3
Phylogenomic Evidence of Adaptive Evolution in the Ancestry of Humans
MORRIS GOODMAN*†‡ AND KIRSTIN N. STERNER*
In Charles Darwin’s tree model for life’s evolution, natural selection adaptively modifies newly arisen species as they branch apart from their common ancestor. In accord with this Darwinian concept, the phylogenomic approach to elucidating adaptive evolution in genes and genomes in the ancestry of modern humans requires a well-supported and well-sampled phylogeny that accurately places humans and other primates and mammals with respect to one another. For more than a century, first from the comparative immunological work of Nuttall on blood sera and now from comparative genomic studies, molecular findings have demonstrated the close kinship of humans to chimpanzees. The close genetic correspondence of chimpanzees to humans and the relative shortness of our evolutionary separation suggest that most distinctive features of the modern human phenotype had already evolved during our ancestry with chimpanzees. Thus, a phylogenomic assessment of being human should examine earlier stages of human ancestry as well as later stages. In addition, with the availability of a number of mammalian genomes, similarities in phenotype between distantly related taxa should be explored for evidence of convergent or parallel adaptive evolution. As an example, recent phylogenomic evidence has shown that adaptive evolution of aerobic energy metabolism genes may have helped shape such distinc-
|
* |
Center for Molecular Medicine and Genetics and |
|
† |
Department of Anatomy and Cell Biology, Wayne State University School of Medicine, Detroit, MI 48201. |
|
‡ |
To whom correspondence should be addressed. E-mail: mgoodwayne@aol.com. |
tive modern human features as long life spans and enlarged brains in the ancestries of both humans and elephants.
Charles Darwin (1859) proposed that natural selection favors inherited modifications that better adapt the organisms of a species to the environment of that species. Darwin also proposed the tree model for life’s evolution. In this model, natural selection adaptively modifies newly arisen species as they branch apart from their common ancestor (Darwin, 1859). Although there is now evidence that symbiotic merges produced the first eukaryotes and that prokaryotic species engage in reticulate evolution (Doolittle, 1999; Margulis and Sagan, 2002; Avise, 2008), Darwin’s model of tree-like branching appears to hold for the evolution of primates and other vertebrates. Having deduced that species share common ancestors, Darwin also reasoned that a truly natural system for classifying species would be genealogical, that is, species should be classified according to how recently they last shared a common ancestor. The hierarchical ranking in such a genealogical system could then be used to indicate how relatively close or distant in geological time extant species are from their last common ancestor (LCA).
In accord with this Darwinian framework, the phylogenomic approach to elucidating adaptive evolution in the ancestry of modern humans involves identifying the changes in genes and genomes on a phylogenetic tree that accurately places humans within the order Primates and, more widely, within the class Mammalia. Viewing the ancestries of many mammals, not just the ancestry of modern humans, could provide examples of convergent adaptive evolution, which may point to specific categories of genetic changes that are associated with important phenotypic changes. This phylogenomic approach could help identify the positively selected genetic changes that shaped such distinctive modern human features as prolonged prenatal and postnatal development, lengthened life spans, strong social bonds, enlarged brains, and high cognitive abilities. In this article, we first briefly sketch out the historical background of ideas and findings that have led to phylogenomic studies of human evolution. We then highlight the concepts that motivate our own efforts and discuss how phylogenomic evidence has enhanced our understanding of adaptive evolution in the ancestry of modern humans.
DARWIN’S VIEWS
In The Descent of Man, and Selection in Relation to Sex, Charles Darwin (1874) suggested that Africa was the birthplace for humankind. The fol-
lowing five passages encapsulate for us Darwin’s thinking about the place of humans in primate phylogeny and about the uniqueness of modern humans.
If the anthropomorphous apes be admitted to form a natural subgroup, then as man agrees with them, not only in all those characters which he possesses in common with the whole Catarhine group, but in other peculiar characters, such as the absence of a tail and of callosities, and in general appearance, we may infer that some ancient member of the anthropomorphous subgroup gave birth to man.
(Darwin, 1874, p. 160)
It is therefore probable that Africa was formerly inhabited by extinct apes closely allied to the gorilla and chimpanzee; and as these two species are now man’s nearest allies, it is somewhat more probable that our early progenitors lived on the African continent than elsewhere.
(1874, p. 161)
In regard to bodily size or strength, we do not know whether man is descended from some small species, like the chimpanzee, or from one as powerful as the gorilla; and, therefore, we cannot say whether man has become larger and stronger, or smaller and weaker, than his ancestors. We should, however, bear in mind that an animal possessing great size, strength, and ferocity, and which, like the gorilla, could defend itself from all enemies, would not perhaps have become social: and this would most effectually have checked the acquirement of the higher mental qualities, such as sympathy and the love of his fellows. Hence it might have been an immense advantage to man to have sprung from some comparatively weak creature.
(1874, p. 65)
As far as differences in certain important points of structure are concerned, man may no doubt rightly claim the rank of a Suborder; and this rank is too low, if we look chiefly to his mental faculties. Nevertheless, from a genealogical point of view it appears that this rank is too high, and that man ought to form merely a Family, or possibly even only a Subfamily.
(1874, p. 158)
Nevertheless the difference in mind between man and the higher animals, great as it is, certainly is one of degree and not of kind.
(1874, p. 130)
Darwin’s application of the theory of evolution by natural selection to discussions of our own origins and place within nature laid the foundation for modern phylogenetic and phylogenomic studies of human evolution. He proposed that humankind originated from man-like apes (first quote) in Africa and that humans are most allied to chimpanzees and gorillas (second quote). Further, Darwin seems to have thought that our progenitors were more like chimpanzees than gorillas (third quote). Darwin challenged the then orthodox view that a whole taxonomic order, the Bimana, should consist of only one species, our own Homo sapiens. Instead, Darwin noted that genealogically, we humans should have no more than a family or even just a subfamily to ourselves (fourth quote), suggesting that Darwin might have been willing to have a family Hominidae that grouped modern humans with man-like apes. Moreover, Darwin also commented on the most widely cited example of human uniqueness, the modern human mind. He postulated that the difference between the modern human mind and the mind of other higher animals was one of degree, not of kind (fifth quote). Nearly a century after Darwin first proposed the theory of evolution by natural selection, molecular evolution emerged as a scientific field and molecular methods began to be used toward the study of human evolution. Molecular evidence inferred from proteins and DNA data generated during the past 50 years have vindicated Darwin’s foresightedness and have decisively established that among living species, modern humans have their closest kinship to common and bonobo chimpanzees.
USE OF MOLECULAR METHODS TO INFER OUR PLACE IN NATURE
More than 100 years ago, Nuttall (1904) observed that rabbit antiserum produced against human whole-blood serum yielded larger precipitates when mixed with serum from human, chimpanzee, or gorilla blood than from orangutan, gibbon, or other mammalian blood. Although Nuttall did not comment on the possible phylogenetic and taxonomic significance of his antihuman serum cross-reacting more strongly with chimpanzee and gorilla sera than with Asian ape sera, he did foresee a promising future for molecular studies of evolution (Nuttall, 1904).
By the middle of the 20th century, molecular biologists had established that DNA contained the genetic information for an organism and that nucleotide sequences in genes encoded the amino acid sequences of proteins. Immunologists could then deduce that a protein’s antigenic divergencies among species reflected amino acid sequence divergencies, the sources of which were nucleotide sequence substitutions. An immunological method that was much improved over Nuttall’s was used to
examine protein divergencies among primate and other mammalian species (Goodman, 1961, 1962a,b, 1963a,b). The observed protein divergencies, interpreted as genetic divergencies, challenged the reigning view (Simpson, 1963) that the human lineage diverged markedly from the ancestral ape state to occupy an entirely new structural-functional adaptive zone. Whereas the then-prevailing view placed chimpanzees and gorillas with orangutans in the family Pongidae, with humans alone among living species in the family Hominidae (Simpson, 1963), the immunologically detected genetic affinities showed humans, chimpanzees, and gorillas to be highly similar and more closely related to one another than to orangutans or other primates, thus supporting chimpanzees and gorillas being grouped with humans in the family Hominidae (Goodman, 1962b, 1963a,b). The indicated genetic kinship between chimpanzees and gorillas was not any closer than the close kinship of either to humans. Indeed some immunological results placed chimpanzees closer to humans than to gorillas [e.g., Fig. 4 in Goodman (1963a)], and they also suggested that rates of molecular evolution had slowed in hominoid lineages (Goodman, 1961, 1962a,b, 1963a). Thus, these first substantial molecular data did not support the claim that the human lineage had diverged radically from an ancestral ape state. Instead, in their proteins, humans, chimpanzees, and gorillas diverged only slightly from one another. The degrees of interspecies antigenic divergence of serum albumin challenged the conventional view that many millions of years of evolution separated modern humans from our nearest nonhuman relatives (Sarich and Wilson, 1967a,b). Instead, when these immunologic divergence data were analyzed by a molecular clock model, the LCA of humans, chimpanzees, and gorillas was placed at only 5 Mya (Sarich and Wilson, 1967a,b).
The determination of the actual amino acid sequences of proteins began in the 1950s (Sanger and Thompson, 1952) and, in the ensuing decades, provided important information about human evolution. Phylogenetic analysis of hemoglobin amino acid sequences pointed to the possibility that chimpanzees and humans were more closely related to each other than either was to gorillas (Goodman et al., 1971, 1982, 1983). This analysis grouped chimpanzee and gorilla with human rather than with orangutan hemoglobin (Goodman et al., 1982, 1983) and showed human and chimpanzee hemoglobin to be identical and slightly divergent from gorilla hemoglobin (Goodman et al., 1971, 1982, 1983).
Estimates of interspecies genetic similarities were also obtained by DNA-DNA hybridization data. An initial set of such data reported in 1972 (Hoyer et al., 1972), similar to the hemoglobin amino acid sequence data, suggested that instead of a human-chimpanzee-gorilla trichotomy, humans and chimpanzees shared the more recent common ancestor. During the 1980s, extensive DNA-DNA hybridization data clearly placed chimpan-
zees closer to humans than to gorillas (Sibley and Ahlquist, 1984; Caccone and Powell, 1989). Direct measurements of interspecies genetic similarities were provided by the actual nucleotide sequences of orthologous DNAs, each set of these DNA orthologues apparently having descended from the same genomic locus in the LCA of the examined contemporary species. By the late 1980s and early 1990s, phylogenetic analysis of such data greatly strengthened the evidence that chimpanzees (common and bonobo) have humans, not gorillas, as their closest relatives (Miyamoto et al., 1987; Bailey et al., 1992; Horai et al., 1992).
With the advent of next-generation sequencing technologies, genomic-level sequence data have provided strong evidence that chimpanzees are our closest living relatives and only slightly diverge from us (Wildman et al., 2003; Chimpanzee Sequencing and Analysis Consortium, 2005; Goodman et al., 2005; Elango et al., 2006; Patterson et al., 2006b; Ebersberger et al., 2007). Large amounts of nucleotide sequence data have also been used to infer the evolutionary relationships among almost all extant primate genera (Goodman et al., 2005; Fabre et al., 2009) and among the major clades of placental mammals (Hallström et al., 2007; Wildman et al., 2007; Prasad et al., 2008). These data reveal that rates of molecular evolution were slower in apes than in Old World monkeys and, within the ape clade, slower in chimpanzees than in gorillas and orangutans and slowest in the human lineage (Elango et al., 2006; Kim et al., 2006). This slowdown can be attributed to the decreased annual mutation rates that must have resulted from lengthened generation times. There may also have been selection for more efficient mechanisms of DNA repair and maintenance of genome integrity (Barja and Herrero, 2000).
A number of recent studies have used fossil calibration points and a variable-rate molecular clock to infer divergence dates across primate phylogeny [e.g., Yoder and Yang (2004), Raaum et al. (2005), Steiper and Young (2006), Fabre et al. (2009)]. Dates inferred from the fossil record and molecular data suggest the human–chimpanzee LCA is more recent than the LCA age for the species of a strepsirrhine genus (either lemuriform or lorisiform) and is close to the LCA age for the species of an Old World monkey genus such as Macaca or Cercopithecus or a New World monkey genus such as Ateles or Callicebus. This objective view of our species recalls Darwin’s vision of our place in a genealogical classification of primates. However, these data suggest that, rather than having a mere subfamily to ourselves, we modern humans should perhaps have no more than a genus or just a subgenus to ourselves; that is, common and bonobo chimpanzees and modern humans would be the only extant members of either subtribe Hominina or genus Homo (Goodman, 1996; Goodman et al., 1998; Wildman et al., 2003). The rules (Hennig, 1966) for such an age-based genealogical classification are that each taxon should represent a clade, and clades at an
equivalent evolutionary age should be assigned the same taxonomic rank. An intragenus sister-grouping of humans and chimpanzees is concordant with the ages of origin of many other mammalian genera (Wildman and Goodman, 2004) and captures the close genetic correspondence of humans to chimpanzees. In accord with this close correspondence, chimpanzees are highly social, have simple material cultures, inhabit a wide range of habitats that range from forests to savannas, and have the ability to use rudimentary forms of language (Fouts and Mills, 1997; Savage-Rumbaugh and Fields, 2000; Whiten et al., 2001; McGrew, 2004; Pruetz and Bertolani, 2007; Sanz and Morgan, 2007).
PHYLOGENOMIC ASSESSMENT OF BEING HUMAN
The extensive sequencing of genomes from primates (Fig. 3.1) and other vertebrates makes possible a phylogenomic search for the genetic basis of modern human traits. The close genetic correspondence of chimpanzees to humans and the relative shortness of our evolutionary separation from chimpanzees suggest that most of the adaptive evolution that produced the distinctive modern human phenotype had already
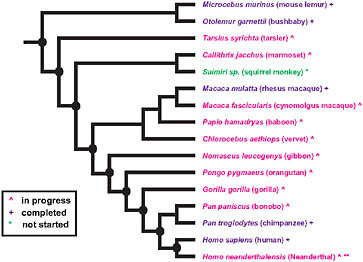
FIGURE 3.1 Summary of primate genome sequencing projects based on the National Human Genome Research Institute list of the status of approved sequencing targets (current as of December 15, 2009). Species with (+) are completed, (^) are in progress, and (*) has not yet been started. **Sequencing of the Neanderthal genome is currently in progress, although not listed by the National Human Genome Research Institute. Nodes of particular interest for examining the evolutionary origins of distinctive human traits are noted by a black oval.
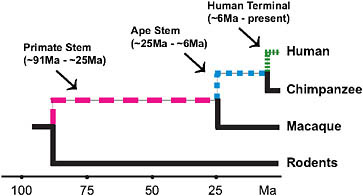
FIGURE 3.2 Lineages of interest when examining the evolutionary origins of distinctly human traits using currently available high-quality genomic data.
occurred by the time of the LCA of chimpanzees and humans. Thus, an assessment of the genetic underpinnings of being human should not just focus on the terminal human lineage but should also encompass earlier periods of human ancestry (Fig. 3.2). Moreover, there are other mammals with aspects of their phenotypes (e.g., enlarged brains) that are similar to aspects of the distinctive modern human phenotype. Examining the ancestries of these mammals is a further way to assess the genetic underpinnings of distinctive modern human phenotypic features and suggests that all such features are not necessarily unique to modern humans.
PHYLOGENOMIC ASSESSMENT OF HUMAN BRAIN EVOLUTION REVEALS ADAPTIVE EVOLUTION IN MULTIPLE STAGES OF HUMAN ANCESTRY
Expanded cognitive abilities are hallmarks of modern humans. Why such abilities were selected for in modern humans and in the human lineage, and how they are maintained, is of great interest. As noted by Darwin more than 135 years ago, differences observed between the modern human mind and the mind of our closest living relatives can be more appropriately characterized as differences in degree, not differences in absolute kind. As such, we expect that the roots of the adaptive evolution that led to the modern human mind trace back to ancient stem lineages in primate and mammalian phylogeny. For example, humans have a phenomenal ability to design and use complex tools, but this ability depends on the opposable thumb, which had evolved in the early primates, as attested to by its presence in slow loris and other primate species.
A striking morphological feature that separates modern humans from other primates is our enlarged cerebral cortex. An initial enlargement of the cerebral cortex in the stem lineage of the anthropoid infra-order Catarrhini was followed by further marked enlargements in the hominid lineage to the chimpanzee/human LCA. After a period of stasis, a further marked expansion occurred during the past 3 million years in the terminal descent to modern humans. This last neocortical expansion resulted from more rapid and prolonged growth of brain mass. Whereas the chimpanzee brain reaches 40% of its adult size by the end of fetal life, the modern human brain at birth has reached only 30% of its adult size (DeSilva and Lesnik, 2008). Nevertheless, although far from its adult size, the newborn human brain is still larger than the newborn chimpanzee brain (DeSilva and Lesnik, 2008).
Anthropoid primates have large brains relative to body size, invasive hemochorial placentation, and long gestations. During this long gestation, the fetal brain consumes approximately 65% of the fetal body’s total metabolic energy (Holliday, 1971). The invasive hemochorial placenta facilitates the transfer of nutrients from mother to fetus. Among anthropoids, modern humans have the largest brain, the most invasive placentation, and the longest gestation. In the earlier ancestry of humans, the threat of destructive maternal immune attacks on the fetus would have necessitated the evolution of mechanisms for immune tolerance at the maternal-fetal interface. Genes that code for galectins, proteins that promote immune cell death, may have provided the anthropoid fetus with an additional immune tolerance mechanism for averting maternal immune attacks (Than et al., 2009). Anthropoid primates have placental-specific galectins that induce apoptosis of T lymphocytes (Than et al., 2009). These genes originated from gene duplications that occurred in the anthropoid stem lineage, and then in that lineage, regulatory evolution of these genes produced placental-specific expression. Moreover, there was also positive selection for amino acid replacements in the placental galectins of the common ancestor of anthropoids, of catarrhines, and of humans. This adaptive evolution contributed to distinctive but not unique modern human features such as lengthened gestation and increased brain-to-body size ratio. Paradoxically, because of the selection that brought about distinctive modern human features, which also include prolonged postnatal development and longer generation times, we are genetically closer to the human/chimpanzee LCA than are chimpanzees (Elango et al., 2006; Kim et al., 2006; Wildman et al., 2007).
Potential genetic correlates to increased brain size in the primate lineage that descended to humans is provided by the evolutionary history of Abnormal Spindle-Like Microcephaly-Associated (ASPM) and microcephalin (MCPH1), two genes that have mutant forms associated with the
severe reduction of brain size that characterizes microcephaly in humans (Bond et al., 2002; Jackson et al., 2002). During descent of the ape stem portion of the primate lineage to humans (approximately 25 to 6 Mya), positive selection acted on microcephalin’s protein-coding sequence, and during the past 6 million years in descent from the chimpanzee/human LCA to modern humans, positive selection acted on ASPM’s protein-coding sequence (Zhang, 2003b; Evans et al., 2004a,b; Kouprina et al., 2004; Wang and Su, 2004).
Language is also considered to be a distinctive human trait. There is evidence of accelerated evolution in the human terminal of the protein-coding sequence of Forkhead Box P2 (FOXP2) (Enard et al., 2002b; Zhang et al., 2002; Spiteri et al., 2007). This gene encodes a transcription factor that influences the expression levels of many brain-expressed genes. FOXP2 mutants have been found in humans with language dysfunction (Lai et al., 2001; MacDermot et al., 2005; Feuk et al., 2006), suggesting that adaptive evolution of FOXP2 may have contributed to the origin of modern human spoken language abilities (Enard et al., 2002b; Zhang et al., 2002). This adaptive evolution may have occurred in archaic humans ancestral to both Neanderthals and modern humans, an inference drawn from the finding that Neanderthal FOXP2 has the same two amino acid replacements that distinguish modern human FOXP2 from the orthologous chimpanzee protein (Krause et al., 2007). The chimpanzee FOXP2 patterns of brain transcriptional regulation differ somewhat from the modern human FOXP2 patterns (Konopka et al., 2009), although there is no direct evidence that the two-amino acid difference of chimpanzee FOXP2 from modern human FOXP2 causes language dysfunction. In addition to evidence suggesting FOXP2 has evolved adaptively in humans, five of the genes that FOXP2 regulates had themselves been under positive selection (Konopka et al., 2009). Several genes involved in the development of the auditory system also show evidence of adaptive evolution in modern humans (A.G. Clark et al., 2003).
A number of recent studies have examined gene expression in the brain transcriptomes of different primate species. More expression changes were observed in the human brain than in other primate brains (Enard et al., 2002a). In general, the majority of these changes involved increased expression (Cáceres et al., 2003). Among the genes found to be up-regulated in modern humans are genes involved in neuronal activity and metabolic processes (Cáceres et al., 2003; Uddin et al., 2004). Genes involved in oxidative phosphorylation (electron transport) are especially up-regulated in humans (Uddin et al., 2004). Most recently, Nowick and colleagues (2009) suggested that major differences in expression of brain-expressed genes observed between human and chimpanzee may be coordinated by a small number of transcription factors that show differential expression
between humans and chimpanzees. Interestingly, many of these transcription factors are associated with pathways involved in energy metabolism (Nowick et al., 2009).
In addition to changes in gene expression level, gene duplication events during human ancestry may have also contributed significantly to our brain evolution. All mammals possess a gene that encodes glutamate dehydrogenase 1 (GLUD1). Whereas GLUD1 is localized to both the mitochondria and cytoplasm where it functions in the metabolism of glutamate, a retrotransposon-mediated duplication event in the hominoid ancestor approximately 18 to 25 Mya (Burki and Kaessmann, 2004) resulted in a second GLUD-encoding gene (GLUD2) that is targeted specifically to the mitochondria (Rosso et al., 2008). Glutamate is the most common neurotransmitter in the brain. It is believed that positively selected amino acid substitutions in GLUD2 allow for more efficient energy metabolism of glutamate in the brain (Plaitakis et al., 2000, 2003; Rosso et al., 2008).
AEROBIC ENERGY METABOLISM GENES AND BRAIN EVOLUTION
Neurons are the most energy-demanding cells of the modern human body (Attwell and Laughlin, 2001). The proliferation and pruning of neurons and their dendrites and the formation of the synaptic connections involved in learning are all energy-intensive processes. Thus it was not unexpected that aerobic energy metabolism (AEM) genes were found to be major targets of positive selection in the adaptive evolution of enlarged brains. This finding was made in a phylogenomic study that examined protein-coding sequence evolution during human ancestry (Uddin et al., 2008). In the time between the Old World monkey–ape LCA and the chimpanzee–human LCA, the most favored targets of positive selection were brain-expressed genes that code for mitochondrial functioning proteins, for example, proteins of the oxidative phosphorylation pathway (Uddin et al., 2008; Goodman et al., 2009). Although not brain-specific, many of these AEM genes are highly expressed in the adult human brain (Uddin et al., 2008). Moreover, these genes not only show evidence of adaptive evolution on the lineage to the LCA of humans and chimpanzees, but also on both the terminal human and terminal chimpanzee lineages. Considering that mitochondria play an essential central role in the aerobic production of energy, it may be inferred that the adaptive evolution of AEM genes improved the molecular machinery that facilitates the functioning of a high-energy-demanding encephalized brain.
Phylogenomic analysis of approximately 15,000 human coding sequences confirmed that AEM genes were favored targets of positive selection in the ape stem period of human ancestry (i.e., between 25 Mya
and 6 Mya), with 52 AEM genes (cellular component GO:0005739; mitochondrion) in the most enriched cluster of genes showing the signatures of positive selection, that is, faster rate of nonsynonymous substitutions (dN) than synonymous (dS) (Goodman et al., 2009). In the human terminal lineage (from 6 Mya to present), there were also many positively selected AEM genes but not significantly more than expected for any category of genes in the human genome. However, when these analyses were confined to only those positively selected genes that also show brain expression levels equivalent to or greater than the median of all modern human brain-expressed genes (Su et al., 2004), the most enriched clusters in the ape stem and human terminal lineages consisted of 20 and 23 AEM genes, respectively. Of these brain-expressed AEM genes, 14 and 10 in the ape stem and human terminal lineage, respectively, were involved in oxidative phosphorylation (Kyoto Encyclopedia of Genes and Genomes pathway map00190; oxidative phosphorylation). For more detailed information about these data and the methods used to infer enrichment for Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes pathways, refer to Goodman et al. (2009).
If the evolutionary origins of enlarged hominid brains depended on adaptively evolved AEM genes, then other large-brained mammals should also have in their ancestry AEM genes as principal targets of positive selection. An opportunity to test this hypothesis was provided by the addition of two afrotherian genomes to the growing set of publically available sequenced genomes. These two afrotherian genomes are from a large-brained mammal, the African savanna elephant (Loxodonta africana) and a small-brained mammal, the lesser hedgehog tenrec (Echinops telfairi). The clade Afrotheria, within which are elephants and tenrecs, is anciently separated from the clade Euarchontoglires, within which are humans and mice (Fig. 3.3A). Although elephants and tenrecs are phylogenetically closer to each other than to humans or mice, elephants resemble modern humans by having such features as large brains, empathetic social bonds, high intelligence, and prolonged development and long life spans [Fig. 3.3B; as discussed in Goodman et al. (2009)]. In contrast, tenrecs, as insectivore-grade mammals, have small, poorly encephalized brains and short life spans. The phylogenomic patterns of adaptive evolution are more similar between elephant and human than between either elephant and tenrec lineages or human and mouse lineages, with adaptively evolved AEM genes being especially well represented in the elephant and human patterns (Fig. 3.3C) (Goodman et al., 2009). In correlation with brain oxygen consumption and brain mass being largest in elephants and next largest in humans, positively selected AEM genes were most evident in the elephant lineage (indeed more overrepresented than any other gene category), next most evident in the human lineage, and not evident (i.e., not overrepresented)
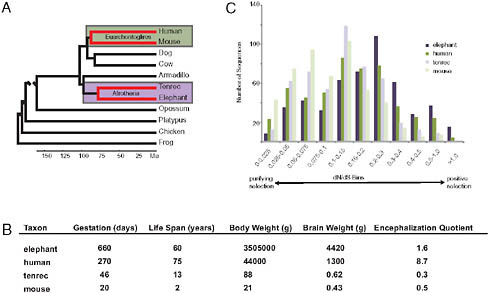
FIGURE 3.3 An example of convergent patterns of adaptive evolution in two large-brained taxa (elephants and humans). All data presented and images are adapted from previously published work (Goodman et al., 2009). (A) Species phylogeny based on DNA and fossil evidence (Hallström et al., 2007; Murphy et al., 2007; Wildman et al., 2007; Prasad et al., 2008). Lineages of interest are highlighted. (B) Life history and phenotype variables among study taxa. Values given are averages. More information and references for these values can be found in Table S1 of Goodman et al. (2009). (C) Lineage protein-coding sequence evolution for 501 mitochondria-related genes.
in tenrec and mouse lineages or in the other examined mammalian pair (the two laurasiatherians Bos taurus and Canis familiaris).
THE HUMAN BRAIN, DIFFERENT BY DEGREE AND NOT KIND
Darwin’s insight that the modern human mind does not differ in kind but rather in degree from other mammalian minds, in our opinion, should serve as the main guidepost for pursuing a phylogenomic search for the genetic roots of the modern human mind. The prospect that high-quality genome sequences will be obtained from thousands of different mammals (Genome 10K Community of Scientists, 2009) promises to make possible such a phylogenomic search. Key to the search will be dense representation of the species and genera in each extant mammalian order. The phylogenetic tree of mammals inferred from genome sequences can then be used to uncover in each evolved lineage the genetic changes that had
occurred between ancestral and descendent genomes. Of particular interest will be those adaptive genetic changes in protein-coding sequences, promoters, and other regulatory sequences.
Phylogenomic research provides an opportunity to identify those parallel or convergent patterns of adaptive genetic evolution that correlate with parallel or convergent patterns of adaptive phenotypic evolution. As an example, brain size increased in parallel in the stem catarrhines and stem platyrrhines (Kay et al., 1997). Encephalization then increased further in ape ancestry, in some Old World monkeys and some New World monkeys (e.g., Cebus) (Marino, 1998). In addition to humans, a number of primate species also exhibit a great deal of phenotypic and behavioral plasticity, including chimpanzees, baboons, macaques, and capuchins. Parallel or convergent patterns of adaptive genetic evolution among these species might help elucidate mechanisms contributing to enhanced brain plasticity in modern humans during childhood when the capacity for learning is greatest. Nevertheless, the search for genetic correlates of distinctive human phenotypic features should explore the possibility that some molecular aspects of modern human brain plasticity might be uniquely human. The hypothesis could be tested that adaptive evolution in our recent ancestry increased the diversity of macromolecular specificities involved in neuronal connectivity and neural plasticity. In testing this hypothesis, genes such as those concerned with cell–cell interaction, adhesion, and receptor–ligand binding and their cis-regulatory motifs should be examined. However, we would not be surprised if phylogenomic studies reveal that the genetic underpinnings for the basic mechanisms of brain plasticity are essentially the same as in other catarrhine primates and that the modern human mind differs from the other species primarily because of the modern human brain’s larger number of neurons and dendritic connections and much longer periods of postnatal development in a social nurturing environment.
A MODERN VOYAGE
With the advent of large-scale sequencing technologies and new bioinformatic tools for processing genomic sequence data, we are poised to embark on a new voyage of exploration and inquiry just as Darwin did in 1831. The last decade in particular has seen exponential growth in genome sequence collection and characterization. As we work toward a more complete understanding of genome structure, biology, and evolution, we become better able to develop and test hypotheses concerning a number of fundamental questions in evolutionary biology and human evolution. At the forefront of our interests are those molecular mechanisms and adaptations that have resulted in the modern human mind.
















