Biomedical research and the practice of medicine, separately and together, are reaching an inflection point: the capacity for description and for collecting data, is expanding dramatically, but the efficiency of compiling, organizing, manipulating these data—and extracting true understanding of fundamental biological processes, and insights into human health and disease, from them—has not kept pace. There are isolated examples of progress: research in certain diseases using genomics, proteomics, metabolomics, systems analyses, and other modern tools has begun to yield tangible medical advances, while some insightful clinical observations have spurred new hypotheses and laboratory efforts. In general, however, there is a growing shortfall: without better integration of information both within and between research and medicine, an increasing wealth of information is left unused.
As illustration, consider the following clinical scenarios;1 in the first example, molecular understanding of disease has already begun to play an important role in informing treatment decisions, while in the second, it has not.
Patient 1 is consulting with her medical oncologist following breast cancer surgery. Twenty-five years ago, the patient’s mother had breast cancer, when therapeutic options were few: hormonal suppression or broad-spectrum chemotherapy with significant side effects. Today, Patient 1’s physician can suggest a precise regimen of therapeutic options tailored to the molecular characteristics of her cancer, drawn from among multiple therapies that together focus on her particular tumor markers. Moreover, the patient’s relatives can undergo
1 These scenarios are illustrative examples describing typical patients. They are not based on individual patients, but reflect current medical practice.
testing to assess their individual breast cancer predisposition (Siemens Healthcare Diagnostics Inc. 2008).
In contrast, Patient 2 has been diagnosed at age 40 with type 2 diabetes, an imprecise category that serves primarily to distinguish his disease from diabetes that typically occurs at younger ages (type 1) or during pregnancy (gestational). The diagnosis gives little insight into the specific molecular pathophysiology of the disease and its complications; similarly there is little basis for tailoring treatment to a patient’s pathophysiology. The patient’s internist will likely prescribe metformin, a drug used for over 50 years and still the most common treatment for type II diabetes in the United States. No concrete molecular information is available to customize Patient 2’s therapy to reduce his risk for kidney failure, blindness or other diabetes-related complications. No tests are available to measure risk of diabetes for his siblings and children. Patient 2 and his family are not yet benefitting from today’s explosion of information on the pathophysiology of disease (A.D.A.M. Medical Encyclopedia 2011; Gordon 2011; Kellett 2011).
What elements of our research and medical enterprise contribute to making the Patient 1 scenario exceptional, and Patient 2 typical? Could it be that something as fundamental as our current system for classifying diseases is actually inhibiting progress? Today’s classification system is based largely on measurable “signs and symptoms,” such as a breast mass or elevated blood sugar, together with descriptions of tissues or cells, and often fail to specify molecular pathways that drive disease or represent targets of treatment.2 Consider a world where a diagnosis itself routinely provides insight into a specific pathogenic pathway. Consider a world where clinical information, including molecular features, becomes part of a vast “Knowledge Network of Disease” that would support precise diagnosis and individualized treatment. What if the potential of molecular features shared by seemingly disparate diseases to suggest radically new treatment regimens were fully realized? In such a world, a new, more accurate and precise “taxonomy of disease” could enable each patient to benefit from and contribute to what is known.
In consideration of such possibilities, and at the request of the Director of the National Institutes of Health, an ad hoc Committee of the National Research Council was convened to explore the feasibility and need, and to develop a potential framework, for creating “a New Taxonomy of human diseases based on molecular biology” (Box 1-1). The Committee hosted a two- day workshop
___________________
2 To clarify, the committee is not suggesting that all diseases would have an equally precise taxonomy, rather each disease should be classified, and treatment provided, using the best available molecular information about the mechanism of disease.
At the request of the Director’s Office of NIH, an ad hoc Committee of the National Research Council will explore the feasibility and need, and develop a potential framework, for creating a “New Taxonomy” of human diseases based on molecular biology. As part of its deliberations, the Committee will host a large two-day workshop that convenes diverse experts in both basic and clinical disease biology to address the feasibility, need, scope, impact, and consequences of defining this New Taxonomy. The workshop participants will also consider the essential elements of the framework by addressing topics that include, but are not limited to:
- Compiling the huge diversity of extant data from molecular studies of human disease to assess what is known, identify gaps, and recommend priorities to fill these gaps.
- Developing effective and acceptable mechanisms and policies for selection, collection, storage, and management of data, as well as means to provide access to and interpret these data.
- Defining the roles and interfaces among the stakeholder communities—public and private funders, data contributors, clinicians, patients, industry, and others.
- Considering how to address the many ethical concerns that are likely to arise in the wake of such a program.
The Committee will also consider recommending a small number of case studies that might be used as an initial test for the framework.
The ad hoc Committee will use the workshop results in its deliberations as it develops recommendations for a framework in a consensus report. The report may form a basis for government and other research funding organizations regarding molecular studies of human disease. The report will not, however, include recommendations related to funding, government organization, or policy issues.
(see Appendix C) that convened diverse experts in both basic biology and clinical medicine to address the feasibility, need, scope, impact, and consequences of creating a “New Taxonomy of human diseases based on molecular biology”. The information and opinions conveyed at the workshop informed and influenced an intensive series of Committee deliberations (in person and by teleconference) over a six-month period. The Committee emphasized that molecular biology was one important base of information for the “New Taxonomy”, but not a limitation or constraint. Moreover, the Committee did not view its charge as prescribing a specific new disease nomenclature. Rather, the Committee saw its challenge as crafting a framework for integrating the rapidly expanding range and detail of biological, behavioral, and experiential information to facilitate basic discovery, and to drive the development of a more accurate and
precise classification of disease (i.e., a “New Taxonomy”), which in turn will enable better medicine.
The vision for a New Taxonomy informed by the proposed “Knowledge Network” shares some similarities with the widely discussed concept of “Personalized Medicine,” recently defined by the President’s Council on Advisors on Science and Technology (PCAST) as “the tailoring of medical treatment to the individual characteristics of each patient … to classify individuals into subpopulations that differ in their susceptibility to a particular disease or their response to a specific treatment. Preventative or therapeutic interventions can then be concentrated on those who will benefit, sparing expense and side effects for those who will not” (PCAST 2008, p. 1). Others have used the related term “Precision Medicine” to refer to a very similar concept (see Glossary). Those who favor the latter term do so in part because it is less likely to be misinterpreted as meaning that each patient will be treated differently from every other patient. However, to be clear, the use of either term in this report refers to the PCAST definition.
A BRIEF HISTORY OF DISEASE TAXONOMIES
One of the first attempts to establish a scientific classification of disease was undertaken by Carolus Linnaeus, who developed the taxonomic system that is still used to classify living organisms. His 1763 publication Genera Morborum (Linné 1763) classified diseases into such categories as exanthematic (feverish with skin eruptions), phlogistic (feverish with heavy pulse and topical pain), and dolorous (painful). The effort was largely a failure because of the lack of an adequate understanding of the biological basis of disease. For example, without a germ theory of disease, rabies was characterized as a psychiatric disorder because of the brain dysfunction that occurs in advanced cases. This illustrates how a classification system for disease that is divorced from the biological basis of disease can mislead and impede efforts to develop better treatments.
Even 100 years ago, the Manual of the International List of Causes of Death, second revision, (Wilbur 1911), which over time would become the International Statistical Classification of Diseases and Related Health Problems (ICD), lumped lung cancer and brain cancer into the category of “cancer of other organs or not specified.” No distinction was made between type 1 and type 2 diabetes, endocrine diseases were categorized under General Diseases, and categories existed for “nervous fever,” “inanition,” and “found dead.”
Today, the ICD, which is currently in its tenth revision, remains the most commonly used categorization of disease (WHO 2007). Published by the World Health Organization (WHO), ICD-10 is used for statistical analyses, reimbursement, and decision support, making it an integral part of health-care systems throughout the world. As will be discussed, the ICD is currently undergoing a
major revision, which will result in the publication of ICD-11 in approximately 2015.
THE TAXONOMIC NEEDS OF THE BIOMEDICAL RESEARCH AND MEDICAL PRACTICE COMMUNITIES
Taxonomies underpin many health-related systems, such as the organization of the curriculum of medical education, the published biomedical literature,3 textbooks, and disease coding systems such as the ICD. Although grounded in a scientific understanding of disease, taxonomies such as the ICD must address the needs of the ever-expanding public health and health-care delivery communities across the globe. Organizations such as WHO must have access to accurate and timely measures of disease incidence and prevalence in multiple continents to make recommendations. Similarly, the health-care industry in the United States depends on an accurate disease classification system to track the delivery of medical care and to determine reimbursement rates. Both of these communities rely on highly robust data collection practices to make decisions that can impact millions of individuals. In this context, a formalized nomenclature is essential for clear communication and understanding. The current practice of updating the ICD nomenclature periodically attempts to balance: (1) the need for a consistent terminology to permit clear communication about diseases that are defined by agreed upon criteria, with (2) the need to ensure that the classification system (i.e., the taxonomy) properly reflects advances in our understanding of molecular pathways and environmental factors that contribute to disease origin and pathology.
However, in part because it must serve the administrative needs of the public health and health-care delivery communities, the current ICD taxonomy is disconnected from much of the biomedical research community (see Figure 1-1). Indeed, few basic researchers know of the existence of ICD, and even fewer use this classification in any way. Thus, two extensive stakeholder groups, represented on one hand by biomedical researchers, and biotechnology and pharmaceutical industries, and on the other by clinicians, health agencies and payers, are widely perceived to be largely unrelated, and to have distinct interests and goals, and therefore taxonomic needs. This is unfortunate because new insights into human disease emerging from basic research and the explosion of information both in basic biology and medicine have the potential to revolutionize disease taxonomy, diagnosis, therapeutic development, and clinical decisions. However, more integration of the informational resources available to these diverse communities will be required before this potential can be fully realized
_________________
3 For example, an information resource used extensively by both clinicians and researchers, PubMed/MEDLINE, is built on the MeSH terminology hierarchy of diseases.
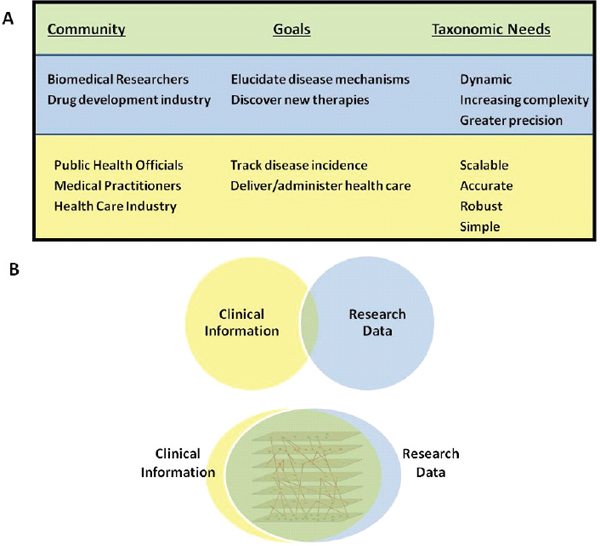
FIGURE 1-1 Integration would benefit all stakeholder communities.
(A) Different stakeholder communities are perceived to have distinct taxonomic and informational needs. (B) Integration of information and a consolidation of needs could better serve all stakeholders.
SOURCE: Committee on A Framework for Developing a New Taxonomy of Disease.
with the attendant benefits of more individualized treatments and improved outcomes for patients.
MISSED OPPORTUNITIES OF CURRENT TAXONOMIES
Currently used disease classifications have properties that limit their information content and usability. Most importantly, current disease taxonomies, including ICD-10, are primarily based on symptoms, on microscopic examination of diseased tissues and cells, and on other forms of laboratory and imaging studies and are not designed optimally to incorporate or exploit rapidly emerging molecular data, incidental patient characteristics, or socio-environmental influences on disease. The ability of current taxonomic systems to incorporate
In 1910 educator Abraham Flexner released a report that revolutionized American medical education by advocating a commitment to professionalization, high academic standards, and close integration with basic science (Flexner 1910). The subsequent rise of academic medical centers with a strong emphasis on research—coupled, after World War II, with greatly expanded merit-based funding of research through the National Institutes of Health and other public and private entities—allowed the United States to capture global leadership in medical research, launch the biotechnology industry, and pioneer countless science-based innovations in health care.
The vast expansion of molecular knowledge currently underway could have benefits comparable to those that accompanied the professionalization of medicine and biomedical research in the early part of the 20th century. Indeed, during his talk at the Committee’s workshop, Dr. Christopher G. Chute, a Mayo Clinic professor and leader in the development of ICD-11, said that the potential of the genomic transformation of medicine “far exceeds the introduction of antibiotics and aseptic surgery.” However, achieving the full potential of the molecular revolution will require—and to an important extent enable—re-thinking both biomedical research and health care on a Flexnerian scale. Creation of a Knowledge Network of Disease that consolidates and integrates basic, clinical, social, and behavioral information, and that helps to inform a New Taxonomy that enables the delivery of improved, more individualized health care, will be a crucial element of this revolutionary change (Chute 2011).
fundamental knowledge is also limited by their basic structure. Taxonomies historically have relied on a hierarchical structure in which individual diseases are successively subdivided into types and subtypes. This rigid organizational structure precludes description of the complex interrelationships that link diseases to each other, and to the vast array of causative factors. It also can lead to the artificial separation of diseases based on distinct symptoms that have related underlying molecular mechanisms. For example, mutations in the LMNA gene give rise to a remarkably diverse set of diseases, including Emery-Dreyfus muscular dystrophy, Charcot-Marie-Tooth axonal neuropathy, lipodystrophy, and premature aging disorders. However, despite their remarkable genetic, molecular, and cellular similarities, these diseases are currently classified as distantly related. While this approach may have been adequate in an era when treatments were largely directed toward symptoms rather than underlying causes, there is a clear risk that continued reliance on hierarchical taxonomies will inhibit efforts—already successful in the case of some diseases—to exploit rapidly expanding mechanistic insights therapeutically.
A further limitation of taxonomic systems is the intrinsically static nature
of their information content. The ICD system is designed to be updated every ten years with minor updates every three years. But many organizations are still working with ICD-9, which was released in 1977, even though ICD-10 was released in 1992. Because of the time it takes to implement ICD revisions in administrative systems, the current taxonomic system is perpetually outdated. Moreover, the static structure of current taxonomies does not lend itself to the continuous integration of new disease parameters as they become available. This is particularly troublesome given that new data regarding the molecular nature of disease are becoming available at an ever-increasing rate.
Current efforts to revise the ICD classification attempt to address these limitations. ICD-11 will be based on a foundational layer from which “linearizations” will be derived (Tu et al. 2010). While the linearizations will be relatively static and hierarchical, the foundational layer is being designed to support multi-parent hierarchies and connections, and to be updated continuously.4 Importantly, the new classification will combine phenomenological characterization of phenotype with genomic factors that might explain or at least distinguish phenotypes.5 Different lung cancers, for example, could be explicitly differentiated by genomic characterization. This is important because knowledge about the specific molecular pathways contributing to the biology of particular types of lung cancer can be used to guide selection of the most appropriate treatment for such patients.
Although the release of ICD-11 will mark an important step forward, the Committee thinks that the amount of information available for this effort can be vastly increased by a two-stage process leading to a Knowledge Network of Disease. As discussed in detail in following sections of this report, the first stage in developing this Knowledge Network would involve creating an Information Commons containing a combination of molecular data, medical histories (including information about social and physical environments), and health outcomes for large numbers of individual patients. The Committee envisions this stage occurring in conjunction with the ongoing delivery of clinical care to these patients, rather than in specialized settings specifically crafted for research purposes. The second stage, the construction of the Knowledge Network itself,
_____________
4 The ICD-11 revision process is closely coordinated with SNOMED—the Systematized Nomenclature of Medicine developed by the International Health Terminology Standards Development Organization (IHTSDO). SNOMED is a large, clinically focused ontology that uses high-level nodes to aggregate more granular data. The WHO and IHTSDO have signed a memorandum of understanding so that the two systems will be complementary rather than competing. The intent is to harmonize the two systems so that the aggregation layer of SNOMED corresponds to ICD-11 and the extensions of ICD-11 become elements of SNOMED.
5 It should be noted that the International Classification of Diseases for Oncology (ICD-O) already attempts to capture genomic data relevant to disease definitions. The third series of the WHO Monographs on the Pathology and Genetics of Tumours sought to integrate genomic data, where available, into disease definitions and indeed today many tumor types are molecularly defined (Vardiman et al. 2009;. Campo et al. 2011; Travis et al. 2011).
would involve data mining of the Information Commons and integration of these data with the scientific literature—specifically with evolving knowledge of the fundamental biological mechanisms underlying disease.
Such a Knowledge Network of Disease would enable development of a more molecularly-based taxonomy. This “New Taxonomy” could, for example, lead to more specific diagnosis and targeted therapies for muscular dystrophy patients based on the specific mutations in their genes. In other cases, it could suggest targeted therapies for patients with the same genetic mechanism of disease despite very different clinical presentations.
As will be described later in the report, the Committee envisions that the Information Commons, which would underlie the Knowledge Network of Disease and the New Taxonomy, would have some analogies with geographical information systems (GISs), which are designed to capture, store, manipulate, and analyze all types of geographically referenced data and make them widely accessible in applications (ESRI 1990) such as Google Maps (Figure 1-2). Most
users would interact with these resources at the higher-value-added levels, the Knowledge Network and the New Taxonomy, rather than at the level of the underlying Information Commons (Figure 1-3). Investigators using the Knowledge Network of Disease could propose hypotheses about the importance of various inter- and intra-layer connections that contribute to disease origin, severity, or progression, or that support the subclassification of particular diseases into those with different molecular mechanisms, prognoses, and/ or treatments, and these ideas then could be tested in an attempt to establish their validity, reproducibility, and robustness. Validated findings that emerge from the Knowledge Network of Disease and are shown to be useful for defining new diseases or subtypes of diseases that are clinically relevant (e.g., which have implications for patient prognosis or therapy) could be incorporated into the New Taxonomy to improve diagnosis and treatment. Whether the “New
Taxonomy” that is informed and refined by the Knowledge Network of Disease would best be realized as a modification of the ICD taxonomy, or should represent an entirely distinct taxonomy that exists in parallel with ICD and other taxonomies, will depend on a number of factors. However, in either case, the goal of basing the New Taxonomy on the Knowledge Network of Disease will be to improve markedly the quantity and quality of information that can be used in biomedicine for the basic discovery of disease mechanisms, improved disease classification, and better medical care.
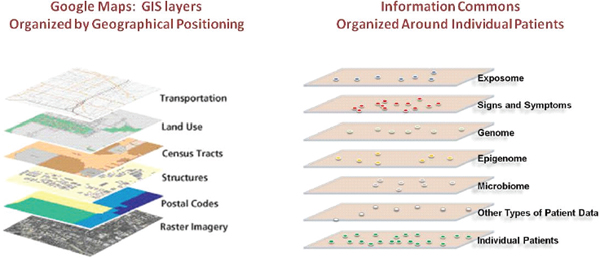
FIGURE 1-2 An Information Commons might use a GIS-type structure.
The proposed, individual-centric Information Commons (right panel) is somewhat analogous to a layered GIS (left panel). In both cases, the bottom layer defines the organization of all the overlays. However, in a GIS, any vertical line through the layers connects related snippets of information since all the layers are organized by geographical position. In contrast, data in each of the higher layers of the Information Commons will overlay on the patient layer in complex ways (e.g., patients with similar microbiomes and symptoms may have very different genome sequences).
SOURCE: FPA 2011 (left panel).
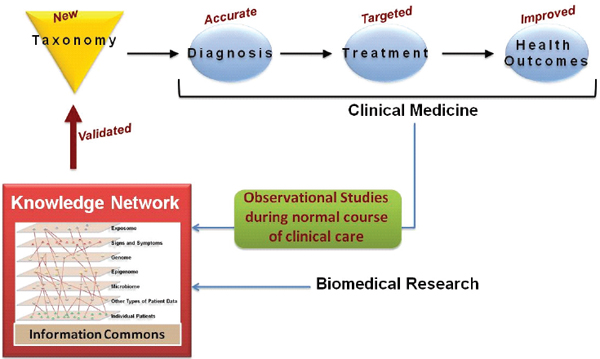
FIGURE 1-3 A knowledge network of disease would enable a new taxonomy.
An individual-centric Information Commons, in combination with all extant biological knowledge, will inform a Knowledge Network of Disease, which will capture the exceedingly complex causal influences and pathogenic mechanisms that determine an individual’s health. The Knowledge Network of Disease would allow researchers to hypothesize new intralayer cluster and interlayer connections. Validated findings that emerge from the Knowledge Network, such as those which define new diseases or subtypes of diseases that are clinically relevant (e.g., which have implications for patient prognosis or therapy) would be incorporated into the New Taxonomy to improve diagnosis and treatment.
SOURCE: Committee on A Framework for Developing a New Taxonomy of Disease.
RATIONALE AND ORGANIZATION OF THE REPORT
Today, historic forces are transforming biomedical research and health care. Information technology, clinical medicine, and the public attitudes that govern the ways that science, medicine, and society interact are all in flux.
A Knowledge Network of Disease could embrace and inform rapidly expanding efforts by the biomedical research community to define at the molecular level the disease predispositions and pathogenic processes occurring in individuals. This network has the potential to play a critical role across the globe for the public-health and health-care-delivery communities by enabling development of a more accurate, molecularly-informed taxonomy of disease.
This report lays out the case for developing such a Knowledge Network of Disease and associated New Taxonomy. Chapter 2 asks “Why now?” It examines basic trends in research, information technology, clinical medicine, and public attitudes that have created an unprecedented opportunity to influence biomedical research and health-care delivery in ways that will benefit all stakeholders.
Chapter 3 asks “What would a Knowledge Network of Disease and New Taxonomy look like?” It describes why the system needs to be dynamic, continuously evolving, integrative, and flexible, and why it needs to enable interrogation by a wide range of users, from basic scientists to clinicians, health-care workers, and the general public.
Chapter 4 asks “How do we get there?” It describes the need for a series of pilot studies to evaluate the feasibility of creating an individual-centric Information Commons and deriving a Knowledge Network and New Taxonomy from it, and to begin to explore the utility of these resources for improving individual health outcomes. This chapter also addresses the impediments that need to be overcome and changes in medical education that will be required before the Knowledge Network of Disease and resulting New Taxonomy can be expected to achieve their full potential for improving human health.
In Chapter 5, the report closes with an epilogue that summarizes the Committee’s rationale for its recommendations and describes how the new resources described in this report could serve the needs of basic scientists, translational researchers, policy makers, insurers, medical trainees, clinicians, and patients.












