8
Genome-wide Patterns of Population Structure and Admixture Among Hispanic/Latino Populations
KATARZYNA BRYC,* CHRISTOPHER VELEZ,† TATIANA KARAFET,‡ ANDRES MORENO-ESTRADA,*§ ANDY REYNOLDS,* ADAM AUTON,*|| MICHAEL HAMMER,‡ CARLOS D. BUSTAMANTE,*§** AND HARRY OSTRER†**
Hispanic/Latino populations possess a complex genetic structure that reflects recent admixture among and potentially ancient substructure within Native American, European, and West African source populations. Here, we quantify genome-wide patterns of SNP and haplotype variation among 100 individuals with ancestry from Ecuador, Colombia, Puerto Rico, and the Dominican Republic genotyped on the Illumina 610-Quad arrays and 112 Mexicans genotyped on Affymetrix 500K platform. Intersecting these data with previously collected high-density SNP data from 4,305 individuals, we use principal component analysis and clustering methods FRAPPE and STRUCTURE to investigate genome-wide patterns of African, European, and Native American population structure within and among Hispanic/Latino populations. Comparing autosomal, X and Y chromosome, and mtDNA variation, we find evidence of a significant sex bias in admixture proportions consistent with disproportionate contribution of European male and Native American female ancestry to present-day populations. We also find that patterns of linkage disequi-
|
* |
Department of Biological Statistics and Computational Biology, Cornell University, Ithaca, NY 14850; |
|
† |
Human Genetics Program, Department of Pediatrics, New York University School of Medicine, New York, NY 10016; |
|
‡ |
Arizona Research Laboratories Division of Biotechnology and Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ 85721; and |
|
§ |
Department of Genetics, Stanford University School of Medicine, Stanford, CA 94305. |
|
|| |
Present address: Wellcome Trust Centre for Human Genetics, Oxford OX3 7BN, UK. |
|
** |
To whom correspondence may be addressed; e-mail: cdbustam@stanford. edu or harry. ostrer@nyumc.org. |
libria in admixed Hispanic/Latino populations are largely affected by the admixture dynamics of the populations, with faster decay of LD in populations of higher African ancestry. Finally, using the locus-specific ancestry inference method LAMP, we reconstruct fine-scale chromosomal patterns of admixture. We document moderate power to differentiate among potential subcontinental source populations within the Native American, European, and African segments of the admixed Hispanic/Latino genomes. Our results suggest future genome-wide association scans in Hispanic/Latino populations may require correction for local genomic ancestry at a subcontinental scale when associating differences in the genome with disease risk, progression, and drug efficacy, as well as for admixture mapping.
The term “Hispanic/Latinos” refers to the ethnically diverse inhabitants of Latin America and to people of Latin American descent throughout the world. Present-day Hispanic/Latino populations exhibit complex population structure, with significant genetic contributions from Native American and European populations (primarily involving local indigenous populations and migrants from the Iberian peninsula and Southern Europe) as well as West Africans brought to the Americas through the trans-Atlantic slave trade (Sans, 2000; S. Wang et al., 2008). These complex historical events have affected patterns of genetic and genomic variation within and among present-day Hispanic/Latino populations in a heterogeneous fashion, resulting in rich and varied ancestry within and among populations as well as marked differences in the contribution of European, Native American, and African ancestry to autosomal, X chromosome, and uniparentally inherited genomes.
Many key demographic variables differed among colonial Latin American populations, including the population size of the local pre-Columbian Native American population, the extent and rate at which European settlers displaced native populations, whether or not slavery was introduced in a given region, and, if so, the size and timing of introduction of the African slave populations. There were also strong differences in ancestry among social classes in colonial (and postcolonial) populations with European ancestry often correlating with higher social standing. As a consequence, present-day Hispanic/Latino populations exhibit very large variation in ancestry proportions (as estimated from genetic data) not only across geographic regions (Sans, 2000; S. Wang et al., 2008), but also within countries themselves (Seldin et al., 2007; Silva-Zolezzi et al., 2009). In addition, the process of admixture was apparently sex-biased and preferentially occurred between European males and Amerindian and/or African females; this process has been shown to be remarkably
consistent among countries and populations including Argentina (Dipierri et al., 1998), Ecuador (González-Andrade et al., 2007), Mexico (Green et al., 2000), Cuba (Mendizabal et al., 2008), Brazil (Marrero et al., 2007), Uruguay (Sans et al., 2002), Colombia (Carvajal-Carmona et al., 2003), and Costa Rica (Carvajal-Carmona et al., 2003).
The rich diversity of variation in ancestry among Hispanic/Latino populations, coupled with consistent differences among populations in the incidence of chronic heritable diseases, suggests that Hispanic/Latino populations may be very well suited for admixture mapping (Smith et al., 2001; González Burchard et al., 2005). For example, differences in relative European ancestry proportions correlate with higher susceptibility in Puerto Ricans to asthma as compared with Mexicans (Salari et al., 2005). Data have also shown an increased risk of breast cancer in Latinas with greater European ancestry (Fejerman et al., 2008) and an interplay between African ancestry and cardiovascular disease and hypertension in Puerto Ricans from Boston (Lai et al., 2009). Hispanic/Latinos are also likely to play an increasingly important role in multi- and transethnic genetic studies of complex disease. Genome-wide scans have identified candidate markers for onset of type 2 diabetes in Mexican-Americans from Texas (Hayes et al., 2007) as well as a region on chromosome 5 associated with asthma in Puerto Ricans (Choudhry et al., 2008).
Quantifying the relative contributions of ancestry, environment (including socioeconomic status), and ancestry by environment interaction to disease outcome in diverse Hispanic/Latino populations will also be critical to applying a genomic perspective to the practice of medicine in the United States and in Latin America. For example, whereas European ancestry was associated with increased asthma susceptibility in Puerto Ricans (Salari et al., 2005), it was also shown that the effect was moderated by socioeconomic status (Choudhry et al., 2006). This suggests that quantifying fine-scale patterns of genomic diversity among diverse U.S. and non-U.S. Hispanic/Latinos may be critical to the efficient and effective design of medical and population genomic studies. A fine-scale population genomics perspective may also provide a powerful means for understanding the roles of ancestry, genetics, and environmental covariates on disease onset and severity (González Burchard et al., 2005).
Here, we introduce a larger, high-density SNP and haplotype dataset to investigate historical population genetics questions—such as variation in sex-biased ancestry and genome-wide admixture proportions within and among Latino populations—as well as provide a genomic resource for the study of population substructure within putative European, African, and Native American source populations. Our dataset includes three Latino populations that are underrepresented in whole-genome analyses, namely, Dominicans, Colombians, and Ecuadorians, as well as Mexicans
and Puerto Ricans, the two largest Hispanic/Latino ethnic groups in the United States. This allows comparison of patterns of population structure and ancestry across multiple U.S. Hispanic/Latino populations. Our dense SNP marker panel is formed by the intersection of two of the most commonly used genotyping platforms, allowing for the inclusion of dozens of Native American, African, and European populations for ancestry inference. Our work expands on high-density population-wide genotype data from the International HapMap Project (HapMap) (International HapMap Consortium, 2005; Frazer et al., 2007), the Human Genome Diversity Panel (HGDP) (Rosenberg et al., 2002), and the Population Reference Sample (POPRES) (Nelson et al., 2008) that have representation of Mexicans but not other Hispanic/Latino groups either from the Caribbean or from South America, with a resulting gap for analyzing admixture in those populations. This project, therefore, represents an important step toward comprehensive panels for U.S.-based studies that can more accurately reflect the diversity within various Hispanic/Latino populations.
RESULTS
Population Structure
We applied the clustering algorithm FRAPPE to investigate genetic structure among Hispanic/Latino individuals using a merged data set with over 5,000 individuals with European, African, and Native American ancestry genotyped across 73,901 SNPs common to the Affymetrix 500K array and the Illumina 610-Quad panel (Materials and Methods). FRAPPE implements a maximum likelihood method to infer the genetic ancestry of each individual, where the individuals are assumed to have originated from K ancestral clusters (Tang et al., 2005). The plots for K = 3 and K = 7 are shown in Fig. 8.1 and for all other values of K in Fig. S1 (available online at www.pnas.org/cgi/content/full/0914618107/DCSupplemental) K = 3. We observed clustering largely by Native American, African, and European ancestry, with the Hispanic/Latino populations showing genetic similarity with all of these populations. However, significant population differences exist, with the Dominicans and Puerto Ricans showing the highest levels of African ancestry (41.8% and 23.6% African, SDs 16% and 12%), whereas Mexicans and Ecuadorians show the lowest levels of African ancestry (5.6% and 7.3% African, SDs 2% and 5%) and the highest Native American ancestries (50.1% and 38.8% Native American, SDs 13% and 10%). We also found extensive variation in European, Native American, and African ancestry among individuals within each population. A clear example could be observed in the Mexican sample, in which ancestry proportions ranged from predominantly Native American to predomi-
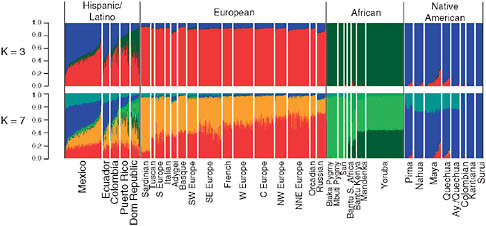
FIGURE 8.1 FRAPPE clustering illustrating the admixed ancestry of Hispanic/Latinos shown for K = 3 and K = 7. Individuals are shown as vertical bars shaded in proportion to their estimated ancestry within each cluster. Native American populations are listed in order geographically, from North to South.
nantly European (with generally low levels of African ancestry). Similar results were found in Colombians and Ecuadorians, whereas Dominicans and Puerto Ricans showed the greatest variation in the African ancestry (Fig. 8.1). Interestingly, at K = 7, we were able to capture signals of continental substructure such as a Southwest to Northeast gradient in Europe and a Native American component that is absent in the two Amazonian indigenous populations (Karitiana and Surui) but that substantially contributes to all other studied Latino populations. We also note that several of the individuals from the Maya and Quechua Native American samples (and to a lesser extent Nahua and Pima) from the Human Genome Diversity Panel (CEPH-HGDP) show moderate levels of European admixture, consistent with previous studies of these populations (Jakobsson et al., 2008). Interestingly, this is not the case for the Aymara and Quechua samples genotyped by Mao et al. (2007).
We also undertook principal component analysis (PCA) of the autosomal genotype data from Hispanic/Latino and putative ancestral populations using the smartpca program from the software package eigenstrat (Fig. 8.2A) (Patterson et al., 2006a). The first two principal components of the PCA strongly support the notion that the three ancestral populations contributing to the Hispanic/Latino genomic diversity correspond exactly to Native American, European, and African ancestry. The Hispanic/Latino populations showed different profiles of ancestry, as exemplified by the fitting of ellipses to the covariance matrix of each population’s first two PCs (Fig. 8.2C). Subsequent PCs showed substructure within Africa, Native
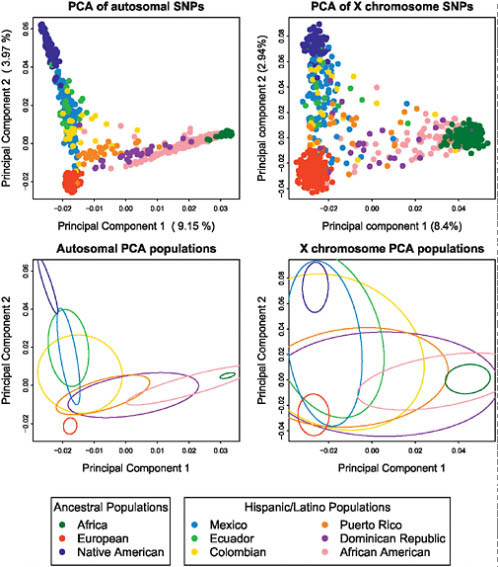
FIGURE 8.2 Principal component analysis results of the Hispanic/Latino individuals with Europeans, Africans, and Native Americans. PC1 vs. PC2 scatterplots based on autosomal markers (Upper Left) and based on X chromosome markers (Upper Right). Ellipses are fitted to the PCA results on the autosomes (Lower Left) and to results from the X chromosome markers (Lower Right).
Americans, and Europeans (Fig. S2, available online at www.pnas.org/cgi/content/full/0914618107/DCSupplemental). PCA on the X chromosome markers (Fig. 8.2B) showed a similar pattern, although because there are only 1,500 markers, this PCA had greater variance, which is illustrated in the fitted ellipses as well (Fig. 8.2D).
We also ran the Bayesian clustering algorithm STRUCTURE in “assignment mode” (Falush et al., 2003), and used a training set of Europeans, Africans, and Native Americans to estimate ancestral allele frequencies and assess admixture proportions within and among the Hispanic/Latino populations. Using STRUCTURE analysis of the autosomes (Fig. 8.3, Upper) and the X chromosome (Fig. 8.3, Lower), we found that, again, Puerto Ricans and Dominicans showed the greatest proportion of African ancestry whereas Colombians, Ecuadorians, and Mexicans showed extensive variation in European and Native American ancestry among individuals. We calculated LD decay curves for all populations with at least
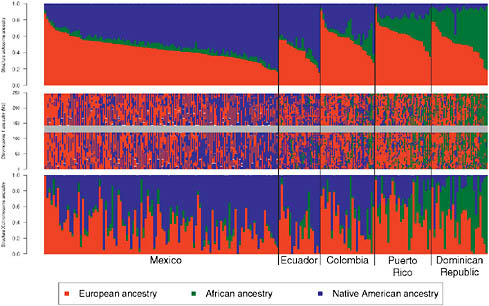
FIGURE 8.3 Genome-wide and locus-specific ancestry estimates for Mexicans, Ecuadorians, Colombians, Puerto Ricans, and Dominicans. Shown for K = 3, clustering of the Hispanic/Latino individuals on the autosomes (Top) and on the X chromosome (Bottom). Individuals are shown as vertical bars shaded in proportion to their estimated ancestry within each cluster. Local ancestry at each locus is shown for each individual on chromosome 1 (Middle). The X chromosome shows greater Native American ancestry and greater variability in African ancestry, with reduced European ancestry.
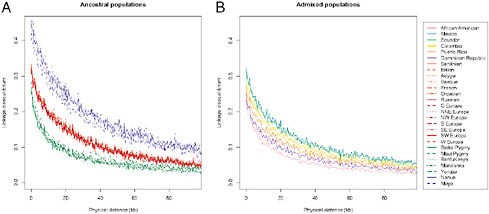
FIGURE 8.4 Linkage disequilibrium, genotype r2 estimated by PLINK, by population as a function of physical distance (Mb). (Left) Native American, European, and African populations. (Right) Hispanic/Latino populations. Scale is the same.
10 individuals, choosing subsets of 10 individuals, and averaging more than 100 random subsets of the data. Patterns of decay of LD were consistent with previously published results (Jakobsson et al., 2008), with Native American populations showing the highest levels of LD and African populations the lowest (Fig. 8.4A). Interestingly, the Hispanic/Latino populations demonstrated rates of decay of LD that correlated strongly with the amount of Native American, European, and African ancestry (Fig. 8.4B). Specifically, the populations with the most Native American ancestry, Mexican and Ecuadorian, exhibited higher levels of linkage disequilibrium among SNP markers, whereas the populations with the highest proportions of African ancestry, the Dominican and Puerto Rican samples, had the lowest levels of LD.
Locus-Specific Ancestry
To reconstruct local genomic ancestry at a fine scale, we used the ancestry deconvolution algorithm LAMP (Sankararaman et al., 2008), allowing for a three-way admixture and focused on the four Hispanic/Latino populations genotyped on the Illumina 610-Quad platform—Dominicans, Colombians, Puerto Ricans, and Ecuadorians (Materials and Methods). Because this same SNP panel had also been genotyped across the HGDP samples (1,043 individuals from 53 populations), the merged dataset containing more than 500,000 markers provided a unique resource for investigating the extent of subcontinental ancestry among diverse Hispanic/Latino populations. We found that individual average ances
tries are in agreement with FRAPPE and STRUCTURE results in which Ecuadorians have the highest Native American proportions, followed by Colombians (showing greater European contribution), and with Puerto Ricans and Dominicans showing the highest African ancestry—specially Dominicans, who show very low contribution from Native Americans (Fig. 8.1). We also used the PCA-based methods of Bryc et al. (2010) to infer ancestry at each locus for the samples genotyped on the Affymetrix 500K, which included more than 100 Mexican samples genotyped by the POPRES project (Nelson et al., 2008) and diverse Native American populations genotyped by Mao et al. (2007). The local admixture tracks for each individual are in large agreement with the genome-wide average ancestry proportions (Fig. 8.3, Middle).
To investigate the genetic relationships among admixed Hispanic/Latino populations and putative ancestral groups, we compared patterns of population divergence among the inferred segments of European, African, and Native American ancestry and corresponding putative source populations using Wright’s FST measure. Specifically, we used LAMP to reconstruct for each individual in our dataset, segments of European, African, and Native American ancestry across both the maximal SNP dataset for all of the admixed and putative source population individuals (i.e., either the 610K Illumina for Puerto Rican, Ecuadorian, Columbian, and Dominican or 500k for Mexicans from Guadalajara) as well as ~70k SNPs common to both platforms. To calculate FST at a given SNP for a given pair of populations, we included only individuals with unambiguous ancestry assignment (i.e., individuals with two European-, two Native American–, or two African-origin chromosomes). One potential confounder for this analysis is that sample sizes differ substantially among subpopulations within major continental regions (e.g., in the Native American set, we have sample sizes that range from n = 7 for Colombian indigenous Americans in HGDP to n = 29 for Nahua from Mexico in the Mao et al. dataset). To minimize the potential bias of differences in sample size, we randomly selected n = 7 individuals from all potential subpopulations and recomputed Wright’s FST. As seen in Table 8.1, we found that consistent with historical records, our results show that African segments of the Hispanic/Latino populations are more closely related to the Bantu-speaking populations of West Africa than other populations. Specifically, we found that the Colombians and Ecuadorians are most closely related to the Kenyan Bantu populations, whereas the Puerto Ricans and Dominicans are closest to the Yoruba from Nigeria. Likewise, European segments show the lowest FST values when compared with Southwest European populations (individuals from Spain and Portugal), as well as French and Italian individuals. Native American segments of the Hispanic/Latino individuals show the least genetic differentiation with Mesoamerican (e.g., Maya and Nahua),
TABLE 8.1 Ancestry-Specific FST Distances Between Hispanic/Latino Populations and Different Putative Source Populations
|
Europe |
Europe |
French |
Italian |
Orcadian |
Russian |
Sardinian |
Tuscan |
|
SW |
W |
|
|
|
|
|
|
|
0.863 |
1.080 |
0.880 |
0.885 |
1.410 |
1.648 |
1.550 |
1.050 |
|
0.537 |
0.730 |
0.613 |
0.610 |
1.093 |
1.413 |
1.270 |
0.825 |
|
0.838 |
1.104 |
0.799 |
0.845 |
1.417 |
1.369 |
1.607 |
0.925 |
|
0.916 |
1.155 |
0.940 |
0.879 |
1.508 |
1.820 |
1.566 |
1.041 |
|
0.122 |
0.265 |
0.270 |
0.271 |
0.793 |
0.882 |
0.852 |
0.336 |
Chibchan (e.g., Colombian), and Andean (e.g., Quechua) populations. The closest relationship is clearly observed between Mexicans from Guadalajara and Nahua indigenous individuals.
Sex Bias in Ancestry Contributions
We used the STRUCTURE ancestry estimates on the autosomes and X chromosome to estimate Native American, European, and African ancestry proportions of each Hispanic/Latino individual. We then compared the estimates of ancestry for each population on the autosomes vs. on the X chromosome [Fig. 8.5 and Figs. S3 and S4 (available online at www.pnas.org/cgi/content/full/0914618107/DCSupplemental)]. Whereas the Native American ancestry was significantly higher on the X chromosome than on the autosomes (including those populations with reduced Native American ancestry, i.e., Puerto Ricans and Dominicans), the autosomal vs. X-chromosome difference was more attenuated with regard to African ancestry. This reduced deviation is present even in those Hispanic/Latino populations analyzed whose non-European ancestry was principally
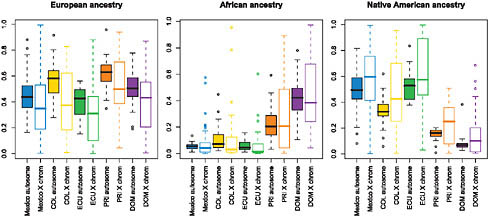
FIGURE 8.5 Boxplots comparing autosomal vs. X-chromosome ancestry proportions by population, shown for European ancestry (Left), Native American ancestry (Center), and African ancestry (Right). Filled boxes correspond to autosomal ancestry estimates; open boxes show X-chromosome ancestry estimates. Median (solid line), first and third quartiles (box) and the minimum/maximum values, or to the smallest value within 1.5 times the IQR from the first quartile (whiskers). For each paired comparison of X chromosomes and autosomes, median Native American ancestries are consistently higher on the X chromosome in all Hispanic/Latino populations sampled, and European ancestries are lower across all populations.
Native American in origin (i.e., Mexicans and Ecuadorians). Furthermore, greater Native American ancestry on the X chromosome in Puerto Ricans did not necessarily imply greater Amerindian ancestry on the autosomes. This finding is similar to those observed by analyzing fine-scale genome pattern of population structure and admixture among African Americans, West Africans, and Europeans (Lind et al., 2007).
Finally, we used SNP and microsatellite genotyping to identify the canonical Y chromosome and mtDNA haplotypes for each of the Hispanic/Latino individuals that we genotyped. Details of the loci and classifications are found in Tables S1 and S2 (available online at www.pnas.org/cgi/content/full/0914618107/DCSupplemental). We found an excess of European Y chromosome haplotypes and a higher proportion of Native American and African mtDNA haplotypes, consistent with previous studies (Fig. 8.6). In addition, we found several non-European Y chromosomal haplotypes with most likely origins from North Africa and the Middle East. We observed that African-derived haplotypes were the predominant origin of mtDNA in Dominicans (17 of 27 individuals), matching the greater African vs. Native American origins of this population on the autosomes and X chromosomes. However, in Puerto Ricans we did not find evidence of a high African female contribution. The predominant Y chromosomal origins in the Puerto Ricans sampled were European and
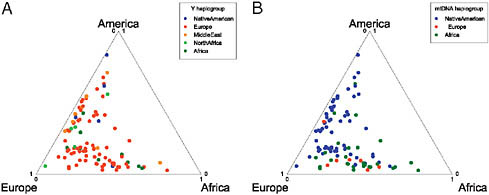
FIGURE 8.6 Comparison of mtDNA and Y chromosome haplotypes. Each individual is represented by a point within the triangle that represents the autosomal ancestry proportions. The most probable continental location for each individual’s haplotype is designated by the shade of the point. The Y chromosome contains a disproportionate number of European haplotypes, whereas the mtDNA has a high proportion of Native American, slightly more African haplotypes, and fewer European haplotypes, consistent with a sex bias toward a great European male and Native American/African female ancestry in the Hispanic/Latinos.
African; but, in contrast, 20 of 27 Puerto Rican individuals had mitochondrial haplotypes of Native American origin, suggesting a strong female Native American and male European and African sex bias contribution. Overall, in all of the Hispanic/Latino populations that we analyzed, we found evidence of greater European ancestry on the Y chromosome and higher Native American ancestry on the mtDNA and X chromosome consistent with previous findings (Dipierri et al., 1998; Green et al., 2000; Sans et al., 2002; Carvajal-Carmona et al., 2003; González-Andrade et al., 2007; Marrero et al., 2007; Mendizabal et al., 2008).
DISCUSSION
Our work has important implications for understanding the population genetic history of Latin America as well as ancestry of U.S.-based Hispanic/Latino populations. As has been previously documented, we found large variation in the proportions of European, African, and Native American ancestry among Mexicans, Puerto Ricans, Dominicans, Ecuadorians, and Colombians, but also within each of these groups. These trends are a consequence of variation in rates of migration from ancestral European and African source populations as well as population density Native Americans in pre-Columbian times (Sans, 2000). We found that Dominicans and Puerto Ricans in our study showed the highest levels of African ancestry, consistent with historical records. European settlers to island nations in the Caribbean basin largely displaced Native American populations by the early to mid-16th century and concurrently imported large African slave populations for large-scale colonial agricultural production (largely of sugar). In contrast, Colombia has wider geographic differences ranging from Caribbean coasts to Andean valleys and mountains, which could explain the enrichment of African ancestry in some individuals and not in others, likely representing the differences in origin within Colombia. Finally, Mexico and Ecuador are two continental countries that had high densities of Native Americans during pre-Columbian times; as expected, the individuals from these two countries show the highest degree of Native American ancestry. Our findings clearly show that the involuntary migration of Africans through the slave trade appears to have left a clear trace in Hispanic/Latino populations proximal to these routes.
From the FST analysis, we found that the high-density genotype data that we have collected is quite informative regarding the personal genetic ancestry of admixed Hispanic/Latino individuals. Specifically, we found that individuals differ dramatically within and among populations and that we can reliably identify subpopulations within major geographic regions (i.e., Europe, Africa, and the Americas) that exhibit lower pairwise
FST (and, therefore, higher genetic similarity) to the inferred European, African, and Native American segments for the 212 individuals studied. We found, for example, that Nahua showed the lowest FST in Mexicans, consistent with the observation that the Nahua are one of the largest Native American populations in this region and are likely to have contributed to the genomes of admixed individuals in Mexico (as opposed, for instance, to the Mexican Pima who fall outside the Mesoamerican cultural region and show considerably higher levels of differentiation). We also found that the lowest FST for the African regions of the Dominican and Puerto Rican genomes are with the Yoruba, a Bantu-speaking West African population that has been shown to be genetically similar to the African segments of African Americans sampled in the United States (Bryc et al., 2010). Although we have limited Native American populations and Hispanic/Latino sample sizes and, thus, the differences in FST with different subcontinental populations suggest that there exists a reasonably strong signal of which present-day populations are most closely related to the ancestral populations that contributed ancestry to each of the Hispanic/Latino populations.
When comparing inferred continental ancestry of the X and Y chromosomes and mitochondrial vs. the autosomal genome, we observed an enrichment of European Y-chromosome vs. autosomal genetic material, and a greater percentage of both Native American and African ancestry on the X-chromosomes and mtDNA compared with the autosomes for the Hispanic/Latino individuals in this study. This suggests a predominance of European males and Native American/African females in the ancestral genetic pool of Latinos, consistent with previous studies. A particularly interesting observation from our work on sex-biased admixture is that the pattern exists not only within populations but among Hispanic/Latino populations as well. In all populations studied, there is an enrichment of Native American ancestry both on the X chromosome and mtDNA compared with the autosomes. This would suggest a greater female Native American contribution to the genome of Latinos. A different result was obtained in relation to African ancestry. We found a smaller difference between mean African ancestry on the X chromosome and the autosomes, compared with the difference in Native American ancestry. Furthermore, unlike in Native American ancestry, we found an overwhelming representation of Native American mtDNA haplogroups in Puerto Ricans, even though non-European ancestry on the autosomes was largely African.
It is important to note that this observation does not necessarily undermine the model of sex-biased admixture among European male and African females in the founding of Hispanic/Latino populations, especially when one considers the predominance of European Y chromosomes in all groups studied. However, it suggests that admixture between Euro-
pean males and Amerindian/African females has been a complex process in the formation of the various Hispanic/Latino populations. Specifically, a reduced X vs. autosome mean African ancestry compared with Native American ancestry suggests a more balanced gender contribution in the Hispanic/Latino genome by individuals of African ancestry. In the case of Puerto Ricans, the only way that one can reconcile greater African ancestry on the X chromosome vs. what would be expected on mitochondrial data would be through transmission of X chromosomes independent of mitochondrial transmission, which is plausible biologically only via males. Caution, however, should be exercised before considering such conclusions as concrete; unlike X chromosomes, which can recombine and thus represent haplotypes derived from thousands of individuals, mitochondrial DNA represents just a sole distant ancestor among these thousands. Thus, a larger mtDNA sample would be necessary compared with X chromosomes to have similar confidence that a cohort would accurately reflect the presumed diversity of ancestry in the population as a whole.
The Y chromosomal results also demonstrate the insufficiency of the paradigm of European males and Native American/African females to capture the complexity within the Latin American populations. For example, we find Y chromosomal haplotypes in Hispanic/Latinos with presumed origins in the Middle East and Northern Africa. Given that historical documentation suggests that most of the non-African and non–Native American contribution to admixed Hispanic/Latino populations is from Southwest Europe, this suggests that the contemporary populations inherited these Y chromosomes from Europeans who, in turn, were descended from Middle Eastern or North African men. Several historical events could have led to the acquisition by Europeans of non-European haplotypes, perhaps during the period of the Roman Empire when the Mediterranean Sea behaved as a conduit (not a physical barrier) between Europe, the Middle East, and North Africa or by Sephardic Jews or Moorish Muslims during the European Middle Ages/Islamic Golden Age. Alternatively, the presence of non-European Y chromosomal haplotypes originating from the Middle East and North Africa could represent the result of Iberian Jews and Muslims (themselves admixed) fleeing the peninsula for New World territories in response to discriminatory policies that strongly pressured both communities at the termination of the Reconquista. Essentially, the diversity of haplotypes in the Y chromosomes in Latinos reflects not only population dynamics from the 15th century onward, but also the historical trends of population movement occurring across the Atlantic during centuries prior.
The marked genetic heterogeneity of Latino populations shown in this study, as previously suggested by other surveys of genetic ancestry (Mao et al., 2007; Price et al., 2007; S. Wang et al., 2008) has important
implications for the identification of disease-associated variants that differ markedly in frequency among parental populations. In their study of 13 Mestizo populations from Latin America, for example, S. Wang et al. (2008) suggested that admixture mapping in Hispanic/Latino populations may be feasible within a two-population admixture framework, since the mean African ancestry in Mestizo populations is typically low (<10%) (S. Wang et al., 2008). Although this is true for Hispanic/Latino populations with origins in the continental landmass of the Americas (such as the populations studied by S. Wang et al.), our results show that this may not apply to Latino populations with origins in the Caribbean, as their African ancestry proportion is considerably higher and is highly variable among individuals, suggesting an extensive three-way admixture and representing additional challenges for admixture mapping. Likewise, we find subtle but reproducible differences in subcontinental ancestry among Hispanic/Latino individuals, suggesting that even a three-way admixture model may not be sufficient to accurately model the dynamic population genetic history of these populations.
Another observation with important implications for designing association studies is the large variation in individual admixture estimates within certain Latino populations (e.g., Mexicans, Colombians, and Ecuadorians). One could expect such outcome when collecting samples from U.S.-based Latino communities, which in turn may come from different locations within their countries of origin (e.g., Colombians and Ecuadorians). However, within the Mexican sample, which has been collected in a single sampling location (i.e., Guadalajara, Mexico), we also observed large variation in European vs. Native American admixture proportions. Our findings are in agreement with previous studies on genetic ancestry from Mexico City (Martinez-Marignac et al., 2007; S. Wang et al., 2008), supporting the idea that such urban agglomerations, in which a large number of epidemiological studies are likely to take place, continue to host a wide range of genetic variability among individuals that may self-identify as individuals from the same population. Therefore, particular attention should be paid to carefully matching representative cases and controls, as well as to carefully control for ancestry when performing association studies using Hispanic/Latino populations. We hope that our dense genome-wide admixture analysis has allowed greater insight into the population dynamics of multiple Hispanic/Latino populations and that it will provide a resource for designing next-generation epidemiological studies in these communities, opening the possibility of better understanding the genetic makeup of this growing segment of the U.S. population.
MATERIALS AND METHODS
Datasets
We genotyped 100 individuals with ancestry from Puerto Rico, the Dominican Republic, Ecuador, and Colombia on Illumina 610K arrays. We extracted 400 European, 365 African American, and 112 Mexican samples from the GlaxoSmithKline POPRES project, which is a resource of nearly 6,000 control individuals from North America, Europe, and Asia genotyped on the Affymetrix GeneChip 500K Array Set (Nelson et al., 2008). We randomly sampled 15 individuals from each European country where possible, or the maximum number of individuals available otherwise, to select the POPRES European individuals to be included in our study. Further description of sampling locations, genotyping, and data quality control are available elsewhere (Nelson et al., 2008). We include 165 and 167 individuals from the HapMap project from the CEU and YRI populations, thinned to the same SNP set (Frazer et al., 2007). We also include all European, Native American, and African individuals from the HGDP genotyped on Illumina 610K arrays (Jakobsson et al., 2008). Finally, we include all Native American populations from the Mao et al. (2007) study genotyped on Affymetrix 500K arrays. For each dataset, we used annotation information to determine the strand on which the data were given and to map all Affymetrix and Illumina marker ids to corresponding dbSNP reference ids [rsids]. SNPs without valid rsids were excluded from analysis. Each dataset was then converted to the forward strand to facilitate merging of the data. Data from the various platforms were merged using the PLINK toolset, version 1.06 (Purcell et al., 2007). Likewise, nonmissing genotype calls that showed disagreement between datasets were omitted. Demographic data for all individuals included in this study are available on GenBank. All samples were approved by institutional review board protocols from their respective studies.
Data Quality Control
The HapMap II release 23, HGDP, Mao et al., and POPRES samples were genotyped and called according to their respective quality control procedures (Frazer et al., 2007; Mao et al., 2007; Jakobsson et al., 2008; Nelson et al., 2008). Our final merged dataset contains 73,901 SNPs with genotype missingness of <0.1 and <0.05 individual missingness across 5,104 individuals.
Population Structure
We used the software FRAPPE, which implements an expectation-maximization algorithm for estimating individual membership in clusters (Tang et al., 2005). This algorithm is more computationally efficient than other MCMC methods, allowing it to analyze many more markers than, for example, STRUCTURE (Falush et al., 2003; Tang et al., 2005). After thinning markers to have r2 < 0.5 in 50 SNP windows, shifting and recalculating every 5 SNPs, we ran FRAPPE on all 64,935 remaining markers for 5,000 iterations. We also assessed admixture proportions for the Hispanic/Latino individuals using STRUCTURE on a reduced dataset of 5,440 markers after thinning for MAF > 0.2 and with a minimum separation of 400 kb between markers. We used the F model with USEPOPINFO = 1 to update allele frequencies using only the ancestral individuals, with 5,000 burn-in and 5,000 iterations (Falush et al., 2003). We also used all 1,518 SNPs on the X chromosome for the same analysis of the X chromosome ancestry. Principal component analysis was conducted using a dataset thinned to have r2 < 0.8 in 50 SNP windows, leaving 69,212 SNPs for analysis using the package smartpca from the software eigenstrat. Ellipses were fitted following the means and 1 SD of the variance–covariance matrix of the PC1 and PC2 scores of each population.
For local ancestry estimation, we used the software LAMP in LAMPANC mode providing allele frequencies for the HGDP West Africans, Europeans, and Native Americans as ancestral populations (Sankararaman et al., 2008). A total of 552,025 SNPs were included in the analysis, and configuration parameters were set as follows: mixture proportions (alpha) = 0.2, 0.4, 0.4; number of generations since admixture (g) = 20; recombination rate (r) = 1e–8; fraction of overlap between adjacent windows (offset) = 0.2; and r2 threshold (ldcutoff) = 0.1. Local ancestry estimation for the Mexican individuals was performed using the two-way PCA-based method described in Bryc et al. (2010) for both the full Illumina 610K and the Affymetrix 500K datasets, in 10 SNP windows. Only Native Americans with <0.01 European ancestry (as estimated from FRAPPE results) were used as the ancestral Native American individuals within their respective datasets. FST was calculated between Native American, European, and African regions of the Hispanic/Latino individuals and the respective continental populations using a C++ implementation of Weir and Cockerham’s (1984) FST weighed equations as previously published. To eliminate bias in estimation of FST due to European ancestry shown in some of the Native Americans, we also removed regions showing European ancestry within any of the Native Americans showing >0.01 European ancestry, using the same local ancestry estimation procedure as described for the Mexican individuals. Furthermore, to avoid any potentially confounding effect of sample size, we used a random sample of 7 (the minimum sample size of
the Native American populations) individuals per non-Hispanic/Latino population to calculate pairwise FST. MAF was set at a threshold >0.1 in the populations compared by FST calculations.
ACKNOWLEDGMENTS
We thank Mariano Rey for support of the project; Peter Gregersen, Carole Oddoux, and Annette Lee for technical assistance; and Marc Pybus for valuable programming support during part of the analyses. This work was supported by the National Institutes of Health (Grant 1R01GM83606) as part of the National Institute of General Medical Sciences research funding programs. Genotype data from 100 Hispanic/Latinos have been deposited in the Gene Expression Omnibus (GEO) series record GSE21248.




















