Molecular Evolutionary Analyses of
Insect Societies
![]()
BRIELLE J. FISCHMAN,*S. HOLLIS WOODARD,*AND
GENE E. ROBINSON*†‡§¶
The social insects live in extraordinarily complex and cohesive societies, where many individuals sacrifice their personal reproduction to become helpers in the colony. Identifying adaptive molecular changes involved in eusocial evolution in insects is important for understanding the mechanisms underlying transitions from solitary to social living, as well as the maintenance and elaboration of social life. Here, we review recent advances made in this area of research in several insect groups: the ants, bees, wasps, and termites. Drawing from whole-genome comparisons, candidate gene approaches, and a genome-scale comparative analysis of protein-coding sequence, we highlight novel insights gained for five major biological processes: chemical signaling, brain development and function, immunity, reproduction, and metabolism and nutrition. Lastly, we make comparisons across these diverse approaches and social insect lineages and discuss potential common themes of eusocial evolution, as well as challenges and prospects for future research in the field.
The social insects are exemplars of cooperative group living. Within their complex societies, there is a reproductive division of labor in which only a small number of individuals reproduce, whereas all other individuals belong to a functionally sterile worker caste that specializes in tasks important for colony growth and develop-
___________________
*Program in Ecology, Evolution, and Conservation Biology, †Department of Entomology, ‡Institute for Genomic Biology, and §Neuroscience Program, University of Illinois, Urbana, IL 61801. ¶ To whom correspondence should be addressed. E-mail: generobi@illinois.edu.
ment (Wilson, 1971). Although there has been much theoretical research on the evolutionary forces that may select for eusociality (Strassmann and Queller, 2007; Nowak et al., 2010), less is known about the actual molecular mechanisms involved in transitions from solitary to social living and in the maintenance and elaboration of eusociality in insects (C. R. Smith et al., 2008).
The social insects provide a powerful comparative framework for investigating mechanisms involved in eusocial evolution. Eusociality has arisen independently at least 12 times in the insects (Cameron and Mardulyn, 2001; Brady et al., 2006; Hines et al., 2007; Cardinal et al., 2010), and eusocial insects have all converged on the following three characteristics: reproductive division of labor, cooperative brood care, and overlapping generations (Michener, 1974). Additionally, despite sharing this core set of traits, there are many differences among eusocial lifestyles, which may be related to ecological, phylogenetic, or other factors specific to particular eusocial lineages (Wilson, 1971). By comparing across social insect lineages, it is possible to both search for common mechanisms of eusocial evolution and explore how eusociality evolves under different conditions.
Analysis of adaptive evolution at the molecular level can yield great insights into the mechanisms underlying the evolution of complex phenotypes, such as eusociality. Genomic sequence provides a molecular record of how natural selection has shaped an organism’s evolutionary history (Clark, 2006). Several methods have been developed for comparing genes and genomes to identify molecular signatures of adaptation. These methods were largely developed during the pregenomic era (Li, 1997) but gain enormous power when large genomic datasets are available, particularly for sets of closely related and phenotypically variable species (Clark et al., 2003; Drosophila 12 Genomes Consortium, 2007). For example, comparisons of primate genomes have identified adaptive genetic changes involved in the evolution of brain size in humans (Pollard et al., 2006), and comparisons of drosophilid genomes have shed light on the ecological pressures that shaped speciation in this group (Clark et al., 2003).
Here, we review some of the first contributions of molecular evolutionary research to our understanding of eusocial evolution in insects. This research has focused on the most well-studied social insects, which include several eusocial lineages within the order Hymenoptera, the ants, bees, and wasps, and the one eusocial lineage in the order Blattodea, the termites (Fig. 8.1). Some studies have performed targeted molecular evolutionary analyses of candidate genes that have been particularly valuable in species for which large amounts of genomic sequence are not yet available. Others have focused on comparative analyses of
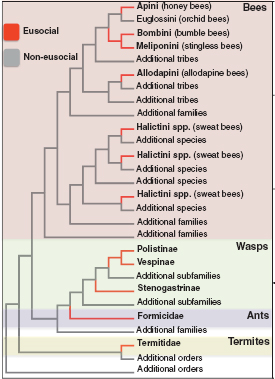
FIGURE 8.1 Cladogram showing the origins of eusociality in insects. Topology and reconstruction of evolutions of eusociality are based on multiple studies (Cameron and Mardulyn, 2001; Brady et al., 2006; Hines et al., 2007; Cardinal et al., 2010).
whole-genome sequence, which is currently available for six social insects, the honey bee, Apis mellifera (Honeybee Genome Sequencing Consortium, 2006), plus five ant species (Bonasio et al., 2010; C. D. Smith et al., 2011; C. R. Smith et al., 2011; Wurm et al., 2011), and for many solitary insects, including three solitary hymenopterans in the parasitoid jewel wasp genus, Nasonia (Werren et al., 2010).
We also draw heavily from our own recent genome-scale study of protein-coding sequence evolution in bees (“bee molecular evolution study”). This study analyzed ~3,600 genes from a set of 10 social and nonsocial bee transcriptomes; these species encompass three independent origins of eusociality (Woodard et al., 2011). Hundreds of genes were identified that exhibit a molecular signature of rapid evolution associated with sociality, defined as a higher ratio of nonsynonymous-to-synonymous nucleotide substitutions (dN/dS) in social relative to nonsocial bee lineages (Woodard et al., 2011). Throughout this review, evidence for rapid
evolution is based on relative dN/dS, and positive selection is defined as dN/dS> 1, unless otherwise specified.
Genes identified in these studies are listed in Table 8.1. The insights gained from these studies have implications for understanding how evolutionary changes in the following five major biological processes might be involved in the evolution of eusociality: chemical signaling, brain development and function, immunity, reproduction, and metabolism and nutrition. We discuss evidence and predictions for the putative functional effects of identified molecular changes in these processes on social phenotypes. We also speculate on the potential adaptive significance of these molecular changes and consider whether these changes evolved in response to the origin, maintenance, or elaboration of eusociality, because each case likely involved a distinct set of selective forces. For the purposes of interpreting and synthesizing results across multiple studies, we present each process separately, but it is important to recognize that these biological processes may evolve in concert and that some molecular
TABLE 8.1 Genes Implicated in the Origin or Maintenance of Insect Society by Molecular Evolutionary Research
|
|
|||
| Gene | Function | Evidence | Type of Changea |
|
|
|||
| Chemical signaling | |||
| decapentaplegic | Gland development (Bradley et al., 2003; Harris et al., 2007) | Rapid evolution in eusocial bees (Woodard et al., 2011) | 1 |
| thickveins | Gland development (Bradley et al., 2003; Harris et al., 2007) | Rapid evolution in eusocial bees (Woodard et al., 2011) | 1 |
| PDGF- and VEGF-related factor 1 | Gland development (Bradley et al., 2003; Harris et al., 2007) | Rapid evolution in eusocial bees (Woodard et al., 2011) | 1 |
| AmOr11 | OR (Wanner et al., 2007) | Responds to main component of queen honey bee pheromone, 9-ODA (Wanner et al., 2007) | 2 |
| Neofem 2 | β-Glycosidase-like (Korb et al., 2009; Weil et al., 2009) | Involved in signaling queen termite presence (Korb et al., 2009; Weil et al., 2009) | 3 |
| Gene | Function | Evidence | Type of Changea |
| GP-9 | Putative OBP (Keller and Ross, 1998; Krieger and Ross, 2005; Gotzek et al., 2007; Leal and Ishida, 2008; Gotzek and Ross, 2009) | Allelic variation associated with fire ant queen number (Keller and Ross, 1998; Krieger and Ross, 2005; Gotzek et al., 2007; Leal and Ishida, 2008; Gotzek and Ross, 2009) | 1,2 |
| Brain development and function | |||
| dunce | cAMP/CREB signaling pathways (Silva et al., 1998) | Rapid evolution in primitively eusocial bees (Woodard et al., 2011) | 1 |
| nejire | CREB binding protein (Silva et al., 1998) | Rapid evolution in primitively eusocial bees (Woodard et al., 2011) | 1 |
| Immunity | |||
| defensin | Antimicrobial protein (Viljakainen and Pamilo, 2008) | Positive selection in ants (Viljakainen and Pamilo, 2008) | 1 |
| termicin | Antimicrobial protein (Bulmer and Crozier, 2004; Bulmer et al., 2010) | Gene duplication, positive selection in termites (Bulmer and Crozier, 2004; Bulmer et al., 2010) | 1,2 |
| GNBP 1 and 2 | Pattern recognition receptors (Bulmer and Crozier, 2006) | Gene duplication, positive selection in termites (Bulmer and Crozier, 2006) | 1,2 |
| relish | Transcription factor, induces production of antimicrobial peptides (Bulmer and Crozier, 2006) | Positive selection in termites (Bulmer and Crozier, 2006) | 1 |
| Reproduction | |||
| tudor | piRNA pathway (Siomi et al., 2010) | Rapid evolution in primitively eusocial bees (Woodard et al., 2011) | 1 |
| Gene | Function | Evidence | Type of Changea |
| capsuleen | piRNA pathway (Siomi et al., 2010) | Rapid evolution in primitively eusocial bees (Woodard et al., 2011) | 1 |
| vasa | piRNA pathway (Siomi et al., 2010) | Rapid evolution in primitively eusocial bees (Woodard et al., 2011) | 1 |
| csd | Sex determination (Beye et al., 2003; Hasselmann et al., 2008a,b) | Gene duplication, positive selection in honey bees (Beye et al., 2003; Hasselmann et al., 2008a,b) | 1,2 |
| Metabolism and nutrition | |||
| MRJPs | Main components of royal jelly (Drapeau et al., 2006) | Gene family expansion, novel feeding-related functions in honey bees (Drapeau et al., 2006) | 2 |
| Hex-1 and Hex-2 | Storage proteins (Zhou et al., 2006, 2007) | Unique insertions in termites (Zhou et al., 2006, 2007) | 1 |
| phosphofructokinase | Key regulator of glycolysis (Kunieda et al., 2006) | Rapid evolution in eusocial bees (Woodard et al., 2011) | 1 |
| hexokinase | Regulator of glycolytic flux (Kunieda et al., 2006) | Rapid evolution in eusocial bees (Woodard et al., 2011) | 1 |
| pyruvate kinase | Regulator of glycolytic flux (Kunieda et al., 2006) | Rapid evolution in eusocial bees (Woodard et al., 2011) | 1 |
|
|
|||
| Note: Although many genes in this table are presumably involved in multiple biological processes, they are classified in one of five processes with known links to insect sociality: chemical signaling, brain development and function, immunity, reproduction, and metabolism and nutrition. | |||
| aType of change: 1, protein coding sequence change; 2, novel gene; 3, change unknown. | |||
changes could potentially affect multiple processes. We end with a discussion of future prospects and challenges for this young field.
CHEMICAL SIGNALING
Social insects use pheromones to coordinate the behavior and physiology of colony members, such as directing the foraging activity of nest-mates, reinforcing dominance status, and inhibiting ovary development in workers (Le Conte and Hefetz, 2008). It is unknown whether chemical signaling was important during the origins of eusociality, because other mechanisms to mediate social interactions, such as physical interactions, serve similar functions in some social insect societies (Wilson, 1971). However, chemical signaling is certainly involved in the maintenance and elaboration of eusociality because it is crucial for the coordination and control of colony members. In humans, in whom vocalization is a major component of social communication, molecular signatures of adaptation have been detected in genes underlying both the production (Enard et al., 2002) and perception (Clark et al., 2003) of vocal signals. Early studies in social insects suggest that analogous changes have occurred in the molecular machinery underlying the production and perception of chemical signals.
Gland Development
Our bee molecular evolution study identified ~200 genes evolving more rapidly in social relative to nonsocial bee lineages (Woodard et al., 2011). Gene ontology enrichment analysis revealed that this set of genes was enriched for genes involved in gland development. This supports a role for these genes in chemical signaling, because glands are the primary organs involved in pheromone production in insects. Moreover, the evolution of complex chemical signaling in the social insects has been associated with the diversification of the gland repertoire (Wilson, 1971).
In other organisms, modular evolution, in which semiautonomous genetic pathways evolve as a functional unit and are reused in multiple contexts, appears to be a common evolutionary mechanism involved in morphological diversification (Wagner et al., 2007). The sequence changes identified in genes involved in gland development in social bees may have caused modular changes to the gland development program, resulting in functional changes to existing glands or the appearance of entirely new glands. This is supported by the evidence that several of these genes (decapentaplegic, thickveins, and PDGF- and VEGF-related factor 1) have specific roles in gland patterning during early development in Drosophila (Bradley et al., 2003; Harris et al., 2007).
Because diversification of gland function is a common characteristic shared by all social insects, it would be fruitful to investigate the sequence evolution and function of these genes in other social insect groups. It is possible that molecular changes in the same or similar genes were involved in gland evolution across other independent eusocial lineages.
Odorant Receptors
Given the diversity of chemical signals used by social insects, odorant receptor genes (ORs) have been predicted to be important targets of selection during eusocial evolution (Robertson and Wanner, 2006). Early support for this prediction was found in the genome of the honey bee, A. mellifera, which, at the time of its publication, contained the largest number of ORs yet found in an insect genome (Honeybee Genome Sequencing Consortium, 2006). However, as more insect genomes have been sequenced, it has been discovered that A. mellifera has an intermediate number of ORs, there is significant variation in OR number between the five ant genomes (Bonasio et al., 2010; C. D. Smith et al., 2011; C. R. Smith et al., 2011; Wurm et al., 2011), and several solitary insect genomes have among the most ORs found in insects so far (Engsontia et al., 2008; Robertson et al., 2010). Thus, the evidence no longer supports an association between sociality and expansion of the OR repertoire. Furthermore, studies in other organisms have revealed that ORs can function combi-natorially and that bioinformatically predicted ORs may not all produce functional proteins, which, together, suggest that the number of ORs in a genome may not scale with the complexity of chemical communication in a species (Nei et al., 2008).
As a result of their functional specificity, ORs are particularly good targets for candidate gene studies, because the adaptive significance of OR evolution may be easier to interpret than for genes with broader functions (Nei et al., 2008). A functional genomics approach was used to identify a novel OR in the A. mellifera genome, AmOr11, which responds to the main component of the honey bee queen pheromone, (E)-9-oxo-2-decenoic acid (9-ODA) (Wanner et al., 2007). The queen pheromone attracts workers to the queen, partially inhibits worker ovary development, and acts as a sex pheromone, among other functions (Wanner et al., 2007). The specific molecular characteristics of AmOr11 that are involved in the perception of 9-ODA are not yet known, but it appears that it arose early in Apis evolution (Plettner et al., 1997; Cruz-López et al., 2005; Urbanová et al., 2008).
Termite Queen Pheromone
Neofem2 is the first gene discovered in termites that is involved in signaling queen presence to workers. It was originally identified as being up-regulated in female neotenic “replacement” reproductives relative to other colony members in two species of Cryptotermes termites (Weil et al., 2009). Knocking down Neofem2 in Cryptotermes secundus queens using RNAi caused an increase in aggressive behavior among workers, which is typically only exhibited under queenless conditions (Weil et al., 2009). Based on sequence similarity, Neofem2 is most closely related to a ?-glycosidase expressed in the salivary glands of the termite Neotermes koshunensis (Korb et al., 2009). ?-glycosidases are enzymes that break down polysaccharides; in wood-dwelling termites, such as N. koshunen-sis and C. secundus, whose diet primarily consists of rotting bark, these enzymes are important for breaking down cellulose (Tokuda et al., 2002). It has thus been suggested that Neofem2 evolved from a wood-digesting enzyme to pheromone (Korb et al., 2009). Supporting this speculation, ?-glycosidases exhibit pheromonal activity in other insects, including the production of an egg recognition signal in another termite species (Korb et al., 2009). The specific molecular changes that have occurred in Neofem2 as it evolved this new social function remain to be discovered. The story of Neofem2 highlights the importance of considering the ecological context of social evolution in a given lineage, because the origin of a social pheromone from a wood-digesting enzyme is almost certainly a phenomenon specific to the wood-dwelling termites.
General protein-9 in Fire Ants
General protein-9 (Gp-9) alleles are strongly associated with variation in queen number in fire ants (genus Solenopsis). In monogynous (single queen) colonies, all females are homozygous for B-type alleles and will not tolerate the presence of multiple queens, whereas in polygynous (multiple queens) colonies, some individuals possess b-type alleles and do accept multiple queens but only if those queens also possess the b-type allele (Gotzek and Ross, 2009). Gp-9 has been called a “greenbeard gene” (Keller and Ross, 1998), because workers carrying one allele favor queens that share the same allele. Molecular phylogenetic analyses of Gp-9 both within and across Solenopsis species have revealed that the b-like alleles form a monophyletic clade, suggesting that monogyny was the ancestral condition in the genus and that polygyny arose once and has been maintained through multiple speciation events (Krieger and Ross, 2005; Gotzek et al., 2007).
At the protein sequence level, Gp-9 most closely resembles odorant-binding proteins (OBPs), which are expressed in chemosensory sensilla
lymph and bind and transport soluble odorants (Gotzek and Ross, 2009). These results have led to the suggestion that Gp-9 is an OBP that plays a role in pheromonal communication in fire ants (Gotzek and Ross, 2009). However, Gp-9 is ubiquitously expressed in the hemolymph, suggesting it may be involved in functions that are unrelated to chemosensation (Leal and Ishida, 2008). In addition, Gp-9 is found in a genomic region with a low recombination rate; therefore, other linked genes in the region may potentially have more influence on the regulation of queen number (Krieger and Ross, 2005; Gotzek and Ross, 2009). Gp-9 alleles are also associated with variation in several life history traits in Solenopsis queens, including body fat and dispersal behavior (Gotzek et al., 2007), suggesting that Gp-9 either acts pleiotropically or with other genes in the region. Although the function of Gp-9 is unresolved, molecular evolutionary analyses suggest that this gene is evolving adaptively, implying that Gp-9 played an important role in fire ant evolution. A signature of positive selection was detected in the branch leading to the b-like allele clade (Krieger and Ross, 2005), suggesting that this allele had an adaptive benefit when it arose. In addition, all b-like alleles share the same amino acid residues at three diagnostic codon positions, and two of these positions show evidence of positive selection in Solenopsis invicta, the species in which it has been best studied (Gotzek et al., 2007).
BRAIN DEVELOPMENT AND FUNCTION
Some of the most striking differences between social and solitary insects are behavioral. Several social insect behaviors appear to be truly novel, such as symbolic dance communication in honey bees and slave making in ants (Wilson, 1971). Other behaviors exhibited by social insects appear to be modified forms of behaviors performed by solitary insects, for example, social foraging, which resembles nest provisioning in solitary insects. It is likely that molecular changes affecting nervous system development and function were important in the evolution of social insect behaviors, but very little is currently known.
Brain Evolution in Primitively Eusocial Bees
Our bee molecular evolution study detected a strong signal of rapid evolution in brain-related genes in primitively eusocial, but not highly eusocial lineages across two independent origins of each lifestyle (Woodard et al., 2011). Among these rapidly evolving genes were dunce and nejire, two genes that mediate learning and memory in invertebrates and vertebrates through cAMP/CREB signaling pathways (Silva et al., 1998).
The detection of molecular changes in brain-related genes exclusively in primitively eusocial bee lineages is perhaps surprising, given that this finding is not what may have been predicted by a prominent hypothesis about the relationship between sociality and brain evolution in vertebrates, the social brain hypothesis (SBH). Originally developed to explain the evolution of the enlarged neocortex in many social vertebrates, the SBH posits that the cognitive demands of social living are a strong selective force in brain evolution (Dunbar and Shultz, 2007). Given that highly eusocial bee societies have larger colony sizes, greater social complexity, and novel behaviors (i.e., dance communication in honey bees) relative to primitively eusocial bees, one might have assumed that the cognitive demands of social living are strongest in highly eusocial species and lead to stronger selection on brain-related genes.
Unique features of insect sociality and the primitively eusocial lifestyle may help to explain why selection on brain evolution appears to have been stronger in the primitively eusocial bees. First, unlike in vertebrate social evolution, where there has been an emphasis on increased individual cognitive abilities, there appears to have been an emphasis on increased connectedness among colony members in insect social evolution, often accompanied by a reduction of individual behavioral repertoires (Oster and Wilson, 1979; Gronenberg and Riveros, 2009). Therefore, individual cognitive abilities may not be correlated with group size in social insects, as has been found in vertebrates. There are also several distinguishing features of the primitively eusocial bee lifestyle that may have placed unique selective pressure on brain evolution in these lineages. Social structure in primitively eusocial bee colonies is typically maintained through fluid and dynamic dominance hierarchies (Michener, 1974; O’Donnell et al., 2007), which can be an especially cog-nitively challenging form of social interaction (O’Donnell et al., 2007; Salvador and Costa, 2009). In addition, a primitively eusocial bee queen is capable of behaving both solitarily, as she does during the colony-founding phase of her life cycle, and socially, as she does once she has reared her first brood of workers (Michener, 1974).
In both ants and wasps, which each evolved eusociality independent of bees, there are some species in which queens exhibit a similar “solitarylike” phase during colony founding and other species that found colonies in swarms, like highly eusocial bees do (Wilson, 1971). A comparison of brain-related genes and/or brain structure in ant and wasp species that do or do not establish colonies solitarily may provide clues as to whether this trait is a strong force in social insect brain evolution. One study in paper wasps reported brain region volume differences between swarm and independent-founding species, suggesting that these differences in colony founding can affect brain evolution (Molina et al., 2009).
IMMUNITY
Pathogens and parasites are thought to have been a strong selective force challenging the maintenance of sociality in a variety of organisms, including social insects (Wilson-Rich et al., 2009). Crowded living conditions, often with closely related individuals, facilitate pathogen transmission (Wilson-Rich et al., 2009). Social insects appear to have responded to this potentially dissolutive selective pressure in three main ways (Viljakainen and Pamilo, 2008). The first way is through “social immunity,” which refers to group-level defenses, such as hygienic behaviors and the use of collected antimicrobial resins for lining nest cavities (Wilson-Rich et al., 2009). The second way is through increasing intracolonial genetic diversity via multiple mating by queens (Tarpy and Seeley, 2006) and high rates of genetic recombination (C. R. Smith et al., 2008) to enhance colony-level disease resistance. The third way is through adaptive evolution of immune genes (Viljakainen and Pamilo, 2008).
Molecular evolutionary analyses of immune genes have provided some of the best examples of positive selection acting in social insect genomes. This may be partly attributable to the fact that immune systems, in general, are often at the forefront of an ongoing evolutionary arms race with pathogens; thus, selection pressure on immune-related genes is typically quite strong (Lazzaro, 2008). In addition, many immune-related genes are functionally well characterized (Hoffmann, 2003), facilitating interpretations of the adaptive significance of sequence changes.
Immune Gene Evolution in Hymenoptera
When the first social insect genome was sequenced, that of A. mel-lifera, researchers were intrigued by the low number of immune genes found in A. mellifera relative to other fully sequenced insect genomes: those of the Diptera, Drosophila melanogaster, and Anopheles gambiae (Honeybee Genome Sequencing Consortium, 2006). Although the main components of canonical immune pathways are conserved, the A. mel-lifera genome contains smaller numbers of gene family members at all points along these pathways (Evans et al., 2006). It was hypothesized that the loss of immune genes was facilitated by novel forms of social immunity in social insects, resulting in relaxed constraint on immune genes (Evans et al., 2006). However, as more insect genomes have been sequenced, it has become apparent that sociality is not necessarily predictive of immune gene number. Rather, it seems that dipterans have unusually large immune gene repertoires, whereas the recently sequenced ant genomes (Bonasio et al., 2010; C. D. Smith et al., 2011; C. R. Smith et al., 2011; Wurm et al., 2011), the solitary wasps Nasonia (Werren et al., 2010), and the solitary pea aphid Acyrthosiphon pisum (International
Aphid Genomics Consortium, 2010) have similar numbers of immune genes as A. mellifera (Evans et al., 2006).
By contrast, molecular evolutionary analysis of individual immune genes in social Hymenoptera has provided evidence that sociality has driven immune gene sequence evolution. One study revealed that some immune genes are evolving more rapidly in species of honey bees, bumble bees, and ants relative to Drosophila (Viljakainen et al., 2009). This study also showed that immune genes are evolving more rapidly than non-immune genes in several honey bee species. Similarly, genes related to innate immunity and humoral immunity were among the fastest evolving (based on branch lengths in phylogenetic trees inferred from protein sequence) in A. mellifera in a comparison of over 3,000 genes among A. mellifera, Nasonia, and their common ancestor (Werren et al., 2010). Additionally, evidence for positive selection has been detected in the antimicrobial protein defensin in a study comparing the sequence of 27 ant species (Viljakainen and Pamilo, 2008). This study revealed that the signal and propeptide regions of defensin, which are cleaved off to activate the mature peptide, are evolving neutrally, whereas the active region of the peptide is under positive selection, including one amino acid site thought to mediate antimicrobial activity. Our bee molecular evolution study did not detect a strong signal of selection on immune genes, but that was likely because these classes of genes were underrepresented in our dataset (Woodard et al., 2011).
Immune Gene Evolution in Termites
A study of the termite defensin-like gene, termicin, in 11 Nasutitermes termite species revealed that this gene has duplicated repeatedly during Nasutitermes radiation and that positive selection has driven a divergence in the molecular charge of the gene copies (Bulmer and Crozier, 2004). Insect defensins are known to function by disrupting bacterial plasma membranes, and experimental evidence suggests that molecular charge may be a crucial component of this activity (Bulmer and Crozier, 2004). It was hypothesized that there is a selective advantage to having two termicins with different charge properties at specific sites (Bulmer and Crozier, 2004). In support of this hypothesis, results from this study suggest that ancestral termicins had relatively high positive charges and that in species in which there has been a gene duplication event, positive selection has driven a decrease in charge for one of the copies. Sequence analysis revealed a strong positive correlation between the strength of selection (dN/dS) and the change in molecular charge along different termicin lineages. Additionally, three amino acid sites that show a signature of positive selection have substitutions at these sites
that contribute to a charge change, and they fall on the external surface of the predicted protein structure, suggesting that these sites may interact with a fungal membrane receptor (Bulmer and Crozier, 2004). A different study of 13 Nasutitermes termite species also found evidence that gene duplication and positive selection are involved in termite immune gene evolution (Bulmer and Crozier, 2006). This study focused on genes encoding Gram-negative bacterial-binding protein 1 and 2 (GNBP1 and GNBP2), which are thought to have duplicated early in termite evolution, and the transcription factor relish, which induces production of antimicrobial peptides in Drosophila. All three genes show evidence of positive selection, with relish showing the strongest signal. Four of the five positively selected sites in relish are in a “spacer” region of the protein that is cleaved by the caspase Dredd. This cleavage is thought to activate relish by generating a DNA-binding Rel homology domain that translocates to the nucleus and binds to promoters of target genes (Stoven et al., 2003). Analysis of the Drosophila simulans ortholog also found positive selection in this spacer region (Bulmer and Crozier, 2006). It was hypothesized that microbial pathogens may be targeting this region of relish to prevent its activation, sparking an evolutionary arms race as relish evolves counterresponses to maintain its normal function (Bulmer and Crozier, 2006). Another study found evidence of positive selection in termicin but not in GNBP2 in two Reticulitermes termite species, a genus distantly related to the Nasutitermes genus (Bulmer et al., 2010). This study used a population genetics approach to analyze intra-specific polymorphism and interspecific divergence in coding sequence, and results indicated that termicin underwent a selective sweep driven by positive selection for beneficial amino acid changes.
REPRODUCTION
In many insect societies, queens are highly reproductive individuals, whereas workers perform almost no reproduction activity. Worker sterility is achieved through a variety of morphological, behavioral, and physiological mechanisms in social insects (Wilson, 1971). For example, in many social species, workers lack spermatheca for sperm storage. In addition, ovary development is tightly regulated by social cues, and queens and workers typically have grossly over- and underdeveloped ovaries, respectively, relative to solitary insects (Wilson, 1971). Sociality also has strong implications for reproductive behavior, particularly for mating frequency, which can affect genetic variation among colony members.
Ovary Development in Primitively Eusocial Bees
Our bee molecular evolution study identified some genes involved in ovary development evolving most rapidly in primitively eusocial bees (Woodard et al., 2011). Although both highly and primitively eusocial bee societies have a strong reproductive division of labor, the reproductive differences between queen and worker in primitively eusocial species are less extreme, and ovary development appears to be more sensitive to social cues in primitively eusocial species (Wilson, 1971). Perhaps the molecular changes in ovary development-related genes found only in the primitively eusocial lineages underlie some of the unique characteristics of the reproductive biology of this eusocial lifestyle.
Several genes (i.e., tudor, capsuleen, vasa) evolving rapidly in one or both of the primitively eusocial bee lineages interact together in the PIWI RNA (piRNA) pathway. The piRNA pathway is expressed only in gametic tissue, and it is involved in regulating gametic cell division and differentiation (Siomi et al., 2010). Functional PIWI genes have recently been discovered in A. mellifera (Liao et al., 2010), suggesting that the piRNA pathway is present and functional in bees. These genes are particularly good candidates for further study, because the tissue specificity of the piRNA pathway suggests that selection on these genes is specifically directed at changes related to reproductive processes, in contrast to genes with broader ranges of tissue expression, where the functional target of selection is harder to infer. Additional ovary development-related genes unrelated to the piRNA pathway also showed a signature of rapid evolution in these primitively eusocial bees (Woodard et al., 2011).
Sex Determination and complementary sex determiner in Honey Bees
More is known about the evolution of complementary sex determiner (csd) in honey bees than probably any other gene in the social insects. The story of csd involves the origin of entirely new genes and pathways, as well as a classic example of balancing selection. Sex determination in honey bees is based on genotype at the csd locus; individuals heterozygous at the csd locus develop into females, whereas hemizygous individuals develop into males (Beye et al., 2003). Sex in many Hymenoptera is probably determined through a similar single-locus system of complementary sex determination (Cook, 1993), but csd is the first and only locus that has been discovered thus far. The genomic region containing csd was first identified through mapping (Beye et al., 2003), and the function of the gene was confirmed by RNAi, which showed that reducing csd expression in genetically female eggs results in male-like development (Hasselmann et al., 2008a). Complementary sex determination not only regulates sex
determination but influences many aspects of social insect biology that are influenced by kinship and degrees of relatedness, including kin selection and the genetic composition of colonies, which are important for division of labor and colony immunity (C. R. Smith et al., 2008).
The csd gene appears to be a honey bee–specific gene because it has been found in multiple Apis species (Hasselmann et al., 2008b) but not outside of the genus (Hasselmann et al., 2008a). The gene likely evolved through the duplication of an adjacent gene, feminizer (fem). The csd and fem genes are similar (>70%) in amino acid sequence, and both are serine/arginine-rich proteins, a class of proteins involved in RNA splicing (Hasselmann et al., 2008a). Both genes share two major domains, but csd has an additional hypervariable region located between these other domains (Hasselmann et al., 2008a). The fem gene has been found in several non-honey bee species and in Nasonia wasps, but not in any additional insect species, suggesting that it evolved sometime before the split between the hymenopteran superfamilies Apoidea and Chalcidoidia ~140 Mya but after the split from Drosophila ~300 Mya (Hasselmann et al., 2008a). The fem gene shares some functional and sequence similarities to transformer (tra), a gene involved in sex determination in Drosophila, and it perhaps evolved from an ancestral form of tra common to fly and bee lineages (Beye et al., 2003; Hasselmann et al., 2008a). RNAi experiments were used to show that csd acts upstream of fem in the sex determination pathway. Genetically female embryos treated with fem RNAi develop male heads, and RNAi knockdowns of csd cause male-specific fem splicing, suggesting that csd is involved in fem splicing (Hasselmann et al., 2008a).
The csd gene has been subject to rigorous population genetic analysis. Because homozygous males do not reproduce, it was predicted that there would be strong negative frequency-dependent selection at the csd locus (Hasselmann et al., 2008b). This prediction has been upheld, because at least 15 different csd alleles have been found in natural populations around the world in three different Apis species (Hasselmann et al., 2008b) and the gene has accumulated 10- to 13-fold more mutations than the rest of the genome (Hasselmann et al., 2008b). Pairwise nonsynonymous differences between alleles are highest in exons 6 and 7 (Hasselmann et al., 2008b), suggesting that this region is a target of positive selection, and is therefore presumably functionally important. Six fixed amino acid differences between csd and fem are located in the coiled-coil domain, which is important in protein binding (Hasselmann et al., 2008a). Strong positive selection was detected on the branch right after the split between the two genes, suggesting that positive selection played a role in their diversification (Hasselmann et al., 2008a).
METABOLISM AND NUTRITION
Transcriptomic analyses have shown that nutritional and metabolic pathways play an important role in queen-worker caste determination in every eusocial insect lineage thus far studied and also contribute to worker-worker division of labor in many species (C. R. Smith et al., 2008). Given these fundamental connections to eusociality, nutritional and metabolic pathways are well studied in social insects and several molecular evolutionary studies have identified changes associated with their function.
Major Royal Jelly Proteins
The evolution of the Major Royal Jelly Proteins (MRJPs) in honey bees is an excellent example of novel genes playing an integral role in the social biology of a species. In the honey bee, A. mellifera, the developmental fate of female larvae is determined by the amount of royal jelly they consume (Kamakura, 2011). Royal jelly is a protein- and lipid-rich substance secreted from the hypopharyngeal glands of brood-feeding “nurse” bees and fed to larvae, which triggers endocrine and epigenetic events that lead to the development of either a worker or a queen (Lyko et al., 2010; Kamakura, 2011). The main components of royal jelly are the MRJPs. The A. mellifera genome contains 10 mrjp genes, encoding 9 MRJPs (one mrjp is a pseudogene). These genes are arranged in tandem in the genome, have high sequence similarity (~60%) to one another, and have a conserved intron/exon structure, suggesting that they are a fairly young gene family (Drapeau et al., 2006). There is evidence that mrjp genes are also present in other Apis species (Drapeau et al., 2006; Yu et al., 2010).
The mrjp gene family in A. mellifera appears to have evolved via a gene duplication event from a member of the yellow gene family. The cluster of mrjp genes in the A. mellifera genome is flanked by members of the yellow gene family, and one of the flanking yellow genes, yellow-e3, shares the characteristic intron/exon structure of the mrjp genes, suggesting that it is their progenitor (Drapeau et al., 2006). Members of the yellow gene family are involved in pigmentation, reproductive physiology, and courtship behavior in insects (Ferguson et al., 2011).
The use of mrjp genes for larval feeding appears to be a derived social trait that is unique to honey bees. Although mrjp-like genes have been found in other social and nonsocial Hymenoptera species, evidence suggests that the yellow gene family is prone to duplication and that the mrjp-like genes in non-Apis species evolved independently of Apis (Werren et al., 2010; C. D. Smith et al., 2011). Furthermore, there is no evidence of a food-related role for any mrjp-like or yellow-like gene
outside of Apis (Ferguson et al., 2011). Because many other social insect species manipulate larval nutrition for the purposes of caste determination without the use of specialized glandular secretions (Webster and Peng, 1988), the evolution of the mrjp genes in honey bees appears to be associated with the elaboration of eusociality and may have been correlated with or dependent on other evolutionary changes, such as changes in gland function.
Hexamerins
The work done on the termite hexamerins is another excellent example of linking genetic changes to protein function and social phenotype. In the lower termites, workers may develop into either reproductives or soldiers, depending on a number of social and environmental cues, and differentiation into the soldier caste is induced by high juvenile hormone (JH) titers (Zhou et al., 2007). RNAi studies in the termite Reticu-litermes flavipes have shown that two hexamerin genes, Hex-1 and Hex-2, are involved in the regulation of this caste determination (Zhou et al., 2006). In many insects, hexamerins act as storage proteins that sequester substances from the diet and release them when food is scarce or inaccessible, such as during early development (Zhou et al., 2007). It has been hypothesized that Hex-1 and Hex-2 work together to regulate caste differentiation in termites via direct interactions with JH (Zhou et al., 2006); however, elucidating the specific molecular mechanisms involved in JH action is a difficult challenge in insects in general (Riddiford, 2008).
Molecular evolutionary studies of Hex-1 and Hex-2 provide clues as to how these genes may interact with JH. Relative to 100+ known Hex genes in other insects, both termite Hex genes have distinctive insertions in their coding regions; the unique insertion in Hex-1 contains a prenylation motif with a proposed function in JH binding, and the unique insertion in Hex-2 shares sequence similarities to the well-characterized blowfly (Calliphora vicina) hexamerin receptor (Drapeau et al., 2006). Consistent with these predicted functions, follow-up experiments demonstrated that the Hex-1 protein has strong binding affinity for JH and the Hex-2 protein shows strong membrane affinity, as would be expected for a receptor protein (Zhou et al., 2006).
Hexamerins also exhibit novel social functions in other social insect species, suggesting that they may be particularly prone to social co-option. Evidence in honey bees (Martins et al., 2010) and Polistes wasps (J. H. Hunt et al., 2010) suggests that hexamerins may be important in caste determination in these social insect lineages, and in ants, hexam-erins appear to be have been important in the evolution of elaborated life history characteristics (Wheeler and Buck, 1995).
JH, Insulin, and Vitellogenin Axis
In the highly eusocial honey bee, A. mellifera, the JH and insulin/insulinlike growth factor-1 (IIS) signaling pathways, as well as the yolk protein precursor vitellogenin (Vg), interact with one another and function in novel ways that are important in multiple social contexts. JH does not function as a gonadotropin in adult honey bees as it does in most insects; instead, it plays a strong role in caste determination and worker division of labor (Robinson and Vargo, 1997). The IIS signaling pathway interacts with JH and is also involved in worker division of labor. Foragers exhibit higher expression of genes in the IIS pathway in the brain relative to nurses, and down-regulating IIS signaling delays the age-related transition from nursing to foraging (Ament et al., 2008). This represents a reversal of the traditional positive relationship between high nutrition and IIS signaling, because foragers are nutritionally deprived relative to nurses (Ament et al., 2008). Vg also shows novel social functions in honey bees. It is highly expressed in some workers, although they are largely nonreproductive; it may be used by nurses in the synthesis of royal jelly (Amdam et al., 2003); and it functions as an antioxidant that may be involved in promoting longevity in queen bees (Corona et al., 2007).
The molecular changes underlying these novel functions of JH, IIS, and Vg are unknown, but insights from solitary insects may provide clues as to what these changes may be. The relationship between genetic variation and regulation of JH titers has been particularly well studied in crickets and butterflies (Zera et al., 2007), molecular evolution and function of the IIS pathway have been investigated across the complete genomes of 12 Drosophila species (Alvarez-Ponce et al., 2009; Grönke et al., 2010), and insect Vgs and their receptors are well characterized at the molecular level (Sappington and Raikhel, 1998).
Carbohydrate Metabolism
Several studies in bees suggest that the evolution of the highly eusocial lifestyle involved molecular changes in genes related to carbohydrate metabolism. Our bee molecular evolution study revealed that genes involved in carbohydrate metabolism are evolving more rapidly in eusocial relative to noneusocial bee lineages and are evolving most rapidly in highly eusocial lineages (Woodard et al., 2011). In particular, 15 genes encoding glycolytic enzymes showed evidence of rapid evolution in eusocial lineages, including enzymes that play a key regulatory role (e.g., phosphofructokinase) or are involved in glycolytic flux (e.g., hexokinase, pyruvate kinase) (Kunieda et al., 2006). Analysis of protein sequence evolution of genes with queen-biased brain gene expression in A. mel-lifera found that queen-biased genes involved in metabolism, inclu-
ding carbohydrate metabolism, were among the most rapidly evolving (based on branch lengths in phylogenetic trees inferred from protein sequence) relative to orthologs from several solitary insects (B. G. Hunt et al., 2010). Comparative analysis of the genome sequences of A. mellifera, D. melanogaster, and A. gambiae suggest that there may also have been bee-specific changes in gene copy number for carbohydrate-metabolizing genes (Kunieda et al., 2006). Given that carbohydrate metabolism is such a fundamental “housekeeping” process, it is not immediately clear why there has been unique selective pressure on these processes in highly eusocial bee lineages. Here, we offer three speculative hypotheses.
First, increases in the flight demands of highly eusocial bees may have placed strong selective pressure on increasing efficiency of glycolytic enzymes, because carbohydrates are the main fuel for flight in bees (Suarez et al., 2005). The individual foraging activity of highly eusocial bee workers appears to be higher than for solitary bees (Roubik, 1992), although, to the best of our knowledge, no direct comparisons of highly and primitively eusocial bee foraging activity have been performed.
Second, highly eusocial bees are unique in relying exclusively on a diet of modified stored sugars (i.e., honey) for long periods of time. Nest thermoregulation during winter months is completely reliant on honey stores as a fuel source to sustain workers, who shiver to produce metabolic heat to maintain optimal hive temperature (Southwick and Heldmaier, 1987). Perhaps these differences in diet have placed some novel selective pressure on glycolytic enzymes in highly eusocial lineages.
Third, perhaps the greatly extended life span of queens in highly eusocial species evolved through changes in metabolism-related genes, including those involved in carbohydrate metabolism. A connection between reduced metabolic rate and increased life span has been shown in many species (Finkel and Holbrook, 2000). In the honey bee, A. mellifera, queens exhibit an age-related reduction in IIS signaling (Corona et al., 2007) that regulates carbohydrate metabolism. If the molecular changes in carbohydrate metabolism genes in highly eusocial bees were attributable to selection for extended queen life span, it can be predicted that similar molecular changes may also be found in independent social insect lineages that also exhibit extended queen life spans (Wilson, 1971).
PROSPECTS AND CHALLENGES
Recent work on molecular evolutionary changes in social insects has identified specific genes, molecular pathways, and biological processes that appear to have been shaped by natural selection. Some of these
changes can be plausibly associated with the origins, maintenance, or elaboration of eusociality, albeit speculatively.
Two insights emerge from this review. First, it appears that there have been unique genetic changes in different social insect lineages, suggesting that the multiple independent occurrences of eusociality have involved multiple molecular routes. These differences may reflect distinct ecological or other constraints for each lineage. For example, the evolution of a queen pheromone in termites from a wood-digesting enzyme seems fitting, given that many termite societies live in rotting wood (Korb et al., 2009).
Second, genetic changes also have occurred in similar biological functions across diverse species of social insects. This supports the concept of a genetic toolkit for eusociality (Toth and Robinson, 2009). This concept is reasonable, because despite the striking diversity among social insect species, they all have converged on a similar suite of traits, which are the defining characteristics of eusociality (Michener, 1974). Previous research suggesting components of a genetic toolkit for eusociality has focused on genes and molecular pathways that are associated both with solitary and related social behaviors in insects, for example, the foraging gene, which is involved in feeding behavior in Drosophila and a variety of other solitary organisms, and social foraging behavior in honey bees and ants (Toth and Robinson, 2009). Transcriptomic studies have also identified shared sets of genes whose expression patterns are associated with division of labor in independent social insect lineages (Toth et al., 2010).
The molecular evolutionary studies we reviewed identify biological processes and specific genes that may be excellent systems in which to investigate the concept of a genetic toolkit for eusociality further. Among the most promising are the following:
(i) Hexamerins. As discussed above, hexamerins have been shown to be involved in queen physiology and other social traits in a variety of social insects, and the work on Hex-1 and Hex-2 in termites demonstrates how hexamerin sequence evolution can be studied and linked to social traits.
(ii) Gland development genes. The rapidly evolving gland development genes identified in our bee molecular evolution study (Woodard et al., 2011) are also good candidates for further study, because the gene functions are relatively well characterized, and gland diversification is a universal phenomenon in social insect evolution.
(iii) Brain-related genes. The rapidly evolving brain-related genes identified in primitively eusocial lineages in our bee molecular evolution study (Woodard et al., 2011) are prime candidates for further study in
primitively eusocial bees, as well as in ant and wasp species that share the primitively eusocial bee lifestyle feature of solitary nest-founding.
The molecular changes and biological processes highlighted in this review are currently the most well studied in social insects. There are almost certainly other equally important types of molecular changes and biological processes associated with social insect evolution that have not yet been discovered, perhaps because of the limited range of taxa subjected to these types of analyses thus far. This gap in our knowledge is largely attributable to a lack of genomic resources, especially for closely related social and nonsocial species. For example, some types of genetic changes, such as chromosomal rearrangements and patterns of DNA methylation, are not possible to study with only fragments of the genome. In addition, the identification of truly novel genes is limited by the small sample size of available genomes and less well-developed forward genetic analyses in social insects relative to model genetic organisms. As these limitations are overcome, it should be possible to search more broadly for different types of genetic changes associated with the evolution of eusocial traits. These analyses can be guided by several theoretical models that have been proposed to predict the types of genetic changes that are most important in social evolution (Nonacs and Kapheim, 2007; Linksvayer and Wade, 2009; Johnson and Linksvayer, 2010).
Whole-genome scans for molecular signatures of adaptive evolution specific to social insects will be particularly useful for generating new hypotheses and implicating new biological processes in social insect evolution. Candidate gene approaches across a broad sample of social and nonsocial insects will allow for greater accuracy in reconstructing the phylogenetic history of molecular changes and testing their associations with social evolution. Once specific sequence changes are identified, functional analyses are necessary to determine their effect on protein-, organismal-, and group-level phenotypes, as well as the adaptive significance of the phenotype change (Dean and Thornton, 2007).
This leads us to raise one important caveat for most molecular evolutionary studies in the social insects: the lack of species-specific information about gene function. As is often the case, gene function in this chapter is typically inferred from orthology to the fruit fly D. melanogaster, which shared a common ancestor with eusocial insect lineages over 300 Mya (Honeybee Genome Sequencing Consortium, 2006). Although gene function for molecular processes is generally highly conserved over evolutionary time, when interpreting findings, it is important to consider the possibility that a particular gene has evolved a novel function. Furthermore, many genes have multiple functions; thus, the target of selection can be difficult to infer solely from identifying molecular evolutionary changes.
Experimental approaches to determining gene function in social insects, via RNAi and transgenesis, will strengthen the interpretation of molecular evolutionary findings. Additional challenges arise in determining the adaptive or ecological significance of molecular changes, even when their functional significance is understood (Feder and Mitchell-Olds, 2003).
Despite these challenges, molecular evolutionary analysis of social insect societies holds promise for testing venerable theories of social evolution using genomic data. Multiple evolutionary scenarios have been proposed as potential routes to group living in insects. These include the composition of incipient social groups, such as associations between mothers and offspring (the “subsocial” route) or between related and unrelated individuals of the same generation (“semisocial” route) (Michener, 1974); mechanisms through which altruism is achieved, such as kin selection (Strassmann and Queller, 2007); parental manipulation of offspring or voluntary helpers at the nest (Charnov, 1978); and necessary preadaptations for social living, such as a monogamous mating system (Hughes et al., 2008) or progressive provisioning of offspring (Nowak et al., 2010). Wedding this rich theory with genome-scale molecular evolutionary analysis and functional experimentation holds the promise of finally answering the compelling question of how eusociality evolved in insects.
ACKNOWLEDGMENTS
We thank members of the G.E.R. laboratory for comments that improved this manuscript. Our bee molecular evolution study was supported by 454 Life Sciences (Roche Diagnostics Corporation) via the Roche 1GB contest, National Science Foundation Grant DEB07–43154 (to G.E.R., Primary Investigator), National Science Foundation Predoctoral Fellowship NSF DGE 07–15088 FLW (to B.J.F.), and National Institutes of Health–University of Illinois Sensory Neuroscience Training Grant PHS2T32DC006612 (to S.H.W. and A. S. Feng, Primary Investigator).
This page intentionally left blank.
























