4
Frameworks for Collaboration
|
Key Points Raised by Speakers
|
PUBLIC–PRIVATE PARTNERSHIPS WITH NIH OR GOVERNMENT
The mission of the NIH, as steward of medical and behavioral research for the nation, is to advance “science in pursuit of fundamental knowledge about the nature and behavior of living systems … and the application of that knowledge to extend healthy life and reduce the burdens of illness and disability.” Those two separate missions, though closely related, have differing implications for the development of partnerships, said Thomas Insel, the director of the National Institute of Mental Health, one of 27 institutes and centers within NIH.
Insel discussed the government role in partnerships by focusing on biobanks, biomarkers, and drug development, pointing out opportunities and challenges in each area. In the area of biobanks, NIH already supports
a large number of often disease-specific, large-cohort studies that collect biosamples. A preliminary estimate of the number of such studies involving more than 5,000 subjects was 139 altogether—90 domestic and 49 international, with some 5 million people involved. These studies are not integrated or standardized, yet the government investment in them could be of great public interest and use.
The collection, storage, and use of biological specimens in biobanks offers the opportunity to look at risk and exposures prospectively as well as retrospectively in a representative sampling of a large population. Biobanks also can be mined in new ways as technology and concepts develop. However, several barriers exist with regard to biobank development, maintenance, and use, including confidentiality concerns when obtaining consents for broad-based studies or sharing of samples, a lack of standardization for collection, handling, and storage, limitations based on the sampled population, and the overall expense.
By examining the NIH’s efforts in the area of biobanks and large cohort studies, a number of possible opportunities to improve or leverage the investments that have been made present themselves, said Insel. The samples that are collected could be improved by thinking about how to sample a population that is actually representative of the United States. Standardized approaches in terms of consent, the kinds of information collected, and the handling and distribution of samples, according to Insel, all could lower some of the barriers to the greater use of biobanks. Additionally, the experiences of other countries in establishing and using biobanks offer lessons for the United States as it moves forward.
In the area of biomarkers, four years ago the Foundation for NIH launched the Biomarkers Consortium as a joint effort with the Food and Drug Administration (FDA) and pharmaceutical and biotechnology companies. The consortium, which now has a large number of for-profit and not-for-profit partners, is organized around four steering committees in the areas of neuroscience, cancer, metabolic disorders, and other disease or scientific areas. The goals of the consortium are to develop biomarkers for diagnosis and treatment, and there already have been a number of examples of success. Funding has come largely from industry, with NIH providing samples or other support.
The consortium provides a valuable opportunity to share resources, said Insel, and the involvement of FDA has great advantages, but there have been barriers to progress. One has been a clash of cultures. “The academics are looking for papers, the industry reps are looking for products, and the NIH folks are often arguing about whether there’s public health impact,” said Insel. There also have been issues about discovery versus the development of biomarkers and about whether industry representatives can speak for their companies. Garry Neil added that “there’s been a lot of
good progress made … and we need to continue to work together to figure out how to optimize these public–private partnerships … but we haven’t completely figured out how to get the most out of it.”
When asked what he would do differently if he were setting up the collaboration today, Neil highlighted several lessons learned from the experience. More emphasis should be put on the science as opposed to the legal aspects of the venture. The areas and topics to explore should be defined at the beginning of the partnership. Participants should be better prepared for areas where the science is not mature enough for development. A mechanism should be in place to allow funding of individual investigators or labs. The executive committee should be involved early to provide direction to the process. Insel added that the best ideas also need to be identified and brought into the program.
Finally, in the area of drug development, Insel pointed out that NIH is very aware of the problems that exist. The number of new drugs in the pipeline has been dropping, and the biotechnology industry is producing very few new candidate drugs (Figure 4-1). In response, the recent health care reform legislation created the Cures Acceleration Network, which would be a half-billion-dollar effort to create an integrated approach to drug discovery led by NIH. The consortium would look first at neglected diseases and then at more common diseases. “We’re not going to replace pharma,” said Insel, but “NIH could become a different kind of player. We’ve got some challenges not only with where we play, but how we play … we have to re-engineer the pipeline as well as [think] about how we catalyze discovery in that pipeline.” However, using public money for drug development is a
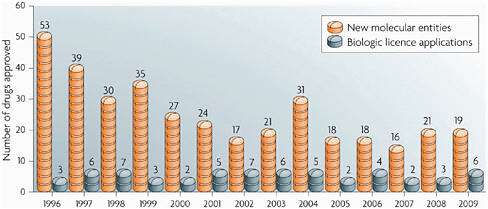
FIGURE 4-1 New molecular entity approvals.
SOURCE: Hughes, Nature Reviews Drug Discovery, 2010.
risky venture, according to Insel. NIH does not have great expertise in this area, it is expensive, and intellectual property issues need to be resolved.
Conflict of interest also poses a problem, said Insel. NIH has been thinking about whether there “are issues about how academia and government scientists interact with industry that need to be managed in a different way going forward, because this has been a source of real despair for the last couple of years, both I think on the industry side and on the NIH side.” Kelly Edwards, associate professor of bioethics and humanities at the University of Washington School of Medicine added that, in terms of conflicts of interest in partnerships between government and industry, people need to trust the institutions set up to develop new knowledge, which may require an “honest broker” for data interpretation and management. Insel agreed that ensuring public trust is “a really serious problem.” NIH has been developing and instituting new regulations governing conflicts of interest, although other issues also have to be resolved. “This needs to involve more than just industry, academia, and NIH and really needs to bring the public into the conversation.” When the research enterprise fails to deliver cures, people begin to wonder who is working for the public good as opposed to personal gain, Insel said.
In all three areas of biobanking, biomarkers, and drug development, NIH plans to emphasize standardization, integration, and sharing. Insel also quoted a recent paper on the pharmaceutical industry’s grand challenge: “Good process will never substitute for good people or good science” (Paul et al., 2010). “We can spend the whole day talking about partnerships,” Insel said, “but unless there’s really very compelling science to drive it, we’re wasting a lot of time in thinking about this just being a process problem.”
ADVANCING TECHNOLOGICAL ACHIEVEMENTS THROUGH COLLABORATION
Christopher Beecher, research professor at the University of Michigan’s Center for Translational Pathology, described the formation of a metabolomics consortium devoted to identifying all of the small molecules in a biological sample in order to discover those which are associated with the presence or progression of a disease. The rationale behind developing a consortium around metabolomics is the low compound identification rates in traditional studies. For example, in a recent study, Beecher and his colleagues tracked approximately 1,200 compounds across 262 samples taken from patients with prostate cancer (Sreekumar et al., 2009). The problem is that he and his colleagues were able to identify only about 37 percent of the compounds that they were tracking and there were many unknowns
that were statistically significant. “That’s the real bugaboo in many of these [studies],” he said.
Beecher decided to confront the problem of low identification by creating the Human Blood Plasma Metabolome Consortium with the goal of isolating and identifying every compound present at a concentration of more than 0.01 nanomoles in a very large quantity of human plasma. Bristol-Myers Squibb, Pfizer, Takeda Pharmaceuticals, Human Metabolome Technologies in Japan, and Agilent Technologies all agreed to fund the project. “The consortium was created to find a solution in which a number of companies could join together to do something that no particular group could do … through any other means,” according to Beecher. The collaboration sought to develop reproducible systems, platforms, and protocols to separate and detect these molecules. All of the results are published after a short embargo period.
All members benefit from participating. In the case of the university, they are “the publishers of the blood plasma metabolome.” The industry members benefit by the direct knowledge they have gained. “If we are successful—and at this moment we are being tremendously successful—we think that this will be a large plus, ultimately to be released to the public, and [will] be for the benefit of science.”
Setting up the consortium, however, took longer than expected—about a year and a half, said Beecher—with universities posing more obstacles than pharmaceutical companies. “The real problem was getting the university to understand and to not put up red flags.”
A major concern in forming the consortium was intellectual property. The organizers of the consortium decided that analyzing normal plasma would reduce the intellectual property issues. The legal experts consulted while the consortium was being formed decided that it would be easier if diseased tissues were not analyzed. “This is not a biomarker discovery attempt,” said Beecher. “This is purely an attempt to characterize, as fully and completely as we can, what a normal, very diverse human population looks like.” Thomas Insel responded that this type of approach would be a serious barrier to progress. “How do you get past that? What are you going to do when you want to study disease?” Beecher replied that the consortium was seeking to produce information that could be used by everyone, just as SEMATECH sought to provide technology that could be used by all of the companies in the semiconductor industry. Stephen Friend observed that “if we as a group were to shy away from working together on disease biology because of IP issues … we’re in real trouble.” Beecher emphasized that universities are risk averse, and to get the consortium funded within an academic environment, it needed to stay away from biomarker development.
OPEN ACCESS PARTNERSHIPS
Partly because of the built-in conservatism of the peer review process, scientific research tends to focus on certain areas and overlook others. For example, 10 years after the human genome was sequenced, 90 percent of the research articles on protein kinases are on just 10 percent of the kinases, even though genetic data indicate that many other kinases have effects that should be investigated. Researchers tend to work in a “tiny universe,” said Aled Edwards, professor of medical biophysics at the University of Toronto and director of the Structural Genomics Consortium, “and that’s a serious problem.”
The Structural Genomics Consortium was established in part to overcome the conservatism of much research. Its goal is to produce 1,000 three-dimensional structures of therapeutically relevant biological targets, along with 100 structures of parasite drug targets. It is funded by the Canadian government, has a board of directors and a scientific committee, and oversees work in laboratories in Toronto, Oxford, and Stockholm. As director of the consortium, Edwards makes many of the managerial decisions. “Don’t run projects by committee or consensus,” he said. “If you can’t find the right person to run it, don’t even start.”
The consortium made an early decision not to pursue intellectual property, which makes negotiations with partners very easy. “If they don’t buy in, we walk away, it takes about two minutes.” The consortium generates data quickly and puts it in the public domain without embargoes and well before the publication of papers describing the results.
The consortium has met its major milestone a year in advance of projections. The consortium now contributes more than 30 percent of the annual global output of human protein structures and accounts for 15 percent of the total output. Its scientists also have published papers in a wide variety of high-profile journals.
The keys to success, he said, are to establish clear and quantifiable objectives, create value for all participants (publications for academics, deliverables for industrial participants), and assume the best in collaborators. “Sometimes if you’re open, you are going to lose and get burned, but 95 out of 100 times, it’s going to work. Just let go of the 5 percent and move on.”
The consortium now has moved the precompetitive boundary by beginning to work on probes for nuclear protein receptors. Whenever reagents and other tools are available to work on a particular receptor, the number of publications on that receptor leaps upward. Yet many interesting genetic targets have received very little attention. As a result, the consortium has organized an effort to make chemical probes available for cell-based assays. The best medicinal chemists are in industry, and the corresponding biolo-
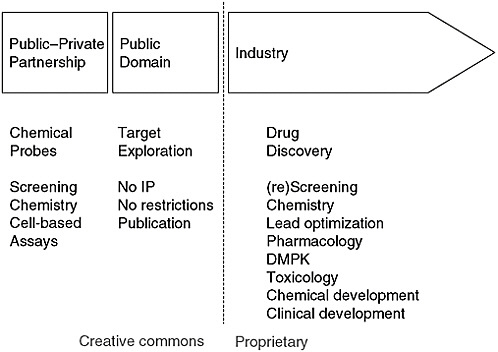
FIGURE 4-2 Model for precompetitive chemistry.
NOTE: DMPK = drug metabolism and pharmacokinetics; IP = intellectual property.
SOURCE: A. Edwards, IOM workshop presentation, July 22, 2010.
gists are in academia, so the consortium created a public–private partnership to produce high-quality tools that can be used for drug discovery and other applications in the proprietary domain (Figure 4-2). Both academic labs and companies are joining the partnership, and tools are starting to be released for use. Edwards cited as an example a G9a methyltransferase probe. Data generated with the probe are freely available on the consortium’s website. The idea, said Edwards, is “to seed this field with papers, and then hopefully more and more people will get at it.”
The final precompetitive barrier Edwards discussed is the failure of novel drug targets in clinical proof-of-concept trials. Data from these failures tend not to be released, which means that patients receive ineffective or even harmful drugs. Edwards suggested that these trials should become precompetitive research. “We should form a precompetitive consortium whose mission is to do open access [research], all the way from inventing the molecule to the phase II [trials]…. All the patients will be involved and know what’s going on, and we’ll make all those data available.” As soon as targets are validated, companies can then use that information to make medicines.
ACCESS TO LARGE-SCALE DATA NETWORKS
Technologies are being developed today that will allow many thousands of patients to have their genomes sequenced. Interactions among vast networks of proteins and metabolites will be mapped. The function of noncoding RNA molecules will be explored and related to the functions of other molecules in the cell. The functions of biological molecules will increasingly be connected to influences in the environment. Yet even these slices of biological function, by themselves, will not be sufficient to understand the mechanisms of disease, said Stephen Friend of Sage Bionetworks. All of this information and more will have to be combined into what he called “more causal or predictable models” of disease (Figure 4-3).
This work will take decades, but already this approach has begun to pay off, said Friend. Genetic association studies have been combined with genome-scale profiling to provide unbiased views of molecular physiology as it relates to disease phenotypes. Pharmaceutical companies are using this approach throughout the drug discovery process. Academics are writing papers on complex networks that have far-flung applications in research and in industry.
Public–private partnerships will be essential to host the data and
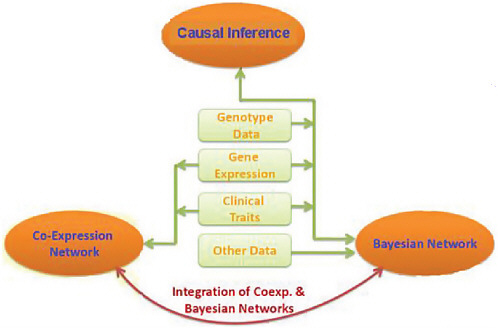
FIGURE 4-3 Integration of genotypic, gene expression, and trait data.
SOURCE: Adapted from Friend, IOM workshop presentation, July 22, 2010.
develop evolving representations of disease, and these partnerships will require significant resources over long time periods. The way to secure this kind of support, said Friend, is to view disease biology as precompetitive research. “We can’t take this on as a single company or single institute.”
That idea is the motivating concept behind Sage Bionetworks, which is based at the Fred Hutchinson Cancer Research Center in Seattle. The organization functions as a commons that produces long-term gains for the entire biomedical community by evolving models of disease. Sage Bionetworks has been pursuing a public-private partnership approach for Alzheimer’s disease and has also been investigating nonresponders to cancer treatments through data on gene expression, proteomics, metabolomics, and other molecular data. Sage Bionetworks is “curating new data sets that should be [available] and . . . putting up tools that allow people to work together to build probabilistic causal models of disease.”
In traditional research, an investigator who gets a grant thinks that he or she can keep the data generated by that grant until postdocs have gotten everything they can out of the data. “That prevents data flow from occurring,” said Friend. Physicists have learned to live in a world where micro-attribution and citation do not rest on publication. He noted that young scientists are already more comfortable living in a more collective world. “In 10 to 20 years, the careers of people will be based on who knows who did what with whom,” said Friend.
Sharing of data and the development of common standards need to occur at a technical level, but they also need to occur at a cultural level, stressed Friend. “Think of a world where interlab communication is equal to intralab communication,” said Friend. “Think of a world where the ability to talk back and forth between labs is the same as it is within labs. To do that, we’ve got to have funders be able to agree that their investigators will be sharing data in certain ways.”
Pilot projects can demonstrate for investigators how to share data. For example, making the data on controls for all clinical trials publicly available could be a model for making data accessible. Another possibility would be to define intellectual property (IP)-free zones, said Friend. “Why don’t we define certain diseases, where everyone who’s working in that disease says, ‘I’m not filing IP’?” New models and fundamental changes in how science is funded and rewarded are necessary to head toward a world in which contributors are more distributed, urged Friend. The patients and their disease foundations will be at the center of this world surrounded by companies, researchers, and government agencies.










