4
Common Themes at the Intersection of Biological and Physical Sciences
In this chapter the committee explores some of the problems and conceptual challenges at the intersection of the biological and physical sciences. Without attempting an exhaustive survey of the subject, it covers a handful of examples that illustrate both the importance of the open scientific problems and some of the breakthroughs that are occurring as a result of inquiries that bridge this intersection.
INTERACTION AND INFORMATION: FROM MOLECULES TO ORGANISMS AND BEYOND
The modern era of biology began when, together, a biologist and a physicist uncovered the nature of the interactions holding together the strands of DNA in the famous double helix. The relatively simple interactions between different pairs of nucleotides immediately revealed the nearly infinite information storage capacity of the DNA heteropolymer and defined, for the first time, the intimate connection between interaction and information that makes up the fabric of living matter. Today, 60 years later, the challenge in studying living matter is to produce a framework for understanding the highly organized and information-rich biological structures that are engaged not only in the acquisition and conversion of metabolic energy but also in the acquisition and transfer of information. Unraveling the complexity of living systems is a challenge that requires not only the creative application of ideas and tools for interacting systems but also the development of new conceptual and mathematical frameworks that can incorporate informa-
tion and information fluxes alongside the existing thermodynamic and statistical principles of physical science. It is at this interface of biology with physics and information theory that the fundamental principles governing living matter are likely to be discovered.
Biological complexity is built on specific interactions between molecules, and these interactions are linked to each other and held in balance through complex networks. These networks underpin the regulation and signaling that govern intracellular function and multicellular behavior all the way to the development of the organism, and their multilayered complexity makes studying the systems challenging.
On the smallest level, interactions are mediated by molecular forces (hydrogen bonding, electrostatics, hydrophobicity), which form the physicochemical basis of molecular recognition between polynucleotide and polypeptide structures. Although we understand the basic laws governing these forces, using these laws to reliably predict specific, complex intermolecular interactions and tracking the effect of the intermolecular interaction to the behavior of a whole organism remain a challenge.
To deal with some of these challenges, ab initio approaches are now frequently complemented by data-driven bioinformatics that analyze and compare empirical data to untangle the interactions between numerous related interacting pairs of molecules. This combination of approaches weaves together ideas and methods from computer science, statistics, physics, and biology, and researchers use them to reveal the patterns (called “code” by some) underpinning the interaction (see Figure 4-1).
Bioinformatic studies provide supramolecular-level descriptions in which, instead of the basic interatomic forces, one works with interaction profiles—that is, the strengths of interactions with different possible partners that de facto define biological molecules. Indeed, characterization of such interaction profiles allows an understanding of interactions at the level of the whole organism and bioinformatic approaches are now extensively used for identifying regulatory targets of transcription factors—proteins that control gene expression. This approach is delivering ingenious quantitative tools that enable us to extract enhanced knowledge from existing biological data in new ways.
The story does not end with describing the specific interactions, however. As mentioned above, the interactions between biomolecules are the building blocks of molecular and genetic networks, and the networks must also be studied and understood. For example, a covalent modification of a specific protein via the enzymatic activity of another (e.g., phosphorylation catalyzed by a kinase) might trigger enzymatic activity of the target protein or cause its re-localization within the cell. In genetic networks, regulation of gene expression and protein synthesis are controlled through the action of transcription factors and recently discov-
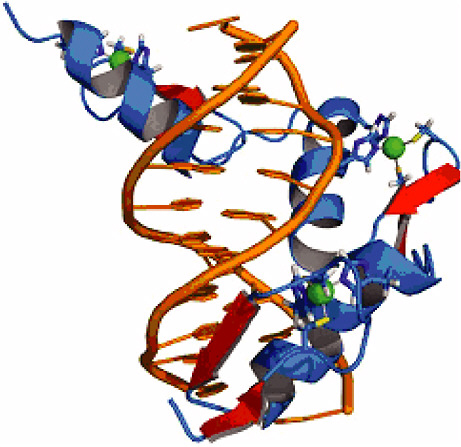
FIGURE 4-1
Zinc Fingers
Zinc finger proteins form a ubiquitous family of transcription factors—proteins that bind DNA in a sequence-specific manner and regulate gene expression. They have a remarkable modularity, which allows mixing and matching of up to six DNA binding domains, fostering highly specific targeting of a diverse set of DNA motifs. Because of this feature, zinc fingers hold great promise as tools for the precise control of gene regulation and have potentially numerous and profoundly important medical applications (Klug, 2005). The design of engineered zinc fingers for practical applications requires a thorough understanding of the interaction “code” that defines transcription factor/DNA binding specificity. Deciphering this code brings together structural analysis with bioinformatics and ab initio computational modeling studies (Paillard et al., 2004). The figure shows the structure of the transcription factor protein Zif268 (blue) containing three zinc fingers in complex with DNA (orange). SOURCE: Pavletich and Pabo, 1991; reprinted with permission from the American Association for the Advancement of Science.
ered micro-RNAs. These and other methods of regulating protein abundance and activity are the links in the causative chains forming cascades and networks that propagate and modulate the effects of different stimuli.
Networks mediated by a mitogen-activated protein (MAP) kinase are another excellent example of this type of network. This family of signaling proteins controls the regulation of diverse processes, ranging from the expression of genes required for a yeast cell to adapt to the carbon sources in the environment to the transcription of proto-oncogenes in the development of cancer, to programmed cell death. Over a dozen MAP kinase family members have been discovered in mammals alone. Each MAP kinase cascade consists of a minimum of three kinase enzymes that, in response to particular stimuli, are activated in series by the MAP kinase above it in the signaling cascade. MAP kinase pathways transfer information to particular effectors that perform a number of functions, including integrating information channeled from other regulatory pathways into the MAP kinase pathway, amplifying particular signals under particular conditions, and precisely directing an array of discrete response patterns. The inherent complexity of the system challenges our ability to characterize and understand the functions and mechanisms of the individual pathways. MAP kinases are only one example of the interconnectedness of the networks of cellular regulatory pathways, and studying such systems provides interesting challenges for both physical and life scientists, because new quantitative concepts and approaches will be necessary to disentangle these interacting network phenomena.
Interaction networks not only exist within cells but also extend outward to coordinate cellular responses to the environment and to coordinate the behavior of groups of cells through intercellular interactions. Paradigmatic examples include bacterial chemical sensing of the environment (chemotaxis) and cell-to-cell interactions through chemical signaling (quorum sensing), illustrated in Figure 4-2. The two bacterial behaviors are unified by the key role played by information transfer. Chemotaxis refers to the ability of bacteria to swim in a biased direction—either toward a gradient of a nutritious compound or away from a gradient of a noxious one. The chemotaxis sensory circuit relies on receptors mounted on the membrane, which bind to attractant or repellant molecules. These binding events elicit a protein phosphorylation cascade, which causes motor proteins to switch the rotation of the flagellar apparatus such that, depending on which direction it is spinning, the bacterium either tumbles or is propelled forward. Quorum sensing is a process of bacterial cell-to-cell communication that involves production and detection of threshold concentrations of signal molecules called autoinducers. The accumulation of a stimulatory concentration of an extracellular autoinducer occurs only when a sufficient number of cells—a “quorum”—is present. Thus, the process is a mechanism that allows bacteria to collectively regulate gene expression and thereby function as multicellular organisms. In both chemotaxis and quorum-
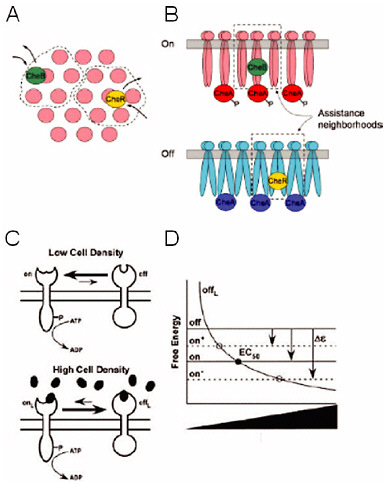
FIGURE 4-2
Chemotaxis and Quorum Sensing
The proteins responsible for eliciting both chemotaxis and quorum sensing are homologous and carry out identical biochemical reactions, yet newly acquired quantitative data reveal a stark difference in their function (see text for details). Experimental data and modeling suggest that chemotaxis receptors are poised to rapidly change signaling strength and the receptors cluster to amplify the signal. This is depicted by top (Panel A) and side (Panel B) views of receptor clusters, whereby transiently bound stimulus enzymes CheR and CheB act not only on the receptor to which they are bound but also on those receptors in their vicinity, or assistance neighborhoods. By contrast, the quorum-sensing receptors require a significant threshold signal concentration to switch activity, and they do not cluster. This is illustrated in Panel C by the single receptor and the heavy arrow, suggesting that the receptors greatly prefer the unbound state in the absence of ligand (low cell density). Panel D shows the corresponding free-energy diagram for the quorum-sensing model. SOURCES: Panels A and B, Hansen et al., 2008; Panels C and D, Swem et al., 2008, reprinted with permission from Elsevier.
sensing, information is passed internally by a series of phosphorylation events that ultimately change the activity of a DNA binding transcriptional regulator that induces/represses genes required for individual or group behaviors.
While the proteins that carry out the chemotaxis and quorum-sensing responses are homologous and, in fact, perform identical biochemical reactions, the two circuits have been shown to possess distinct design properties suggesting that the two signaling systems evolved to optimally solve very different biological problems. As a result of work by both physical scientists and life scientists, we now know that in chemotaxis, bacterial cells must respond rapidly to small, differential changes in ligand concentration. Consistent with this, chemotaxis receptors are poised to rapidly change signaling strength, spending nearly half their time in the on state. Moreover, the chemotaxis receptors cluster, which promotes signal amplification. By contrast, the quorum-sensing receptors have dramatically different signaling properties. First, in the absence of ligand, the quorum-sensing receptors are nearly always in the on state and thus require a significant threshold ligand concentration to switch off. Second, the quaternary arrangement of receptors precludes higher order complexes and thus clustering. This arrangement excludes chemotaxis-style signal amplification. Therefore, the quorum-sensing apparatus appears designed to respond slowly to the accumulation of ligand. This dramatic difference in the output of two seemingly homologous systems selects for high-sensitivity differential signaling accompanied by amplification for chemotaxis, while selecting against exactly those features in quorum-sensing signaling.
These two sensory relay systems of bacteria provide a striking example of insight into the function and design principles of biological networks that can be gained by a comparative study. They also serve as classic examples in which the close collaborations between life scientists and physical scientists, and others (biologists examining mutant phenotypes, chemists synthesizing agonist and antagonist molecules, physicists modeling network properties, and engineers studying the functioning of simplified synthetic circuits) have brought about a fundamentally new understanding of how cells process information and how cooperative behaviors evolve.
Intercellular interaction and information transfer are, of course, also central to processes of development in multicellular organisms—a subject to which we return later in discussing the problem of biological self-assembly. Indeed, the theme of specific interactions and encoding and transfer of information are ubiquitous at all levels of biological organization. It provides a natural interface with physical sciences, which can lend quantitative ideas and tools to advance our understanding of living matter. This interface is rich with fundamental questions, and we expect much progress in the near future.
DYNAMICS, MULTISTABILITY, AND STOCHASTICITY
The dynamics of simple systems are the bread and butter of physics, taught in all introductory physics courses. More sophisticated dynamical systems analysis is central to many fields in physical sciences and engineering. Dynamical systems theory provides a body of mathematical concepts and methods that allow us to describe complex dynamics in systems with many degrees of freedom (Jackson, 1991). In particular, it provides a systematic approach for identifying generic, parameter-independent behaviors as well as for reducing dimensionality by identifying the variables most essential for the dynamics. It also addresses questions of stability and multistability and provides methods for dealing with temporal changes unfolding on widely disparate timescales. Dynamical systems theory has been particularly valuable for understanding the behavior of nonlinear systems, and so the discipline has made a natural expansion into biology, the quintessential “nonlinear science.”
Dynamics plays a ubiquitous role in living systems. Some of the most obvious examples are provided by rhythmic behaviors: cell cycle and circadian rhythms, respiration and heartbeat, locomotion and neural oscillations. But rhythmic behaviors are not the only manifestations of dynamics. Adaptation, growth, and evolution are also dynamical processes that cover a broad range of timescales, from seconds to millions of years. The dynamics are not always orderly and deterministic: randomness and stochasticity play important roles both on molecular and evolutionary timescales. Insight into dynamic behavior cannot be achieved without recourse to quantitative analysis. The latter provides an immediate connection to physical sciences and mathematics, fields that have developed powerful tools and concepts for studying deterministic and stochastic dynamical processes.
One common manifestation of dynamics in living systems is adaptation. Cells and organisms interact with their environment, more often than not by adaptation, a response that mitigates the effects of change. Biological examples of adaptation include the very familiar experience of our eyes needing time to adapt to darkness when the light is switched off. This phenomenon involves multiple layers of feedback operating on the molecular and cellular levels in the retina. A generally similar “negative feedback” mechanism underlies homeostasis of blood glucose level (disrupted by diabetes) and many other processes. An opposite type of feedback—positive feedback—amplifies the effect of perturbations, resulting in excitable behavior. Positive feedback plays the key role in the generation of action potentials in neurons, which was elucidated in the classic work of biophysics by Hodgkin and Huxley in 1952. Positive feedback is often associated with the existence of alternative steady states and the possibility of switching between them. Epigenetic phenomena that control cell differentiation provide another example of this behavior. In fact, it is characteristic of living systems to possess many possible
states, a situation that in mathematics and physics is associated with multistability. In this case, the state of the system depends on the past history of its dynamics, meaning that the system has memory. Physical sciences provide many elegant examples of systems in which multistability and memory play important roles in dynamics, and many ideas and mathematical approaches have been developed to accurately describe such situations.
An important set of ideas bridging living and physical systems pertains to relaxational dynamics, energy landscapes, and fluctuations. Thermodynamically stable states are the minima of (free) energy: Near-equilibrium dynamics tends to drive systems into these states, while thermal fluctuations oppose this convergence. Relaxational dynamics generalizes to a broad class of nonequilibrium processes and plays an important role in the way we understand the stability and robustness of many living systems. It is also central to control theory and to the design of complex engineered systems (Freeman and Kokotovic, 2008). Evolved or designed energy landscapes can be used to understand the control of protein structure and dynamics (Figure 4-3) (Onuchic and Wolynes, 2004), while multistable dynamical landscapes in a system of interacting neurons provide a compelling hypothesis for the nature of memory (Figure 4-4) (Hopfield, 2007).
Energy landscape ideas provide powerful insight into the dynamics of complex biological molecules that catalyze chemical reactions or, in the case of molecular motors, perform mechanical work. Similar to man-made machines, “molecular machines” are built from many heterogeneous components whose interactions are carefully orchestrated. Understanding the design principles behind these machines remains a key issue for theoretical biological physicists. An excellent example of these issues is provided by a molecular motor kinesin. Our understanding of the motility of kinesin motors has advanced since the discovery of kinesin’s unidirectional transport of cellular organelles along the microtubule (Vale et al., 1985). For example, force-adenosinetriphosphate (ATP) velocity relationships measured via single-molecule assays (Schnitzer et al., 2000) and kinetic ensemble experiments have enabled scientists to decipher the phenomenological energy landscape of kinesin motor dynamics. Importantly, the ideas pioneered in these studies are not restricted to the kinesins.
Many other molecular components related to cellular function, indeed, are molecular motors that utilize molecular fuels such as ATP, oxygen, cyclic adenosine monophosphate (cAMP), guanosine 5’-triphosphate (GTP), and Ca2+ to form a cycle of conformational switches, in the process performing work essential to maintain cellular life. It will be fascinating as new physical developments (such as nonequilibrium fluctuation theorems) shed light on the type of nanoscale dynamics that seem to have been captured so efficiently by evolution of these magnificent machines.
The ideas of multistable landscapes are also relevant in a very different context: that of formation and maintenance of memories in the brain. A range of experi-
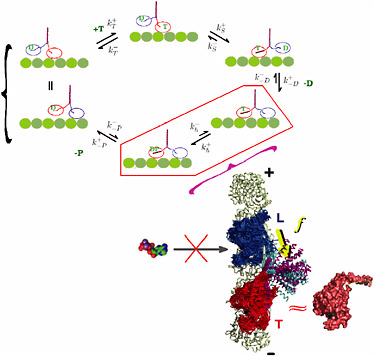
FIGURE 4-3
Dynamics of Molecular Motors
The dynamics of molecular motors directly couples protein folding to protein function. The prototypical kinesin motor, which ferries cargo throughout the cell, consists of a pair of motor domains (“heads”) coupled by a flexible “neck” to a coiled-coil “stalk.” The kinesin steps along the microtubule filament in a hand-over-hand fashion. Motor action occurs through an ATP-powered mechanochemical cycle (shown in the figure) involving sequential binding and unbinding of the motor heads to the microtubule along which the motor “walks.” What is the mechanism coordinating large changes in the conformation of different domains—the physical “steps” that the motor takes—with the chemical activity that powers the motion? Recent modeling efforts have approached this problem using the landscape ideas for protein folding and have identified the critical role of the intermediate state (highlighted in the figure) during which both motor heads are transiently bound to the microtubule. The topological constraint of such binding introduces mechanical stress into the structure and results in an asymmetric straining of the two motor heads. Therefore the variation of kinesin structure itself is indispensable for the coordinated dynamics of the motor. The internal tension built on the neck-linker (yellow arrow) prevents the premature binding of ATP molecule to the leading head by deforming its catalytic site from its nativelike environment. The tension on the neck-linker is maintained as long as both head domains of the motor are bound to the microtubule surface. This asymmetry provides the directionality and, ultimately, the high processivity of kinesin motor action. The role of internal stress in protein conformation dynamics is likely to be central to the function of many if not all molecular machines and, as a subject of study, provides a direct example of the link between physics, chemistry, and biology. SOURCE: Changbong Hyeon and José Onuchic, University of California at San Diego.
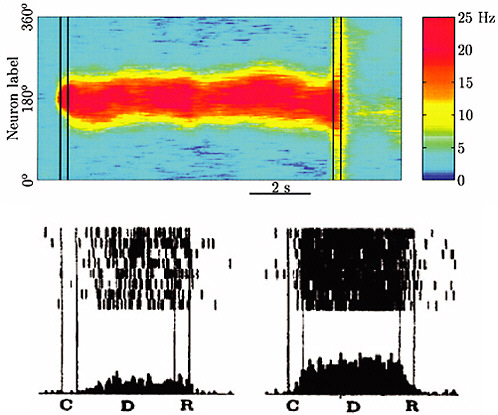
FIGURE 4-4
Brain Dynamics and Memory
Multistability in cortical networks has been proposed as a mechanism for spatial working memory. The top panel of the figure shows the spatiotemporal map of action potential firing rate in a computational model of monkey prefrontal cortex. The model network is presented with a cue at a 180-degree angle at time C and retains a memory in the form of a stable “bump” state during a delay period D until the memory is recalled at time R. The action potential firing pattern and spike rate histogram of an individual model neuron in the network (bottom right) closely resemble those of a cortical neuron in a monkey performing a cue-delay-recall task (bottom left). Action potential firing patterns in the bottom plots are represented as rastergrams, where each line shows the activity recorded during one experimental trial and each action potential is represented by a tick mark. SOURCE: Compte et al., 2000, reprinted with the permission of Oxford University Press.
mental, computational, and theoretical results suggest that memory relies at least in part on multistability phenomena. Multiple stable states can coexist in neuronal systems at several levels, from the molecular machinery at individual synapses to large-scale network activity states, and states can be stable on widely different timescales, from a few milliseconds to an entire lifetime. The basic mechanism is that a neural system becomes attracted to one of multiple stable states, which can be thought of as local minima in an energy landscape. An example is spatial working memory in a cortical network (Compte et al., 2000) where physical models reproduce the results from experiments in which monkeys are presented with visual cues at different locations (0-360 degrees) along a circle on a screen and are required to remember that location for a delay period after the cue disappears. The monkey subsequently makes an eye movement to the cued location. Recordings of electrical activity from prefrontal cortical neurons during this task show that the spike-firing rate increases in a subset of neurons when the cue is presented and remains high throughout the delay period, thus appearing to represent a memory of the cue. A computational (theoretical) network model consisting of thousands of excitatory and inhibitory neurons whose synaptic connection pattern is mimicking the cellular and synaptic organization of prefrontal cortex reliably reproduces such memory-related activity states (see Figure 4-4). This example shows that the synaptic architecture of neuronal systems can support multistability as a memory mechanism; it also illustrates the power of computational modeling and system analysis tools from the physical sciences when applied to complex biological systems.
A very important common concept equally relevant to the dynamics of physical and biological systems is stochasticity. The study of stochastic (i.e., random) behavior in biological systems provides an excellent recent example of scientific progress driven by the cross-fertilization of ideas from different disciplines, aided by technological advances enabling quantitative measurements. With the advent of fluorescent reporters and the capacity to image and individually track large numbers of single cells over extended periods of time, it became possible to observe and quantify fluctuations in different genes in single cells, even in bacteria, the smallest cells on Earth. The design of the experiments to study fluctuating biological behavior and the interpretation of the data were built on the fundamental understanding of stochastic processes in physics and mathematics. The results of the studies are shedding light not only on the molecular biology of the underlying processes but also on the possible importance of stochasticity in cell “decision making” (Suel et al., 2007; Losick and Desplan, 2008).
Studies of stochasticity show that genetically identical organisms can exhibit substantial phenotypic differences. During the process of gene expression a single gene is transcribed by RNA polymerase to produce about one hundred to several hundred mRNAs, each of which is translated to produce tens to thousands of proteins. The small number of molecules involved in this process, especially at
the transcription step, can lead to substantial fluctuations in protein abundance between genetically identical cells. Because protein concentrations control the rates of many biochemical processes, stochastic fluctuations in transcription have the capacity to induce differences in physiological states. Stochastic effects of this type are, for example, suggested to play a role in tripping the switch between the dormant and active replication modes of viral infection in bacteria (Arkin et al., 1998). Elowitz and collaborators (2002) pioneered an elegant experimental design and analysis framework to measure variability, or “noise,” in gene expression and to distinguish between variability that arises from intrinsic or extrinsic noise sources (see Figure 4-5). Their insight was to realize that if they used two fluorescent reporters, the difference in the levels of two reporters defined the noise intrinsic to the process of expression of the two genes while the common component of fluctuation represented the extrinsic noise arising from variability that affects the expression of all genes in a cell. Using this methodology Elowitz et al. demonstrated that substantial intrinsic noise exists in bacterial gene expression. Since this landmark study many groups have utilized this approach to measure noise in gene expression in other microbial and animal systems as well as to understand cellular individuality (Ghaemmaghami et al., 2003; Kaufmann and van Oudenaarden, 2007).
To illustrate the vast range of dynamical phenomena and the vast range of tem-
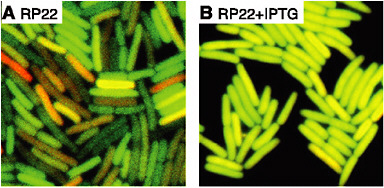
FIGURE 4-5
Molecular Noise and Stochasticity of the Cell
Single-cell imaging of genetically encoded fluorescent proteins (CFP and YFP, shown in orange and the darker green color channels) enables quantitative measurements of molecular noise in E. coli. Panel (A) exhibits strong fluctuations, which appear when genes (in this case CFP and YFP) are expressed at low levels because of repression by the LacI transcription factor. In panel (B), repression is eliminated and both fluorescent proteins are expressed at higher levels and the cells exhibit reduced noise. SOURCE: Adapted from Elowitz et al., 2002; reprinted with permission from the American Association for the Advancement of Science.
poral and spatial scales on which dynamics may unfold, we conclude this section with some important questions concerning the dynamics of ecosystems, biomes, and the Earth system. All of these systems exhibit properties that depend on the physiology, biochemistry, and development of individual organisms, but particular characteristics of the systems emerge depending on the interactions among organisms or interactions between organisms and their environments. These so-called emergent properties have many dimensions, and they link evolutionary, ecological, and Earth-system processes in a network of interactions that operate on diverse timescales. Emergent properties play central roles in determining the suitability of a habitat and have contributed to Earth and its life-support systems.
Nitrogen, one of the key building blocks of life, is but one example where mutually dependent interactions on many scales create an amazingly complex network, the understanding of which will require applying knowledge from both the physical and life sciences. Although nitrogen is the most abundant element in the atmosphere, the low reactivity of N2 gas makes the atmospheric pool generally unavailable and contributes to widespread constraints on plant and animal growth by biologically available nitrogen (Vitousek, 2004). On the other hand, excess nitrogen can be a source of profound problems. Excess biologically available nitrogen plays a role in the dead zone at the mouth of the Mississippi and other large rivers. In the atmosphere, nitrous oxide (N2O), a gas released from fertilizer, contributes to the greenhouse effect about 300 times as much, on a molecule by molecule basis, as carbon dioxide. The flow of biologically available nitrogen through the Earth system has been dramatically increased by human activity, having more than doubled in the last two centuries.
Every transformation in the global nitrogen cycle at every level of organization depends on interactions between physical and biological processes, with implications that feed back to alter the characteristics of the biological and physical environment. At the cellular scale, the assembly of nitrogen-fixing symbioses involves a complicated set of biochemical, physical, and physiological interactions between plants and micro-organisms, leading to the assembly of multiorganism “factories” that provide the delicately regulated conditions necessary for high rates of N2 fixation. The roles of climate, nutrients released from rocks, pH in the soil, and enzymes cast into the external environment all highlight the role of the physical environment in regulating this process. Unknown variables in the area of nitrogen transformations include the importance of microbial diversity, the flexibility of the environmental constraints, and the interaction of large-scale, climate-driven processes with small-scale microbe-driven processes.
As with many of the great challenges in the environmental end of science at the interface of the biological and physical sciences, the unknowns in the global nitrogen cycle involve processes that interact across a vast range of temporal and spatial scales. Understanding nitrogen fixation or gas loss depends on effectively integrat-
ing processes that range in temporal scale from seconds for the chemical signaling among plants and micro-organisms to millennia for the evolution of microbial genomes and the development of soil texture. Relevant spatial scales range from organism to organism signaling at the scale of microns to climate change at the global scale. The committee reiterates that some of the grandest challenges in science involve understanding how emergent properties link physical and biological properties, from the molecular to the global scale, and how their properties can be managed to achieve specific effects.
SELF-ORGANIZATION AND SELF-ASSEMBLY
In physics, crystalline structure is explained in terms of energy and thermodynamics, and the dynamics of crystallization can create a multitude of forms, as, for example, in snowflakes. Assembly is another fundamental issue in the natural sciences that, like the issues of interaction and dynamics discussed above, brings complexities not encountered in the physical sciences.
Life itself is the ultimate paradigm of self-organization. What does it take to transition from inanimate to animated, living matter? The answer to this question involves issues of complexity in interacting systems, energy and information fluxes, memory, and other ingredients of self-organizing and self-replicating systems. Theoretical foundations for thinking about self-organizing and replicating systems have been laid down by von Neumann and Burks (1966). Yet, unlike Schrödinger’s vision of aperiodic solids as the seat of molecular memory and genetic heritability (Schrödinger, 1944), which materialized when the structure and function of DNA were uncovered, von Neumann’s ideas about self-organization have yet to bear fruit. Biological systems provide material that is ripe for applying these ideas and for the development of new ideas. Continuing interdisciplinary efforts to explore artificial life systems are likely to yield deep insights into the question of the origin of life itself.
Even the narrowest interpretation of “self-organization” as biological self-assembly and development encompasses a wide range of fascinating and important phenomena, ranging from the self-assembly of multiprotein nanoscale structures such as viral capsids and bacterial flagella to the growth and differentiation of animal tissues in the process of organogenesis. These phenomena involve dynamical processes that can be superficially compared to the execution of a program by a computer, yet all of the program “instructions” and contingencies are resident in the components themselves. How do the slight variations in viral capsomer proteins encode the structure of the capsid? How does the assembling flagellum “know” when it reaches the correct size? What are the mechanisms that ensure the proper size and proportions of a fly wing?
Lessons from biological self-assembly will advance our ability to build artificial
nanostructures and advanced materials. Conversely, answers to the general questions of engineered self-assembly, such as How much information must be encoded in a set of parts to assemble a desired structure with high fidelity? may help to uncover some of the fundamental principles of living matter.
A fascinating example of molecular self-assembly in biology is provided by the bacterial flagellar motor (Chevance and Hughes, 2008) (see Figure 4-6). This remarkable molecular machine is a proton motive force (PMF) driven motor that rotates the flagellum, a flexible filament that propels swimming bacteria. Its assembly involves the coordinated sequential expression of over 50 genes encoding different protein components (Kalir et al., 2001). Protein products of genes expressed early initiate the self-assembly process and build an integral membrane ring structure, which will become the rotor, followed by the assembly onto this ring of the PMF-driven secretion system, which plays the key role in the timely export of the specific proteins that assemble the rod-and-hook structure. The assembly follows the same strategy as construction work on a skyscraper: The partially completed structure acts as a conduit for transporting building materials up to the moving “front” of construction at the top level. As the rod-and-hook structure is completed, the specificity of the secretion system switches and the structure exports protein components for the flagellar filament. We now have tantalizing insights into the molecular mechanisms that control the assembly process. For example, the length of the hook appears to be controlled by a “molecular ruler” (Journet et al., 2003) protein, which trips the specificity switch of the export system once the rod-and-hook assembly reaches a certain length. Another feedback mechanism, export of a specific transcriptional inhibitor, couples completion of the rod-and-hook assembly to the initiation of the late gene expression program (Kutsukake, 1994). The ability of scientists to dissect a molecular process of this complexity is truly inspiring and sets the stage for addressing fundamental questions concerning biological self-assembly. Can we understand evolutionary connections between different but related molecular machines? Can we distill the general principles of biological self-assembly and implement them to construct artificial nanostructures of increasing complexity? Progress in this field will clearly require a joint multidisciplinary effort among biological scientists who have a detailed understanding of these natural systems and physical scientists who are developing nanoscaled structures and advanced materials with some self-assembly or self-repair capability.
The diversity and complexity of physical forms in the living world is striking. Shapes are the most readily observable phenotypes, and it is not surprising that it was the observation of finch beak shapes that inspired Darwin’s theory of natural selection. Nearly 100 years ago Darcy Thompson took the first steps to apply mathematics to the description of living shapes, but the real progress in understanding morphogenesis came with advances in genetics and molecular biology that brought us to the point where we now know the genetic factors that control the shape of
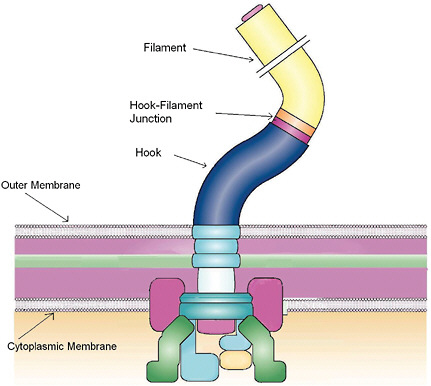
FIGURE 4-6
Flagellar Motor Self-Assembly
The flagellar apparatus is assembled within the cell wall with the filament extending outside the cell. Self-assembly of this complex structure is aided by the just-in-time expression, production, and secretion of the numerous protein components as they become required for the assembly. The assembly is regulated by multiple feedback loops that control the order of protein secretion. Switching off the supply of components determines the length of the rod, the hook, and the filament shown in the figure. Quantitative characterization of biological self-assembly in model systems such as the flagella will uncover the mechanisms of assembly of complex structures and their evolution. SOURCE: Adapted from Chevance and Hughes, 2008; reprinted and adapted with permission from Macmillan Publishers Ltd.
bird beaks (Abzhanov et al., 2004). Nontrivial structure and shape of organs, limbs, and whole bodies are the products of carefully controlled cell proliferation, rearrangement, and migration processes that constitute the developmental program of organisms. These fundamental aspects of morphogenesis are controlled by the genetically determined molecular interaction networks of the type mentioned above. Yet, morphogenesis ultimately involves the physical organization of tissues.
To relate molecular factors to macroscopic phenotypes such as the shape and structure of organs, one must understand the dynamical mechanisms and intercellular interactions that bridge the molecular and whole-organ scales. Understanding these complex phenotypes will require increasingly complex and quantitative hypotheses and the development and adoption of new quantitative tools and ideas. The problems of morphogenesis link molecular genetics with the field of pattern formation studied by physicists and applied mathematicians (Turing, 1952; Cross and Hohenberg, 1993) (Figure 4-7) and constitutes an exciting scientific frontier where interdisciplinary collaboration is likely to be the main driver of progress.
Layered onto shapes are patterns. Some familiar ones include spiral patterns of seeds in a sunflower or spines on a pineapple. Leonardo da Vinci and Johannes Kepler were among the first to ponder the remarkable mathematical regularity of these patterns: The number of visible right- and left-handed spirals is invariably a pair of successive Fibonacci numbers (5-8, 8-13, 13-21, etc.) (Adler et al., 1997). Understanding the logic of the phyllotaxis pattern has remained a challenge since that time. In 1870, Hofmesiter formulated the mathematical rules that describe phyllotaxis. More recently, Levitov showed how phyllotaxy can arise as the lowest energy state in a system of particles with long-range repulsion (1991), while Douady and Couder devised an experimental demonstration showing that sequential addition of ferrofluid droplets (which repel each other) as a function of the rate of addition naturally reproduces the full range of possible phyllotaxis patterns (1992) (Figure 4-8). Yet, the connection with plant morphogenesis was missing: What was the relation between the physical packing of repelling objects and the process of plant growth that produced beautiful patterns in biology?
The answer had to wait until molecular genetic and fluorescent imaging techniques were developed, thereby allowing investigators to determine the distribution and transport of auxin, a plant hormone promoting leaf primordia growth in the shoot apical meristem (SAM). Specifically, auxin is actively transported within the surface layer of cells by pumps that transport auxin in the direction of increasing concentration (Reinhardt et al., 2003; Jonsson et al., 2006). There is positive feedback: The steeper the auxin concentration gradient, the greater the pumping activity that further increases the gradient. This positive feedback gives rise to an instability and, in turn, to the formation of spots of high auxin concentration, which cause local outgrowth of leaf primordia. The high auxin regions forming the primordia effectively repel each other because the periphery of each high auxin region is depleted of auxin. Thus, as happens with ferrofluid droplets, a pattern, spiral in this case, emerges in the plant. To fully understand the morphogenetic implications of this molecular mechanism, plant biologists had to team up with computational modelers (Jonsson et al., 2006; Smith et al., 2006); this partnership led to a quantitative model that not only demonstrated the emergence of phyllotaxis but also made detailed, falsifiable predictions concerning the spatial
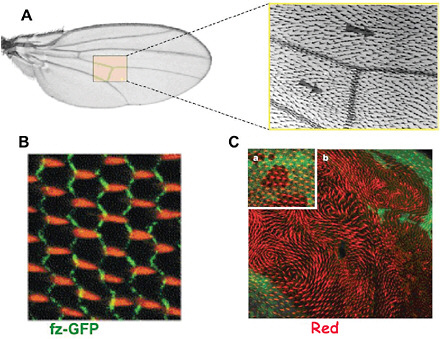
FIGURE 4-7
Planar Cell Polarity in Drosophila Wing Development
A close-up view of a fly wing (A) reveals an ordered array of distally pointing short “hairs”; (B) An array of actin bundles (orange) forming the prehairs emanates from the distal edge of each cell during the early pupal stage of wing development. Polarization of actin prehairs depends on intercellular interactions that involve the formation of asymmetric ligand-receptor complexes bridging neighboring cells. (C) Mutants defective in certain genes (in this case, a gene called Fat) show a swirling pattern of prehairs (red), which, while losing global orientation, retain local alignment. This behavior strongly suggests an analogy with ferromagnetism, which also involves formation of a globally ordered magnetization under the action of short-range interactions. We thus have a striking illustration of how physics ideas can be relevant to biological phenomena far removed from their original physical context. SOURCES: (A) and (B) Strutt, 2001, reprinted with permission from Elsevier; (C) Ma et al., 2003, reprinted with permission from Macmillan Publishers Ltd.
distribution of auxin pumps. The model for phyllotactic pattern formation is similar in spirit, although not in letter, to the Turing model in providing a bridge between the molecular and morphological scales. The phyllotactic model is now being used in close collaboration with experiment to further explore the dynamics of SAM growth and its regulation. The model also has interesting mathematical aspects that are being examined and generalized by applied mathematicians.
Ultimately, the analogy between self-assembly and developmental processes in
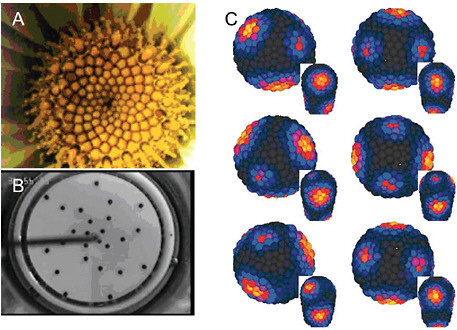
FIGURE 4-8
Phyllotaxis
Panel (A) presents a typical Fibbonacci spiral pattern of floral phyllotaxis. Panel (B) shows phyllotaxis in a physical system: a phyllotactic pattern generated by sequential addition of ferrofluid droplets in the center of a dish with a slow radial flow of viscous fluid. Panel (C) shows the early stages of the spiral formation in a computational model of auxin-driven patterning of outgrowth primordia (yellow/red) in a shoot apical meristem. SOURCES: (A) and (B) Douady and Couder, 1992, reprinted with permission from the American Physical Society; (C) Jonsson et al., 2006, courtesy of the Proceedings of the National Academy of Sciences.
biology suggests that not only is there no limit to the complexity of assembly, but also that only the ideas and concepts underlying the different assembly protocols remain to be discovered.
CONCLUSION
This chapter outlines just a few of the systems in which biological and physical scientists have collaborated on new concepts for viewing and solving issues in their fields. Among these, the multifaceted study of interactions and information transfer, studies of dynamical behavior, and studies of self-assembly and devel-
opmental processes provide a glimpse of how interdisciplinary approaches could bring us closer to understanding the fundamental principles of nature and the process of life itself.
REFERENCES
Abzhanov, A., M. Protas, B.R. Grant, P.R. Grant, and C.J. Tabin, 2004. Bmp4 and morphological variation of beaks in Darwin’s finches, Science 305: 1462-1465.
Adler, I., D. Barabé, and R.V. Jean, 1997. A history of the study of phyllotaxis, Annals of Botany 80: 231-244.
Arkin, A., J. Ross, and H.H. McAdams, 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells, Genetics 149: 1633-1648.
Chevance, F.F., and K.T. Hughes, 2008. Coordinating assembly of a bacterial macromolecular machine, Nature Reviews Microbiology 6: 455-465.
Compte, A., N. Brunel, P.S. Goldman-Rakic, and X.J. Wang, 2000. Synaptic mechanisms and network dynamics underlying spatial working memory in a cortical network model, Cerebral Cortex 10: 910-923.
Cross, M., and P. Hohenberg, 1993. Pattern formation outside of equilibrium, Modern Physics 65: 851.
Douady, S., and Y. Couder, 1992. Phyllotaxis as a physical self-organized growth process, Physical Review Letters 68: 2098-2101.
Elowitz, M.B., A.J. Levine, E.D. Siggia, and P.S. Swain, 2002. Stochastic gene expression in a single cell, Science 297: 1183-1186.
Freeman, R.A., and P.V. Kokotovic, 2008. Robust Nonlinear Control Design: State-Space and Lyapunov Techniques, Basel, Switzerland: Springer Verlag, Modern Birkhäuser Classics.
Ghaemmaghami, S., W. Huh, K. Bower, R.W. Howson, A. Belle, N. Dephoure, E.K O’Shea, and J.S. Weissman, 2003. Global analysis of protein expression in yeast, Nature 425: 737-741.
Hansen, C.H., R.G. Endres, and N.S. Wingreen, 2008. Chemotaxis in Escherichia coli: A molecular model for robust precise adaptation, Public Library of Science Computational Biology 4: 14-27.
Hodgkin, A., and A. Huxley, 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve, Journal of Physiology-London 117: 500-544.
Hopfield, J.J., 2007. Hopfield Network, Scholarpedia 2: 1977.
Jackson, E. Atlee, 1991. Perspectives of Nonlinear Dynamics, Cambridge, England: Cambridge University Press.
Jonsson, H., M.G. Heisler, B.E. Shapiro, E.M. Meyerowitz, and E. Mjolsness, 2006. An auxin-driven polarized transport model for phyllotaxis, Proceedings of the National Academy of Sciences of the United States of America 103: 1633-1638.
Journet, L., C. Agrain, P. Broz, and G.R. Cornelis, 2003. The needle length of bacterial injectisomes is determined by a molecular ruler, Science 302: 1757-1760.
Kalir, S., J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M.G. Surette, and U. Alonl, 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria, Science: 292, 2080-2083.
Kaufmann, B. B., and A. van Oudenaarden, 2007. Stochastic gene expression: from single molecules to the proteome, Current Opinion in Genetics & Development 17: 107-112.
Klug, A., 2005. The discovery of zinc fingers and their development for practical applications in gene regulation, Proceedings of the Japan Academy Series B-Physical and Biological Sciences 81: 87-102.
Kutsukake, K., 1994. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium, Molecular & General Genetics 243: 605-612.
Levitov, L.S., 1991. Phyllotaxis of flux lattices in layered superconductors, Physics Review Letters 66: 224-227.
Losick, R., and C. Desplan, 2008. Stochasticity and cell fate, Science 320: 65-68.
Ma, D., C.H. Yang, H. McNeill, M.A. Simon, and J.D. Axelrod, 2003. Fidelity in planar cell polarity signalling, Nature 421: 543-547.
Onuchic, J.N., and P.G. Wolynes, 2004. Theory of protein folding, Current Opinion in Structural Biology 14: 70-75.
Paillard, G., C. Deremble, and R. Lavery, 2004. Looking into DNA recognition: zinc finger binding specificity, Nucleic Acids Research 32: 673-682.
Pavletich, N.P., and C.O. Pabo, 1991. Zinc finger-DNA recognition—Crystal-structure of a Zif268-DNA complex at 2.1-A, Science 252: 809-817.
Reinhardt, D., E.R. Pesce, P. Stieger, T. Mandel, K. Baltensperger, M. Bennett, J. Traas, J. Friml, and C. Kuhlemeier, 2003. Regulation of phyllotaxis by polar auxin transport, Nature 426: 255-260.
Schnitzer, M.J., K. Visscher, and S.M. Block, 2000. Force production by single kinesin motors, Nature Cell Biology 2: 718-723.
Schrödinger, E., 1944. What is Life? Cambridge, England: Cambridge University Press.
Smith, R.S., S. Guyomarc’h, T. Mandel, D. Reinhardt, C. Kuhlemeier, and P. Prusinkiewicz, 2006. A plausible model of phyllotaxis, Proceedings of the National Academy of Sciences of the United States of America 103: 1301-1306.
Strutt, D.I., 2001. Asymmetric localization of frizzled and the establishment of cell polarity in the drosophila wing, Molecular Cell 7: 367-375.
Suel, G.M., R.P. Kulkarni, J. Dworkin, J. Garcia-Ojalvo, and M.B. Elowitz, 2007. Tunability and noise dependence in differentiation dynamics, Science 315: 1716-1719.
Swem, L.R., D.L. Swem, N.S. Wingreen, and B.L. Bassler, 2008. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harvey, Cell 134: 461-473.
Turing, A.M., 1952. The chemical basis of morphogenesis, Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 237: 37-72.
Vale, R.D., T.S. Reese, and M.P. Sheetz, 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility, Cell 42: 39-50.
Vitousek, P., 2004. Nutrient Cycling and Limitation: Hawai’i as a Model System. Princeton, N.J.: Princeton University Press.
von Neumann J., and A.W. Burks, 1966. Theory of Self-Reproducing Automata. Champaign, Ill: University of Illinois Press.





















