3
Digital Health Data Uses: Leveraging Data for Better Health
KEY SPEAKER THEMES
Leenay
• Shared information in health care must evolve to include all stakeholders as active constituents in the health care conversation.
• Data included in current digital record systems must be more accurate, timely, and standardized to support actionable decisions.
• ACOs represent an alignment of incentives for the collection of higher-quality data, with greater completeness and accuracy, and the increased liquidity of this data.
Kush
• Data standards are key to improving data quality.
• Quality must be built into research methods from the beginning.
• Regulated clinical research requires high-quality, or “sushi-grade,” data, which can be obtained from EHRs if certain processes are applied and requirements met.
• Information from patient diagnosis, treatment, and treatment response should be aggregated and transformed to computable, standardized data for improved and more effective clinical decision support.
• EHR data quality issues may be mitigated through triangulation of multiple sources.
• Genome-directed cancer treatment is a driving-use case for learning cancer systems.
Buehler
• Data quality requirements depend on the purpose of those data.
• Public health surveillance systems must be prepared to take full advantage of the data influx resulting from implementation of Meaningful Use.
• Linking public health and direct health care services research through data will serve to strengthen the population-level approach to surveillance.
LaVenture
• Requirements for public health data quality vary by the specific program needs.
• Greater EHR use with improved standards and quality checks will increase the prevalence of better-quality data to improve care and public health.
• Incomplete records from EHRs with limited standards, specifications, and certification criteria create obstacles for the use of that data for surveillance.
• Value of data, and quality improvement, must be taught and encouraged as a standard of practice.
• A reliable bidirectional exchange of data with public health requires a shared responsibility for achieving high levels of data quality.
Different data uses have different requirements and priorities. This chapter includes presentations and discussions focused on data uses and quality requirements from the perspectives of various stakeholders in the field. Mark Leenay, Chief Medical Officer and Senior Vice President at OptumHealth Care Solutions, discussed challenges and opportunities specific to practice management and the clinical care digital data utility. Rebecca Kush, Founder and the current President and CEO of Clinical Data Interchange Standards Consortium (CDISC), built on this topic in her discussion of data quality requirements, challenges, priorities and enabling standards/processes for the clinical research enterprise. Later, Mia Levy, Director of Cancer Clinical Informatics for the Vanderbilt-Ingram Cancer Center, detailed the case example of Vanderbilt’s experience and successes in implementing a translational informatics data management system for cancer diagnosis, treatment, and care. James Buehler, Director of the Public Health Surveillance & Informatics Program Office at the Centers for Disease Control and Prevention (CDC), focused on data quality for public health surveillance at the national level, while Marty LaVenture, Director of the Office of Health Information Technology and e-Health at the Minnesota Department of Health, spoke to the local and state levels.
In his discussion of the digital data utility and its role in clinical practice management, Mark Leenay emphasized the requirements necessary to enable sustainable private health information exchanges while ensuring data are connected, intelligent, and aligned. Actionable data at the point of care, increased data liquidity, and integration of data across the care continuum, as well as across different types of data, are all integral to incorporating digital health data into practice management.
While the quality of health data is important to their use, Leenay said, flow of data also plays a major role in supporting population management. Currently, data platforms are not integrated into routine care, rendering the flow of digital health information incomplete, and leaving the many different stakeholders managing care with only a partial view of the situation. Without a central repository of digital health information, from which each stakeholder is able to extract information to make decisions, this ineffective communication stream is difficult to rectify. The current lack of information fluidity, Leenay concluded, warrants continued efforts designed to bring data to the point of care for individuals.
In conjunction with the challenge of fluidity, identity resolution continues to be a barrier to providing integrated, longitudinal data. Without national
member IDs, for both patients and providers, effective use of digital health data for practice management will continue to be a struggle. As an example of this challenge, Leenay cited that out of people using health care exchanges, it is predicted that 50 percent will be eligible for Medicaid at some point during the year. As these individuals presumably will alternate between a private exchange and Medicaid, integrating their data presents a challenge. Further complicating the issue, National Provider IDs (NPIs) are used inconsistently, which causes difficulties for data aggregation across provider and hospital groups.
Content within data systems, both administrative and clinical, presents additional challenges. Leenay suggested that 85 percent of the information in EHRs is administrative rather than clinical data. Administrative claims data typically are designed for fee-for-service billing as opposed to pay-for-performance. Historically, incentives have been designed to reward complexity of service; the more complex the service, the more the provider will be paid. However, as the system shifts to pay-for-performance, incentives will need to be structured so that physicians are incentivized to enter more clinical data in order to be reimbursed appropriately. Additionally, the limited link between claims data and clinical data, and provider resistance to efforts to forge that link, presents a challenge to supporting the sorts of analyses that require insight into both cost or utilization and clinical outcomes. Citing an area for potential short-term progress, Lennay mentioned that from a clinical data perspective, there is minimal use of national registries. Such registries could provide a way to look at clinical outcomes that are not necessarily as complex as EHRs. As a cautionary note, Lennay pointed out that from an administrative dataset perspective, migration from ICD-9 to ICD-10 will involve a transition from fewer data points to more, a complicated extrapolation that will be an ongoing challenge.
Lastly, Leenay emphasized, the data actually included in EHRs are often inconsistent and incomplete. However, in light of the changing health care environment, primarily the development of accountable care organizations (ACOs), data quality requirements are changing. He pointed out that ACOs represent an alignment of incentives for the collection of higher quality data, with greater completeness and accuracy, and the increased liquidity of this data.
In summary, Leenay underscored several priorities to improve integration of the digital data utility into clinical care moving forward: identity resolution, information exchange standards, registries, and attention to disparities. Identity resolution will be critical to increase the accuracy of digital record use for patient care; strategies to develop and improve current dataset systems must include a focus on standards and normalization to facilitate coordinated information exchange. Organizations and clinicians should make greater use of national registries. And given the current
socioeconomic disparities in health care, it is important for stakeholders in the digital movement to guard against worsening such disparities through the digital divide.
In her discussion of clinical research, Rebecca Kush emphasized that different types of analyses require different grades of data quality, likening the data quality requirements for clinical decisions and regulated research to “sushi-grade” data, or the highest quality available. As depicted in Figure 3-1, Kush laid out these requirements on a sliding scale, dependent
on the type of activities supported. Pointing to the extremes, she suggested that those projects investigating nonmedical sales and marketing have far different data quality requirements than those involving regulated research and clinical decisions.
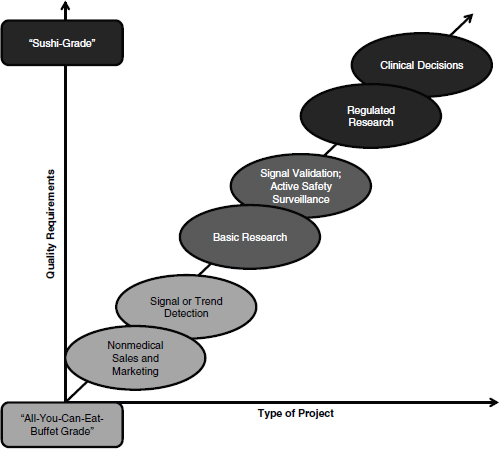
FIGURE 3-1 Spectrum of data quality requirements based on intended use.
SOURCE: Reprinted with permission from Rebecca Kush.
Currently, Kush said, clinical research (especially regulated clinical research) presents a plethora of logistical challenges to clinical investigators. The average active study site has 3 or more disparate data entry systems; 50-60 percent of trials involve paper data collection on 3- or 4-part forms, while the remaining 40-50 percent of trials involve electronic data capture tools. Data are entered 4-7 times total on average, including 2-3 times by the clinicians or study coordinators. Thus, there is plenty of opportunity to introduce transcription errors. In addition, reporting an unexpected or serious adverse event does not fit into normal clinical care workflow and takes excessive time, so that researchers often refrain from doing so. Given these inefficiencies and labor-intensive procedures, Kush emphasized, most clinicians do one regulated clinical research study and no more. Further exacerbating the data quality issue, efforts to ensure that study data are clean can involve significant resources and financial consequences; depending on the point of the research process in which the error is identified, correction of a single error in the database can cost upward of $8,000.
However, promise lies in the growing industry appreciation of the power of standardization. Kush described CDISC’s progress in this field through development of integration profiles with the capability to enable the extraction of a standardized, clinical research dataset (CDASH, Clinical Data Acquisition Standards Harmonization) from EHRs. The resulting interoperability specification (a set of standards) meets global regulations for collection of electronic research data and produces the minimum dataset needed on any clinical trial for regulated purposes. This combination of workflow enablers and standards has been used in safety reporting, regulatory reporting, and Phase 4 trials; and presents an opportunity to support research with EHRs and contribute to the process of research informing clinical decisions faster with higher quality information.
Kush then pointed to the Coalition Against Major Diseases (CAMD), an effort initiated by the Critical Path Institute (C-Path) to pool Alzheimer’s-related trial data from multiple sources with the goal of generating better information from a larger, aggregated database. By standardizing this data into the CDISC format and then pooling it across sources, C-Path was able to create a database of more than 6,000 patients and has now made this database available to researchers around the world. A standard guide has been developed for researchers moving forward, so that they can collect Alzheimer’s data in the CDISC format from the start and it can be easily compared with the current database. As such, this standardization effort has allowed researchers the capability to easily break out different cohorts
and better identify trends in their patient populations in order to identify personalized treatments or to populations most likely to respond.
With CAMD as an exemplar of the opportunities available, Kush delineated priorities for clinical research to bring such successes to scale across the field. Data quality should be built into the clinical research system from the beginning, and those individuals involved in the research process (including site personnel, the project team, reviewers, and auditors) should be trained and educated to incorporate data quality measures, including standards for data collection, into their work. Data collection should be simplified with well-defined requirements for the necessary data set and standardized formats. Data should be handled only the minimum amount throughout the process, thereby reducing potential errors due to transcription or reentry. Additionally, Kush noted, data quality measures should be considered and incorporated throughout the postmarketing process.
In her final comments, Kush emphasized that greater standardization offers considerable promise for clinical research moving forward, particularly in leveraging EHRs for research. As exemplified by CAMD and similar efforts, standardization facilitates both data sharing and data aggregation, presenting the opportunity for groundbreaking research efforts to identify new treatments and therapies with larger, standardized datasets.
In her discussion of translational informatics, Mia Levy focused on her experience at the Vanderbilt-Ingram Cancer Center, where genome-directed cancer treatment is the focus of the Center’s work. Currently, Levy noted, genomics is playing an ever more important role in the care of patients across the cancer continuum; cancer diagnosis, treatment selection, and care are all experiencing an era of genomics.
Traditionally, cancers have been categorized and treated according to the site of their origin and their histology. Now, the molecular subtypes of cancers are determining the course of care, and the molecular variance being discovered in these subtypes is vast. Levy noted that for those patients with characterized molecular subtypes, their mutations are considered actionable. Either an FDA-approved, standard-of-care therapy is available to treat the subtype of their cancer or a medication for their specific mutation is in the clinical research pipeline. However, in this genomic era, even patients for whom a mutation has not been identified are also considered to be actionable, in that they are spared from receiving ineffective, costly, and potentially harmful treatments. These developments hold great promise for the field of cancer treatment, but the process of implementing a system capable of processing and managing this information poses an entirely new range of challenges to those involved in translational informatics.
Reporting molecular diagnostic results in an EHR is typically unstructured and unwieldy she stated. Text is entered into a reporting template, and that form is then scanned and uploaded into the EHR as an image file, rendering it noncomputable. Another challenge associated with this type of reporting is the sheer amount of data to be reported. In her example, Levy highlighted that variance on 40 different mutations had to be reported at the same time. Not only does that require an increase in data points, but the complexity associated with this variance information must also be reflected in the system. Information must be reported in a way that is clinically useful for physicians, in order to help inform them of the findings’ clinical significance. Levy emphasized that much of this information is actionable only through its ability to link a patient’s results to clinical trial eligibility, and traditional reporting mechanisms do not possess this ability.
Levy noted that approaches to addressing these challenges are varied. Visualization of test results, complete with color coordination and coding, has proved very helpful at Vanderbilt, allowing researchers and clinicians to quickly scan information and identify positive findings. Findings are reported in a structured way, so that there is an entity, an attribute, and a value behind each piece of data. Moreover, information and results recorded in the EHR are linked directly to a database that provides information on the clinical significance of a patient’s particular mutation variant, thereby identifying potential targeted therapies. Further guidance is provided through inclusion of relevant, summarized clinical trial literature, which links clinicians to full, PubMed sources should they need to see additional information on the significance of the trial to their patient’s care. The data management system also links the EHR to a clinical trials database, providing clinicians with the means to identify relevant trial eligibility criteria. All of these strategies, Levy emphasized, offer promise for the effective and efficient incorporation of complex and varied digital data into the process of cancer care.
Levy finished her discussion by looking to the future, contemplating how to make systems like Vanderbilt’s sustainable and scalable with respect to content generation as well as content dissemination. Aggregation of institutional data, she suggested, is critical for rendering the data clinically useful. Information from patient diagnosis, treatment, and treatment response should be aggregated and transformed to computable, standardized data for improved and more effective clinical decision support. Moreover, the records incorporated into this type of database should be combined with other data, including patient-reported outcomes as well as cost information, both of which would be beneficial to understanding treatment comparative effectiveness. Given the complexity of genome-directed cancer treatment and translational informatics on the whole, Levy underscored her experiences with the importance of triangulation of data from multiple sources
to better approximate the probability of an event and use this as a basis for learning processes. EHR data can be useful for learning systems, but it must be of high quality and mitigated through triangulation of multiple resources.
SUPPORTING PUBLIC HEALTH AND SURVEILLANCE AT THE NATIONAL LEVEL
In the context of public health surveillance, data quality has varying definitions. As James Buehler of the CDC explained in his comments, quality requirements depend on the public health purpose the data are serving. For those working to prevent and contain specific diseases or adverse health events, the required data includes information about disease characteristics and severity, where and when it is occurring, its antecedents, its evolution over time, and its consequences. Moreover, public health professionals need data on those who are affected, individuals’ risk factors and whether certain groups of people are affected more than others, outcomes, and disease susceptibility to treatment. All of this information, often generated by individuals’ utilization of health care services, provides insight into what can be done to craft, target, and direct and redirect public health interventions. As such, public health surveillance is not simply about collecting information; it is about analyzing and using that data for a purpose, and that purpose can vary from disease surveillance, to situational awareness of a community’s status, to local, sometimes individual, interventions. While the data-quality requirements vary for each of these different purposes, Buehler continued, some apply to the broad range of public health surveillance uses. The data should be complete, reliable, timely, and inexpensive, and they should provide accurate insights into the local surveillance context. In practice, it is often not possible for a surveillance system to achieve all of these desirable attributes, requiring balance of desirable and sometimes competing attributes to maximize utility and value.
In order to meet these requirements, current public health surveillance data sources and systems are becoming progressively more automated. Attention is increasingly directed toward integrating EHRs into both the reporting and feedback arms of surveillance, so that individuals’ direct interactions with the health care system can serve as an additional source of electronic public health data. However, the process of moving this automation and integration forward faces a number of challenges Buehler noted, outlining several priorities for addressing those challenges. It is critical that public health surveillance systems are prepared to take full advantage of the data influx resulting from implementation of meaningful use, he said. The public health workforce likewise must be equipped to make the best use of this information, as it presents a great opportunity for more effective and
efficient public health surveillance. Finally, he noted, linking public health and direct health care services research in this way will serve to strengthen the population-level approach to surveillance.
SUPPORTING PUBLIC HEALTH AND SURVEILLANCE AT THE LOCAL LEVEL
In line with Buehler’s discussion of public health surveillance at the national level, Martin LaVenture shifted the focus to public health at the state and local (city and county) level. LaVenture reinforced the earlier assertion that necessary data quality attributes vary and depend on the context of the local public health activity. For example, timeliness is of particular importance for newborn screening, acute disease surveillance, and outbreaks, while completeness is especially critical for maintenance of immunization records. Accuracy is crucial for monitoring cancer clusters, while currency, comprehensiveness, and access to the primary data source all are relevant for public health surveillance and clinical decision support.
These quality characteristics all contribute to the usability of public health surveillance data today. Currently, surveillance data is collected from many sources, and health facilities increasingly are adopting EHRs for patient information management and decision support. However, frequent miscoding and mismapping of this information can result in loss of trust in both the data and the providers using those data; in which case the value of those data suffers. Moreover, the limited EHR standards and specifications and certification criteria lead to incomplete and invalid records, which create obstacles to efficient use of that data for clinical and disease surveillance purposes. This can lead to additional work by providers and public health officials and delay important public health intervention, prevention, and policy decisions.
In the face of these challenges, LaVenture said, the public health digital environment is changing. Greater EHR use with standards and quality checks built in will increase the prevalence of better quality data, thereby creating the opportunity for quality information exchange for care and public health and improved point-of-care decision support. To facilitate this progress, LaVenture proposed a number of priorities. Health information systems need to move beyond information management to rapid, accurate knowledge creation with support from public health information systems such as an immunization information system. The EHR certification process, he suggested, should include more comprehensive, structured content requirements for data quality, including thresholds at the point of capture, review, and exchange thus helping ensure higher-quality outputs for broader use. Standards for quality checks and improvement are needed to ensure updates and corrections can be completed quickly and propagation
of errors to other settings can be minimized. LaVenture went on to suggest that health care professionals should be educated on the value of quality data to encourage further focus on and enthusiasm for high-quality data, and incentives should encourage use of this data to increase value and quality. Public health agencies need similar incentives and support to modernize state and local systems in order to enable bidirectional flow of this information. Additionally, LaVenture noted that better use of existing standards and adopting new standards for the content and quality of data will reduce variability and increase usability for multiple purposes, and continuous improvement of data sources will ensure that their output is of the highest quality possible. It is also critical that information generated from these sources, and the knowledge from its analysis, is brought back to the source, to foster continuous improvement at the source level. Finally, LaVenture concluded by emphasizing that continued innovation with the public health case in mind will lead to better-quality data and better surveillance through improved adoption, use, and exchange of health information.












