Potential Pathways and Models for Moving Forward
DISCUSSION POINTS HIGHLIGHTED BY INDIVIDUAL PRESENTERS
• Volumes of molecular, clinical, and epidemiological data already exist in thousands of data repositories. Integrating data that are already in the public domain to generate new hypotheses for testing can help to identify new diagnostics, therapeutics, and disease mechanisms.
• An integrated knowledge ecosystem that supports moving data from discovery to actionable intelligence could drive better decision making and a learning health care enterprise.
• The overarching biomedical informatics challenges are systems issues; scale, standards, and sharing; software, storage, and security; sustainability; and social issues (changing mindsets and behaviors).
• Convenience and personal empowerment drive the disruptive innovation in service industries, and this will be the case for health care as well, with the patient or consumer as a primary disrupter.
• Numerous end-user applications can be developed on a core enterprise analytics platform, allowing researchers, clinicians, administrators, and others to analyze high-quality data. A common infrastructure can also support secondary uses of data.
To set the stage for discussion in the third panel session, John Mendelsohn, Forum chair, highlighted some of the key needs identified thus far in the speaker presentations. One main area of concern was the collection, structure, storage, and analysis of big datasets, including the need for interoperability and standardization of systems. In addition, observational research requires that the data be de-identified and pooled. Mendelsohn noted that the more technical issues of computer power, software, and interconnectivity did not seem to be major concerns.
Another main area of discussion was data scrutiny and use, which are affected by ethical and social issues more than scientific issues. Key issues raised by individual participants were patient privacy and trust. Many stakeholders need or want access to the data, including patients themselves, investigators, universities, pharmaceutical companies, the government, and others. There are questions of whether there is ownership of the data and, if so, by whom.
With these needs and concerns in mind, panelists discussed a variety of approaches for moving the field of cancer informatics forward.
PUBLIC DATA-DRIVEN SYSTEMS AND PERSONALIZED MEDICINE
Atul Butte, chief of the Division of Systems Medicine at Stanford University and Lucile Packard Children’s Hospital, shared examples of how public data can drive science and enable personalized medicine. There are tremendous volumes of data already in the public domain. For example, a DNA microarray or “gene chip” can quantitate every gene in the genome. This high-throughput genome technology is now widely used in research laboratories and has led to massive volumes of microarray data. One of the repositories tasked with holding these data is the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO). This United States–based repository currently contains more than 625,000 publicly available microarray datasets. Together with a comparable European repository, there are more than 900,000 microarray datasets in the public domain. At the current pace, the content doubles every 2 years (Butte, 2008).
Data generation has been commoditized, Butte said. He offered the example of Assay Depot, an online marketplace for scientific research
services. One can search for anything from assays to animal models and can purchase services from vendors “as easily as finding a song on iTunes.” These are laboratories around the world with excess capacity that are ready to do business with “dry bench” researchers, as well as laboratories in need of a specific service or laboratories that find they get faster results, or higher-quality results, by outsourcing these types of services via the Internet than by using the local university facility.
At this point, most of the steps along the translational pipeline can be commoditized, including clinical and molecular measurements, statistical and computational methods, and validation. The step that cannot be commoditized or outsourced, Butte said, is asking good questions. Given all of the public data, what are the new kinds of questions we should be asking? Given this commoditization of data, one no longer needs a wet lab to conduct academic research or launch commercial research ventures. All one needs is a place to formulate questions (and the means to pay for the studies).
Integrative Genomics to Identify Novel Targets
The first example Butte described involves using integrative genomics on public data to find causal factors for complex diseases, factors that can be targets for new drugs. Butte argued that the only way forward for some complex diseases will be to develop algorithms to integrate genetic, genomic, proteomic, preclinical model, and clinical data.
A decade ago, Butte and colleagues conducted a microarray study on type 2 diabetes and identified 187 genes that were differentially expressed in diabetes and non-diabetes muscle tissue (Patti et al., 2003). Now, intersecting those data with the results of 130 similar independent microarray experiments looking at muscle, fat, beta cells, and liver from rat, mouse, and human, Butte sought to identify common genes across all of these tissues and species (Kodama et al., 2012). Most of the genes in the genome were positive in just one of those studies. One gene, which Butte referred to as Gene A, was differently expressed between diabetes and control samples in 78 of the diabetes microarray experiments but, remarkably, had never been pursued for elucidation of the pathophysiology of type 2 diabetes. Further study by Butte and his group showed that Gene A codes for a functioning cell-surface receptor. Another common differentially expressed gene, which he referred to as Gene C, codes for the ligand for that receptor. Subsequent studies in mice (done through collaboration or purchased services, Butte noted) showed that the receptor is upregulated in mice fed a high-fat diet
and is expressed on inflammatory cells in adipose tissue. Gene A turns out to be a well-known receptor, and a knockout mouse is available from Jackson Laboratories. Further testing showed that the knockout mouse had increased insulin sensitivity (i.e., does not die from diabetes and fares better than the wild type), which Butte suggested is why this gene was never studied further for diabetes.
Knocking out this receptor now makes this an interesting therapeutic target, Butte said. Because a soluble form of Gene A protein can be detected in the blood, serum from patients was assayed for the diabetes marker, hemoglobin A1c (HbA1c), and Gene A protein. The data show that the lower the level of the receptor, the lower the level of HbA1c. Treatment for 7 days with therapeutic antibody to Gene A protein lowered the blood sugar of mice fed a high-fat diet.
In summary, Butte said, by using publicly available data that anyone can access, a protodrug, an antireceptor antibody, and a serum companion diagnostic have been developed, all in about 18 months. There are also human pathology data, mouse models, and human genetics data. Butte and colleagues have used the same approach for type 1 diabetes, small-cell lung cancer, and other diseases.
Genomic Nosology and Drug or Diagnostic Discovery
In another approach, Butte sought to find every microarray experiment that has looked at normal and disease samples in the same experiment. Many cancer researchers study metastatic versus nonmetastatic disease, but very few studies actually include normal controls. The first challenge was discovering that there were 200 words for “normal” in the repository (e.g., normal, vehicle, wild type, control, time zero, margins).
From the searches, a systematic classification of disease based on similarities in gene expression was assembled. Butte highlighted the fact that colon cancer and colon polyps clustered together based on molecular profiles (as would be expected since certain polyps are associated with cancer); however, cervical cancer was most similar to type 1 autoimmune polyglandular syndrome. In other words, cervical cancer was more similar to a very rare pediatric genetic disease that is not a cancer than it was to another cancer, colon cancer.
Around the same time, the Broad Institute released the Connectivity Map, a repository of genomewide transcriptional expression data from human cell lines treated with more than 1,500 different drugs at varying
doses. Matching the disease gene expression data with the drug gene expression data, Butte identified hundreds of correlations and is currently pursuing two: a seizure drug, topiramate, that may be effective for inflammatory bowel disease and an ulcer drug, cimetidine (Tagamet), that may be effective on lung adenocarcinoma (Sirota et al., 2011). Both have shown efficacy in animal models.
Again, Butte stressed, this entire work was done using publicly available data, and he urged investment in building these types of repositories, keeping them updated, and facilitating access to them.
In conclusion, Butte said, bioinformatics is more than just building tools. There are plenty of tools published every month, and the molecular, clinical, and epidemiological data already exist. There are thousands, perhaps tens of thousands, of repositories today. We can identify new diagnostics, therapeutics, and disease mechanisms by integrating datasets. We just need to demonstrate what can be done, he said.
Finally, there is a need for investigators who can imagine the basic questions to ask of these clinical and genomic repositories and are willing to make a career of studying publicly available data. Investigators need to move beyond the mindset that “if it’s not your own data you can’t trust it.” The data are just sitting there, Butte said, waiting for people to use them.
ADAPTING TO DATA-INTENSIVE, DATA-ENABLED BIOMEDICINE
Data represent the fastest-growing resource on Earth, said George Poste, chief scientist for the Complex Adaptive Systems Initiative at Arizona State University. The volume, variety, and speed with which new data are being generated in biomedicine are staggering, and the current computational power may not be sufficient. Data are global and range across multiple users and scales, from the molecular level to the patient in the clinic. The central challenge is how to integrate these data.
Data Production, Analysis, and Utilization in Biomedicine
Poste summarized the overarching themes in meeting the biomedical informatics challenges as the following:
• systems,
• scale, standards, and sharing,
• software, storage, and security,
• sustainability, and
• social issues (changing mindsets and behaviors).
With regard to data production in biomedicine, Poste said, there are more data, but many reported findings have not been validated, and there are issues with replication, suitability for a specified purpose (e.g., regulatory), and authenticity (e.g., information on the Internet). There are more powerful, high-throughput analytic tools, but we deploy them against small sets of samples, resulting in inadequate analytic and statistical rigor. Technology convergence and the creation of multidisciplinary datasets are handicapped by single-specialty silos. There are more participants, locations, and distributed data, but there is a pervasive lack of interoperable exchange formats and standards for data annotation, analysis, and curation. There is also a poor record of sharing.
Data analysis and utilization in biomedicine, Poste said, require more rapid, real-time data access, but data are often trapped in isolated and hierarchical databanks. There is a need for more quantification and precision analytics, but insufficient numbers of personnel are trained for large-scale data analysis. More complexity and uncertainty exist, but there are escalating gaps in institutional and individual cognitive and analytic capabilities to handle it. Finally, the rate of change in data is increasing, as is the rate at which our knowledge and competencies depreciate.
Most of the current approaches to bioinformatics and health care informatics lack the agility and extensibility to meet projected needs, whether in basic or clinical research. We need much more sophisticated approaches for end-to-end system design, Poste said. Systems must meet the needs of a multiplicity of end-user communities without creating new silos. Ours is a data-driven, data-enabled society. Most data are now fundamentally networked, and an increasing fraction of data is digital from the outset. But as datasets become ever larger, they become increasingly unmovable with the existing infrastructure. Sophisticated simulations and meta-analytics can amplify the data streams.
Poste referred to the “fourth paradigm” of scientific discovery espoused by computer scientist Jim Gray, which states that we are now in a period of data-driven knowledge, intelligence, and actionable decisions (following the earlier paradigms of experiment, theory, and simulation). The nature of discovery has moved from hypothesis driven to hypothesis generating, based upon the analysis of large datasets, and explanation
involves complex statistical probabilities instead of simplistic, unitary (particularly binary) values.
Having reviewed the gaps and challenges, Poste asked if we are still building systems and infrastructure that merely support the collection of data or are working toward an integrated knowledge ecosystem that supports moving the data from discovery to actionable intelligence that can drive a learning health care enterprise.
Importance of Having the “Right” Data in the System
Poste stressed the importance of pre-analytical variables such as rigorous selection of specimen donors, standardized specimen collection, and annotated health records. Most researchers, however, do not have access to highly standardized, stringently collected, and phenotyped patient samples. He quoted Carolyn Compton, former director of the NIH Office of Biorepositories and Biospecimen Research and now president and CEO of C-Path, who has stated in several venues that “the technological capacity exists to produce low-quality data from low-quality analytes with unprecedented efficiency . . . we now have the ability to get the wrong answers with unprecedented speed.” This is a pervasive problem in biomarker identification and validation, which suffers from a “small N” problem that leads to bias and overfitting. It has been suggested that more than 50 percent of the data from academic labs cannot be replicated by industry for new drug target validation and submission to the FDA (Ioannidis and Panagiotou, 2011; Mullard, 2011).
The ability of each individual to have his or her genome sequenced inexpensively will add to the complexity. He said that having this genome information will allow for modulation of gene expression that can be transmitted transgenerationally, altering the epigenome in the progeny. The 95 percent of the genome that is noncoding is also turning out to be profoundly important in regulating the other 5 percent, Poste said.
What is going to represent a complete and accurate analysis of genome sequence, architecture, and regulation, Poste asked, for the purposes of informing regulatory and clinical decisions? He cited a recent publication describing only 88 percent concordance of single-nucleotide variants when the same samples were analyzed on two different sequencing platforms (Lam et al., 2011). The FDA is currently reviewing validation issues for the clinical use of genome sequencing.
Researchers also can access numerous protein–protein interaction and
pathway databases, based on the critical assumption that the databases are accurate, he said, citing Schnoes and colleagues (2009), who describe inaccuracies and misannotations in large primary protein databases (including GenBank NR and TeEMBL).
As discussed by Leroy Hood (Chapter 3), mapping the dysregulation of biological networks in disease is a rational foundation for targeted drug discovery. The goal is to target an intervention where the network is being perturbed. However, these are complex, adaptive systems, and a unifocal point intervention in a network will almost certainly be compensated for or have a bypass circuit available. One approach is to try to define network choke points as targets, subverting the alternate compensatory pathways as well. He said a challenging question in cancer drug development is, “At what point does the level of network dysregulation eclipse any feasible approach to achieve a homeostatic reset with drugs?” In this regard, Poste suggested that diagnostic technologies for early detection may be the more prudent option for research investments.
In summary, Poste said that a huge amount of data being put into the public domain is not accurate. Moving forward, Poste listed the need for controlled vocabularies and ontologies; minimal information checklists and open source repositories; algorithms and source code for analytic tools; exchange formats and semantic interoperability; and cross-domain harmonization, integration, migration, and sharing. Ultimately, the only valuable data are validated, actionable data.
Computational Capabilities for Large Datasets
Other disciplines are skilled at handling big datasets, but Poste suggested that insularity makes us reluctant to look at, learn from, and import these approaches into biomedicine.
Most of us, he said, have been trained in “static world” biomedicine that involves conventional collaborations, traditional social and professional preferences and hierarchies, minimum patient input, and a general reluctance to share data. The world we live in, however, is increasingly dynamic. This includes web-based collaborations, fluid populations of diverse participants with many unanticipated productive inputs, the ability to mine huge amounts of public data, open source networks, and extended communities.
Despite this overall movement toward increased access, a 2009 review of the top 500 papers published in the 50 journals that had the highest impact factor found that only 9 percent had deposited the primary raw data
into a public database (Alsheikh-Ali et al., 2011). Of the portion of the 500 papers that were covered by a journal or funding agency data access policy, 59 percent were not compliant.
Access to the raw data and computer code is absolutely essential, Poste stressed, and new incentives are needed to encourage people to share data. There is a need for ways to ensure due credit, attribution, and citation of the original dataset when it is used by others. He said the greatest challenge, however, is how to drive molecular medicine and IT-centric capabilities in routine clinical medicine. Designing next-generation health IT systems that will comprehensively capture the genetic, biological, behavioral, social, environmental, and ecological factors relevant to disease risk, progression, and outcomes is extremely challenging. Electronic health records need to be thought of in a dynamic sense, rather than simply a digital version of the original fixed paper format. Most EHRs, however, are not designed to support secondary use of data. A comprehensive clinical data integration system would include, for example, current and planned clinical trials, observational data from the provider as well as patient-reported information, SEER data, mobile health or remote sensor data, and payer datasets.
The final reckoning for actionable data is regulatory science, Poste said. Yet while there are many references to personalized medicine in FDA strategic planning documents, Poste noted that there is scant reference to how the challenges of personalized medicine will be met, including the informatics needs.
We have the capability to create a new health care ecosystem from the convergence of technologies and markets, Poste said (Figure 5-1), but this depends upon cyberinfrastructure for both e-science and e-medicine. He referred participants to the recently released report from the National Science Foundation (NSF, 2012) on cyberinfrastructure needs for 21st century science and engineering. If we do have standardized, validated data, how do we move them? Most academic data remain isolated in laboratories or centers. Contemporary academia does not have the necessary connectivity (e.g., optical networks with 10,000 megabit per second transfer) or the computational capacity to gain access to the data it needs. There is a growing imbalance between the ability of the end-user population to access data and embrace their complexity and the ability (or lack thereof) of institutions to access and analyze large sets of data. Poste suggested that institutions that cannot harness big datasets will suffer “cognitive starvation” and relegation to competitive irrelevance in the scientific and engineering domains.
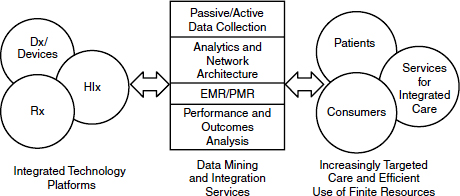
FIGURE 5-1 A new health care ecosystem arising from convergence of technologies and markets.
NOTE: Dx = diagnostics; EMR = electronic medical record; HIx = health information; PMR = personal medical record; Rx = (bio)pharmaceuticals.
SOURCE: Poste presentation (February 28, 2012).
Whether one has access to a high-performance computing center internally or participates with others in a consortium to create a cluster that provides high-performance computing capability, the cloud is a ubiquitous option. There are numerous commercial cloud computing services, and there is no single business model for cloud computing adoption at this stage. Poste noted that although the cloud provides on-demand access to large-scale, economically competitive computing capacity and flexibility, there are concerns about security, reliability, intellectual property, and regulatory compliance.
Moving forward begins with changing minds and changing behaviors to transition from informational silos to integrated systems. Technology is only the enabler, Poste said. We must embrace new organizational structures and must engage and educate multiple constituencies. The health care space will become increasingly decentralized with regard to how data are generated and increasingly centralized for data analytics and decision support. Data flows will increase as patient encounters with the health care system evolve from being episodic to more continuous, real-time monitoring.
In developing a new framework that can adapt to the scale and logistical complexity of modern biomedicine, Poste suggested that research
sponsors (e.g., NIH) focus less on single-investigator awards that offer incremental progress and instead seek to fund high-risk and high-reward projects with the potential for radical, disruptive innovation; that a single-discipline career focus be replaced with obligate assembly of diverse expertise for multi-dimensional engagement; that new study sections with broader expertise, including industry, be assembled; and that siloed datasets be abandoned for large-scale, standardized, interoperable open source databases with professional annotation, analytics, and curation.
Government has a vital role to play, Poste said, in the promulgation of standards, centralized coordination of resources, enforcement of data sharing, and proactive design of regulatory frameworks to address new technologies. Industry plays an important role by participating in pre-competitive private–public partnerships and by taking a proactive role in shaping new transdisciplinary education, training, and employment opportunities.
Ours is a world of massive data, Poste summarized, and to manage these data we need disruptive change and new products, services, and partnership models. He emphasized that moving forward will require courage to declare that radical change is needed; resilience to combat denial and deflection by entrenched constituencies; competitiveness and new participants who drive disruption at the margins or at convergence points (the voice of patients, payers, and new industrial participants will drive e-science and health IT); and accountability and responsibility, providing improved return on investment of public and private funding and addressing urgent societal and economic imperatives.
BIG DATA AND DISRUPTIVE INNOVATION: MODELS FOR DEMOCRATIZING CANCER RESEARCH AND CARE
More so than in any other area of health care, cancer research and cancer care are especially overloaded by data, said Jason Hwang, executive director of Health Care at the Innosight Institute. Before the advent of next-generation sequencing techniques, the doubling time of DNA sequencing data was about 19 months, slightly slower than the doubling time of hard-disk storage capacity (about 14 months). After the uptake of new sequencing technologies, however, the doubling time of sequencing data decreased dramatically to around 5 months (Stein, 2010). Sequence data are just one component of biomedical data. Combined with all of the data being generated in the clinic and in research, the volume of biomedical data, especially cancer-related data, is growing exponentially.
As an internist who subsequently earned a master of business administration, Hwang said that many of the problems facing biomedicine are common challenges that other industries have solved or are also trying to solve. The challenges of big data are not unique to health care, and studying the approaches used by industries (e.g., retail, airlines, banking) can be very informative, he said.
Learning from Users of Big Data in Diverse Non-Health Venues
The idea of using big data to make better decisions is not new and predates computing by a number of years, Hwang said. He provided one example of big data before the existence of computers that comes from the U.S. Navy. Matthew Fontaine Maury (1806–1873), referred to as the Pathfinder of the Seas and the Scientist of the Seas, is also considered the father of modern oceanography and naval meteorology. In his time, each ship charted its own course across the ocean, and in general, the experience of any given ship that made it across was not shared. Maury realized that data collected on a voyage (e.g., meteorological data, currents, winds) were trapped in silos (the ship’s log) and that these data could be integrated to create optimal routes for any ship on any given day. He created a standardized reporting mechanism that included a reward for any ship captain that submitted his log books along with the maps that Maury had provided. He then compiled these data and revised his maps for the next journey.
Another more recent example that Hwang described involves a group of mathematicians and computer enthusiasts who were dubbed “the gang that beat Las Vegas.” Until that time, betting lines and odds were set by individual bookkeepers who made predictions based on the (generally limited) information to which they had access, adjusting their betting lines based on trends and statistics and the bookkeeper’s intuition. Members of the computer group realized that if they had access to computing power, they could plug in all of the available data, more than any individual bookkeeper could ever hope to assemble and process, and probably do much better on the odds, Hwang explained. They collected data not just on last week’s game results, but on all of the opponent’s games, the weather, the latest injury reports, etc. Ultimately, the group fared so well that no one in Las Vegas would take its bets anymore, and a syndicate of people had to be created to place these bets. According to Federal Bureau of Investigation (FBI) records, between 1980 and 1985, the main members of the group amassed close to $14 million in profits (about $20 million to $25 million
in today’s dollars), with a return on investment of more than 10 percent. Because this number included only members of the core computer group and did not include many other bets placed by family members and friends with whom they shared odds data, the actual profits are likely quadruple that amount. Although they were indicted for other crimes (related to placing bets over the phone), the FBI was never able to charge them with the most serious crime of bookkeeping.
No matter the industry, data abound. It is what we are able to do with the data that is important, Hwang said. Many modern success stories of the use of big data are not particularly representative of their traditional industries, for example, Amazon.com (retailer, bookseller) and Netflix (video distribution). They excel at data collection and use it to drive decisions that improve their business model day after day.
For example, Hwang said that Netflix realized that a video rental service by mail is easily commoditized and has a low barrier to entry and that any number of start-ups could have entered the same space. What Netflix also realized was that it could collect data on people’s preferences and start making customized recommendations, making it the service of choice. Two-thirds of video selections made on Netflix are driven by the recommendations its software makes. Amazon collects data on what people decide to purchase, as well as what they clicked on and decided not to purchase, and uses these data to make recommendations of “you may also like. . . .” These data on unchosen clicks (or “data exhaust”) that were processed and incorporated into Amazon’s decision making are often thrown away by other companies, Hwang noted.
Another big data user, Google, developed its search engine in a way that was very different from anybody else, Hwang said. Google realized that if it gave you a list of ranked search results and you clicked on number four instead of number one, and dozens of other people did that as well, this was an opportunity to improve its search results (e.g., rank that number four result higher next time). Using this data exhaust, Google was able to create the best spell-checkers for nearly every language in the world. Every time someone misspells something, Google will suggest what it thinks they mean, but it gives them the opportunity to say, “No, I meant to spell it that way.” If enough people say that is how they intended to spell it, Google takes the opportunity to learn and improve its spell-checker. Google has built its translators in a similar learning fashion. Google is rapidly expanding its translation engines and capabilities in a way that is far cheaper, because it is essentially crowdsourcing its software construction, Hwang explained.
The real impact of big data, Hwang suggested, is that the data will allow better decision making based on computer algorithms, rather than relying upon just one expert’s individual opinion (which can vary widely among experts, especially in health care). Data-based algorithms could aid in choosing the most appropriate treatment when there are multiple to choose from, for example. This is not unlike the transformation in banking, Hwang said, where previously a loan officer had to base a decision about people’s ability to repay on factors such as the type of clothes they wore and the car they drove, their job and how long they worked there, marital status, and so forth. With better analytics, computerized credit reporting and monitoring services now help make that decision, in many cases replacing the human decision maker. This opens up the door to other opportunities, Hwang said, such as allowing consumers to see the criteria on which their creditworthiness is based and to optimize their credit score in order to get the best loans possible.
In the ability to use big data there is opportunity to create new economies. If we think of the marketplace as concentric circles, Hwang said, customers who have the most money and the most expertise are at the center. These are the people who are the early adopters of any new product or service in an industry. Moving outward in the rings, the amount of money and expertise diminishes, until the outermost circle represents the people with the least amount of money and the least amount of expertise, who are the last adopters of any product or service.
Using service industries as an example, Hwang explained that solutions have classically been very centralized. That is, if there is a problem that needs to be solved, we go to a source of the solution and pay that source for the expertise to help solve the problem. In essence, technologies have now extracted that expertise from the brain of the expert and embedded it in a piece of software, a tool, or a technology, such that anyone can use it. This commoditizes that expertise and democratizes access to industries by making it convenient and affordable. When booking travel, for example, no longer does one need to always call a travel agent. Most people are capable of making a simple booking using the tools available online. There are sources of decision support to help people make the right decision for their needs. While travel agents are still there to help with more complicated needs, there is choice in the marketplace, granted by technologies that have
commoditized travel expertise. This can be observed in many different industries. One can go to an accountant or choose to use TurboTax or some other type of software support that commoditizes the expertise of tax accountants. One can choose to go to a bank teller during banking hours or to an automated teller machine (ATM) whenever one wants.
Convenience and personal empowerment drive the disruption that we see in service industries, and Hwang said that this will be the case for health care as well. Think now of the centralized source not as a travel agent, an accountant, or a real estate agent, but as the local general hospital. Many health care services are either embedded within or revolve around the big general hospital in every local district. In this case, the goal of disruptive innovation is to suggest that the expertise and tasks being done in general hospitals today could be shifted outside such that people do not have to travel to the hospital for care. What services can be provided in an out-patient setting? What tools and technologies could equip a general practitioner to do what only groups of specialists would normally do in the wards of a hospital? Further, what is being done in the outpatient setting that could be delivered in a doctor’s office or even a kiosk and could be delivered not just by doctors but by nurse practitioners, pharmacists, physician assistants, and allied health professionals? Ultimately, what can be done in patients’ homes and managed by patients themselves? Disruptive innovation is about where health care can exist in a sustainable and affordable fashion in this country, Hwang said.
Big data can help to facilitate the decentralization of our highly centralized and expensive health care structure and help to emphasize prevention and wellness. Hwang listed some of the many different types of data-enabled business models that could help to achieve decentralized health care, including telehealth and e-visits, automated kiosks, home monitoring, wireless health devices, retail and worksite clinics, and others. All of these are enabled by technologies such as telecommunications and precision diagnostics as well as better decision-making tools that are derived from big data (Figure 5-2).
The goal is not to put hospitals and doctors out of business, Hwang stressed; however, health care delivery is a scarce resource that could well be supplemented by this approach. He likened the potential for decentralization in health care to what has happened in the legal world. Simple actions that used to be done by lawyers, such as trademark registration, small business incorporation, simple real estate leases, and legal discovery, for example, fall into a category that economists call “automating the automatable.”
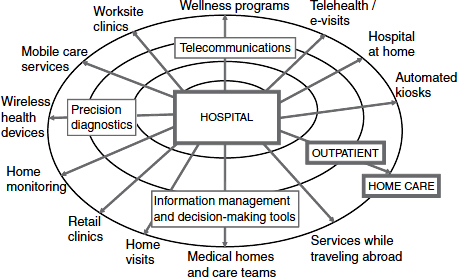
FIGURE 5-2 A new ecosystem of disruptive business models.
SOURCE: Hwang presentation (February 28, 2012).
For example, software programs can now assemble documents that used to require hours of work. When software can manage some of the tasks that used to be reserved only for professionals, it frees up the professionals so that they can spend far more time talking to their clients and focusing on higher-value work, where their expertise is really needed.
In summary, Hwang predicted that big data will transform cancer care and research first, because the data deluge in the field outpaces anything else in health care. Building EHRs and other data repositories is just the beginning; the truly sustainable value will be provided by enterprises capable of extracting wisdom from the data. The ultimate goal, he said, is to use big data to create the tools that will commoditize expertise and make care more accessible to more people.
Democratizing Big Data Informatics for Cancer and Other Therapeutic Areas
Kris Joshi, global vice president of health care for Oracle, expanded on the concept of democratization of informatics. With so many sources of data currently or soon to be available (e.g., the anticipated $100 genome, mobile health devices, imaging), the challenge is providing people with
tools that are affordable, readily available, and easily manageable so they can derive value from the data. Patients are interested in a value-based health system that consistently delivers new therapies and better care at a cost they can afford, but they do not want affordability at the expense of innovation, Joshi said. Similarly, patients want privacy protection, but they understand that collaboration across the life sciences and health care is necessary to achieve this innovation, and they do not want collaboration to be stopped under the guise of privacy protection.
Joshi likened informatics to an iceberg. The small tip that is visible is the data analysis and presentation that everyone is interested in doing. The challenge is the rest of the iceberg lurking below the surface—the data acquisition from myriad complex clinical, financial, administrative, and research source systems and the attendant cleansing, integration, and warehousing of these data. This is almost always underappreciated in terms of the magnitude of the work involved, Joshi said.
Informatics done right can transform health systems to a point where the insights coming out of EHR data can be translated within the institution into improved processes and procedures that reflect the best knowledge, not from 10 years ago but from 2 weeks ago, because that knowledge was incorporated in a learning health care workflow (Figure 5-3). A closed-loop learning health care system empowers clinicians and nurses who can look at the data and effect change. However, current health IT systems do not have the capability to support this.
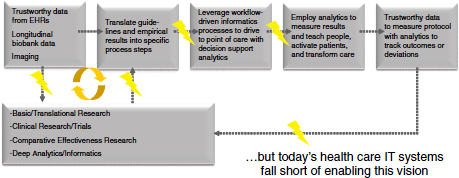
FIGURE 5-3 Learning health care paradigm supported by robust, interoperable informatics.
NOT: EHR = electronic medical record.
SOURCE: Joshi presentation February 28, 2012. Reprinted with permission from Kris Joshi and Brett Davis.
The fundamental problem is turning the growing mass of data into transformational insights. Because biases in the data can lead different individuals to reach different conclusions, transparency in how one derived the insight from the data is critical, Joshi stressed. These challenges are general informatics problems (i.e., not specific to cancer or even specific to research), and they should be solved like general informatics problems, Joshi said. Also, as noted by others, much can be learned from other industries. He added that in trying to solve one problem, you often find that some of the solutions also solve another problem elsewhere in the health care ecosystem (e.g., addressing a research informatics issue also ends up solving a payer issue).
Many decisions are now made using analytics, which must be reproducible if one is to be able to justify the decision. Analytics, Joshi said, is a niche industry where only those who know how to deal with the data can derive value from it, and this keeps analytics from being a highly used tool.
One ongoing challenge in analytics has been integrating and normalizing data from the different source systems (e.g., clinical, financial, administrative, research). A lot of data quality problems start at this point, Joshi noted. The informatics requirements for data integration, validation, and normalization are not trivial and require an enterprise approach. However, once a core enterprise analytics platform is in place, numerous applications can be developed that allow researchers, clinicians, administrators, and others to analyze those high-quality data. A common infrastructure can also support multiple secondary uses of data and thereby lower costs. For example, data can be used for clinical trial optimization, decreasing time lines and enhancing efficiency and accuracy. Joshi suggested that in addition to thinking about secondary uses of data, we should consider secondary uses of IT infrastructure as another way to reduce overall costs. Specifically, creating an entire system focused on cancer runs the risk of spending too much time, effort, and money; not focusing on innovation; and most likely duplicating existing systems to some extent.
In closing, Joshi mentioned one example of the regional initiatives that are gearing up to enable collaboration across the health care ecosystem: the Partnership to Advance Clinical electronic Research (PACeR) initiative, which is a public–private partnership between New York State hospitals and life sciences companies.
Consumers as Disruptive Innovators
The Patient Protection and Affordable Care Act (ACA) put in place incentives for coordination of care and for innovation around patient engagement, explained Farzad Mostashari, national coordinator for health information technology in the Office of the National Coordinator for Health Information Technology (ONC). Insurers no longer want to pay for care as piecework, he said. Per the ACA, if hospitals can provide care for Medicare patients that is more coordinated, they can share in any resulting savings.
Mostashari listed three elements that will aid in the successful coordination of care: (1) the technology infrastructure for care coordination exists, (2) there is a strong business case for care coordination and patient engagement, and (3) there is movement toward the democratization of information and information tools. The consumer is the ultimate disrupter here, he said.
As a case example, Mostashari offered his thoughts on some of the reasons Google Health failed. While certain elements are specific to Google, others are more generalizable, and we can learn from them. First, people had to spend hours typing in their own information. Per HIPAA, patients have a right to get a copy of their own medical information (and certain legislative provisions may require that it be provided electronically and within a specified time period). It is legally possible for people to get access to their own records, but it should also be easy to do. People are often uncomfortable asking for their records, worrying that asking is in some way challenging the doctor, Mostashari added. This attitude needs to change, he said. Knowing your information is part of what being a good patient is about.
Another problem with Google Health, Mostashari said, was that once people typed all of their information in, they could not do much more with it except read or print it. People want to be able to find a clinical trial, learn what the side effects of their medications are, get a second opinion, see their images, share their data, or keep abreast of the latest research on their cancer. They want to find resources and find other people like themselves.
The progress in health IT for doctors and hospitals is exciting, Mostashari said, but what is more exciting is the disruptive possibilities of consumers and their caregivers as the nexus where information comes together and is used to generate new knowledge.
THE EHR AND CANCER RESEARCH AND CARE
Mostashari said that when he joined ONC in 2009, about 10 percent of hospitals and 20 percent of primary care providers used a basic EHR system. The Health Information Technology for Economic and Clinical Health Act (HITECH) of 2009 put in place financial incentives for doctors and hospitals to adopt and meaningfully use EHRs and created some of the digital infrastructure to help small practices. By 2011, 37 percent of hospitals were using an EHR system. Mostashari anticipated that by 2013, it would be more than 50 percent.
The EHR is not just an office system, he said. ONC has developed interoperability standards to facilitate sharing and has a program to certify EHR systems that conform to these standards. Per the standards, information on clinical care, medications, procedures, and other data will be in a standard XML format with tags to tell users what the data elements are and where they go. Data will be shared when patients transition from care setting to care setting. Data will also be shared with the patient, because this is one of the requirements for meaningful use of an EHR, Mostashari noted.
The EpicCare System as a Model for the Uses of EHRs in Cancer Research and Care
Sam Butler, a physician and member of the clinical informatics team at software developer Epic, described the features of the EpicCare system as a model for the uses of EHRs in cancer research and care. He estimates that EpicCare has been used for somewhere between 108 million and 142 million patients in the United States, across 270 health care clients. An available add-on module to the main EHR is Beacon, Epic’s oncology information system. Currently, Butler explained, Beacon is primarily for chemotherapy management. Functions include staging, problem lists, protocols, treatment planning, review and release, pharmacy verification, electronic medication administration record (eMAR), flow sheets, and reporting. Of the 270 current EpicCare EHR users, 140 of them are actively installing Beacon.
The system is extensible, and it is easy for an institution to add information to a set of problems. The model system includes 250 protocols, both standardized regimens and research protocols. These are not meant to be a review of the literature, Butler noted, but are representative protocols
using standard protocol language to serve as starting points. Customers are required to review the protocols, validate them, and make them their own protocols. A protocol can be adapted as a treatment plan for a particular patient. Once a treatment plan is created, nurses review the information when a patient arrives, and the nurse or pharmacist releases the orders, which then go through a process of pharmacy verification. Administration of treatment is instantly documented in the eMAR, often by using bar coding. Everything is captured in the flow sheet, which informs and supports physician decision making regarding the next round of treatment. Then the cycle of treatment plan, pharmacy verification, eMAR documentation, and flow sheet begins again. Beacon can also capture discrete data such as reasons for changing a treatment plan or discontinuing it.
This cyclical system works well in a large institution where the oncologists, pharmacists, and nurses all are in the same organization. However, if the physicians are in their community practice and they have another EHR, or another program to create their treatment plan, there is no standard to communicate that treatment plan to a hospital, infusion center, or pharmacist with a different system.
Application and workbench data are exported nightly or more frequently and deposited into a reporting database that is relational and that can be accessed by analytic tools. The data can also be combined with other data warehouses or biorepositories in other systems. The database can be used to create custom data marts that are customer developed and owned.
In the future, the system will allow more patient-entered data. Epic is working on ways to engage patients, such as sending out surveys, aggregating the data that are reported, and displaying that information in graphs and reports to help patients make an informed decision about their treatment by showing them what other patients have experienced. The challenge, Butler noted, will be getting enough patient data to make those graphs meaningful and significant. Epic is also working on incorporating genomics and adding the ability to document the care a patient received before entering this system, for a complete oncological history.
Butler noted several challenges in moving forward, including encouraging physicians to see the value of using an EHR (he said physicians are very concerned that using an EHR is going to slow them down), interoperability, and management of big datasets.
CANCER CENTER–BASED NETWORKS FOR HEALTH RESEARCH INFORMATION EXCHANGE
Following on his discussion of the informatics challenges facing cancer centers (Chapter 2), William Dalton, of the Moffitt Cancer Center, offered an aspirational model for a research and health care information exchange network that would allow many different partners and stakeholders to participate, contribute, and benefit (Figure 5-4). Expanding on the research information exchange he described earlier (see Figure 2-3), the data warehouse would become a federated network information system.
The development of personalized cancer care relies on the efforts of many people contributing to the continuous cycle of discovery, translation, and delivery of health care. Data networks allow for the discovery of associations between specific molecular profiles and clinical information from individual patients, leading to new knowledge that can be translated into more personalized cancer care.
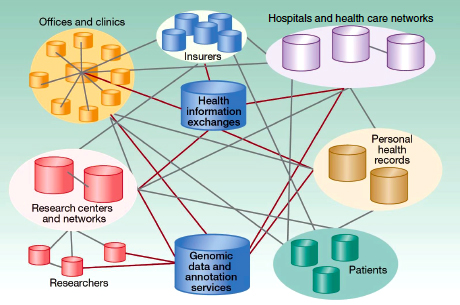
FIGURE 5-4 Designing a new federated research and health care network model.
SOURCE: Dalton et al., 2010. Reprinted with permission from the American Association for Cancer Research.
One approach to facilitate personalized care is to ask patients to participate as partners in the care journey. As an example, Dalton described the Moffitt “Total Cancer Care Protocol.” This IRB-approved observational study protocol includes three critical questions for the patient:
1. Can we follow you throughout your lifetime? (With the goal of entering any health care data into a central data warehouse)
2. Can we study your tumor using molecular technology? (With the goal of entering genetic and genomic data into a central data warehouse)
3. Can we recontact you? (For purposes of sharing information that might be of importance to you, such as a clinical trial designed for patients like you)
As part of a public–private partnership with Merck, the protocol is currently open in a consortium of 18 sites in 10 states, all using the same protocol and consent and following standard operating procedures for tumor collection and data aggregation, and many using the same central IRB. Now finishing its sixth year, more than 85,000 people have been enrolled to date. More than 32,000 tumors have been collected, all clinically annotated according to standard operating procedures, and almost half have been profiled.
Dalton noted that in its early stages, the database was more of a repository than a warehouse, with some of the queries taking weeks, if not months, and Moffitt sought expert assistance from Oracle, TransMed, and Deloitte. Through this strategic partnership, an integrated health research information platform was developed that creates the real-time relationships and associations from disparate data sources that are needed to create new knowledge for improved patient treatments, outcomes, and prevention. Critical to this endeavor was harmonization of the data through creation of a data dictionary. In addition to defined elements, the dictionary also incorporated a means of measuring the quality and veracity of the data. Because patient information resides in many sources, often with different identification numbers, the first challenge was to create a means of cohort identity. Harmonization of the data takes place in the data factory before the data are placed in a warehouse, where they can then be queried for different uses by different partners and stakeholders. The data warehouse is a robust, scalable dataset of oncology patients, Dalton said, and queries are done in real time.
As one example, Dalton described how the data warehouse could be queried for cohort identification. One could, for example, query for females diagnosed and/or treated at Moffitt between 2005 and 2009 with breast cancer, with the histology of infiltrating ductal carcinoma, not otherwise specified, who were stages III to IV at diagnosis and who were estrogen receptor positive, with a documented family history of breast cancer. Out of more than 212,000 female patients in the database, the successive real-time queries identified a cohort of 25. In a second example, Dalton demonstrated how queries could identify patients meeting select clinical trial criteria who were eligible and available at a particular consortium site and who also had banked tissue samples that could be assayed.
It is one thing to be able to do health research information exchange at a single institution or within a defined consortium, but it is quite another, Dalton said, to manage this on a national scale. He proposed a national health and research information exchange, incorporating regional “hub and spoke” platforms, with cancer centers as the hub and their individual colleagues within the community contributing data and having access to those data.
This federated framework could facilitate many aspects of cancer research, including basic science, translational research, drug discovery and development, clinical trials, companion diagnostics, comparative effectiveness and outcomes research, postmarketing surveillance, and others. The goal, as with the other models discussed earlier, is a rapid-learning information system. Each patient added iteratively improves the learning process.
In summary, Dalton said that the guiding principles for developing cancer center collaboration are inclusiveness, accessibility of data (especially real-time access through a research information exchange), and public–private partnerships to achieve long-term sustainability.
During the discussion, panelists offered additional comments on pathways forward and other examples of instructive models in other domains.
Brandon Hayes-Lattin, cancer survivor, cancer researcher, and senior medical adviser for the Lance Armstrong Foundation, said that the foundation’s Share Your Story campaign has transformed cancer survivorship through individual patients posting personal stories so that others might be helped by them. Along these same lines, he supported developing tools where patients could contribute their clinical data to a shared resource and also access aggregated clinical data to guide their personal decision making. He noted that the Livestrong constituency was surveyed regarding sharing their data, and 87 percent of the 8,500 respondents agreed that researchers should have the ability to review their information as long as it is not directly linked to them. Further, 71 percent felt that their data was safer when stored electronically than on paper.
Patients are looking for a range of things, Hayes-Lattin said. They want to trust their physician, but they want to double-check, too. Getting a second opinion requires their raw data, not just the interpretations. Patients also say that they want to be able to find resources. There are a lot of resources available for cancer patients, but it can be hard to find them, he said. It is important for patients to be able to put their situation into context in the larger world of cancer, to learn from the experiences of others.
Bradford Hesse, chief of the NCI Health Communication and Informatics Research Branch, added that a significant portion of traffic to government health websites such as NCI or the Centers for Disease Control and Prevention (CDC) involves patients. Patients are engaged and activated, and they need to have a health system that is prepared to help them. He also referred to the SHARP initiatives (Strategic HIT Advanced Resource Projects), some of which focus on the security and privacy of health IT.
Providing a Substrate for Innovation
Hwang mentioned the role of government in creating a substrate for innovation in the private sector. For example, no private-sector company or start-up was likely going to invent the Internet, but a government agency with the will and the resources did create such a network and opened it up to the private sector for use, essentially launching a new economy. Another example was the launch of global positioning system (GPS) satellites and allowing the private sector to use those data to generate whatever products and services it could imagine. Most companies would not try to launch
their own satellites, but many have created innovative products using the data. Poste added that the Internet predecessor, Advanced Research Projects Agency Network (ARPANET), and GPS technology were both products of the military sector. A primary factor driving military technology is a perception of an existential threat. There are similar threats in biomedicine, Poste said, including the economics and viability of the health care system at large and the threat at the level of the individual facing a terrible disease.
In these and other instructive precedents, government has taken the lead in recognizing the threat, recognizing the need for a coherent systems-based approach, and allowing the mobilization of the creativity community (i.e., innovation from the bottom up). Poste argued that what is needed is a combination of national leadership, the courage to acknowledge that much of the system is broken, and the willingness to allow individual, bottom-up contribution and participation in the larger infrastructure.
Mining Data to Assess the Quality of Cancer Care
Allen Lichter, chief executive officer of the American Society of Clinical Oncology (ASCO), noted that ASCO is interested in informatics from a quality-of-care perspective. Quality monitoring is especially important in oncology. As mentioned earlier, most cancer patients are treated in community settings where the vast majority of oncologists are generalists. In addition, diagnostic and therapeutic options are increasing rapidly, and physicians need to make sure that they are up-to-date.
The ASCO Quality Oncology Practice Initiative (QOPI) was designed to monitor the quality of cancer care and has been in place for about 10 years. One concern, Lichter said, is that it assesses quality retrospectively (reviewing cases from 6 months or more prior), uses sample cases, has a limited number of measures, and is manual (taking close to an hour per chart). ASCO is looking to evolve this into a real-time electronic system, reviewing consecutive cases, monitoring the full spectrum of care, and providing decision support.
Butler noted that researchers make their living from their data and they are understandably hesitant to give it up. However, over the course of the workshop there was wide support for the concept that the data should be used to benefit society and should be made available for broad use. Butler
suggested starting with data sharing efforts for just a few diagnoses. For example, in pediatric gastroenterology, 30 institutions provide all of their data on inflammatory bowel disease patients to one institution that then aggregates the anonymized data. They did not try to start too big. He added that the United Network for Organ Sharing (UNOS) has done the same with transplant data.
Murphy offered pediatric oncology as a model for collaborating. The field of pediatric oncology has a long history of collecting data and using the information in a virtuous cycle to inform and improve the next generation of care. The field is less competitive and more geared toward sharing.
Education, Training, and Funding
Mia Levy, director of cancer clinical informatics at the Vanderbilt-Ingram Cancer Center, said that there is a real need for more people who are computationally oriented in medical schools. There is also a need for career paths and leadership positions for people who are trained in both medicine and informatics. In funding these researchers, Levy said that review committees need to be receptive to the potential of informatics and the value of observational data.
If Data Are Available, Users Will Come
Levy referred to how Butte and others have made use of publicly available data repositories of cancer information. If data are available, people will start using them for a variety of purposes. To facilitate more use of the data sources that are already available, she said there has to be increased accessibility, more sharing of data, and an invitation to the broader informatics community to come to the table and start working on problems that are important to cancer.
Alsheikh-Ali, A. A., W. Qureshi, M. H. Al-Mallah, and J. P. Ioannidis. 2011. Public availability of published research data in high-impact journals. PLoS ONE 6(9):e24357.
Butte, A. J. 2008. Translational bioinformatics: Coming of age. Journal of the American Medical Informatics Association 15(6):709-714.
Dalton, W. S., D. M. Sullivan, T. J. Yeatman, and D. A. Fenstermacher. 2010. The 2010 Health Care Reform Act: A potential opportunity to advance cancer research by taking cancer personally. Clinical Cancer Research 16(24):5987-5996.
Ioannidis, J. P., and O. A. Panagiotou. 2011. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. Journal of the American Medical Association 305(21):2200-2210.
Kodama, K., M. Horikoshi, K. Toda, S. Yamada, K. Hara, J. Irie, M. Sirota, A. A. Morgan, R. Chen, H. Ohtsu, S. Maeda, T. Kadowaki, and A. J. Butte. 2012. Expression-based genome-wide association study links the receptor CD44 in adipose tissue with type 2 diabetes. Proceedings of the National Academy of Sciences 109(18):7049-7054.
Lam, H. Y., M. J. Clark, R. Chen, R. Chen, G. Natsoulis, M. O’Huallachain, F. E. Dewey, L. Habegger, E. A. Ashley, M. B. Gerstein, A. J. Butte, H. P. Ji, and M. Snyder. 2011. Performance comparison of whole-genome sequencing platforms. Nature Biotechnology 30(1):78-82.
Mullard, A. 2011. Reliability of “new drug target” claims called into question. Nature Reviews Drug Discovery 10(9):643-644.
NSF (National Science Foundation). 2012. Advanced computing infrastructure: Vision and strategic plan. NSF Document nsf12051. http://www.nsf.gov/pubs/2012/nsf12051/nsf12051.pdf (accessed April 27, 2012).
Patti, M. E., A. J. Butte, S. Crunkhorn, K. Cusi, R. Berria, S. Kashyap, Y. Miyazaki, I. Kohane, M. Costello, R. Saccone, E. J. Landaker, A. B. Goldfine, E. Mun, R. DeFronzo, J. Finlayson, C. R. Kahn, and L. J. Mandarino. 2003. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proceedings of the National Academy of Sciences 100(14):8466-8471.
Schnoes, A. M., S. D. Brown, I. Dodevski, and P. C. Babbitt. 2009. Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLoS Computational Biology 5(12):e1000605.
Sirota, M., J. T. Dudley, J. Kim, A. P. Chiang, A. A. Morgan, A. Sweet-Cordero, J. Sage, and A. J. Butte. 2011. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science Translational Medicine 3(96):96ra77.
Stein, L. D. 2010. The case for cloud computing in genome informatics. Genome Biology 11(5):207.




























