1
Introduction
|
Key Points Raised by Speakers
|
The field of genomics has the potential to revolutionize our interactions with the health care system, from increasing awareness of our risk for common diseases to individualized drug design. However, even with the significant increase in basic discoveries since the sequencing of the human genome 10 years ago, there has been a troubling paucity of new disease therapeutics, diagnostics, and preventive measures based on these discoveries.
This gap between discovery and application results in part from the complexity of human biology and disease processes. The growth of large databases in genomics, proteomics, metabolomics, and other dimensions of biological systems has helped reveal the inherent complexities of mul-
tiple gene-environment interactions, incomplete gene penetrance, and the dynamics of biological networks. However this gap also results from a lack of funding to substantiate the clinical relevance of basic discoveries and the increasing time and cost required to convert discoveries into commercial applications.
One way to bridge this growing divide in the translational pathway is through partnerships that distribute the risks involved in research and development (R&D) among multiple parties in a precompetitive manner. Industry, academia, and government all hold tools, knowledge, and biological materials that could be used collaboratively to speed the development of new drugs, diagnostics, and preventive measures. In particular, the sharing of biological specimens (biospecimens) and the data derived from those biospecimens through large-scale precompetitive collaborations could offer substantial benefits to all parties involved and the public at large.
However, numerous issues such as intellectual property (IP) protections and funding can be cumbersome or completely inhibitory to establishing collaborative ventures and must be overcome to facilitate this process and realize the potentially immense benefits. To explore these issues and develop potential solutions, the Roundtable on Translating Genomic-Based Research for Health held a workshop on July 22, 2010, entitled “Establishing Precompetitive Collaborations to Stimulate Genomics-Driven Drug Development” (see Appendix A for the full agenda). Representatives of government, industry, academia, and nonprofit organizations participated. Speakers and workshop participants sought to:
-
Examine specific examples from other industries that have engaged in precompetitive collaborations;
-
Identify how the best practices from other successful collaborations can be applied to genomics;
-
Clarify the rules of engagement for each stakeholder that would allow for genomics-based collaboration; and
-
Elucidate a conceptual framework for the precompetitive sharing of biological resources from many different stakeholders—academia, industry, government, and others.
While the workshop had a particular focus on stored biospecimens and the data derived from those samples, most of the observations could apply much more broadly in the development of drugs, diagnostics, and preventive measures in biomedicine. However, the goal of the workshop was not to solve the problems surrounding the establishment of precompetitive collaborations but rather to foster discussions that could clarify the issues and identify potential solutions.
EXPANDED USE OF PRECOMPETITIVE COLLABORATIONS
Recent years have seen a substantial expansion in the number of precompetitive, collaborative, multistakeholder consortia or relationships that have been established to address bottlenecks within the drug development process. Several examples of these include the Innovative Medicines Initiative in Europe, which is focused on bottlenecks in safety, efficacy, and preclinical toxicity; the Biomarkers Consortium, which is developing predictive biomarkers for such diseases as breast cancer, sarcopenia, and atherosclerosis; the Predictive Safety Testing Consortium, which is looking at liver, muscle, vascular, and renal diseases and carcinogenicity; and the Coalition Against Major Diseases, which is developing a shared Alzheimer’s database and quantitative disease models.
However, these collaborative arrangements, many of which are relatively new, fall far short of meeting current needs, said Geoff Ginsburg, professor of medicine and director of the Center for Genomic Medicine at Duke University, during his introductory remarks. Approval rates for new molecular entities in the pharmaceutical industry have been flat or declining over the past two decades (Booth and Zemmel, 2004; CBO, 2009). The return on investment has dropped precipitously for companies seeking to develop new drugs (Figure 1-1) and the problem of declining returns for increasing effort cannot be solved by any one organization working alone, suggested Ginsburg. Companies, government, and academic researchers need to combine their complementary skills and resources if they are to take advantage of the wealth of new information and knowledge made available through genomics-based research.
A wide variety of topics and tools are potential targets for precompetitive collaboration according to Ginsburg, including biospecimens, model systems, drug targets, probes, clinical and molecular data, preclinical models, software, and clinical trials data.
IDENTIFYING NEW APPROACHES TO PRECOMPETITIVE COLLABORATION
Stephen Friend, president and chief executive officer (CEO) of Sage Bionetworks, summarized the key points from an earlier Institute of Medicine (IOM) workshop on precompetitive collaboration in oncology research as background for the discussion (IOM, 2010).
For collaborations to succeed, related Friend, people need to work together in very different ways than they have in the past. Partners have to be able to share not just information but materials and personnel. They need to be willing to develop innovative tools, activities, and infrastructures for collaboration. Taking these steps requires a commitment from key
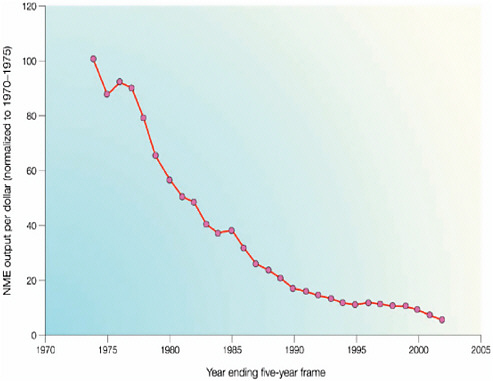
FIGURE 1-1 Normalized new molecular entity output per dollar expended.
SOURCE: Booth and Zemmel, Nature Reviews in Drug Discovery, 2004.
leaders. “You can dream all you want about how beautiful it is to have public-private partnerships, but if you don’t have leaders who are willing to build those tools and an infrastructure that allows sharing, it’s very hard,” said Friend.
Successful collaborations may require particular inputs, recounted Friend. One likely element of success is a neutral convener that can combine the relevant stakeholder perspectives and provide a legal safe harbor for collaboration. It does not need to be government, “it doesn’t need to be a nonprofit, it doesn’t need to be industry,” said Friend. “It has to be someone who actually can look at what are the consequences for the many groups that are coming together.”
Successful collaborations also can require governance structures that allow collaborators to develop strategies that are economically and intellectually sustainable. Sufficient incentives may have to be put in place to overcome existing incentives and to motivate the creation of policies that reward the development of an infrastructure for effective collaboration. These incentives can take the form of support for process, participation, or
reporting; support for results; or support for data exchange and meaningful analysis. As Garry Neil, corporate vice president for Johnson & Johnson, pointed out, building a robust national infrastructure to conduct and disseminate the data from clinical trials could bring tremendous cost savings to industry, government, and consumers.
Finally, effective collaborations may require the identification and prioritization of “bottleneck” knowledge gaps that can be addressed more effectively through precompetitive collaboration, the development of information “utilities” such as data standards and infrastructures, less regulatory uncertainty, and more head-to-head evaluations of collaborative models to identify key features and best practices. For example, Friend suggested that Clinical and Translational Science Awards could provide a mechanism for greater collaboration, as the institutions that have received these awards are working on ways to share data that could provide a template for many other kinds of partnerships.
In some collaborations, the use of open source principles can enable distributed innovation. “Tasks do not have to be built out of centralized efforts run from a single point,” said Friend. “The real power in the twenty-first century will come from distributing tasks.” Open source collaborations can require novel intellectual property provisions, a resource model that can support such work, and the establishment of conditions for entering, exiting, and ending a collaborative effort.
Another possible approach, said Friend, is the use of prizes to motivate collaborative research. “You do not have to have two years of going through a grant review process and then pay people to do certain tasks. Prizes [offer] a much more effective way to fund things.” Any organization could use such a mechanism, said Friend, including the National Institutes of Health (NIH). He contended that “the sole emphasis on the grant structure the way it is misses an opportunity to have prizes drive some of those opportunities.” Friend also suggested that perhaps one-third of R01 funds could be distributed in different ways than through traditional grants. For example, large, infrastructure-driven projects could produce faster results for patients and reduce repetitive work. “I think it’s going to go in that direction … as the government has less money,” said Friend. Though he added that he did not know whether alternative funding mechanisms would be easy to institute, he said they are “worth considering.”
In addition, patients can drive collaborations. For example, patients can organize initiatives to collect samples associated with phenotypes and share those samples and information widely with researchers. Patients are extremely aware of what their diseases are doing to them, said Friend, and they can gather information about their diseases that would be impossible for physicians to gather.
Importantly, summarized Friend, all the participants in a collaboration
need to benefit. Patients need to have an opportunity to contribute to the development of more effective and ultimately personalized treatments. FDA needs to receive data and other input for evidence-based regulatory policy. Pharmaceutical companies need to see opportunities for more efficient drug development and approval. The device industry needs to benefit from larger markets and less risk. Academic researchers need to receive better clinical data and be able to work toward more effective treatments. “Altruism is a great philosophical concept, but someone’s got to feel there’s something in it for them,” said Friend.
Friend ended his talk by discussing the concept of “extending the spectrum” of precompetitive biomedical research. The sharing of non-compound data and disease models occupies the middle of the spectrum. However, the middle of the spectrum is surrounded by many other activities that can be done collaboratively, including sharing analytical tools, revenue, scientists, consultants, clinical compound data, toxicity data, and compounds themselves. “We should mark our progress over time, where we are along that spectrum,” Friend said.
Three themes to keep in mind in considering the precompetitive space, said Friend, are the following:
-
Diverse approaches are needed for diverse goals.
-
The cultural barriers are often higher than the technical barriers.
-
Collaboration requires neutral and well-funded coordination with incentives for all participants.
THE POTENTIAL FOR PRECOMPETITIVE SHARING OF BIOBANKED SPECIMENS
As an example of an area where collaboration could be of substantial benefit to all parties, biospecimens represent a significant untapped resource of genetic and genomic information that can be used for target discovery, target validation, biomarkers for decision making, pharmacodynamics, pharmacogenomics, predisposition, prognosis, efficacy and monitoring, and replication.
Much more widespread use of biospecimens and their derived data could benefit multiple stakeholders, according to Ginsburg. Patients could receive better treatments, diagnostics, and use of their samples and data. Regulatory bodies could create and streamline standards for policy making. Pharmaceutical companies could use biomarkers to investigate drug targets, biological pathways, and disease mechanisms, leading to more efficient drug discovery and development, targeted therapies, and faster and less costly innovation cycles. Diagnostic companies could expand their portfolio of potential diagnostic products and reduce the risks in the discovery process.
Universities and academic health systems would have access to new data, funding, and hypotheses. Also, Ginsburg said, the efforts of all of these stakeholders would produce benefits for the broader society.
As an example of potential benefits, Ginsburg cited a research project by David Goldstein and John McHutchison at Duke University who used data from a clinical trial to discover a genetic variant that has a dramatic effect on the efficacy of a drug combination in patients with hepatitis C (Ge et al., 2009). In a second study, Goldstein, McHutchison, and their colleagues used access to six cohorts of hepatitis C patients around the world to demonstrate that the genetic variant could predict rapid clearance of the virus. Within months, the company LabCorp announced the availability of a genotype test to support individualized treatment decisions for patients with hepatitis C infection. “This is an example of access to large clinical data sets, applying genomic technologies, and, at least in my experience, one of the most rapid translations to a product that’s on the market,” said Ginsburg.
More than 300 million biospecimens are currently stored in the United
TABLE 1-1 Biospecimen Storage in the United States, 1999
|
Type of Repository |
No. of Cases |
No. of Specimens |
Cases per Year |
|
Large tissue banks, repositories, and core facilities |
>2.8 million |
119.6 million |
390,790 |
|
Longitudinal studies |
>340,088 |
508,088 |
|
|
Pathology specimens |
>160 million |
>160 million |
>8 million |
|
Newborn screening laboratories |
>13.5 million |
>13.5 million |
<10,000 to >50,000 |
|
Forensic DNA banks |
1.4 million |
1.4 million |
|
|
Sperm, ovum, and embryo banks |
>>200 |
>9,900 |
>9,900 |
|
Umbilical cord blood banks |
>18,300 |
>18,300 |
|
|
Organ banks |
|
>75,500 |
>75,500 |
|
Blood banks |
|
~12 million |
~12 million |
|
Total |
>178 million |
>307.1 million |
>20.5 million |
|
SOURCE: E. Eiserman and S. Haga, RAND Handbook on Human Tissue Sources: A National Resource of Human Tissue Samples, MR9540ST, 1999. |
|||
States (Eiseman and Haga, 1999) (Table 1-1) and an informal survey Ginsburg conducted among colleagues before the workshop found that one major pharmaceutical company enrolls about 32,000 patients per year in clinical trials and an academic health center reported 45,000 patients in trials and registries in 2010, although each collects specimens from only a fraction of those patients. Additionally, a recent survey by Willett and colleagues (2007) revealed that roughly a million specimens are associated with large U.S. cohort studies for which clinical data exist. Based on these surveys, Ginsburg estimates that millions of biospecimens in the United States are linked to at least some phenotypic information, but access to these samples is limited, and the biobanks tend to be fragmented and isolated. “Samples are widely distributed and nobody actually knows where they are [and], for a lot of them, what they are,” said Ginsburg, “with limited access even among investigators in the academic health center enterprise.” Thus, immense benefit could be realized if a framework were developed that would allow these samples to be pooled together and shared precompetitively in a collaborative venture.








