References
Denny FW, Glezen WP, Karzon DT, Katz SL, Krugman S, McIntosh K, Parrott RH. Commentary. J Pediatr. 1976;88:1057.
Department of Health and Human services [sic]. Annex 11: Pandemic influenza response and preparedness plan. Washington: The Department; 2003 Aug 26.
Dowdle WR. Influenza pandemic periodicity, virus recycling, and the art of risk assessment. Emerg Infect Dis. 2006;12:34–9.
Dowdle WR, Hattwick MAW. Swine influenza virus infections in humans. J Infect Dis. 1977;136: S386–9.
Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, Sharrar RG, et al. Legionnaires’ disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–97.
Hodder RA, Gaydos JC, Allen RG, Top FH Jr, Nowosiwsky T, Russell PK. Swine influenza A at Fort Dix, New Jersey (January–February 1976). III. Extent of spread and duration of the outbreak. J Infect Dis. 1977;136:S369–75.
Kilbourne ED. Flu to the starboard! Man the harpoons! Fill with vaccine! Get the captain! Hurry! New York Times. 1976 Feb 13. p. 32, col. 4.
Obama B, Lugar R. Grounding a pandemic. Op-ed section. New York Times. 2005 Jun 6. [cited 1 Nov 2005]. Available from http://www.nytimes.com/2005/06/06/opinion/06obama.html?ex=1130994000&en=1b199f715505a19c&ei=5070.
Schmeck HM Jr. More deaths reported after shots but no link to flu vaccine is found. New York Times. 1976 Oct 14; Sect. A:1+.
Schoenbaum SC, McNeil BJ, Kavat J. The swine-influenza decision. N Engl J Med. 1976;295:759–65.
Schonberger LB, Hurwitz ES, Katona P, Holman RC, Bregman DJ. Guillain-Barré syndrome: its epidemiology and associations with influenza vaccination. Ann Neurol. 1981;9(Suppl):31–8.
Schwartz H. Swine flu fiasco. New York Times. 21 Dec 1976:33.
Unprepared for a flu pandemic [editorial]. New York Times. 2005 Jul 17; Sect. 4:11 (col. 1).
A12
SOUTHERN HEMISPHERE, NORTHERN HEMISPHERE: A GLOBAL INFLUENZA WORLD
Kennedy F. Shortridge, Ph.D.79
The University of Hong Kong
Comment
The global response to the 2009-H1N1 influenza A pandemic has been heartening, drawing in no small measure from experience gained from the 1997 influenza H5N1 “Bird Flu” and 2002/2003 severe acute respiratory syndrome (SARS) outbreaks. In each case, recognition of the virus’s principal source, chicken in
the case of the H5N1/HK/97 virus (Shortridge, 1999a) and civet cat80 for the SARS coronavirus (Guan et al., 2003), prevented further zoonotic spread. The Bird Flu incident provided the foundation for dealing with SARS. There is still much to be learned about influenza toward improving pandemic preparedness. This will require renewed vigor across a range of influenza studies and public health measures. It cannot be denied that our goal must be to the vision of no more pandemics. The 2009-H1N1 influenza A pandemic has rekindled this vision and is seen, in part, against a background of 30 years in Hong Kong, a place that in effect functioned as an influenza sentinel post. This informal designation followed the hypothesis that southern China is an epicenter for the emergence of pandemic influenza viruses (Shortridge and Stuart-Harris, 1982), a hypothesis that was the anchor for subsequent thinking and goals considered here.
Another H1N1 Virus
Should we have been surprised?
Simple epidemiological information based on the ages of those initially infected by the pandemic H1N1 virus in 1918 in Canton (now Guangzhou) in southeastern China suggests that an H1N1-like virus had been active in the area for about 11 years prior to 1918 (Shortridge, 1999b). Molecular evolutionary studies indicate that such a virus had circulated from as early as 1911 (Smith et al., 2009a). These data, seroarcheological studies, and virus isolation suggest that H1N1 or H1-like viruses circulated in humans four times over the last 120 or so years. Indeed, the H1N1 lineage may have extended back to the 1830s (Gammelin et al., 1990), a dating in accord with the emergence of a pandemic in China, possibly southern China, in 1830-1831 (Patterson, 1985). Thus, it is clear that this subtype of virus has an affinity for the human host. This includes the 1950-like H1N1 virus that reappeared in 1977 (Kendal et al., 1978; Nakajima et al., 1978) whether or not it is a rogue virus. And now, in 2009 another H1N1 virus has been able to establish a niche in humans—and doing so with pandemic gusto—in the face of a global population seemingly well protected with H1 antibodies. Whereas the pandemic viruses of the last century arose in southern China (see below about the 1918 virus) and were of Eurasian genetic extraction, the H1N1 2009 virus arose in North America and was of mixed geographical and host extractions.
The answer to the question posed is both “no” and “yes.” There is much to learn about the epidemiology, ecology, and science of H1N1 viruses.
China H1N1—Then and Now
While the 1918 pandemic seemingly manifested in Western Europe and the United States in late winter and early spring, it did not apparently do so in China
until June in Canton (Table A12-1; Cadbury, 1920; Chun, 1919; Shortridge, 1983). Infection there was mild, and no influenza deaths were recorded at the Canton Hospital. Consonant with the time of the Canton outbreak, more solid data come from Hong Kong 110km to the southeast, where influenza deaths abruptly started in June, continuing thereafter throughout the year (Table A12-2; Shortridge, 1983). The effects of the outbreak were most severe from 1918 to
TABLE A12-1 China H1N1—Then and Now
|
1918 |
H1N1-like virusa |
||
|
South |
Virus smoldering ~11 years pre-1918? |
||
|
South |
June, outbreaks in pigs following northward human spread |
||
|
North-east |
October, pigs infected at same time as humans |
||
|
South |
Hong Kong, June, human deaths |
||
|
2009-H1N1 influenza A human cases virologically confirmed (publicly available data)b |
|||
|
Mainland |
5,592 |
September 9 |
|
|
Hong Kong |
15,357 |
September 9 |
1st case May 1 |
|
Taiwan |
94 |
September 9 |
|
|
Macao |
1,435 |
September 9 |
|
|
Porcine infection not known to this time. |
|||
|
aAdapted from Cadbury (1920), Chun (1919), and Shortridge (1983). bCHP (2009). |
|||
TABLE A12-2 Deaths Due to Influenza in Hong Kong for Each Month from 1918-1928
1922, exacting a heavy toll on a territory already coping with other infectious disease problems including malaria, tuberculosis, plague, smallpox, cholera, and measles (e.g., Starling, 2007) (Box A12-1).
The 2009-H1N1 influenza A virus was recognized in the United States and Mexico in mid-April. Airplane travel could quickly seed the virus worldwide, more so than in previous pandemics, catching the Southern Hemisphere winter and summer in the Asian tropics at the usual time of peak influenza activity there (see below). Hong Kong, with its experience of dealing with influenza, was able to react with sound public health measures and diagnostic services. The first case was recognized on May 1st and 4 months later there were around 15,000 cases, a figure that is probably an underestimate. The vast majority of cases were mild. At the time of the Institute of Medicine (IOM) workshop (mid-September 2009), the numbers of cases in mainland China, Taiwan, and Macao were trickling in. Elsewhere in tropical Asia, reported numbers were also low (SEARO/WHO 2009).
|
BOX A12-1 Brief Overview of the Origin of the 1918 Pandemic H1N1 Virus and the Classical H1N1 Swine Flu Virus Origin of the 1918 Pandemic Virus Differing times of yearly influenza occurrence in temperate and tropical zones may be a key factor, something not appreciated until the late 1980s (see later discussion). Economic migrants from the southestern part of the influenza epicenter would have carried the H1N1 virus to Western Europe and the United States prior to 1918. Outbreaks there occurred in late winter and early spring 1918 before later doing so that year in the Canton area and in Hong Kong in summer, the usual time of peak occurrence (Shortridge, 1999b). Origin of Classical Swine Flu Virus Outbreaks of influenza in pigs followed the spread of the virus northward of Canton and were recorded at the time of human infection in the far northeast (Chun, 1919), suggesting initial human-to-pig transmission. Could there have been a similar occurrence in pigs in the United States around 1918 resulting in endemicity? Infection of piglets experimentally with the reconstructed 1918 pandemic virus is of interest (Weingartl et al., 2009). Phylogenetic studies on the hemagglutinins (HAs) from a range of H1N1 viruses suggest that the classical swine/Iowa/15/30 virus was not a direct descendant of the 1918 pandemic virus, rather from a common ancestral virus around 1905 (Kanegae et al., 1994). It seems that much activity was taking place in southern China involving an H1 or H1-like virus from around the turn of the century until 1918. While much must be conjectured, we may overlook it to our peril. |
Temporal Occurrence
Attention has been drawn to the influence of season on times of influenza occurrence. Influenza had been thought to be a winter infection. It clearly occurs in temperate zones with a recognizable disease burden, but in the tropics and subtropics? A meeting was convened in Singapore in late 1988 with influenza colleagues from around the Pacific Basin with an emphasis on those from Asia. Although much of the data were rudimentary or incomplete, generalized patterns of occurrence were discernible and are shown in Figure A12-1 (Reichelderfer et al., 1989). The winter occurrence in northern Asia was consistent with winter elsewhere, but in tropical and subtropical zones the influenza virus was present for much of the year or year-round, often with a peak at the hottest, weather-
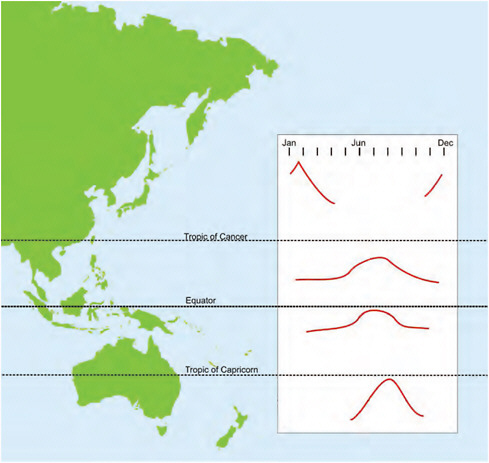
FIGURE A12-1 General patterns of temporal occurrence of influenza A and B viruses in eastern Asia and Australasia.
SOURCE: Adapted from Reichelderfer et al. (1989).
affected times usually midyear. An important fallout from this was the recognition that the disease burden in the Asian tropics and subtropics is just as significant as that in temperate zones, warranting inter alia recommendation for influenza vaccination (Chiu et al., 2002, 2009; Wong et al., 2004, 2006).
The ability of seemingly the same influenza viruses to manifest in a cold, dry, or damp winter and year-round in a hot, humid climate is more than a little curious. Experimental guinea pig studies suggest that aerosol transmission predominates during winter in temperate regions and through contact in the tropics (Lowen and Palese, 2009). No doubt, a variety of factors are involved, for example, host susceptibility, virus transmissibility including quantity and quality of virus spread, and, perhaps, hand hygiene in different zones given that it seemingly contributes to preventing household infections in subtropical Hong Kong (Cowling et al., 2009). The writer takes the view that stress is also probably a factor. He considers that the most stressful time in a temperate zone is winter, whereas the tropics are stressful year-round, particularly so with seasonal climatic changes (Chan et al., 2009). In either case, the lining of the upper respiratory tract may be compromised, facilitating influenza infection and infections by other respiratory viruses. It is noteworthy that epidemics of respiratory syncytial virus pneumonia in children in equatorial Indonesia are associated with weather changes (Omer et al., 2008).
Patterns of Occurrence of 2009-H1N1 Influenza A in Australasia and Hong Kong
The time of appearance of the 2009-H1N1 influenza A virus provided the opportunity to see how the Southern Hemisphere would behave as a guide to dealing with the forthcoming outbreak in the Northern Hemisphere winter. As a bonus, the virus would be expected to be present in tropical East Asia in the Northern Hemisphere around the same time (Figure A12-1).
In New Zealand, the outbreak followed the usual seasonal pattern starting in May and declining from mid-July onward (Baker et al., 2009). The situation was generally similar in Australia, with a little interstate variation (Australian Influenza Surveillance Summary Report, 2009). In both countries, the 2009-H1N1 influenza A virus soon predominated the seasonal virus. Its morbidity was probably lower than might have been expected. While health services coped well, those most affected usually had underlying conditions. Meanwhile, in Hong Kong a similar situation prevailed: 2009-H1N1 influenza A started to rise in mid-June, peaking in July and beginning to fall in August. The resumption of school in September saw a big rise in cases, and then falling off by early October (CHP, 2009). By contrast with Australia and New Zealand, the seasonal H3N2 was less easily displaced and comprised as much as about 50 percent in the peak period falling to less than 5 percent by early October (Table A12-3). Other respiratory viruses were isolated during midsummer including medically important paramyxoviruses,
which occur variably throughout the year, and respiratory syncytial virus in the summer (Sung et al., 1987, 1992).
Thus, the experience in Australasia and Hong Kong in the first round indicates that the pandemic is mild. Experimental ferret infection studies by Perez et al. (2009) showed that the 2009-H1N1 influenza A virus “is more transmissible than, and has a biological advantage over, prototypical seasonal H1 or H3 strains.” The authors also point out the possibility of dual infections by H1N1 2009 and H3N2 viruses and clinical implications. In this sense, the isolation of a high percentage of H3N2 viruses at the time of the summer peak in Hong Kong (Table A12-3) is of relevance. Ideally, as many isolates of the three categories of variants as practicable from the Northern Hemisphere winter should be sequenced for evidence of reassort ment, as should those obtained from tropical East Asia throughout the year.
Recycling of H Subtypes
The appearance of the 2009-H1N1 influenza A virus has rekindled the notion that there is a limited range of H subtypes that can cause pandemics, namely H1, H2, and H3. This does not exclude the possibility that other H subtypes have expressed this trait in earlier centuries or that we are in a period of transition of human susceptibility in which other H subtypes, say H5, H7, and H9, are possible pandemic candidates.
With the caveat of the reliability of seroarcheology and interpretation of the historical record, H1N1 or H1N1-like viruses have occurred singly or have coexisted with H2 or H3 viruses (Shortridge, 1992, 1999b). This raises concern that a similar situation could arise with the 2009-H1N1 influenza A virus.
Immediate concerns are that (1) it usurps one, the other, or both prevailing H1N1 and H3N2 variants and (2) it coexists with one, the other, or both of the variants. A medium- to long-term concern is that an H2-like virus will emerge, usurp, or coexist with prevailing variants.
Based on the evidence that H1 and H3 viruses cocirculated before the 1918 pandemic, it seems a reasonable possibility that the prevailing variants will be usurped. Preliminary ferret infection experiments suggest that H1N1 2009 viruses will do this (Perez et al., 2009). A range of such experiments must be pursued.
And what might the future hold for the influenza type B virus in the face of the new H1N1 2009 competitor or a triad of type A variants?
H5N1 Virus
While the 2009-H1N1 influenza A virus may have overtaken the H5N1 virus in pandemicity, occasionally reported human cases and continued virus presence in avians are cause for concern (WHO, 2009). The virus has spread
TABLE A12-3 Respiratory Pathogens Isolated in Hong Kong at Selected Times During the 2009-H1N1 Influenza A Outbreaka
|
Agent |
July 13-19, 2009 |
August 10-16, 2009 |
September 27 to October 3, 2009b |
|||
|
Number |
Type/Subtype |
Number |
Type/Subtype |
Number |
Type/Subtype |
|
|
Adenovirus |
16 |
Untyped: 3 Type 1: 3 Type 2: 2 Type 3: 1 Type 4: 1 Type 5: 2 Type 6: 1 Type 8: 2 Type 14: 1 |
15 |
Untyped: 8 Type 1: 2 Type 2: 5 |
|
|
|
Influenza A |
1022 |
Untyped: 4 H1: 65 H1 2009: 549 H3: 404 |
1691 |
Untyped: 1 H1: 88 H1 2009: 846 H3: 756 |
1001 |
Untyped: H1: 21 H1 2009: 910 H3: 54 |
|
Influenza B |
13 |
|
30 |
|
14 |
|
|
Parainfluenza |
38 |
Type 1: 22 Type 2: 3 Type 3: 12 Type 4: 1 |
17 |
Type 1: 4 Type 2: 1 Type 3: 12 |
|
|
|
Respiratory syncytial virus |
10 |
|
37 |
|
|
|
|
Rhinovirus |
6 |
|
22 |
|
|
|
|
Mycoplasma pneumoniae |
7 |
|
3 |
|
|
|
|
aData abstracted from reports from Virology Division, Centre for Health Protection, Department of Health, Hong Kong SAR (CHP, 2009). bBlank spaces in this timeframe denote no isolation reported. |
||||||
from eastern Asia across the Eurasian land mass to northern Africa. The pattern of human infection in Egypt is different from that recorded elsewhere (Dudley, 2009), and asymptomatic infection is increasingly appreciated (Dudley, 2008). The virus appears to be endemic in poultry in southern China (Chen et al., 2006) and recent studies have shown its presence in wild birds in 10 provinces (Kou
et al., 2009) and in mammals, namely raccoon dogs (Qi et al., 2009) and pikas (Zhou et al., 2009). Continued virus surveillance and responsible reporting are necessary. International human and animal health authorities now face exceedingly complex influenza issues.
Food, Flu, and the Future
With the big increase in global population in the past 60 or so years has come the need to supply dietary meat protein on an unprecedented scale, bringing with it infectious agents arising from intensified animal production (Greger, 2006). The 2009-H1N1 influenza A virus most likely arose in the United States and Mexico region through intensive pig production (Greger, 2009), distant from the epicenter of southern China. This suggests the prospect of new influenza pandemic epicenters elsewhere in the future.
A similar situation applies to poultry meat and egg production in Asia generally and China in particular, where chicken has become a “near-daily staple.” This was facilitated by the control of Newcastle disease, the major poultry disease of the region, through vaccine improvements and distribution (Shortridge, 1982; Copland, 1987; Higgins and Shortridge, 1988), benefits that have extended to minor poultry. The industrialization of poultry production probably played a major role in the genesis and spread of the H5N1 virus as pig production seemingly has done for the 2009-H1N1 influenza A virus. A more serious problem could arise through lapses in biosecurity, resulting in an avian or porcine reassortant capable of great human devastation.
In a land noted for consumption of wildlife and where nothing is wasted (Parham, 2006), wildlife farming in China poses new threats. The H5N1 virus was isolated from farmed Bar-headed Geese at Qinghai Lake, western China (Butler, 2006), a location from which migratory birds may have started the virus’s westward spread across Eurasia. (Commercially raised civet cats were the source of the SARS virus; see Guan et al., 2003.)
Industrial avian and animal production have wider implications for human health (Nierenberg and Garces, 2005). At this point it is hard to see how a projected human population of 9 billion in 2050 will have adequate food security (Adams, 2009). The time is now ripe to put microbes, biotechnology and reconstituted ribosomes to good use for the large-scale production of dietary protein.
Some Areas for Investigation
The appearance of the 2009-H1N1 influenza A virus and its ability to give rise to a pandemic prompt a number of areas for follow-up investigation. Some of these intersect. The areas are listed in Table A12-4 and briefly discussed.
TABLE A12-4 Some Areas for Investigation
|
Category 1: 2009-H1N1 influenza A virus |
|
|
Category 2A: Recycling of HA antigens |
|
|
Category 2B: Clinical |
|
|
Category 3: The HA |
|
|
Category 4: A rock and a hard place |
|
2009-H1N1 Influenza A Virus
The role of the pig as a “mixing vessel” is central, a hypothesis set rolling by the isolation of early human H3N2 variants in pigs from China (Shortridge et al., 1977; Scholtissek et al., 1985). Events surrounding the genesis of the 2009-H1N1 influenza A virus need not be reiterated. Suffice it to ask whether the acquisition of Eurasian “avian-like” swine genes through reassortment in the pig could have facilitated the transition of a virus of North American geographical lineage into a pandemic virus? The Eurasian genetic composition of the pandemic viruses of the past century prompts this question. While there does not seem to be a common genetic template for a pandemic virus (save for a change of HA), the recognition of likely progenitors of a pandemic virus some years earlier in the case of the 2009-H1N1 influenza A virus puts a new spin on the concept of pre-pandemic “bridging strains” (Shortridge et al., 1979). Moreover, the virus had seemingly transmitted to humans several months before the recognition of the outbreak (Smith et al., 2009b). Maybe George Orwell in his satire on farm animals got it right—pigs rule (Orwell, 1945).
That the 2009-H1N1 influenza A virus was able to squeeze through a human “firewall” of H1 antibodies signals that its HA antigen intrinsically has an unrecognized propensity for antigenic change or that an adaptation process has taken
place over a number of years in the massive high-density pig populations of North America. While it is not possible to predict the number of antigenic variants that the virus is capable of producing, current information suggests that its life expectancy may be relatively short, akin, say, to the 11-year reign of the H2N2 Asian virus. In addition, the 2009-H1N1 influenza A virus may behave as a short-term third-party or facilitating virus undergoing further reassortment in a pig or human with a novel HA leading to a new round of pandemicity.
In addition to ramping up virus surveillance of pigs, it is essential that recommendations on industrial pig production be pursued (Greger, 2009). This is now an inescapable global issue.
Recycling of HA Antigens
The reappearance of an H1N1 pandemic virus in 2009 revives the concept of recycling of H subtypes. Furthermore, its presence in humans along with prevailing H3N2 variants may pave the way for an H2 virus, perhaps sooner rather than later, either as a totally “new” virus (as was the case in 1957) or as a reassortant of the H1N1 or H3N2 virus or both. If it were a totally “new” H2N2 virus, matching its gene constellation with that of the 1957 pandemic virus would be illuminating toward understanding the genetic factors that engender pandemicity. The apparent cocirculation of H1- and H2-like viruses at least in southern China from 1888 to 1898 (Cantlie, 1891; Shortridge, 1999b) raises the possibility of a repeat of this in the early part of this century. H2 or H2-like viruses do not appear to have cocirculated with H3-like viruses. Evidence of H2 virus infection of humans must be sought.
That being said, it is important that all unidentified human and porcine isolates be checked for H2 gene sequence or HA antigen and for, say, H5, H7, and H9 viruses (CHP, 2009). Recent estimates suggest that the pandemic H2N2 1957 virus entered the human population two to six years before the pandemic (Smith et al., 2009a). The isolation of avian/swine reassortant H2N3 viruses from diseased pigs in the United States is of concern (Ma et al., 2007). While the isolation of H2 viruses from pigs or humans may prove elusive, it would be wise to set about the detection of H2 antibody in humans born after 1968, particularly in children. The interpretation of serological studies for the detection of presumptive antibody to avian influenza viruses in mammalian sera by widely used hemagglutination inhibition is more complex than the simple test belies. Whatever the diagnostic approach, it is suggested that investigation be carried out through an international collaborative effort using standardized reagents and techniques. Furthermore, there should be an international group to set about this task as a matter of urgency.
Clinical
An epidemic of encephalitis lethargica (EL) followed the 1918 pandemic, claiming the lives of an estimated half-million people. The appearance of an
H1N1 pandemic virus partly of swine origin nine decades later raises the possibility of EL in this one.
It is not known whether the 1918 H1N1 virus was the cause of the EL epidemic. von Economo, who initially described it, apparently thought not. Recent investigation of EL and other disorders suggests that EL may be the outcome of an autoimmune response to streptococcal infection (Dale et al., 2004; Vincent, 2004; Vilensky and Gilman, 2006). The extent of post-H1N1 influenza streptococcal pneumonia in survivors of the 1918 pandemic is not known. Nonetheless, it would seem reasonable to keep the possibility of EL-like illness in clinical purview now, even though the 2009-H1N1 influenza A pandemic is generally mild and with limited antibiotic cover.
The HA
Although there is accumulating evidence indicative of recycling of H subtypes H1, H2, and H3, the possibility that the other 13 H subtypes have pandemic capabilities cannot be excluded irrespective of their avian hosts and ecological backgrounds. There is no simple answer, but it might initially be approached in two parts.
-
HA structure and function through determination of three dimensional structure, antigenic epitope mapping, and receptor binding site.
-
Biologically through the detection of virus escape mutants in culture under pressure of using extensive panels of monoclonal antibodies as a gauge of a virus H subtype’s ability to escape the host’s immune response (e.g., Kaverin et al., 2004).
It could be reasoned that, unless an HA has intrinsic hierarchic antigenic capabilities to produce a succession of antigenic variants in humans, it is unlikely to progress beyond initial infection. The detection of presumptive antibody to all avian H subtypes examined in rural southern China contributes to this view (Shortridge, 1992). The ability of H3N2 viruses to have a 41-year presence thus far in humans might be inferred from the coexistence of many antigenic variants in the domestic duck population of southern China (Shortridge et al., 1990). In this sense, H1 and H2 viruses are an enigma. With so few H1 and H2 viruses in domestic ducks and wild waterfowl, it will be as illuminating as it is critical to measure the extent of the H1N1 2009 virus’s ability to produce escape mutants—even more so for the H2 subtype because of the increasing possibility of its being recycled.
Intrinsic to understanding the outcome of infection is understanding the earliest stages of pathogenesis in the upper respiratory tract. Might it be possible to distinguish among events that take place in, say, the nasal turbinates following infection by a seasonal flu virus, a precursor pandemic virus, and the earliest detected pandemic virus? Whatever the situation, it would be beneficial to know
much more about the overall process of pathogenesis given that human defenses have been breached since 1997 by H5, H9, H7, and H1 viruses.
While the role of cell receptor type in determining the outcome of virus infection now seems less clear (Nicholls et al., 2008), the strength of cell-virus binding may be influenced by secondary factors such a torsion in the HA molecule and flexibility of the subterminal carbohydrate side chain. This is a complex area that will take a long time to resolve.
A Rock and a Hard Place
A universal influenza vaccine is a long way off and, even with the best of intentions, it is unrealistic to expect sufficient vaccine to be available for yearly needs, a pandemic such as that of 2009-H1N1 influenza A let alone confront a full-blooded pandemic. Factors here include vaccine manufacturing problems, cost, distribution, and administration of the vaccine. Nor will there be sufficient antiviral agents available of an order of magnitude. Many of the world’s population will be affected, particularly the poor. Improved understanding of the outcome of severe influenza infection indicates that the response of the host is more important than the amount of virus generated (La Gruta et al., 2007). Dysregulation of the cytokine response is the basis of much of this; its control through immunomodulatory agents offers an indirect approach to dealing with infection. Cheap generic agents such as statins, and fibrate and glitazone PPAR agonists used to treat a variety of medical conditions are attractive propositions (Fedson, 2009a,b). Statins, for example, which are used to treat high cholesterol levels can reduce the levels of pro-inflammatory cytokines and chemokines in influenza infection. The prophylaxis/therapy envisaged for severe influenza is simple, logical and imaginative (Clark and Alleva, 2009); rigorously explored experimentation beckons.
No More Pandemics
Having observed the changing tide of progress in influenza over the years from the Hong Kong influenza sentinel post, I feel the time is appropriate in the early phase of the 2009-H1N1 influenza A pandemic to rethink our view of pandemics. There is now better understanding of influenza virus ecology, the zoonotic dimension of pandemics, reaction to threats and outbreaks in incipient and pandemic phases, and special features of the virus and its disease to allow transition from a reactive or defensive position to a more positive one, namely, no more pandemics (Figure A12-2). This transition derives largely from almost four decades of consolidation of the hypothesis that pandemic influenza is a zoonosis that is consequent upon studies on the origin of the 1968 H3N2 (Hong Kong) pandemic virus (Webster and Laver, 1972). This signifies the nonhuman virus’s
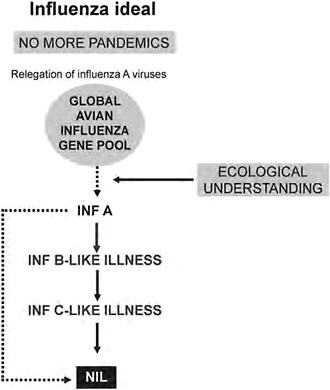
FIGURE A12-2 Long-term steps for the prevention of influenza pandemics.
ability to cross the species barrier to humans and the most likely H subtype of the time to do so. A recognition of these properties provides the type and level of preventative action appropriate for birds and mammals to break the chain of zoonotic transmission. There is still a long way to go; it will require inter alia global mapping of avian influenza viruses and the influenza viruses of other hosts bearing in mind that pandemic influenza by its very nature is a zoonosis that is noneradicable.
If pandemics can be stopped, it might be reasoned that over time, without renewal of genetic vigor through cycles of pandemicity, prevailing type A variants may be of declining pathogenicity, doing so to the level of the type B virus. Ideally, they disappear or, more likely, reduce to the level of type C virus pathogenicity. After all, as pointed out by Gammelin et al. (1990), influenza B and C viruses have a common root with A viruses and may have developed their own type under selection pressures specific for humans. These views represent a mindset that is really an influenza vision well into the future.
Virus Surveillance
Virus surveillance is the cornerstone of influenza ecology, epidemiology, disease occurrence, and public health measures through to pandemic preparedness. Yet surprisingly little is done; when it is, it is often in response to a disease outbreak. It is not often appreciated that dangerous or potentially dangerous influenza viruses can be carried by subclinically infected birds and mammals and be spread from population to population and across different populations. One might reasonably ask if, in hindsight, long-term systematic virus surveillance of pig populations, on the one hand, and concerted action on the part of farm operators and civil authorities, on the other, would have averted the 2009-H1N1 influenza A pandemic. Whatever the case, 2009-H1N1 influenza A is another wake-up call indicating the need for long-term surveillance, especially of domestic birds and mammals. This requires long-term scientific will, political will, and financial commitment. With the increasing movement toward industrial agribusiness operations for meat protein production, surveillance for influenza viruses (and other recognized and unrecognized agents) is imperative. Surveillance must be increased globally (Figure A12-3). The time is nigh for international and other agencies to formulate policies and recommendations to this end.
FIGURE A12-3 Emphasizing the need for increasing influenza virus surveillance for the prevention of pandemic influenza.
Global Need
Understanding the origin of influenza pandemics has moved from superstition to science (Anonymous, 1920). The scientific advances in influenza and their potential for global benefit must be clearly enunciated to national and international decision makers. This approach will particularly require international collaboration, cooperation and collective action for the common good. Nations must work as one global community (Figure A12-4). This approach is definitely possible and not without precedent. All nations worked together in the wake of World War II in spite of the difficulties of the Cold War for the eradication of smallpox. It is anticipated that poliomyelitis will be eradicated within a decade, in no small measure due to substantial funding and effort from Rotary International.
A pandemic requires panglobal effort and is best dealt with through the decision-making process of the United Nations. This may well bring with it the need for structural and organizational changes to deal effectively with the many complexities of influenza and its pandemic viruses. Similar principles should apply to all emerging infectious diseases (Morse, 2009).
FIGURE A12-4 Toward a unified, global effort for the prevention of pandemic influenza.
Guiding Principle
The basic principle that emerges through this report is the prevention of influenza pandemics, taking prevention further to a vision: no more pandemics. The foundation for such a vision, especially in the case of influenza pandemics and their historical association with China, is not new. It goes back about 2,500 years to what Joseph Needham called the Chinese Hippocratic Corpus—The best medicine is preventive medicine—later consolidated to—A skillful doctor cures illness when there is no sign of disease, and thus the disease never comes (Figure A12-5; Needham, 1980; Needham and Lu, 2000).
Appreciation
Appreciation for core support to the writer and others is due to the World Health Organization, Geneva, in the 1970s for promoting and fostering studies on the ecology of influenza viruses through its Veterinary Public Health Unit; the National Institutes of Health, Bethesda, Maryland, for supporting such studies through St. Jude Children’s Research Hospital, Memphis; the Centers for Disease Control and Prevention, Atlanta, for studies in moving the animal-human influenza link forward and, later The Wellcome Trust, London and the Li Ka Shing

FIGURE A12-5 Fundamental principles still apply.
SOURCE: Adapted from Needham (1980) and Needham and Lu (2000).
Foundation, Hong Kong, for supporting further studies in these areas. It would be fair to say that the generous support from these and other organizations to so many across the field has contributed much to the global pool of understanding about influenza virus ecology and epidemiology that provided the foundation for dealing with the H5N1 virus in 1997, SARS in 2003, and now the 2009-H1N1 influenza A pandemic. These experiences will be stepping stones to future influenza and other infectious disease issues that lie ahead in a changing world.
References
Adams, D. 2009 (October 21). GM research is needed urgently to avoid food crisis, says Royal Society. The Guardian, http://www.guardian.co.uk/environment/2009/oct/21/gm-research-food/print (accessed November 17, 2009).
Anonymous. 1920 (March). How influenza started in China! The Far East Review March:170.
Australian Influenza Surveillance Summary Report. 2009. No. 13. Reporting period: 7 August 2009-14 August 2009. Department of Health and Aging, Australian Government.
Baker, M. G., N. Wilson, Q. S. Huang, S. Parine, L. Lopez, D. Bandaranayake, M. Tobias, K. Mason, G. F. Mackereth, M. Jacobs, C. Thornley, S. Roberts, and C. McArthur. 2009. Pandemic influenza A (H1N1)v in New Zealand: the experience from April to August 2009. Eurosurveillance 14(34):1-5.
Butler, D. 2006. Blogger reveals China’s migratory goose farms near the site of the flu outbreak. Nature 441(7091):263.
Cadbury, W. W. 1920. The 1918 pandemic of influenza in Canton. China Medical Journal 34:1-17.
Cantlie, J. 1891 (August 29). The first recorded appearance of the modern influenza epidemic. British Medical Journal:491.
Chan, P. K. S., H. Y. Mok, T. C. Lee, I. M. T. Chiu, W. Y. Lam, and J. L.Y. Sung. 2009. Seasonal influenza activity in Hong Kong and its association with meteorological variations. Journal of Medical Virology 81(10):1797-1806.
Chen, H., G. J. D. Smith, K. S. Li, J. Wang, X. H. Fan, J. M. Rayner, D. Vijaykrishna, J. X. Zhang, L. J. Zhang, C. T. Guo, C. L. Cheung, K. M. Xu, L. Duan, K. Huang, K. Qin, Y. H. C.. Leung, W. L. Wu, H. R. Lu, Y. Chen, N. S. Xia, T. S. P. Naipospos, K. Yuen, S. S. Hassan, S. Bahri, T. D. Nguyen, R. G. Webster, J. S. M. Peiris, and Y. Guan. 2006. Establishment of multiple sublineages of H5N1 influenza viruses in Asia: implications for pandemic control. Proceedings of the National Academy of Sciences 103(8):2845-2850.
Chiu, S. S., Y. L. Lau, K. H. Chan, W. H. Wong, and J. S. Peiris. 2002. Influenza-related hospitalizations among children in Hong Kong. New England Journal of Medicine 347(26):2097-2103.
Chiu, S. S., K. H. Chan, H. Chen, B. W. Young, W. Lim, W. H. S. Wong, Y. L. Lau, and J. S. M. Peiris. 2009. Virologically confirmed population-based burden of hospitalization caused by influenza A and B among children in Hong Kong. Clinical Infectious Diseases 49(7):1016-1021.
CHP (Centre for Health Protection). 2009. Flu express/notifiable infectious diseases/swine and seasonal flu monitor/update on human swine influenza/virus isolation and serology testing (respiratory pathogens). Centre for Health Protection, Department of Health, The Government of the Hong Kong Special Administrative Region.
Chun, J. W. H. 1919. Influenza including its infection among pigs. National Medical Journal, China 5:34-44.
Clark, I., and L. Alleva. 2009. Invited commentary on David Fedson’s article Influenza and Other Respiratory Viruses 3(5):199-201.
Copland, J. W., editor. 1987. Newcastle disease in poultry: a new food pellet vaccine. ACIR Monograph No.5. Canberra, Australia: Australian Centre for International Agricultural Research.
Cowling, B. J., K. H. Chan, V. J. Fang, C. K. Y. Cheng, R.O.P. Fung, W. Wai, J. Sin, W. H. Seto, R. Yung, D. W. S. Chu, B. C. F. Chui, P. W. Y. Lee, M. C. Chiu, H. C. Lee, T. M. Uyeki, P. M. Houck, J. S. M. Peiris, and G. M. Leung. 2009. Facemasks and hand hygiene to prevent influenza transmission in households: a cluster randomized trial. Annals of Internal Medicine 151(7):437-446.
Dale, R. C., A. J. Church, R. A. H. Surtes, A. J. Lees, J. E. Adcock, B. Harding, B. G. R. Neville, and G. Giovannoni. 2004. Encephalitis lethargica syndrome: 20 new cases and evidence of basal ganglia autoimmunity. Brain 127(Pt. 1):21-33.
Dudley, J. P. 2008. Public health and epidemiological considerations for avian influenza risk mapping and risk assessment. Ecology and Society 13(2):21.
———. 2009. Age-specific infection and death rates for human A (H5N1) avian influenza in Egypt. Eurosurveillance 14(18):19198.
Fedson, D. S. 2009a. Confronting the next influenza pandemic with anti-inflammatory and immunomodulatory agents: why they are needed and how they might work. Influenza and Other Respiratory Viruses 3(4):129-142.
———. 2009b. Meeting the challenge of influenza pandemic preparedness in developing countries. Emerging Infectious Diseases 15(3):365-371.
Gammelin, M., A. Altmuller, U. Reinhardt, J. Mandler, V. R. Harley, J. P. Hudson, W. M. Fitch, and C. Scholtissek. 1990. Phylogenetic analysis of nucleoprotein suggests that the human influ enza A viruses emerged from a 19th century avian ancestor. Molecular Biology and Evolution 7(2):194-200.
Greger, M. 2006. Bird flu. A virus of our own hatching. New York: Lantern Books.
———. 2009. CDC confirms ties to virus first discovered in U.S. pig factories, http://www.humanesociety.org/news/news/2009/04/swine_flu_virus_origin_1998_042909.html (accessed November 17, 2009).
Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C .L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J .S. M. Peiris, and L. L. M. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302(5643):276-278.
Higgins, D. A., and K. F. Shortridge. 1988. Newcastle disease in tropical and developing countries. In Newcastle Disease, edited by D. J. Alexander. Boston: Kluwer Academic Publications. Pp. 273-302.
Kanegae, Y., S. Sugita, K. F. Shortridge, Y. Yoshioka, and K. Nerome. 1994. Origin and evolutionary pathways of the H1 hemagglutinin gene of avian, swine and human influenza viruses. Archives of Virology 134(1-2):17-28.
Kaverin, N. V., I. A. Rudneva, N. A. Ilyushina, A. S. Lipatov, S. Krauss, and R. G. Webster. 2004. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. Journal of Virology 78(1):240-249.
Kendal, A. P., G. R. Noble, J. J. Skehel, and R. G. Webster. 1978. Antigenic similarity of influenza A (H1N1) viruses from epidemics in 1977-1978 to “Scandinavian” strains isolated in epidemics of 1950-1951. Virology 89(2):632-636.
Kou, Z., Y. D. Li, Z. H.Yin, S.Guo, M. L.Wang, X. B. Gao, P. Li, L. J.Tang, P. Jiang, Z. Luo, Z. Xin, C. Q. Ding, Y. B. He, Z. Y. Ren, P. Cui, H. F. Zhao, Z. Zhang, S. Tang, B. P. Yan, F. M. Lei, and T. X . Li. 2009. The survey of H5N1 flu viruses in wild birds in 14 provinces of China from 2004-2007. PLoS ONE 4(9):e6926.
La Gruta, N. L., K. Kedzierska, J. Stambas, and P. C. Doherty. 2007. A question of self-preservation: immunopathology in influenza virus infection. Immunology and Cell Biology 85(2):85-92.
Lowen, A., and P. Palese. 2009 (August 18). Transmission of influenza viruses in temperate zones is predominantly by aerosol, in the tropics by contact. PLoS Currents: Influenza, http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2762697/ (accessed January 4, 2010).
Ma, W. J., A. L. Vincent, M. R. Gramer, C. B. Brockwelll, K. M. Lager, B. H. Janke, P. C. Gauger, D. P. Patnayak, R. J. Webby, and J. A. Richt. 2007. Identification of H2N3 Influenza A viruses from swine in the United States. Proceedings of the National Academy of Sciences 104(52):20949-20954.
Morse, S. S. 2009. Emerging infections: condemned to repeat? In Microbial evolution and co-adaptation. Washington, DC: The National Academies Press. Pp. 191-208.
Nakajima, K., U. Dresselberger, and P. Palese. 1978. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature 274(5669):334-339.
Needham, J. 1980. China and the origins of immunology. Centre of Asian Studies occasional papers and monographs, No.41. University of Hong Kong, Hong Kong.
Needham, J., and G. D. Lu. 2000. The origins of immunology. In Science and civilization in China. Biology and biological technology, Part VI: Medicine. Vol.6, edited by N. Sivan. Cambridge: Cambridge University Press. Pp. 114-174.
Nicholls, J. M., R. W. Y. Chan, R. J. Russell, G. M. Air, and J. S. M. Peiris. 2008. Evolving complexities of influenza virus and its receptors. Trends in Microbiology 16(4):149-157.
Nierenberg, D., and L. Garces. 2005. Industrial animal agriculture. The next global health crisis? London: World Society for the Protection of Animals.
Omer, S. B., A. Sutanto, H. Sarwo, M. Linehan, I. G. G. Djelantik, D. Mercer, V. Moniaga, L. H. Moulton, A.Widjaya, P. Muljati, B. D. Gessner, and M. C. Steinhoff. 2008. Climatic, temporal and geographic characteristics of respiratory syncytial virus disease in a tropical island population. Epidemiology and Infection 136(10):1319-1327.
Orwell, G. 1945. Animal farm. Harmondsworth, England: Penguin Books.
Parham, W. 2006. South China’s taste for wildlife, http://fas.org/blog/china/reports-and-data/south_chinas_taste_for_wildlife (accessed November 17, 2009).
Patterson, K. D. 1985. Pandemic and epidemic influenza, 1830-1848. Social Science and Medicine 21(5):571-580.
Perez, D., E. Sorrell, M. Angel, J. Q. Yi, D. Hickman, L. Pena, G. Ramirez-Nieto, B. Kimble, and Y. Araya. 2009. Fitness of pandemic H1N1 and seasonal influenza A viruses during co-infection: evidence of competitive advantage of pandemic H1N1 influenza versus seasonal influenza. PLoS Currents: Influenza, http://knol.google.com/k/daniel-perez/fitness-of-pandemic (accessed November 17, 2009).
Qi, X., X. H. Li, P. Rider, W. X. Fan, H. W. Gu, L. T. Xu, Y. H. Yang, S. W. Lu, H. Wang, and F.Y. Lui. 2009. Molecular avian influenza A viruses isolated from raccoon dogs in China. PLoS ONE 4(3):e4682.
Reichelderfer, P., A. P. Kendal, K. F. Shortridge, A. Hampson, et al. 1989. Influenza surveillance in the Pacific Basin. Seasonality of virus occurrence: a preliminary report. In Proceedings: First Asia Pacific Congress of Medical Virology, Singapore, 1988, edited by Y. C. Chan, S. Doraisingham, and A. E. Ling. Singapore: World Scientific. Pp. 412-444.
Scholtissek, C., H. Burger, O. Kistner, and K. F. Shortridge. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147(2):278-294.
SEARO/WHO (World Health Organization Regional Office for Southeast Asia). 2009. Pandemic H1N1-2009 influenza situation in SEA region, http://www.searo.who.int/en/section10/Section2562.htm (accessed November 17, 2009).
Shortridge, K. F., editor. 1982. Newcastle disease and its control in Southeast Asia. Hong Kong: Hong Kong University Press.
———. 1983. Pandemic influenza: application of epidemiology and ecology in the region of southern China to prospective studies. In The origin of pandemic influenza viruses, edited by W. G. Laver. New York: Elsevier Science Publishing Company, Inc. Pp. 191-200.
———. 1992. Pandemic influenza: a zoonosis? Seminars in Respiratory Infections 7(1):11-25.
———. 1999a. Poultry and the H5N1 outbreak in Hong Kong, 1997: abridged chronology and virus isolation. Vaccine 17(Suppl 1):S26-S29.
———. 1999b. The 1918 “Spanish flu:” pearls from swine? Nature Medicine 5(4):384-385.
Shortridge, K. F., and C. H. Stuart-Harris. 1982. An influenza epicentre? Lancet 2(8302):812-813.
Shortridge, K. F., A. Cherry, and A. P. Kendal. 1979. Further studies of the antigenic properties of H3N2 strains of influenza A viruses isolated from swine in southeast Asia. Journal of General Virology 44(1):251-254.
Shortridge, K. F., P. A. Underwood, and P. A. King. 1990. Antigenic stability of H3 influenza viruses in the domestic duck population of southern China. Archives of Virology 114(1-2):121-136.
Shortridge, K. F., R. G. Webster, W. K. Butterfield, and C. H. Campbell. 1977. Persistence of Hong Kong influenza virus variants in pigs. Science 196(4297):1454-1455.
Smith, G. J. D., J. Bahl, D. Vijaykrishna, J. X. Zhang, L. L. M. Poon, H. L.Chen, R. G. Webster, J. S. M. Peiris, and Y. Guan. 2009a. Dating the emergence of pandemic influenza viruses. Proceedings of the National Academy of Sciences 106(28):11709-11712.
Smith, G. J. D., D. Vijaykrishna, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. M. Peiris, Y.Guan, and A. Rambaut. 2009b. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459(7250):1122-1125.
Starling, A. E. 2007. Plague, SARS and the story of medicine in Hong Kong. Hong Kong Museum of Medical Sciences Society 2006. Hong Kong: Hong Kong University Press.
Sung, R. Y. T., H. G. S. Murray, R. C. K. Chan, D. P. Davies, and G. L. French. 1987. Seasonal patterns of respiratory syncytial virus infection in Hong Kong. Journal of Infectious Diseases 156(3):527-528.
Sung, R. Y. T., R. C. K. Chan, J. S. Tam, A. B. F. Cheng, and H. G. S. Murray. 1992. Epidemiology and aetiology of acute bronchiolitis in Hong Kong infants. Epidemiology and Infection 108(1):147-154.
Vilensky, J. A., and S. Gilman. 2006. Encephalitis lethargica: could this disease be recognized if the epidemic recurred? Practical Neurology 6(6):360-367.
Vincent, A. 2004. Encephalitis lethargica: part of a spectrum of post-streptococcal autoimmune disease? Brain 127(1):2-3.
Webster, R. G., and W. G. Laver. 1972. The origin of pandemic influenza. Bulletin of the World Health Organization 47(4):449-452.
Weingartl, H. M., R. A. Albrecht, K. M. Larger, S. Babiuk, P. Marszal, J. Neufeld, C. Embury-Hyatt, P. Lekcharoensuk, T. M. Tumpey, A. Garcia-Sastre, and J. A. Richt. 2009. Experimental infection of pigs with the human 1918 pandemic influenza virus. Journal of Virology 83(9):4287-4296.
WHO (World Health Organization). 2009. Situation update avian influenza, http://www.who.int/csr/disease/avian_influenza/updates/en.index.html (accessed November 17, 2009).
Wong, C. M., K. P. Chan, A. J. Hedley, and J. S. M. Peiris. 2004. Influenza-associated mortality in Hong Kong. Clinical Infectious Diseases 39(11):1611-1617.
Wong, C. M., L. Yang, K. P. Chan, G. M. Leung, K. H. Chan, Y. Guan, T. H. Lam, A. J. Hedley, and J. S. M. Peiris. 2006. Influenza-associated hospitalization in a subtropical city. PLOS Medicine 3(4):e121.
Zhou, J. Y., W. B. Sun, J. H. Wang, J. Q. Guo, W. Yin, N. P. Wu, L. J. Li, Y. Yan, M. Liao, Y. Huang, K. J. Luo, X. T. Jiang, and H. L. Chen. 2009. Characterization of the H5N1 highly pathogenic avian influenza virus derived from wild pikas in China. Journal of Virology 83(17):8957-8964.





















