely response to emerging diseases. Early media reports of respiratory illness in Mexico were among the first signs of the 2009-H1N1 influenza A pandemic and unofficial information sources are a critical mechanism for following the pandemic (Brownstein et al., 2009b). Studies to evaluate the sensitivity and specificity of informal sources and to improve the detection of emerging disease outbreaks are under way.
Introduction
The work of ProMED-mail (the International Society for Infectious Diseases Program for Monitoring Emerging Diseases) and that of the Institute of Medicine (IOM) have intersected in several ways since the well-known IOM (1992) publication of Emerging Infections: Microbial Threats to Health in the United States. This report ended a period of complacency in medicine where we had seen terms like the “conquest of infectious diseases” or “the end of infectious diseases,” and
|
70 |
Editor, ProMED-mail, International Society for Infectious Diseases; Department of Medicine, University of Massachusetts Medical School, Boston, Massachusetts. To whom correspondence should be addressed at Hinton State Laboratory Institute, 305 South Street, Jamaica Plain, MA 02130, Phone: 617-983-6800, e-mail: lmadoff@promedmail.org. |
|
71 |
Director, HealthMap; Children’s Hospital and Harvard Medical School, Boston, Massachusetts. |
we were brought into the present day by the emergence of several important diseases. The IOM has done a great deal in bringing these issues to the forefront.
A Tale of Two Emerging Diseases
Let us consider two emerging diseases, one before the birth of the Internet and one after it became a mature entity. The first was detected in 1981 (CDC, 1981). An article in Morbidity and Mortality Weekly Report (MMWR) was published recognizing a cluster of Pneumocystis pneumonia in gay men in Los Angeles. This outbreak became apparent only because treatment for Pneumocystis carinii was only available from the Centers for Disease Control and Prevention (CDC). CDC investigators noted this cluster of an unusual pathogen in an unusual patient population and that was the first recognition of what came to be known as HIV/AIDS.
However, we know well that HIV infection did not start in 1981 and that the epidemic had been going on for many years, largely in Africa, but also in other parts of the world. Certainly, HIV was widespread throughout Africa at the time it became apparent in the United States and yet, although this was not a subtle disease that could easily be missed, it was essentially unknown in the West.
What if we had known earlier? Two trends were visible in the early 1990s. One was the growth and popularization of the Internet, as it was coming out of the exclusive domain of the military and a few academic centers. This was the age, for example, when America Online (AOL) was born. The second was the recognition of the importance of emerging infectious diseases as HIV, Legionnaire’s disease, and resurgent rheumatic fever became evident. Many clinicians and scientists soon had access to electronic mail and some began to wonder whether this medium could be used as a way of speeding the transmission of information about emerging diseases.
A group of very prescient individuals, Steve Morse, Jack Woodall, and Barbara Hatch-Rosenberg, met at a UN-sponsored conference on detection of the use of biological weapons and began an email list among the attendees at the meeting. This mailing list became the nidus of the forum, the beginning of what was to become ProMED-mail (for Program for Monitoring Emerging Diseases). They began sending each other reports of emerging diseases, some of which might have involved the accidental or intentional release of biological weapons material. Soon many people wanted to share this information and join this mailing list, and from the initial group of about forty individuals, ProMED-mail was born (Madoff, 2004; Madoff and Woodall, 2005).
The outbreak referred to in this report and reproduced in Box A9-1 on the following page, of course, was the beginning of the pandemic that would become known as severe acute respiratory syndrome (SARS), the second disease in our tale. The report was in many ways typical of a ProMED report: an email from a reader who had overheard a rumor of what was going on in Guanzhou. Comments in an informal online source said that there were hospitals that had been
|
BOX A9-1 PNEUMONIA - CHINA (GUANGDONG): RFI ******************************************** Date: 10 Feb 2003 From: Stephen O. Cunnion, MD, PhD, MPH International Consultants in Health, Inc Member ASTM&H, ISTM This morning I received this email and then searched your archives and found nothing that pertained to it. Does anyone know anything about this problem? “Have you heard of an epidemic in Guangzhou? An acquaintance of mine from a teacher’s chat room lives there and reports that the hospitals there have been closed and people are dying.” SOURCE: ProMED (2003). |
closed and people who were dying. Since there were H5N1 avian influenza cases in Hong Kong at around this time, there were many questions about whether this could have been avian influenza. All ProMED reports are moderated and the moderator comment questioned whether this outbreak in fact was due to flu. It was not clear at the time.
SARS traveled quickly. The first Canadian death occurred in March 2003, raising the issue of who needs to know what and when (Poutanen et al., 2003). One of the problems with the traditional public health system is that it often presumes to know who needs access to information. But the ethos of ProMED has always been for transparency—that we cannot predict who is going to need to know. Who would have guessed that doctors in an emergency room in Toronto were going to be seeing cases of SARS so quickly after this mysterious illness appeared in Asia?
Hierarchical Surveillance Systems Versus Informal-Source Surveillance Systems
If we look at how traditional public health works, we can see that there is a flow of information from the ground up (Figure A9-1). Laboratories, practitioners, and members of the general public report to local officials, who then report to regional officials, and they to national officials. These, in turn, report to world bodies, who will publicize or convey back information to others as they deem necessary and inform the people who need to be involved in response to an outbreak or who need to be aware of it.
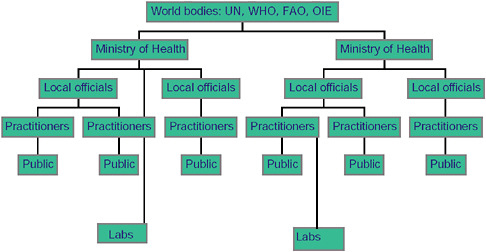
FIGURE A9-1 Hierarchichal nature of traditional public health reporting.
This is a powerful system and one that is very good at capturing much information and funneling it towards people who can collect, digest, and process it. However, it is also a system that takes time and one in which any break in the chain can lead to the loss of information.
In contrast, the idea behind informal biosurveillance systems (Figure A9-2) is that they not only deal with a hierarchical system but also can communicate in both directions with many levels of the system, such as local health officials, laboratories, ministries, and the World Health Organization (WHO), in addition to healthcare workers in the field, the public, and the media. This kind of process can speed the flow of information and can improve our ability to detect outbreaks.
Automated and Manual Biosurveillance Systems
Shortly after ProMED began operating, it became clear that the space on the Internet was becoming larger and larger and that it really was not possible for a person to look at everything and see everything. The idea of web crawling, or using automated search systems to mine the Internet for early warnings of emerging diseases, was born.
One of the first systems in the public health domain was the Global Public Health Information Network (GPHIN), established by the Public Health Agency of Canada and still operated by this entity (Mykhalovskiy and Weir, 2006). GPHIN remains a large and robust system that alerts public health officials and agencies such as CDC and the WHO that use it on a paid subscription basis to find out information about emerging diseases.
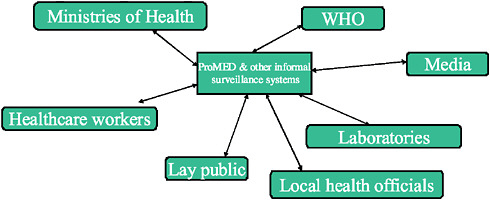
FIGURE A9-2 Informal-source surveillance.
HealthMap, based at Children’s Hospital in Boston and with whom ProMED has been recently collaborating, is another type of automated tracking system for mining the Internet for reports infectious diseases (Brownstein and Freifeld, 2007). HealthMap uses automated systems similarly to GPHIN and goes a step further in trying to place reports in a geographical context, that is, on a map. They use multiple sources, including ProMED reports, WHO reports, a variety of news media sources, and other freely available sources. HealthMap places these reports in a geographical context and makes them available to everyone at no cost. Through their collaboration, HealthMap and ProMED are developing innovative ways to look at how emerging disease information is collected and distributed.
There are several automated or partially automated “biosurveillance” systems, terminology used by Reis and others (2003). Veratect is a commercial project that makes use of a variety of automated and human sources that was begun by personnel from the ARGUS system. Other biosurveillance systems include MedIsys, a web crawling tool organized by the European Union and focused on that region, and Biocaster, based in Japan. A number of these systems have been extensively reviewed in a recent publication (Walters et al., 2009).
There are also several human-based or manual biosurveillance systems, including the well-known Epi-X run by CDC, which is a closed and confidential network open only to public health officials. Other systems include an emerging infections network (EIN) run by the Infectious Disease Society of America (IDSA) (Polgreen et al., 2008), the Geo-Sentinel system run by the International Society of Travel Medicine (ISTM) with involvement of CDC (Freedman et al., 1999), and WHO’s Global Outbreak Alert and Reporting Network (GOARN). Other systems focus on particular diseases or particular regions, or have a specialized focus.
An important point is that redundancy in this setting is good. These systems do not always pick up exactly the same signals, and having a variety of systems
in place helps keep checks and balances on the other systems, and helps fill in and recognize gaps.
Automated and Manual Biosurveillance Collaboration
The collaboration between ProMED and HealthMap began with the simple ideas that ProMED reports would feed HealthMap and that HealthMap would automatically parse the text-rich and unstructured ProMED reports, categorize them by disease, and place them on their map of the world with each map point containing a link back to the ProMED report.
Soon it became apparent that there were other areas for collaboration. One of them was that ProMED could exploit HealthMap’s automatic systems for finding and reporting on disease outbreaks in the news media and other sources. HealthMap could generate alerts for ProMED staff so that, for example, a ProMED virology moderator could receive reports up to several times a day on a particular set of viral disease search terms.
This would clearly help in the discovery of new content and, we hoped, improve the timeliness of reporting because it would not depend on a reader seeing a report and sending it to ProMED, or on a ProMED rapporteur or staff member finding the report and posting it. The improved timeliness would also improve the capture.
HealthMap uses automated systems to place events geographically (so-called “geotagging”), to categorize events by disease type and by location. These were not always accurate; for example, a news story that talks about a “plague of cheating” on the football field could be detected as a plague outbreak, or a report of a disease outbreak in guinea pigs could be placed in Guinea on the map.
These types of errors can be captured and corrected by what we call “ curation” of the HealthMap data, which is done manually by the ProMED staff. This kind of interaction, or community input from the ProMED staff into the HealthMap administration, would help improve the precision of the report. Also, the geotagging of HealthMap reports, which is automated, could place events at the country level, sometimes at the province or state level, but not more precisely than that. ProMED and HealthMap have developed tools so that its staff can virtually take a pin and place it on the precise location or locations of an outbreak.
This is essentially the marriage of an automated system with a human-based system in a way that strengthens both. Figure A9-3 shows a specialized map developed by HealthMap with the locations in ProMED reports. Each point is a clickable link that leads back to a full report of the outbreak.
Other Types of Informal Surveillance: Google Flu Trends
There are some other creative ways for monitoring Internet data for disease outbreak events.
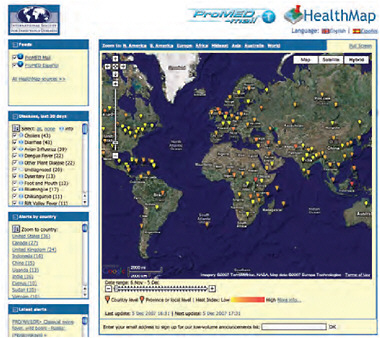
FIGURE A9-3 HealthMap screen shot.
SOURCE: See http://healthmap.org/promed/en.
One can monitor the “searchstream,” that is, the terms that people use to search on the Internet with the premise that if an outbreak is going on, people will search using terms related to that outbreak (Polgreen et al., 2008). If there is a flu epidemic, for example, people will search “flu,” or “fever,” or “Tamiflu,” or various similar terms. This approach has been refined and validated by Google Flu Trends (Ginsberg et al., 2009).
Google Flu Trends show that indeed if they monitor this searchstream it would detect influenza peaks at times before influenza-like illness (ILI) was actually detectable by other methods. Figure A9-4 is a screen shot of Google Flu Trends that shows the slow rise in ILI activity at this time of year. Also shown are the previous year’s peaks and the spring peak associated with the 2009-H1N1 influenza A outbreak here in the United States. This can be monitored at the national level and then also at the state level.
H1N1 and Informal Source Surveillance
What clues to the ongoing 2009-H1N1 influenza A outbreak could be found in informal sources? A weblog from the journal Science (excerpted in Box A9-2) (Cohen, 2009) has recorded the outbreak of swine flu day by day and, according
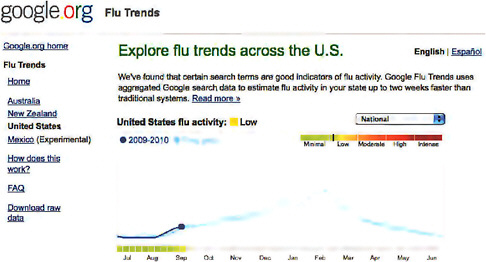
FIGURE A9-4 Google Flu Trends screen shot.
SOURCE: See http://www.google.org/flutrends/us/.
to its May 5, 2009, entry, the first documented cases were in Mexico City, later found to be confirmed with swine flu.
Retrospectively, one of the earliest reports appeared on HealthMap on April 1, 2009, from the Veracruz region of Mexico, showing an outbreak of pneumonia in that region.
Other blog entries discuss the role of the traditional public health system, its detection of the swine flu outbreak through laboratory findings, and its response to it. But at the same time informal sources were used, including HealthMap and Veratect, for recording information on this outbreak early on.
The April 11, 2009, entry discusses the revised IHRs, which became effective in 2007. The revised regulations recognize and to some extent codify the use of informal sources as valid sources of information for world public health and allow countries to report on reportable diseases of potential public health and international importance.
This was a key event that took many years to accomplish and WHO and the World Health Assembly deserve great credit for allowing it to happen. ProMED’s first report on April 22, 2009, followed the MMWR publication of the swine flu cases in California.
Box A9-2 is a brief summary of the use of informal information sources in the context of this current outbreak. Informal surveillance systems played a relatively minor role and the traditional public health system worked quite well in terms of this outbreak. The systems that have been in place for what we all thought would probably be an avian flu outbreak functioned quite effectively.
|
BOX A9-2 Swine Flu Day by Day 11 March: First documented symptoms (as of 5 May) in a Mexico City resident who later would be found to have confirmed infection with A(H1N1) swine flu. 30 March: A 10-year-old boy with fever, cold, and vomiting goes to the Naval Medical Center San Diego in California. As part of a clinical study, a nasopharyngeal swab is sent across town to the Naval Health Research Center (NHRC). 1 April: NHRC researchers determine that the boy is likely infected with influenza A, but they cannot subtype the strain. As per protocol, the sample is sent to Marshfield Labs in Wisconsin. HealthMap, a global disease alert system run by academics, flags a news story from Mexico about a strange respiratory outbreak in the state of Veracruz that has claimed two lives. 6 April: Veratect, a Kirkland, Washington-based company that scours news reports for emerging threats, reports in its subscription-only database that local Mexican health officials have declared an alert because of respiratory disease outbreak in La Gloria, Veracruz state, Mexico. 11 April: As per the International Health Regulations (IHR), the World Health Organization (WHO) has a pandemic alert and response network, which relies on designated people or institutions in each member country to report unusual disease patterns. PAHO, a regional office of WHO, asks the Mexican IHR “focal point” to verify the outbreak reported in the news. 12 April: Mexico’s director general of epidemiology confirms to PAHO the existence of acute respiratory infections Studies continue. Mexico’s focal point considers outbreak to be a “potential public health event of international importance” because it meets IHR criteria: severe public health impact and an unusual event. 21 April: Samples from Mexico arrive at PHAC. 22 April: CDC publishes first dispatch in the Morbidity and Mortality Weekly Report (MMWR) about two cases in California. Mexico reports atypical influenza behavior associated with severe pneumonia in various cities. InDRE [Instituto de Diagnóstico y Referencia Epidemiológicos] ships samples to PHACs National Microbiology Laboratory in Winnipeg and CDC. ProMED’s first report on human cases citing CDC report. 23 April: Samples from Mexico arrive at CDC. PHAC and CDC confirm Mexico cases are the same A(H1N1) of swine origin. SOURCE: Excerpted and adapted from Cohen (2009). |
It is important to mention, however, that informal sources have played a role in monitoring the progress of this outbreak and in keeping tabs on what is happening. To date, ProMED has posted more than 230 reports about it, many of them long, multipart reports, since April 2009.
Establishing a Baseline to Evaluate Informal Source Disease Surveillance
How well do informal sources work in detecting and reporting public health events? How can we look at this, evaluate it, and improve it? One of the activities that ProMED has pursued in collaboration with HealthMap is to take the archive of ProMED data, which consists of more than 40,000 free text reports dating back to 1994, and put them into a structured database. This was done by extracting, in a mostly automated way, the information from reports based on disease occurrence, type of disease, location, numbers of cases, dates of onset, dates of detection, dates of lab confirmation, and so forth, and putting them into a structured database and combining these data with external informal sources (such as news media). For the first time ProMED was able to see clearly what it had been doing.
Each circle in Figure A9-5 represents a particular disease. Many of the diseases that ProMED reports on are undiagnosed or unidentified, but there are quite a few others that are; avian flu is again at the forefront.
Through this approach, diseases can be followed over time. An individual disease can be observed and in some ways a bit of the history of emerging infectious diseases over the past 15 years can be seen. The system provides the ability to look at ProMED reports, the numbers of ProMED reports over time, and to track disease occurrence. It also shows that it is important to look not just for what is expected, but for that which is not.
From the structured database, it is possible to visualize the locations of ProMED reports over the period. It was noted that reports tend to occur most frequently in the northern hemisphere, in the information-rich and media-rich regions of the world, the United States and Western Europe in particular. Thus, the Southern Hemisphere, South America, Africa, and Asia are much less well covered by ProMED, a problem that ProMED is aware of and striving to solve.
One of the ways in which ProMED is addressing this issue is through regional programs. One of its oldest and best-established programs is in Latin America, which Eduardo Gotuzzo helped form in collaboration with the Panamerican Infectious Disease Association (API). ProMED has established relationships with the Mekong Basin Disease Surveillance Group in the countries that border the Mekong River in Southeast Asia in collaboration with WHO and the Rockefeller Foundation. With funding provided by Google.org it has established two new networks, in East Africa and in Francophone Africa, particularly West Africa; and the Nuclear Threat Initiative (NTI) provided funding for a Russian-language system based in the former Soviet Union.
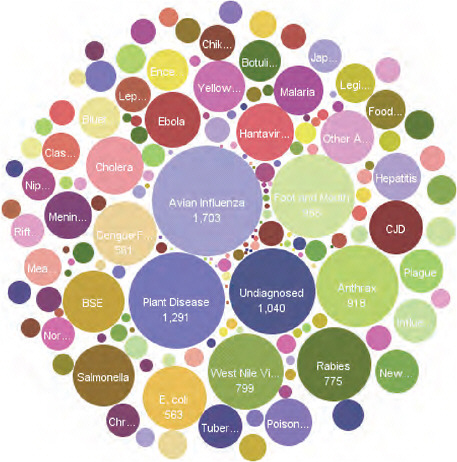
FIGURE A9-5 Quantitation of subjects in ProMED reports from 1996-2008. The size of each circle corresponds to the number of reports on a given topic.
These are all areas where disease surveillance was and continues to be relatively poor, so the regional networks serve two functions: one is to improve regional collaboration to help providers and public health within these regions, and the other is to help inform the broader world about problems within these regions. They serve that function nicely and ProMED expects to see further growth within these networks.
ProMED and HealthMap have studied the structured global baseline of its reports, news reports, and other reports by comparing the dates of detection of a series of outbreaks and establishing a group of distinct outbreaks based on geographic region and time period. We first assessed the sensitivity and scope of these datasets with a descriptive analysis of ProMED reports. Next, analyzing the WHO’s Global Alert and Response (GAR) reports between 1996 and 2008, we
selected human outbreaks of infectious, non-food-borne diseases that were not considered seasonal or endemic to the region, and were not isolated, imported cases, then extracted the corresponding ProMED and HealthMap reports. WHO reports are not necessarily the first report or the first time that WHO becomes aware of or works on an outbreak. However, they are a gold standard, a publicly available record that could be used to look at these data.
We subsequently created timelines of the progression and reporting for each outbreak, compared the timing in reporting by official and informal sources, and attempted to identify factors that may contribute to differences in the timing of reporting. A qualitative analysis of the ProMED data set revealed a sensitive increase in the number of ProMED reports for specific diseases and locations around the time the WHO reported corresponding outbreaks. Figure A9-6 is a timeline showing time differences between official WHO reports, informal reports, and various “outbreak milestones.” The line at 0 days represents no lead/lag over the WHO report. The line represents the date of the EPR report and each blue dot represents the date of something else, a date identifiable within a ProMED report. For the earliest ProMED report, the dates of symptom onset, hospitalization, death, or lab confirmation are recorded and the black diamonds represent the median time before the publication, or the official verification of the report. For 355 WHO-confirmed outbreaks, ProMED reported on average 18 days (95 percent C.I.: 12.2-23.8) earlier than WHO’s GAR reports, while HealthMap reported 12 days (5.4-18.2) earlier (Figure A9-1). A further analysis revealed country- and disease-dependent differences in reporting. Sensitivity was 0.946 (0.923-0.970) for ProMED (n = 355), and 1.000 (1-1) for HealthMap (n = 39). This preliminary work shows that informal online disease reporting can facilitate both sensitive and timely detection of disease outbreaks. An examination of the finer-grained differences in reporting depending on the disease and location reveals the most informatively valuable areas in which efforts for monitoring the vast amount of informal online reports should be targeted. Early and accurate recognition of outbreaks is crucial for expediting the initiation of appropriate interventions.
It is also possible to look at these data by country and see clear differences between individual countries and regions in the speed of early reporting according to disease type. Some diseases are reported in a much more timely way than others, both by ProMED and by WHO. The structured database can be used to try to identify the gaps in ProMED’s and other informal surveillance systems too, by disease type, by geography, and by language; find the most effective signals, both in terms of accuracy and timeliness; and learn ways to reduce noise. Signal-to-noise ratio is a major problem with biosurveillance systems.
It is possible that one of the reasons more alarm bells did not go off when outbreaks of pneumonia were being reported in Mexico is that outbreaks of pneumonia are frequent. How do we know which ones matter and how do we know which ones are worth our attention? Hopefully an analysis of these data will help
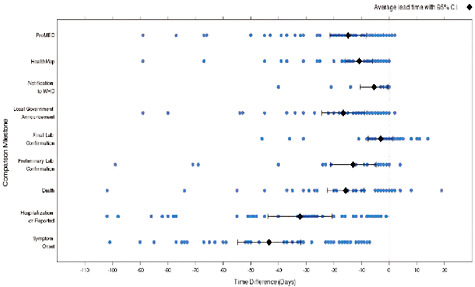
FIGURE A9-6 Timeline showing time differences between official WHO reports, selected informal reports, and various “outbreak milestones.”
identify the right signals and make it possible to shorten the interval between outbreak and detection. Using this analysis, we can prospectively evaluate these findings using ProMED and HealthMap as we go forward.
Conclusions
The monitoring of informal sources of information or rumors is an important tool in public health. It is free of political constraints, it is transparent, and it allows for clinicians and other observers to have a role in reporting on emerging diseases.
Informal sources of information can complement and assist the traditional public health systems rather than attempt to replace them. Multiple systems are complementary and enhance the ability to detect outbreaks. Those using informal surveillance systems need to maintain a broad view and not focus on a particular disease, region, or type of disease. They need to keep their eyes on the horizon and need to work to improve the signal-to-noise ratio and to improve geographic coverage.
Acknowledgments
We are grateful to the following people and groups: ProMED and HealthMap participants, subscribers, and staff; Google.org; the Oracle Corporation; Harvard
School of Public Health; Nuclear Threat Initiative; the Rockefeller Foundation; The Bill & Melinda Gates Foundation; and the Robert Wood Johnson Foundation. We thank Maria Jacobs for editorial assistance.
References
Brownstein, J. S., and C. C. Freifeld. 2007. HealthMap: the development of automated real-time Internet surveillance for epidemic intelligence. Eurosurveillance 12(11):E071129.5.
Brownstein, J. S., C. C. Freifeld, and L. C. Madoff. 2009a. Digital disease detection—harnessing the Web for public health surveillance. New England Journal of Medicine 360(21):2153-2155, 2157.
———. 2009b. Influenza A (H1N1) virus, 2009—online monitoring. New England Journal of Medicine 360(21):2156.
CDC (Centers for Disease Control and Prevention). 1981. Pneumocystis pneumonia—Los Angeles. Morbidity and Mortality Weekly Report 30(21):1-3.
Cohen, J. 2009. Swine flu outbreak, day by day, http://blogs.sciencemag.org/scienceinsider/special/swine-flu-timeline.html (accessed November 15, 2009).
Freedman, D. O., P. E. Kozarsky, L. H. Weld, and M. S. Cetron. 1999. GeoSentinel: the global emerging infections sentinel network of the International Society of Travel Medicine. Journal of Travel Medicine 6(2):94-98.
Ginsberg, J., M. H. Mohebbi, R. S. Patel, L. Brammer, M. S. Smolinski, and L. Brilliant. 2009. Detecting influenza epidemics using search engine query data. Nature 457(7232):1012-1014.
IOM (Institute of Medicine). 1992. Emerging infections: microbial threats to health in the United States. Washington, DC: National Academy Press.
Madoff, L. C. 2004. ProMED-mail: an early warning system for emerging diseases. Clinical Infectious Diseases 39(2):227-232.
Madoff, L. C., and J. P. Woodall. 2005. The Internet and the global monitoring of emerging diseases: lessons from the first 10 years of ProMED-mail. Archives of Medical Research 36(6):724-730.
Mykhalovskiy, E., and L. Weir. 2006. The Global Public Health Intelligence Network and early warning outbreak detection: a Canadian contribution to global public health. Canadian Journal of Public Health 97(1):42-44.
Polgreen, P. M., Y. Chen, D. M. Pennock, and F. D. Nelson. 2008. Using Internet searches for influenza surveillance. Clinical Infectious Diseases 47(11):1443-1448.
Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Bruham, A. J. McGreer, National Microbiology Laboratory Canada, Canadian Severe Acute Respiratory Syndrome Study Team. 2003. Identification of severe acute respiratory syndrome in Canada. New England Journal of Medicine 348(20):1995-2005.
ProMED. 2003. Pneumonia—China (Guangdong): RFI, http://www.promedmail.org/pls/pm/pm?an=20030210.0357 (accessed November 20, 2009).
Reis, B. Y., M. Pagano, and K. D. Mandl. 2003. Using temporal context to improve biosurveillance. Proceedings of the National Academy of Sciences 100(4):1961-1965.
Walters, R., P. Harlan, N. Nelson, and D. Hartley. 2009. Data sources for biosurveillance. In Wiley handbook of science and technology for homeland security, edited by J. Voeller. Hoboken, NJ: John Wiley and Sons, Inc. Pp. 1-17.














