Using Emerging Science and
Technologies to Address Persistent
and Future Environmental Challenges
Chapter 2 discussed some of the broad drivers and challenges that are inherent to the mission of the US Environmental Protection Agency (EPA) today and in the future. Remarkable progress has been made in the last several decades in the development of new scientific approaches, tools, and technologies relevant to addressing those challenges. The purpose of this chapter is to highlight new and changing science and technologies that are or will be increasingly important for science-informed policy and regulation in EPA.
New tools and technologies can substantially improve the scientific basis of environmental policy and regulations, but it is important to remember that many of the tools and technologies need to build on and enhance the current foundation of environmental science and engineering in the United States. In addition, addressing the complex “wicked problems” facing EPA today and in the future requires not only new science and technology but a more deliberate approach to systems thinking, for example, by using frameworks that strive to integrate a broader array of interactions between humans and the environment. From the perspective of scientific advances relevant to the future of EPA, it will be increasingly important that all aspects of biologic sciences and environmental sciences and engineering—including human health risk assessment, microbial pathogenesis, ecosystem energy and matter transfers, and ecologic adaptation to climate change—be considered in an integrated systems-biology approach. That approach must also be integrated with considerations of environmental, social, behavioral, and economic impacts.
A SIMPLE PARADIGM FOR DATA-DRIVEN, SCIENCE-INFORMED DECISIONS IN THE ENVIRONMENTAL PROTECTION AGENCY
New scientific advances, including the development and application of new tools and technologies, are critical for the science mission of EPA. Effec-
tive science-informed regulation and policy aimed at protecting human health and environmental quality relies on robust approaches to data acquisition and to knowledge generated from the data. For science to inform regulation and policy effectively, a strong problem-formulation step is needed. Once a problem is formulated, EPA scientists can evaluate what types of data are needed and then determine which available tools and technologies are appropriate for gathering the most robust data (see Figure 3-1). As described in detail in this chapter, management and interpretation of “big data” will be a continuing challenge for EPA inasmuch as new technologies are now capable of quickly generating huge amounts of data. Senior statisticians are needed in the agency to help analyze, model, and support the synthesis of that data. In many instances, large amounts of data are directly acquired as a component of hypothesis-driven research. However, many new technologies are also used for discovery-driven research— that is, generating large volumes of data that may not be a derivative of a clear, hypothesis-driven experiment, but nevertheless may yield important new hypotheses. In both instances, the data themselves do not become knowledge that can be applied as solutions to problems until they are analyzed and interpreted and then placed in the context of an appropriate problem or scientific theory. As depicted in Figure 3-1, there are iterations and feedback loops that must exist, particularly between data acquisition and data modeling, analysis, and synthesis.
The generation of knowledge, which can take many forms depending on the question being addressed and the nature of the data, ultimately serves as the basis of science-informed regulation and policy (see Figure 3-1). The committee recognizes that scientific data constitute one—albeit important—input into decision-making processes but alone will not resolve highly complex and uncertain environmental and health problems. Ultimately, environmental and health decisions and solutions will also be based on economic, societal, and other considerations apart from science. They need to take into account the variety and complexities of interactions between humans and the environment. But with better scientific understanding, regulations and other actions can be more effective and can have better and more cost-effective outcomes, such as improved human health and improved quality of ecosystems and the environment.
In accordance with the above discussion, it is imperative that EPA have the capacity and knowledge to take advantage of the latest science and technologies, which are always changing. The remainder of the chapter highlights a number of scientific and technologic advances that will be increasingly important for state-of-the-art, science-informed environmental regulation. It also includes several examples of how emerging science, technologies, and tools are transforming the way in which EPA will use data to address important regulatory issues and decision-making, and they demonstrate the need for a systems approach to addressing these complex problems. The chapter has been organized in parallel to the challenges identified in Chapter 2. The main topics that will be discussed are tools and technologies to address challenges related to 1) chemical exposures, human health, and the environment; 2) air pollution and climate
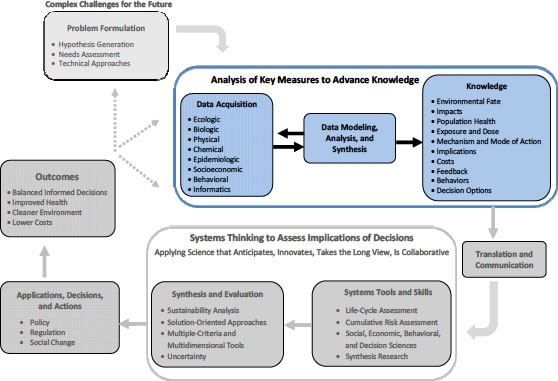
FIGURE 3-1 The iterative process of science-informed environmental decision-making and policy. The process starts with effective problem-formulation, which drives both the experimental design and the selection of data to be acquired. Modeling, synthesis, and analysis of the data are necessary to generate new knowledge. Only through effective translation and communication of new knowledge can science truly inform policies that can generate actions to improve public health and the environment. An evaluation of outcomes is an essential component in determining whether science-informed actions have been beneficial, and it, in turn, adds to the knowledge base.
change; 3) water quality and nutrient pollution; and 4) shifting spatial and temporal scales. The chapter ends with a section on, “Using New Science to Drive Safer Technologies and Products”, which discusses ways in which EPA can prevent environmental problems before they arise.
The examples in this chapter are not intended to be comprehensive; rather, they are provided to illustrate from different perspectives the many ways in which new advances in science, engineering, and technology could be embraced by the agency, its scientists, and regulators to ensure that the agency remains at the leading edge of science-informed regulatory policy to protect human health and the environment. Having assessed EPA’s current activities, the committee notes that EPA is well equipped to take advantage of most of the new scientific and technologic advances and that, in fact, its scientists and engineers are leaders in some fields.
New technologies will be important to EPA for identifying chemicals in the environment, understanding their transport and fate in the environment, assessing the extent of actual human exposures through biomonitoring, and identifying and predicting the potential toxic effects of chemicals. Current and emerging tools and technologies related to these topics are discussed in the sections below.
Identifying Chemicals in Environmental Media
Analytic chemistry continues to improve at breakneck speed, and analytic determinations for both metals and organic chemicals have improved exponentially. Chemicals can now be detected at ever lower concentrations. For some organic chemicals, such as chlorinated dioxins, standard EPA methods include the routine measurement of samples in parts per quadrillion (ppq) or picograms per liter (pg/L) (EPA 1997), which allows risk managers to characterize lifetime uptake of exposure to various carcinogens and daily uptake rates in chronic hazard quotient assessments of chemicals that were not previously detectable. Simply being able to measure concentrations of chemicals in environmental media or blood confronts EPA with new decisions on whether to set maximum contaminant levels in drinking water or allowable daily intakes in food or whether to allow states to do so independently if health effects are uncertain.
As the public learns about new methods of detection of chemicals in, for example, their blood, their children’s blood, and the environment (water, air, and soil), questions arise as to what such occurrences mean. Of course, the simple detection of chemicals in relevant receptors does not necessarily imply any human health or ecologic effects. To evaluate the health implications of chemical
exposures throughout the range of exposure levels, sufficiently large epidemiologic studies that incorporate state-of-the-art analytic methods are needed (see the section “Applications of Biomarkers to Human Health Studies”). But, even when biologic effects are not evident (and in special cases of hormesis when there are potentially beneficial effects), the challenge for EPA is to provide meaningful and relevant information to potentially affected parties.
It is now possible, while testing for emerging contaminants of interest and their metabolites, to monitor the effluent of a publicly owned wastewater-treatment plant and determine trace quantities and metabolites of substances— such as pharmaceuticals (licit and illicit), personal-care products, and hormones (natural and synthetic)—that are being used and disposed of or excreted by people in each town (Zegura et al. 2009; Jean et al. 2012; Neng and Nogueira 2012). The mass emission factors per capita can be calculated for the chemicals without determining individual household use. However, without better knowledge of the environmental and human health risks of such low-dose exposures, the advanced detection capabilities do not necessarily help the agency to interpret the results or to protect human health and the environment more effectively. One example is mercury. On one hand, from a toxicologic standpoint, mercury is one of the most studied elements (Schober et al. 2003; Jones et al. 2010). On the other hand, it is still difficult to make a conclusive assessment of the health effects of mercury emitted into the environment (EPA 2011a). Finding cost-effective research opportunities for connecting data on environmental chemicals with environmental and health outcomes can contribute to an increase in knowledge and can inform policy.
Fate and Transport of Chemicals in the Environment
EPA has long been recognized as a leader in developing computer models of the fate and transport of chemical contaminants in the environment, a key component in constructing models of human exposure and health outcomes, as well as in source attribution for ecologic and human endpoints. It develops and supports models for both scientific purposes and application in environmental management. Although many of its models are well established and now backed by years of application experience, EPA and the broader environmental-modeling community face challenges to improve spatial and temporal resolution, to account for stochastic environmental behaviors and for modeling uncertainties, to improve the characterization of transfers between environmental media (air, surface water, groundwater, and soil), and to account for feedback between contaminant concentrations and environmental behavior (for example, the effects of such short-lived radiative-forcing agents as ozone and aerosols have on climate change). Furthermore, sources, properties, and behaviors of some contaminants remain poorly understood, even after years of study. EPA also faces significant challenges and opportunities for integrating models with data from new monitoring systems through data assimilation and inverse model
ing techniques. Specific examples of ways in which new approaches to environmental fate and transport modeling are enhancing the understanding of health and ecologic impacts of pollutants are provided in the section on “Tools and Technologies to Address Challenges of Air Pollution and Climate Change”.
Assessing the Extent of Human Exposures Through Biomonitoring
Historically, exposure research in EPA has focused on discrete exposures—in external or internal environments, concentrating on effects from sources or effects on biologic systems, and on human or ecologic exposures— one pollutant or stressor at a time. Tools and methods have evolved for undertaking those specific challenges, but targeted approaches have led to sparse exposure data (Egeghy et al. 2012).
The broader availability and ease of use of advanced technologies are resulting in a profusion of data and an overall democratization of the collection and availability of exposure data. The US Centers for Disease Control and Prevention (CDC) National Health and Nutrition Examination Survey (NHANES) alone has provided one of the most revealing snapshots of human exposures to environmental chemicals through the use of biomonitoring (CDC 2012). The collaboration between CDC and national and international organizations quickly expanded the breadth and depth of data available at the population and subpopulation level. That rapid progress was predicated on the availability of better analytic methods and a national commitment to generate baseline data.
Scientific and technologic advances in disparate fields—including computational chemistry, climate change science, health tracking, computational toxicology, and sensor technology—have provided unprecedented opportunities to address the needs of exposure research. Many of the tools are more accessible and easier to use than earlier ones and are slowly being deployed by researchers and stakeholders, such as state agencies and public-interest groups. For example, advances in personal environmental monitoring technologies have been enabled because people around the world routinely carry cellular telephones (Tsow et al. 2009). Those devices may be equipped with motion, audio, visual, and location sensors that can be controlled through wireless networks. Efforts are underway to use them to create expanding networks of sensors to collect personal exposure information.
As discussed in Chapter 2, biomonitoring for human exposure to chemicals in the environment has provided a new lens for understanding population exposures to toxicants. Although the analytic and technical methods discussed to measure human exposure to environmental toxicants will continue to improve, without better information to understand whether the dose is of sufficient magnitude to cause an effect, simply identifying the presence of a toxic substance may raise more questions than it answers. Therefore, there are continuing advances needed to measure and understand the burden of chemicals and their metabolites in the human body.
Recent advances in microchip capillary electrophoresis for separation and identification of nucleotides, proteins, and peptides and advances in spectrometrics, such as nuclear magnetic resonance imaging and mass spectrometry, have changed the nature of health effects monitoring. These technologic advances— especially in genomics, proteomics, metabolomics, bioinformatics, and related fields of the molecular sciences (referred to here collectively as panomics)— have transformed the understanding of biologic processes at the molecular level and should eventually allow detailed characterization of molecular pathways that underlie the biologic responses of humans and other organisms to environmental perturbations. Advances in “–omics” technologies provide EPA with a better understanding of mechanistic pathways and modes of action that can support the risk assessment process. Also, the integration of those technologies with population-based epidemiologic research can contribute to the discovery of major environmental determinants, dose-response relationships, mechanistic pathways, susceptible populations, and gene-environment interactions for health effects in human populations. Appendix C discusses some of the recent advances in -omics technologies and approaches, their implications for EPA, where EPA is at the leading edge of applying the technologies to address environmental problems, and where EPA could benefit from more extensive engagement.
New high-throughput -omic and biomonitoring technologies are providing a greater number of potential biomarkers to assess multiple exposures simultaneously over the course of a lifetime. The biomarkers address exposures to a wide variety of stressors, including chemical, biologic, physical, and psychosocial stressors. The exposome is now being presented as a unifying concept that can capture the totality of environmental exposures (including lifestyle factors, such as diet, stress, drug use, and infection) from the prenatal period on by using a combination of biomarkers, genomic technologies, informatics, and environmental exposures (Figure 3-2) (Wild 2005; Rappaport and Smith 2010; Lioy and Rappaport 2011). The exposome, in concert with the human genome and the epigenome, holds promise for elucidating the etiology of chronic diseases and relevant contributions from the environment (Rappaport and Smith 2010). The concept of the exposome will be of particular value to EPA in assessing and comparing potential health and environmental consequences of individual chemical exposures against previously identified risks. It may also allow for more carefully designed and rational experiments to evaluate potential chemical interactions that contribute to the exposome of individuals or populations.
Exposure information is a key component of prediction, prevention, and reduction of environmental and human health risks. Exposure science at EPA has been limited by the availability of methods, technologies, and resources, but recent advancements provide an unprecedented opportunity to develop higher-throughput, more cost-effective, and more relevant exposure assessments. Research in this field is funded by other federal agencies and international programs, such as the National Institute of Environmental Health Sciences Expo-
sure Biology Program; the National Science Foundation Environmental, Health, and Safety Risks of Nanomaterials Program; and the European Commission’s exposome initiative. Those organizations provide valuable partnership opportunities for EPA to build capacity through strategic collaborations. Moreover, an integral need for EPA in the future will be to develop processes and procedures for effective public communication of the potential public health and environmental risks associated with the increasing number of chemicals, both old and new, that will undoubtedly be identified in food, water, air, and biologic samples, including human tissues. Risk communication strategies should include the latest approaches in social, economic, and behavioral sciences, as discussed in Chapter 5.
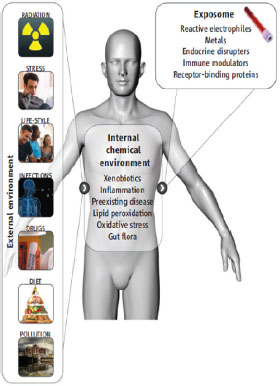
FIGURE 3-2 Characterizing the exposome. The exposome represents the combined exposures from all sources that reach the internal chemical environment. Examples of toxicologically important exposome classes are shown. Biomarkers, such as those measured in blood and urine, can be used to characterize the exposome. Source: Adapted from Rappaport and Smith 2010.
Applications of Biomarkers to Human Health Studies
Epidemiologic research plays a central role in assessing, understanding, and controlling the human health effects of environmental exposures. In 2009,
the National Research Council (NRC) report Science and Decisions: Advancing Risk Assessment recommended that EPA increase the role of epidemiology, surveillance, and biomonitoring to support cumulative risk assessment (NRC 2009). The most successful and current epidemiologic studies leverage multiple resources and use highly collaborative and multidisciplinary approaches (Seminara et al. 2007; Baker and Nieuwenhuijsen 2008). In the United States, a number of high-quality prospective cohort studies funded mostly by the National Institutes of Health have followed millions of people and have collected bio-specimen repositories (blood, urine, nails, and DNA) and sociodemographic, genetic, medical, and lifestyle information (Seminara et al. 2007; Willett et al. 2007; NHLBI 2011). Major prospective cohort studies have also been undertaken in other countries (Riboli et al. 2002; Ahsan et al. 2006; Elliott and Peak-man 2008).
With some exceptions, current prospective cohort studies generally lack information on environmental exposures. EPA can contribute to closing this gap by, for instance, adding high-quality environmental measures to studies that already have good followup and outcome measures. Examples of collaborations in which EPA plays a critical role are the Agricultural Health Study (NIH 2012), the Multiethnic Study of Atherosclerosis and Air Pollution (MESA Air) (University of Washington 2011), and the National Children’s Study (NRC/IOM 2008). In the National Children’s Study, the linkage of monitoring data on toxicants in air, water, food, and ecosystems to individual participant data has already been explored in depth in Queens, New York, one of the Vanguard National Children’s Study sites (Lioy et al. 2009). Budgetary and implementation challenges for the National Children’s Study will require innovative strategies for recruitment, examination, and followup without compromising the quality of the science (Kaiser 2012).
Alternatively, EPA could add followup and outcome measures to studies that have good measures of exposure, although this is likely to be more time-consuming and expensive. At a minimum, EPA should ensure that environmental indicators, including country-wide air-monitoring and water-monitoring data, meet quality and accessibility criteria, for example, through a public data-access system. The indicators can then be merged with individual and community-level data in population-based studies by using geographic and temporal criteria. Biomonitoring and modeling approaches to predict exposure and dose and other advances in exposure science—including the exposome (Weis et al. 2005; Sheldon and Cohen Hubal 2009; Rappaport and Smith 2010; Lioy and Rappaport 2011), -omic technologies, and complex systems approaches (Diez Roux 2011)—could be incorporated into the prospective studies. By building expertise and leadership in exposure assessment and by working in collaboration with other national and international efforts, EPA can play a principal role in the incorporation of environmental exposures into prospective cohort studies and thus contribute to the discovery of major environmental determinants, dose— response relationships, mechanistic pathways, and gene—environment interactions for chronic diseases in human studies.
Environmental informatics plays an important role in the human-population—based studies described above. Although environmental informatics received much of its momentum from central Europe in the early 1990s (Pillmann et al. 2006), EPA has recognized its importance and has played a role in shaping its direction. The agency helped to establish the Environmental Data Standards Council, which was subsumed in 2005 by the Exchange Network Leadership Council (Environmental Information Exchange Network 2011), an environmental-data exchange partnership representing states, tribes, territories, and EPA. The council’s mission includes supporting environmental information-sharing among its partners through automation, standardization, and real-time access. The scope of data exchange covers air, water, health, waste, and natural resources, and covers multiple programs. Cross-program data include data from the Department of Homeland Security, the Toxics Release Inventory, pollution-prevention programs, the Substance Registry Services System, and data obtained with geospatial technologies. The council is an example of useful and productive national efforts to generate environmental informatics data. On the basis of technologic advances and new environmental challenges discussed throughout this report, it will be necessary for EPA to begin to make data standards flexible and adaptable so that it can use data that are less structured and less groomed.
Health informatics has a strong history in the United States. There are numerous national and state data registries on chronic and nonchronic diseases, such as the Surveillance, Epidemiology, and End Results cancer registry and the National Birth Defects registry. The Agency for Healthcare Research and Quality of the Department of Health and Human Services maintains a national hospital discharge database and, as previously mentioned, CDC’s National Center for Health Statistics conducts the NHANES annually to study health behaviors, dietary intake, environmental exposure, and disease status of the US population. EPA could also work with CDC’s National Center for Health Statistics and the National Center for Environmental Health to facilitate the merging of environmental-monitoring data (on air, water, and ecosystems) with national databases that have biomarker and health data, such as NHANES. Such merging, following the NHANES model of public access, could constitute a major advance in the understanding of environmental exposures and their health effects and in informing policy regulation and the prevention and control of environmental exposures. Collaborating with other epidemiologic research efforts, EPA will have the opportunity to identify the optimal population-based prospective cohort study protocol to answer environmental-health questions, to ensure that high-quality data on environmental exposures are incorporated into large epidemiologic studies, and to contribute to the analysis and interpretation of exposure and health-effect associations. In addition, there are proprietary databases owned by healthcare providers and insurers, including Medicare and Medicaid. These databases lay out the foundation of health informatics in the United States and have been successfully used in environmental health research.
Identifying and Predicting the Potential Toxic Effects of Chemicals
In 2007, NRC convened a panel of experts to create a vision and strategy for toxicity testing that would capitalize on the -omics concepts described in Appendix C and on other new tools and technologies for the 21st century (NRC 2007a). Conceptually, that vision is not very different from the now classic four-step approach to risk assessment—hazard identification, exposure assessment, dose—response assessment, and risk characterization—that was laid out in the NRC report Risk Assessment in the Federal Government: Managing the Process (commonly referred to as the Red Book) (NRC 1983) and that has been widely adopted by EPA as its chemical risk assessment paradigm (EPA 1984, 2000). However, the vision looks to new tools and technologies that would largely replace in vivo animal testing through extensive use of high-throughput in vitro technologies that use human-derived cells and tissues coupled with computational approaches that allow characterization of systems-based pathways that precede toxic responses. The computational approach to predictive toxicology has many advantages over the current time-consuming, expensive, and somewhat unreliable paradigm of relying on high-dose in vivo animal testiXng to predict human responses to low-dose exposures.
Although there is generally widespread agreement that the new panomics tools (that is, genomics, proteomics, metabolomics, bioinformatics, and related fields of the molecular sciences), coupled with sophisticated bioinformatics approaches to data management and analyses, will transform the understanding of how toxic chemicals produce their adverse effects, much remains to be learned about the applicability and relevance of in vitro toxicology results to actual human exposures at low doses. With the fundamental mechanistic knowledge, it should be easier to distinguish responses that are relevant to humans from responses that may be species-specific or to identify responses that occur at high doses but not low doses or vice versa. That knowledge would contribute to a reduction in the frequency of false-positive and false-negative results that sometimes plague high-dose in vivo animal testing.
A key issue in the use of such technologies is phenotypic anchoring,1 which is an important step in the validation of an assay. It is essential to validate treatment-related changes observed in an in vitro –omics experiment as causally associated with adverse outcomes seen in the individual. A single exposure to one dose of one chemical can result in a plethora of molecular responses and hundreds of thousands of data points that reflect the organism’s response to that exposure. Quantitative changes in gene expression (transcriptomics), protein content (proteomics), later enzymatic activity, and concentrations of metabolic
![]()
1 The concept of phenotypic anchoring arose from studies that examined the effects of chemical exposures on gene expression in tissues (transcriptomics). In that context, the term is defined as “the relation[ship between] specific alterations in gene expression profiles [and] specific adverse effects of environmental stresses defined by conventional parameters of toxicity such as clinical chemistry and histopathology” (Paules 2003).
substrates, products, cofactors, and other small molecules (metabolomics) can all be measured. But which of those signals, if any, are quantitatively predictive of the ultimate adverse response of interest is the key. Changes in the profiles are dynamic, tissue-specific, and dose-dependent, so the results may be drastically different depending on the tissue that was examined, the time when the sample was taken, and the dose or concentration that was used. Sophisticated bioinformatic analyses will be required to make biologic sense out of such massive amounts of data. Tremendous advances have been made in this field in the last 5 years, and it is now possible to coalesce such information into pathway analyses that may have utility in toxicity assessment. Indeed, EPA’s ToxCast program has begun to examine approaches discussed above to predictive in vitro toxicity assessment (Judson et al. 2011).
Example of Using Emerging Science to Address Regulatory Issues and Support Decision-Making: ToxCast Program
In 2006, EPA began a new computational toxicology program aimed at developing new approaches to assess and predict toxicity in vitro (Judson et al. 2011). Agency scientists in the computational toxicology program have been substantial contributors to the development of new approaches to toxicity testing. They have collectively published over 130 peer reviewed articles since its inception, including 38 publications from ToxCast (EPA 2012a). Although the use of an array of high-throughput in vitro tests—focused on different putative toxicity endpoints and pathways—to predict in vivo outcomes is attractive from both a cost-savings and time-savings perspective, it entails many challenges, including the following:
• Chemical metabolism and disposition may differ between the in vitro and in vivo situations. A principle tenet of toxicology is that the concentration of a toxicant at a specific target site is a key determinant of toxicity. If a metabolite of a toxicant, not the parent molecule, is responsible for toxicity, the in vitro systems must be able to form that metabolite—and other metabolites that might modify the response (for example, alternate detoxification pathways)—in a ratio similar to what occurs in vivo. If an in vitro system fails to form the toxicant or if it forms one that does not occur in vivo, the test system will generate a false-negative or false-positive response. The large amounts of data that can be generated from -omics experiments may be useful in identifying putative pathways of toxicity, but the relevance of the pathways to human exposures depends on a reasonably accurate simulation of the metabolic disposition of the substance that would occur in vivo.
• The time course of effects observed in vitro may be very different from what occurrs in vivo. Many chemical treatments of cells result in immediate changes in gene expression, and the nature and magnitude of the changes are highly dynamic. Initial responses may be largely adaptive in nature, and not necessarily reflective of an ultimate toxic effect. Adaptive responses can indi-
cate the potential for future toxicity, but many intervening biologic processes may abrogate downstream responses, so the fact that a particular pathway is activated by a chemical does not necessarily mean that the same will occur in vivo. It will be important for high-throughput screening approaches to consider multiple time points for analysis.
• Dose-response assessment determined in vitro may be difficult to correlate with in vivo responses and administered doses. Relating dose rate (in milligrams per kilogram per day) in vivo at specific tissues to cell-culture concentrations tested in vitro is extremely difficult and requires detailed knowledge of the absorption, distribution, metabolism, and excretion of a xenobiotic after in vivo exposure. It also requires knowledge about protein binding to plasma and intracellular proteins, lipid portioning, tissue-specific activation, and detoxification for interpretation of the relevance of an in vitro cell concentration to a target-tissue concentration after in vivo administration. Thus, physiologically based pharmacokinetic modeling, which will require some in vivo data, will continue to be an important part of hazard evaluation and risk assessment for chemicals that are identified as being potentially of concern on the basis of in vitro screening assays. Although advances in in vitro toxicity assessment continue to improve and will certainly decrease the number of animals required for in vivo testing, it is unlikely that in vitro tests will fully replace the need for in vivo animal testing for understanding the pharmacokinetics and pharmacodynamics of toxic substances because of the complex interplay between tissues and organs that are ultimately critical determinates of a toxic response.
The importance of those concepts was recently illustrated in some modeling studies of EPA ToxCast data. In the first phase of the ToxCast program, EPA scientists used hundreds of in vitro assays to screen a library of agricultural and industrial chemicals to identify cellular pathways and processes that were modified by specific chemicals; they intended to use the data to set priorities among chemicals for further testing (Judson et al. 2010). However, the potency of a chemical in an in vitro assay may or may not reflect its biologic potency in vivo because of differences in bioavailability, clearance, and exposure (Blaauboer 2010). Scientists at the Hamner Institute, in collaboration with EPA scientists, recently developed pharmacokinetic and pharmacodynamic models that incorporate human dosimetry and exposure data with the ToxCast high-throughput in vitro screening data (Rotroff et al. 2010; Wetmore et al. 2012). Their results demonstrated that incorporation of dosimetry and exposure information is critically important for improving priority-setting for further testing and for evaluating the potential human health effects at relevant exposures.
EPA scientists have played a leading role in the new approaches, and it will be important for them to continue to lead the way in both computational and systems toxicology in the future. With further improvements, such as inclusion of human dosimetric and exposure data, high-throughput in vitro assays for screening of new chemical entities for potentially hazardous properties will
probably become widely used for toxicity testing. Although the new technology-driven approaches to in vitro toxicity testing and high-throughput screening constitute an important advance in hazard evaluation of new chemicals, they are not yet ready to replace traditional approaches to hazard evaluation because of inherent limitations of extrapolation from in vitro to in vivo findings, as discussed above. But they will be very useful in setting priorities among new chemicals for more thorough toxicity testing. Additionally, the new technologies will greatly augment traditional approaches to in vivo toxicity evaluation by providing mechanistic insights and more detailed characterizations of biologic responses at doses well below those shown to produce toxicity. That will be especially important in evaluating endocrine-active chemicals and chemically induced alterations that may occur during early life.
McHale et al. (2010) have discussed the importance of new –omics technologies and of a systems-thinking approach to human health risk assessment of chemical exposures, or systems toxicology. EPA has already begun to examine such approaches to predictive in vitro toxicity assessment through the ToxCast program (EPA 2008a). It is evident that new approaches to data management and analysis will be critical for the success of computational approaches to predictive toxicology. The statistical and modeling challenges are immense in addressing the large volumes of data that will come from systems-toxicology experiments, which are an essential element of EPA’s computational-toxicology effort. It will be critical for the success of this and other efforts that involve large amounts of data for EPA to have access to the best available tools and technologies in informatics.
Example of Using Emerging Science to Address Regulatory Issues and Support Decision-Making: Predicting the Hazards of a New Material
Nanotechnology is an emerging technology that poses new challenges for EPA. Deemed the next industrial revolution, nanotechnology is predicted to advance technology in nearly every economic sector and be a major contributor to the nation’s economy. The rationale of that prediction is that nanoparticles, with dimensions of 1-100 nm, have properties that are useful in a wide variety of applications, including electronic, photovoltaic, structural, catalytic, diagnostic, and therapeutic.
A potential concern is that some of the properties of nanoparticles might pose risks to human health or the environment. The challenge for EPA is to use or develop the science and tools needed to assess and manage the widespread use of nanoscale materials that have unknown hazards. That includes assessing potential risks associated with an emerging technology and, if necessary, monitoring potential exposures and hazards. Using nanotechnology as an example, the committee identified several questions that can be used to better understand the risks associated with new science and tools. Many of these issues regarding the environmental, health, and safety aspects of nanotechnology are addressed in
a 2012 NRC report, A Research Strategy for the Environmental, Health, and Safety Aspects of Nanotechnology (NRC 2012).
First, do nanoparticles present different properties and inherent risks from smaller molecules or larger particles? To answer this question, it will be important for EPA to adapt and develop new science and tools that strengthen the correlation between the structure and identity of a nanomaterial and the hazard posed by it. That means that new analytic tools or approaches that permit reliable and rapid assessment of engineered-nanomaterial structure and purity are needed. Rapid tests to screen for hazards and set priorities among materials for further testing are essential to keep pace with the development of new materials and to make efficient use of resources available to test materials. To model and predict the properties of the new materials, it will be necessary to develop precisely defined reference materials to ensure that inputs to predictive models and informatics efforts are robust and reliable. The measurement tools, rapid screening approaches, defined reference materials, and modeling and informatics approaches, advanced in an integrated fashion, can determine more rapidly what, if any, unique hazards are associated with this emerging technology.
Second, what are the likely routes and venues of exposure to engineered nanomaterials? Consumer-use patterns, production methods, and life-cycle effects of emerging technologies are unknown. To identify likely ways in which exposure can occur, it will be important for EPA to use physical science, engineering, and social science tools in a multidisciplinary approach that seeks to understand the life cycle of the materials, the supply chains that incorporate them, the projections for market growth, and consumer behaviors in using nanomaterial-containing products. By identifying the intersection between the most likely exposures and unique hazards, EPA can focus on further characterizing the potential risk and using science to inform policies needed to monitor and manage the risk.
Third, how can nanomaterials be detected, tracked, and monitored in complex biologic and environmental media? To complement the science to assess unique hazards and realistic exposures described earlier in this section, EPA will require tools to monitor the distribution of and potential exposures to nanomaterials. The characterization of pristine nanomaterials has been a challenge given the lack of specialized tools for detecting and measuring them. Once distributed, nanomaterials pose even greater challenges to detection, tracking, and monitoring than small molecules or micron-scale particles. This is because nanomaterials tend to have distributions of sizes and surface coatings, their high surface area leads to agglomeration or deposition, their surface chemistry has been shown to be dynamic, and their speciation can be complex. EPA and its collaborators and contractors will need to invent, develop, or refine tools to detect, track, and monitor nanomaterials. In some cases, the solution may be to integrate the use of existing tools. In others, new tools will be required. In addition to direct detection of the materials, strategies that exploit the use of biomarkers as described earlier in this chapter may prove essential for understanding exposures.
The three questions posed in this section may be similarly applied to any emerging material to identify concerns surrounding new hazards, exposure routes, and material tracking. The case of nanotechnology is an example of how EPA will need to approach many emerging tools, technologies, and challenges in general in the future. In order to have the capacity to address those tools, technologies, and challenges, it will need to have enough internal expertise to identify and collaborate with the expertise of all of its stakeholders in order to ask the right questions; determine what existing tools and strategies can be applied to answer those questions; determine the needs for new tools and strategies; develop, apply, and refine the new tools and strategies; and use the science to make recommendations based on hazards, exposures, and monitoring.
TOOLS AND TECHNOLOGIES TO ADDRESS CHALLENGES RELATED TO AIR POLLUTION AND CLIMATE CHANGE
As discussed in Chapter 2, EPA’s first goal in its 2011–2015 strategic plan is “taking action on climate change and improving air quality” (EPA 2010). Improved modeling capabilities are integral to attaining that goal inasmuch as models are needed to test the understanding of sources, environmental processes, fate, and effects of airborne contaminants and to investigate the effects of potential mitigation measures. Examples of the many areas in which new technologies will impact air quality and climate change are discussed in the following sections on air-pollution modeling; carbon-cycle modeling, greenhouse-gas emissions, and sinks; and air-quality monitoring.
Air-Pollution Modeling
EPA has a strong history of leadership in air-quality modeling. Its Community Multi-scale Air Quality (CMAQ) model is used both domestically and internationally as a premier platform for “one atmosphere” modeling of the chemistry and transport of ground-level ozone, particulate matter, reactive nitrogen, mercury, and dozens of other materials. In recent years, EPA researchers have worked with other government and university scientists to develop capabilities to run the CMAQ model in a real-time forecast mode (Eder et al. 2009); to couple the CMAQ model to an advanced meteorologic model, the Weather Research and Forecasting system (Appel et al. 2010); and to build advanced sensitivity analysis and inverse modeling capabilities (Napelenok et al. 2008; Tian et al. 2010).
In coming years, investments in modeling efforts will advance the understanding of sources and environmental processes that contribute to particulate-matter loadings and health and environmental effects. Modeling efforts will also improve the understanding of interactions between climate change and air quality with a special focus on relatively short-lived greenhouse agents, such as ozone, black carbon, and other constituents of particulate matter. Improved
modeling capabilities will enable EPA to evaluate actions that have dual benefits for reducing radiative-forcing agents (such as ozone and aerosols) and improving air quality, and it will also enable EPA to understand better how tropospheric particulate matter may have masked some global warming in the past. The committee has identified several efforts that will likely be important for EPA in the future. They include, working with other federal and university scientists to improve the use of global climate model predictions to inform air-quality management and other climate-adaptation decisions; working toward a better understanding of the global mass balance of mercury and other biologically active metals, including the role of natural sources and re-emission, chemical and biologic processing, and interregional transport; improving its understanding of physical and chemical processes; leading the integration of models and observations (including satellite and other remote sensing techniques2) to help to estimate emissions of greenhouse agents and conventional air pollutants, especially from dispersed or fugitive sources; and expanding its efforts to integrate socioeconomic and biophysical systems models for integrated assessment, including examination of air and climate effects of changing agriculture, energy, information, land-use, and transportation systems.
Carbon-Cycle Modeling, Greenhouse-Gas Emissions, and Sinks
EPA is engaged in a variety of science, engineering, regulatory, and policy development activities related to greenhouse-gas emissions, the global carbon cycle, and impacts of resulting changes on human health. The agency is responsible for the national-level inventory of greenhouse gases in the context of the Framework Convention on Climate Change. Under the Clean Air Act, the agency has authority to regulate greenhouse gases, including carbon dioxide, methane, nitrous oxide, and hydrofluorocarbons. Much attention is also focused on estimating ecosystem uptake and sequestration of carbon as a quantifiable (and monetizable) ecosystem service.
Fossil-fuel emissions can be estimated with relatively high precision, and the science of monitoring and modeling of their uptake by terrestrial and marine ecosystems is evolving rapidly. National-scale and continental-scale estimates of carbon fluxes are now produced through several approaches. In one approach, atmospheric-inversion models rely on regional measurements of atmospheric carbon dioxide coupled to surface ecosystem fluxes and atmospheric circulation
![]()
2 Remote sensing—the study of Earth processes and phenomena without direct physical contact—will be discussed several times throughout this chapter. It includes both passive sensors, which measure electromagnetic radiation that is emitted or reflected by the object or area being observed, and active sensors, such as synthetic-aperture radar or light detection and ranging systems, which emit energy and measure its return to infer properties of the scanned surfaces. Remote sensing complements expensive and slow data collection on the ground and provides local-to-global areal coverage of many key environmental processes.
(Gurney et al. 2002). In another approach, which is more direct, biomass inventories (for example, forest and cropland inventories) are used for estimating uptake by monitoring changes in biomass stocks. A third approach involves spatially explicit modeling of ecosystem processes on the basis of weather, soil, land use, and land cover (Schwalm et al. 2010). Each of those approaches has limitations and uncertainties, and derived estimates show only moderate agreement (Hayes et al. 2012). Hayes et al. (2012) demonstrate the value of the inventory approach, which relies on stock estimates obtained from EPA reports (for example, EPA 2011b), for subcontinental-scale estimates of carbon fluxes.
Integrated modeling of greenhouse-gas sources and sinks3 will continue to develop rapidly given continuing advances in remote sensing of ecosystem properties and understanding of the carbon cycle. To meet its regulatory mandate and to support policies that address climate change, EPA could benefit from increased science and engineering capacity in ecosystem ecology and Earth-system science.
Air-Quality Monitoring
Advances in atmospheric remote sensing have created a new paradigm for air-quality monitoring and prediction from regional to global scales (NRC 2007b). Research and applications have focused on fine particulate aerosols, tropospheric ozone, nitrogen dioxide, formaldehyde, sulfur dioxide, and carbon monoxide, but have also included other compounds, such as benzene, ethylbenzene, and 1,3-butadiene (NRC 2007b, Fishman et al. 2008, Hystad et al. 2011). Active sensors, such as satellite and aircraft-mounted light detection and ranging systems (LiDAR) (for example, the cloud-aerosol LiDAR with orthogonal polarization), can provide information on the vertical distribution of clouds and aerosols on the basis of the magnitude and spectral variation in backscatter of the vertical beam. However, most remote sensing of air quality has relied on passive sensors, for example, measurements of pollution in the troposphere, the moderate-resolution imaging spectroradiometer and multi-angle imaging spectroradiometer on the National Aeronautics and Space Administration’s (NASA) Terra platform, and the ozone-monitoring instrument and tropospheric emission spectrometer on NASA’s AURA platform (Martin 2008). Those collect radio-metric data on solar backscatter or thermal infrared emissions that are then used in retrieval algorithms that incorporate other geophysical information and radiative-transfer models. The reliability of results depends on the surface reflectivity or emissivity, clouds, the viewing geometry, and the retrieval wavelength (Martin 2008). Estimating ground-level concentrations, which are of greatest relevance to EPA, requires additional information on the vertical structure of the
![]()
3 The ocean is the largest sink, inasmuch as carbon dioxide is dissolved in seawater and is in equilibrium with the atmosphere (in freshwater bodies, it can change the water pH to some extent).
atmosphere, especially for ozone and carbon monoxide. Inverse modeling is required to infer pollutant source strength from observed concentration patterns.
Although it is not a substitute for ground-based air-quality measurements, satellite-derived data provide important spatial, temporal, and contextual information about the extent, duration, transport paths, and distances of pollution from a source, which is generally not possible with in situ ground-based measurements. For example, Morris et al. (2006) linked increases in surface ozone in Houston to wildfires in Alaska and western Canada, and Heald et al. (2006) traced an increase in springtime surface aerosols in the northwestern United States to anthropogenic sources in Asia. As retrieval algorithms and the spatial and spectral quality of satellite data have improved, remote sensing has provided a means of obtaining relatively consistent estimates of air-pollutant exposure over large areas for health-effects assessments (van Donkelaar et al. 2010; Hystad et al. 2011), which has facilitated large-scale epidemiologic investigations in settings where monitoring data are inadequate to determine spatial contrasts (Crouse et al. 2012). Another important trend is the assimilation of concurrent data from multiple sensors with ground data; that has proved especially useful in improving estimates of ground-level ozone (Fishman et al. 2008).
Example of Using Emerging Science to Address Regulatory Issues and Support Decision-Making: Remote Sensing to Monitor Landfill Gas Emissions
Great progress has been made in reducing or eliminating releases of toxic substances from concentrated sources (also known as point sources), but monitoring and mitigating emissions from so-called area sources has been technically difficult and remains one of the persistent challenges faced by EPA. Recent efforts to use emerging technology in monitoring provide a glimpse on a very broad scale of what might be possible with further advances. EPA’s National Risk Management Research Laboratory used a tunable diode laser to perform optical remote-sensing of fugitive methane, hazardous air pollutants (including mercury), volatile organic compounds, and nonmethane organic compounds emitted from three landfills. With multiple measurements of concentrations along different light paths, the system calculates a mass emission flux for the entire area. What had been thought to be an excessively expensive monitoring challenge is now financially and practically manageable (EPA 2012b).
Example of Using Emerging Science to Address Regulatory Issues and Support Decision-Making: Multipollutant Analysis Standard-Setting
Regulation in the United States is predicated on single-pollutant standards or control strategies. Improved understanding of health effects of cumulative and mixed exposures calls for new approaches to standard-setting that consider a multipollutant approach. The shift will require understanding of the joint behav-
ior of multiple stressors, the interactions among them, and their contributions to health outcomes.
The air-pollution health community has been examining the science-readiness of a multipollutant regulatory strategy (Dominici et. al. 2010; Greenbaum and Shaikh 2010). The challenges, opportunities, and future research needs related to multipollutant approaches for the assessment of health risks associated with exposures to air pollution were evaluated in a public workshop held in 2011 (Johns et al. 2012). The workshop highlighted the need for a transdisciplinary research approach for developing more relevant tools and methods in the fields of exposure science, human and animal toxicology, and air pollution epidemiology. More important, it recommended collaboration among science, engineering, and policy communities to develop practical and implementable approaches that could ultimately inform decision-making (D. Johns, EPA, personal communication, May 9, 2012).
Related efforts to characterize toxicity of mixtures of chemicals in chemical risk assessment are under way. A key challenge is to define the universe of possible combinations of mixtures that are representative of real-world exposures. In a recent analysis, EPA researchers investigated methods from the field of community ecology originally developed to study avian species cooccurrence patterns and adapted them to examine chemical co-occurrence (Tornero-Velez et al. 2012). Their findings showed that chemical co-occurrence was not random but was highly structured and usually resulted in specific predictable combinations. Novel application of tools and approaches from a variety of research disciplines can be used to address the complexity of mixtures, advance the scientific communities’ understanding of exposures to the mixtures, and promote the design of relevant experiments and models to assess associated health risks.
TOOLS AND TECHNOLOGIES TO ADDRESS CHALLENGES RELATED TO WATER QUALITY
As discussed in Chapter 2, there are several important drivers of water quality and water-quality policies for which new technologies and approaches can be instrumental in enhancing data-driven regulations. For the purposes of this chapter, examples of the many areas in which new technologies will impact water quality are divided into the following areas: remote sensing technologies for water-quality monitoring; water modeling; and detecting microorganism and microbial products in the environment.
Water-Quality Monitoring
Multispectral imagery has been successfully applied to water-quality monitoring for several decades, notably for monitoring surface temperature and concentrations of suspended sediments and algae (see reviews by Mertes 2002; Matthews 2011). Modern multispectral sensors—such as the moderate-resolution imaging
spectroradiometer and the European medium-resolution imaging spectrometer sensor, which have moderate (about 250m) spatial resolution, 10–15 spectral bands, and high sampling frequency—have accelerated progress in remote sensing of suspended sediments, dissolved organic matter, chlorophyll, phycocyanin, and other water-quality indicators that are extensive enough to suit sensor resolution (Bierman et al. 2011). Satellite-based assessments of water quality will probably be increasingly routine, especially with better integration and assimilation of in situ data and multiscale sensor data via empiric and physically based models (Matthews 2011). As mentioned in the section “Air-Quality Monitoring” above, the new tools and technologies are not a substitute for ground-based water-quality measurements, but they provide important spatial and contextual information about the extent, duration, transport paths, and distances of pollution from a source and should be used to enhance the current water-monitoring infrastructure and related exposure assessment efforts.
Water Modeling
Real-time reporting of water-quality data would complement EPA research programs. Data could be downloaded to a community Website so that other researchers and the general public could understand water-quality and quantity (storm-flow) information better. That type of network would eventually allow analysis of infiltration or inflow problems, including policy options (such as disconnecting storm drains from the sanitary sewer) and the likely effectiveness of infrastructure investment in light of climate change (such as more intense storm events). Figure 3-3 illustrates how a sensor network might be set up.
Spatially detailed high-frequency sensing of water resources that uses an embedded network can provide breakthroughs in water science and engineering by promoting understanding of nonlinearities (the knowledge base to discern mechanisms and basic kinetics of nonlinear water processes) (Ostby 1999; Coppus and Imeson 2002; Nowak et al. 2006); scalability (the ability to scale up complex processes from observations at a point to the catchment basin) (Ridolfi et al. 2003; Sivapalan et al. 2003; Long and Plummer 2004); prediction and forecasting (the capacity to predict events, to model and anticipate outcomes of management actions, and to provide warnings or operational control of adverse water-quantity and water-quality trends or events) (Christensen et al. 2002; Scavia et al. 2003; ASCE 2004; Vandenberghe et al. 2005; Shukla et al. 2006; Hall et al. 2007); and discovery science (the discovery of heretofore unknown and unreported processes) (Jeong et al. 2006; Messner et al. 2006; Loperfido et al. 2009; 2010a,b).
Detecting Microorganisms and Microbial Products in the Environment
Development of detection methods for microbial contamination in water, soil, and air is a critical part of environmental protection. EPA is one of the few federal agencies that oversees a substantial research portfolio that includes new
analytic techniques for environmental assessment. Although more modern biochemical methods are available, coliform bacteria and enterococci continue to be used as indicators for the assessment of safe drinking and recreational waters (EPA 1986, 2002, 2005), and cultivation methods for viability remain the gold standard (Messer and Dufour 1998). In recognition of the inadequacy of the bacterial indicator system over the years, research methods have been developed and improved for measuring enteric viruses (Fong and Lipp 2005; Yates et al. 2006; Pepper et al. 2010) and protozoa (Sauch et al. 1985; Rose 1988; Aboytes et al. 2004) and for expanding the understanding of risk (Slifko et al. 1997, 1999; Aboytes et al. 2004). National surveys of groundwater and surface water have directly influenced important rule-making, including the Surface Water Treatment Rule, the Information Collection Rule, the Long Term Enhanced Surface Water Treatment Rule, the Disinfectants and Disinfection Byproducts Rule, the Ground Water Rule, and final rules for the use or disposal of sewage sludge.
Assessment and control of waterborne diseases still rely on the ability to sample and quantify fecal indicator organisms and pathogens as part of the evaluation of water quality. The most recent advances in the detection of microorganisms in water include quantitative polymerase chain reaction (PCR) methods, which can be designed for any microorganism of interest because they are highly specific and quantitative. The PCR methods can produce information relatively fast and, under the Clean Water Act and the Beaches Environmental Assessment and Coastal Health Act, their adoption has moved quickly toward meeting total maximum daily load requirements and beach safety (see the example below on “Beach Safety”).
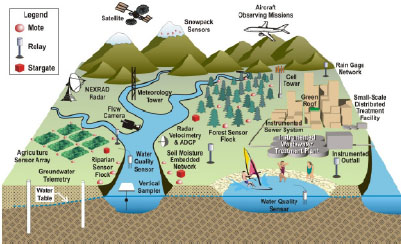
FIGURE 3-3 Schematic of an instrumented watershed in an observatory of the national network. Real-time sensors for meteorology, rainfall, stream velocity, suspended sediment, water quality, soil moisture, groundwater, and snowpack are shown with wireless communications equipment necessary for transmitting the data. Source: WATERS Network 2009.
New approaches to next-generation DNA-sequencing technologies offer the promise of characterizing healthy water by ensuring the absence of harmful biotic organisms, even rare ones (see Appendix C for background information on genomics tools and technologies). Just as the human microbiome studies are examining the diversity and ecology of microorganisms in the intestinal tract, DNA-sequencing methods are being used to explore the water microbiome in polluted, pristine, and unique environments, although finding rare microbial populations that will exhibit genetic characteristics with the potential for harm to humans is difficult. Metagenomics of the wastewater system, and in particular the viral genome, provide insight into the complex world of water microbiotas, but is only being used for exploration. Current efforts are being spent in developing methods and generating large amounts of data (Table 3-2); the methods are able to identify which microorganisms (including potentially pathogenic organisms) are present, but their viability and functional activity are often not known. Finally, genomic data have not been used much to inform microbial risk assessment. In the next decade, environmental microbiome studies and data will need to move toward sophisticated data interpretation and modeling, and substantial investment in bioinformatics will be necessary. With the growing understanding of the ecosystem microbiome and its interaction with human health and the environment, it is becoming evident that the microbiome plays an important role in modulating health risks posed by broader environmental exposures. Understanding such interactions will have important implications for understanding individual and population susceptibility and the observed variability in risks posed by environmental exposures.
Other recent advances that are facilitating the use of molecular tools include new techniques for increasing sample concentration—such as ultrafiltration, continuous filtration, and new types of filters—for improved recovery and automated extraction of nucleic acids with less contamination, less inhibition, and more rapid throughput (Hill et al. 2005; Srinivasan et al. 2011). New quantitative PCR approaches for monitoring the viability of pathogens of concern are of particular interest, and several approaches show some promise. Such dyes as ethidium monoazide and propidium monoazide have been used to distinguish between live cells and heat-killed cells, but the dyes are not able to penetrate apparently killed cells when applied to disinfected treated sewage samples, so the signals that are produced through quantitative PCR methods are comparable with counts made before and after disinfection with or without use of the dyes (Varma et al. 2009; Srinivasan et al. 2011). More work is needed to address the possible presence of viable but nonculturable cells in disinfected effluents. An approach to examining viability associated with bacteria is to use quantitative PCR methods to target the precursors of ribosomal RNA (rRNA). That was done to quantify viable cells of Aeromonas and mycobacteria in water (Cangelosi et al. 2010) and showed promise for both saltwater and freshwater and for post-chlorination monitoring. Those types of methods will require verification in the monitoring of disinfected drinking water and wastewater. There may be a need
| Environment Sampled | Target and Approach | Findings | Reference |
| Wastewater biosolids | Bacterial 16S rRNA genes; PCR, pyrosequencing (454 GS-FLX sequencer) | Most of the pathogenic sequences belonged to the genera Clostridium and Mycobacterium | Bibby et al. 2010 |
| Wastewater (activated sludge, influent, and effluent) | Bacterial 16S rRNA gene (hypervariable V4 region); PCR, pyrosequencing (454 GS-FLX sequencer) | Most of the pathogenic sequences belonged to the genera Aeromonas and Clostridium | Ye and Zhang 2011 |
| River sediment | Bacterial antibiotic-resistance genes; MDA, pyrosequencing (454 GS-FLX sequencer) | Large amounts of several classes of resistance genes in bacterial communities exposed to antibiotic were identified | Kristiansson et al. 2011 |
| Reclaimed and potable water | Viral DNA and RNA; tangential flow filtration, DNase treatment, MDA, pyrosequencing (454 GS-FLX and GS20 sequencer) | Over 50% of the viral sequences had no significant similarity to proteins in GenBank; bacteriophages dominated the DNA viral community; the RNA metagenomes contained sequences related to plant viruses and invertebrate picornaviruses | Rosario et al. 2009 |
| Wastewater biosolids | Viral DNA and RNA; DNase and RNase treatment, reverse transcription for RNA, pyrosequencing (454 GS-FLX sequencer), optimal annotation approach specific for viral pathogen identification is described | Parechovirus, coronavirus, adenovirus, aichi virus, and herpesvirus were identified | Bibby et al. 2011 |
| Lake water | Viral RNA; tangential flow filtration, DNase and RNase treatment, random amplification (klenow DNA polymerase), pyrosequencing (454GS-FLX sequencer) | 66% of the sequences had no significant similarity to known sequences; presence of viral sequences (30 viral families) with significant homology to insect, human, and plant pathogens | Djikeng et al. 2009 |
Abbreviations: DNase, deoxyribonuclase; MDA, multiple displacement amplification; PCR, polymerase chain reaction; RNase, ribonuclease; rRNA, ribosomal RNA.
Source: Aw and Rose 2012. Reprinted with permission; copyright 2012, Current Opinion in Biotechnology.
for a method that combines some type of cultivation with quantitative PCR techniques in real time to address viability. The use of molecular tools that can be used to inform decisions for water treatment and public-health protection will still require substantial investment in sample concentration, hazard characterization, quantification, and assessment of viability.
Example of Using Emerging Science to Address Regulatory Issues and Support Decision-Making: Beach Safety
Shorelines provide benefits to society as a whole and in particular are directly associated with tourism, which remains one of the largest economic sectors around the world. According to the Natural Resources Defense Council (NRDC 2011), beaches in the United States were given advisories or were closed 24,091 times in 2010—the second-highest number of advisories and closures in the 21 years since NRDC began reporting. It was suggested that aging and poorly designed sewage-treatment systems and contaminated stormwater were the main causes of pollution that led to fecal—indicator concentrations that exceeded the state’s health and safety standards. There were also more than 9,000 days of Gulf Coast beach notices, advisories, and closures due to the Deepwater Horizon oil-spill disaster in 2010 (NRDC 2011).
As part of an overhaul of the Clean Water Act, the Beaches Environmental Assessment and Coastal Health Act mandated that research be undertaken to understand coastal pollution, address polluted sediments, decrease response time, and improve protection of public health (EPA 2006a); most of the research programs under this act have yet to be realized, and improving public-health protection has been slow. EPA has begun to update water-quality standards, address health studies and swimmer surveys, and advance the use of new genomic technology for the rapid testing of water quality. The development of the first standardized quantitative PCR method for enterococci is being promoted for recreational-water assessment (Wade et al. 2006). Evaluations based on new quantitative PCR methods for indicators in ambient and recreational waters are being published (Byappanahalli et al. 2010; Noble et al. 2010), but there are challenges to using these methods for regulatory purposes because interpretation of the signals may underestimate or overestimate human health risks and could lead to beach closures that cause unnecessary economic losses (Srinivasan et al. 2011). Continued investment in new methods, applications for surveys, and links to health effects and management strategies are necessary.
Wastewater and stormwater are key culprits in water pollution, and further improvement of water safety cannot occur unless point and nonpoint sources of pollution are elucidated. Research on microbial source tracking has advanced the use of molecular tools for investigating the presence of pathogens in impaired waters and to setting total maximum daily load requirements. EPA is taking a leadership role in the microbial source-tracking research (EPA 2005). In addition, California has organized one of the largest blind studies, the Global
Inter-Laboratory Fecal Source Identification Comparison Study, which involves the evaluation of 39 microbial source-tracking methods by 29 laboratories (Shanks 2011).
To maximize the benefits of clean water, protect the general public, sustain water resources, and restore impaired shorelines, decision-makers will need to rely increasingly on an understanding of the long-term and short-term changes in water quality and aquatic ecosystems. The advanced science and technology are poised to play an increasingly important role in providing forecasts of effects on appropriate temporal and spatial scales. Advances could be made quickly for safe and sustainable water resources in the promotion of methodologic developments and applications in rapid and predictive monitoring; development of and investment in a safe-waters program that links genomic tools with watershed and beach-shed characterizations; continued microbial characterization of stormwater, combined sewage overflows, and wastewater; and development of and investment in innovative engineering designs to reduce pollution loads.
Example of Using Emerging Science to Address Regulatory Issues and Support Decision-Making: Quantitative Microbial Risk Assessment
Quantitative microbial risk assessment had its beginnings in the 1980s; it is associated with the first publication of dose—response models (Haas 1983) and is now an accepted process for addressing waterborne disease risks and management strategies (Haas et al. 1999; Medema et al. 2003). Although great strides have been made in using quantitative microbial risk assessment in EPA’s Office of Homeland Security (including leading an interagency working group and the exchange of information with CDC), EPA has yet to take a leadership role in developing the necessary databases for use in a national risk assessment of wastewater, stormwater, and recreational water.
Linking biology, mathematics, health, the environment, and policy will require substantial interdisciplinary research focused on problem-solving and systems thinking. Quantitative microbial risk assessment has been seen as an important framework for pulling science and data together and can lead to innovative work in decision science. According to the Center for Advancing Microbial Risk Assessment, “ultimately, the goal in assessing risks is to develop and implement strategies that can monitor and control the risks (or safety) and allows one to respond to emerging diseases, outbreaks and emergencies that impact the safety of water, food, air, fomites, and in general our outdoor and indoor environments” (CAMRA 2012). The framework is being promoted by the World Health Organization (WHO 2004), and the international need for data, education, and mathematical tools to assist countries around the world with the implementation of quantitative microbial risk assessment strategies is paramount. More recently, Science and Decisions: Advancing Risk Assessment (NRC 2009) called for more integration with the risk-assessment—risk-management paradigm. This approach will provide a pathway to the integration
of new tools and science for addressing EPA’s goals of safe and sustainable water.
If a quantitative microbial risk-assessment framework were put into practice by EPA, it would need to incorporate alternative indicators based on genomic approaches, microbial source-tracking, and pathogen-monitoring. Also, the complete human-coupled water cycle would need to be explored, including built and natural systems. Implementation of a quantitative microbial risk-assessment framework would require investment in a health-related water microbiology collaborative research network. The network would bring molecular biologists, ecologists, engineers, and water-quality health and policy experts together to build internal capacity, to develop external partnerships, and to foster national collaboration. Regardless of whether EPA decides to systematically use a quantitative microbial risk-assessment framework, the future of science at the agency would benefit from continuing to build exposure databases and support work on the survival and inactivation of pathogens that can feed into quantitative microbial risk assessment. Agency science would also benefit from new informatics and application tools that are based on quantitative microbial risk assessment models to enhance decision-making to meet safe-water goals.
An example of an area in which EPA may be able collaborate to more effectively fill information gaps or address funding overlap in a resource-constrained environment is through microbiology research. There are other organizations that have microbiology programs, but few address the environment. NIH’s Division of Microbiology and Infectious Diseases supports clinical research and basic science for microbes and infectious disease. NIH has recently partnered with the National Science Foundation and the US Department of Agriculture to fund research on the ecology and evolution of infectious disease. The partnership addresses diseases that have an environmental pathway and can include waterborne diseases, but most of the efforts have been related to cholera and little attention has been given to other groups of pathogens. EPA has not yet played a role in the partnership, but it could contribute to filling a gap in knowledge about wastewater treatment and monitoring as it relates to microbes and environmental and human health.
TOOLS AND TECHNOLOGIES TO ADDRESS CHALLENGES RELATED TO SHIFTING SPATIAL AND TEMPORAL SCALES
Chapter 2 noted that current environmental challenges are expanding in both space and time and it emphasized that long-term data are needed to characterize such changes and to characterize the cause and the potential implications of different policy options. To address the challenges of increasing spatial and temporal scales for a variety of environmental problems, new approaches, tools, and technologies in such areas as computer science, information technology (IT), and remote sensing will become increasingly important to EPA. The ability to take full advantage of all the new tools and technologies discussed in the pre-
ceding sections of this chapter will require EPA to have state-of-the-art IT and informatics resources that can be used to manage, analyze, and model diverse datasets obtained from the vast array of technologies.
Computer Science, Informatics, and Information Technology
The future needs for IT and informatics in support of science in EPA are subject to two principal influences: the future directions of EPA’s mission and the underlying science in future directions taken by the IT industry. Science in EPA will increasingly depend on its capability in IT and informatics. IT is concerned with the acquisition, processing, storage, and dissemination of information with a combination of computing and telecommunication (Longley and Shain 1985). The term informatics, as used here, refers to the application of IT in the generation, repository, retrieval, processing, integration, analysis, and interpretation of data obtained in different media and across geographic and disciplinary boundaries that are related to the environment and ecosystem, community and human activities, and human health (see He 2003). Informatics is also concerned with the computational, cognitive, and social aspects of IT. One way in which IT can be used for data acquisition is through public engagement. Taking advantage of expertise outside of EPA (from academia, industry, and other agencies) and considering the general public as a source of new information is a way in which knowledge and resources can be combined in a cost-effective manner. Examples include taking advantage of social media and crowdsourcing. Appendix D provides additional background information on various important and rapidly changing tools and technologies in the field of information technology and informatics.
Example of Using Emerging Science to Address Regulatory Issues and Support Decision-Making: Social Media
EPA does substantial outreach to the public and to other agencies and research communities via such media as blogs and wikis. It also supports mobile, desktop, and laptop collaboration and it clearly sees the role of social media for these outward-facing purposes. The general IT activities are the responsibility of several entities in the Office of Environmental Information and elsewhere in the agency, such as the Office of Solid Waste and Emergency Response (EPA 2012c) and the Office of Water (EPA 2012d). Social media also have a role to play in crowdsourcing and citizen science, as will be discussed in the following section. Another important topic in the near future will be the use of social media for scientific collaboration. The emergence of secure enterprise social networks provides a host of opportunities for greatly enhanced internal and external collaboration, particularly as tighter budgetary circumstances force the dissolution of some departmental and interagency boundaries.
Those networks already securely provide an environment for microblogging, private messaging, profiles, administered groups, directories, and secure external networks of partners. Conversations may be fully archived and are searchable with tags, topics, and links to documents and images. The technology is accelerating rapidly and will surely be part of the expectations for the next generation of EPA scientists. As the new technologies are emerging, consolidating, and maturing, following such changes closely would help EPA to make anticipatory decisions for adopting the appropriate technology that provides the greatest benefit to the agency at the least cost.
Example of Using Emerging Science to Address Regulatory Issues and Support Decision-Making: Crowdsourcing
Massive online collaboration, or crowdsourcing, can be defined as the “sourcing [of] tasks traditionally performed by specific individuals to an undefined large group of people [or community]—the crowd—through an open call. For example, the public can be invited to help develop a new technology, carry out a design task, [propose policy solutions,] or help capture, systematize, or analyze large amounts of data—also known as citizen science” (Ferebee 2011). With a well-designed process, crowdsourcing can help assemble the data, expertise, and resources required to perform a task or solve a problem by allowing people and organizations to collaborate freely and openly across disciplinary and geographic boundaries. The emergence of crowdsourcing, such as citizen science, with widely dispersed sensors will produce vast amounts of new data from low-cost unstructured sources. This can inform multiple domains of environmental science, but may have the greatest potential for monitoring environmental conditions and creating more refined models of human exposure.
The idea behind regulatory crowdsourcing is that many areas of regulation today, from air and water quality to food safety and financial services, could benefit by having a larger number of informed people helping to gather, classify, and analyze shared pools of publicly accessible data. Such data can be used to educate the public, enhance science, inform public policy-making, or even spur regulatory enforcement actions. Indeed, there are many arenas in which experts and enthusiasts, if asked, would help to provide data or to analyze existing data.
EPA is no stranger to crowdsourcing. With such peers as NASA and CDC, EPA is a pioneer and visible leader in collaborative science. One example is its use of broad networks to engage outside environmental problem-solvers, for example, through the Federal Environmental Research Network, InnoCentive challenges, and the Challenge.gov Web site (Preuss 2011). More recent examples of using the public to gather information are discussed in Box 3-1. Broad community participation has led to a wide array of new data sources to provide a baseline for monitoring the effects of climate change on local tree species, for wildlife toxicology mapping, and for real-time water-quality monitoring.
Today, a growing number of regulatory agencies (including EPA, the Securities and Exchange Commission, and the Food and Drug Administration) see social media and online collaboration as a means of providing richer, more useful, and more interactive pathways for community participation. For EPA and its stakeholders, the question is whether the agency can take advantage of this growing social interconnectivity to engage the public in environmental protection better while bolstering both its science activities and its capacity for effective regulatory monitoring and enforcement. There are a number of ways in which crowdsourcing or citizen science could augment or enhance EPA’s scientific and regulatory capabilities. They include harnessing new technologies to engage broader communities along the lines of crowdsourced data collection, especially in the context of environmental monitoring, exposure assessment, health surveillance, and social behaviors; crowdsourced data classification and analysis; and crowdsourced environmental problem-solving. Crowdsourcing also provides an opportunity for EPA to gain a better understanding of the general sentiment of the public on issues that are of concern to EPA.
Crowdsourcing initiatives are typically low in cost because the most expensive resource (people’s time) is supplied voluntarily. Whether classifying galaxies or recording observations of bird species or local environmental quality, participants in a crowdsourcing project are intrinsically motivated to participate. For an agency like EPA, crowdsourcing presents an opportunity to gather and analyze large amounts of data or input inexpensively. That being said, crowdsourcing projects are not free to run either. There are costs involved in supplying the infrastructure for participation (typically a Web site or mobile interface where participants can record observations and discuss issues) and managing the overall effort.
Acquisition of Environmental Data through Remote Sensing
In the 40 years since the launch of Landsat 1—the first Earth-observing satellite-borne sensor designed expressly to study the planet’s land surfaces— there have been enormous advances in remote-sensing systems for environmental mapping and monitoring. They include multispectral digital imaging systems and imaging radar (1970s), hyperspectral imaging systems (1980s), and profiling and imaging LiDAR (1990s to present). In that period, remote sensing has benefited from rapid improvements in instrument capabilities and calibration, positional control and global positioning systems, computer performance, processing algorithms and software, fusion of imagery from multiple sensors, and closer integration with geographic information system and ground measurements and monitoring systems. As a result, remote sensing of the environment has evolved from a narrow research community to a large and diverse user community that is applying remote-sensing products on local, regional, and global scales (Schaepman et al. 2009).
BOX 3-1 Engaging the Public to Gather Information
One example of crowdsourcing is a “computer game” called Fold-IT, in which participants work to fold proteins in different configurations (Foldit 2012). The scoring for the game is based on packing the protein (the smaller the better), hiding the hydrophobic side chains, and folding the protein so that sidechains are not too close together. The information gathered from this program has resulted in the publication of several scientific papers (Cooper et al. 2010a,b, 2011; Gilski et al. 2011; Khatib et al. 2011a,b) and has shown that sometimes human knowledge and intuition can outperform computational methods.
Another example is when EPA set out to produce an action plan for the Puget Sound estuary in Washington state. It launched an information challenge that invited the broader community to assemble relevant data sources and begin to articulate solutions. Over 600 residents, businesses, environmental groups, and researchers contributed 175 new data sources (Tapscot and Williams 2010). Examples include a tree-ring database from 2006 that provides a baseline for monitoring the effects of climate change on local tree species, wildlife toxicology maps of the Puget Sound area, and real-time water quality-monitoring tools, including water measurements taken from local ferries that could complement existing buoy measurement systems. Former EPA Chief Information Officer Molly O’Neill said afterwards, “we can actually use these kinds of mass collaboration tools to transform government, not just add layers to government” (Tapscot and Williams 2010). The kinds of “emergent behavior” observed in cases like the Puget Sound information challenge can be applied in nearly all aspects of the regulatory system and lead to new insights, innovations, and strategies that even the most capable agencies could not produce in isolation (PSP 2011).
EPA has long recognized the scientific value and cost effectiveness of remote sensing for large-area environmental mapping and monitoring, and remote sensing data are increasingly being used to strengthen human exposure characterization for air pollutants and other contaminants. The agency has been a contributing partner in national satellite-based mapping programs such as the Multi-Resolution Land Cover Program, the Coastal Change Analysis Program, and the Gap Analysis Program. It has also supported research and application efforts in remote sensing of water quality and air quality, notably in the use of aircraft-borne sensors for local pollution and hazardous-substance detection and monitoring. For example, from 2002 to 2010, the agency partnered with the Department of Defense and the National Geospatial Intelligence Agency to operate Airborne Spectral Photometric Environmental Collection Technology, an aircraft-borne set of sensors designed to provide emergency-response data on hazardous releases.
Much of the progress in remote sensing has depended on tight integration of imagery with other geospatial data and process models that use appropriate parameters and aircraft and satellite data. A key opportunity for EPA science
lies in extended collaboration with remote-sensing scientists to advance such integrated approaches, especially in fields in which EPA has extensive or nascent expertise in such domains as pollutant fate and transport, landscape ecology, ecosystem service mapping and monitoring, environmental-disaster monitoring, and health impact assessment.
Terrestrial-Ecosystem Monitoring with Remote Sensing
Remote sensing of land surfaces has evolved from technically complex but thematically relatively simple land-use and land-cover mapping and monitoring to technically and scientifically complex monitoring and modeling of surface properties and processes, such as three-dimensional (3D) vegetation structure and net primary production. From a technologic perspective, important trends in remote sensing of terrestrial ecosystems include (Wang et al. 2010)
• Increasing availability of multispectral imagery with very high spatial resolution (0.5-10 m) from satellite systems such as IKONOS, GeoEye-1, SPOT-5, and FORMOSAT-2.
• Increasing availability of imaging spectrometer data with more than 100 narrow (10-20 nm) spectral bands at moderately high (10-500 m) resolution from satellite-borne systems, such as EO-1 Hyperion, and aircraft-borne systems, such as the Compact Airborne Spectrographic Imager.
• Imaging LiDAR from aircraft platforms for regional studies (for example, laser vegetation-imaging sensors) and satellite platforms for global studies (for example, the Geosciences Laser Altimeter System carried on the Ice, Cloud, and Land Elevation Satellite).
• Well-calibrated thermal remote-sensing data at fine spatial resolution (the Advanced Spaceborne Thermal Emission and Reflection Radiometer and the Hyperspectral Infrared Imager), moderate resolution (the Moderate Resolution Imaging Spectrodiameter and the Visible Infrared Imager Radiometer Suite), and coarse resolution (the Geostationary Operational Environmental Satellite and the Meteosat Second Generation Satellite) for monitoring surface-energy balance, evapotranspiration, plant stress, and drought.
• Constellations of small satellites capable of high-frequency global coverage for environmental event and disaster monitoring (for example, the UK Disaster Monitoring Constellation Satellite).
Imaging spectrometers hold special promise for obtaining detailed information about plant-community composition and the physiologic condition of canopies and allowing monitoring of community succession, phenology, species invasions, crop yield, soil chemistry, and nutrient cycling. Issues of data quality and data access are diminishing, and progress is being made in radiative-transfer models, spectral-mixture models, and physically based inversion models for multiscale monitoring of terrestrial ecosystem processes (Schaepman et al. 2009).
Imaging LiDAR is especially powerful for tracking changes in surface elevation, above-ground vegetation biomass, 3D vegetation structure, and 3D distribution of canopy-leaf area. Particularly in forested regions, LiDAR can be used to improve estimates of net ecosystem production and carbon stocks over large areas (Goetz and Dubayah 2011; Hall et al. 2011). Imaging radar has also been an important tool for monitoring vegetation structure, especially in cloud-prone areas and areas subjected to seasonal flooding (Bergen et al. 2009). Image fusion, the combined use of imagery from two or more sensors, can be used to exploit complementary information from very-high-resolution multispectral imagery, hyperspectral imagery, and LiDAR (Koetz et al. 2007).
Long-Term Datasets in Real-Time
Dense, long-term environmental datasets in real-time could create a foundation for informed decision-making. A suite of decision-support tools could be developed for integration with air- and water-quality models at various scales. For example, data in hospital admission forms could be combined with meteorological and air quality models in real time to provide health forecasts and warnings. Real-time sensing and modeling of water-borne pathogens in situ could provide drinking water treatment plants with threat forecasts, alerting them to the need to change source water or treatment techniques. Special research attention could be given to handling uncertainty of both data and models. In addition to deterministic models with uncertainty analyses, probabilistic approaches can be extremely powerful when computational intelligence tools are used.
Data assimilation and data mining approaches provides innovative possibilities. An example is the use of an intelligent real-time cyberinfrastructure-based information system called the Intelligent Digital Watershed to better understand the interactions and dynamics between human activity and water quality and quantity. Such an approach provides “1) novel uses of data mining algorithms in data quality and model construction, 2) development of specialized data mining algorithms for [environmental forecasting] applications, 3) development of data transformation algorithms, [4)] data-driven modeling of non-stationary processes, [such as storm forecasting for by-pass wastewater discharges], and [5)] development of decision-making algorithms for models constructed with data mining algorithms”.4 Using data in a novel way could greatly expand the analysis capability of EPA and provide insights previously impossible to obtain without such innovations.
Already, the Consortium of Universities for the Advancement of Hydro-logic Science, Inc. Hydrologic Information System (HIS) project has a systematic data acquisition network for the publication, discovery, and access of water
![]()
4 NSF-CDI. 2008-2011. CDI-Type II: Understanding Water-Human Dynamics with Intelligent Digital Watersheds. (#0835607). Jerald L. Schnoor (PI), David Bennett, Andrew Kusiak, Marian Muste, and Silvia Secchi.
data (CUAHSI 2012a). HIS has pooled datasets from many sources into a coherent and accessible prototype national system for water resource data discovery, delivery, publication, and curation (CUAHSI 2012b). What is missing is the integration of the data into a modeling or forecasting system, which EPA could provide. Problems could be analyzed and solved by using an intelligent digital environmental data system. A human information system is also needed to archive land use, census, voting, planning, and other socioeconomic data relevant to environmental processes and management. The socioeconomic and environmental data would be referenced to common coordinates for use in cross referencing and to enable testing of hypotheses concerning how to solve problems in innovative ways (such as behavioral incentives vs command-and-control).
A central tenet of an intelligent digital environmental data system is that dense, coherent, accessible, multidisciplinary data will serve as an attractor to bring together a broad range of environmental scientists, social scientists, and engineers to pose research questions and devise solutions to environmental problems. It could encourage a social transformation in how interdisciplinary work is accomplished.
Archives and Repositories
It is essential to characterize the environment in diverse ways, although many of the data that result are of limited use without the ability to detect change over time. The implications of exposure to toxic and harmful materials are understood to some extent, but many of the issues being addressed by EPA are in the context of environmental factors whose effects are best characterized in terms of changing exposures or accumulation of materials. Given the great spatial and temporal variability of those same factors, it is often difficult to understand the importance of measurements at a single point in time or space, so measurements of low spatial and temporal scope can easily lead to spurious conclusions. To ensure that EPA and environmental scientists more broadly can effectively understand the relative importance of any single environmental data-set, it is critical to develop and maintain long-term records that are composed of multiple parameters (Lovett et al. 2007). The challenge is to ensure that enough environmental data are collected and preserved to support understanding of long-term trends among the key parameters now identified as important, while providing a high likelihood of providing the data necessary to understand emerging issues. Making data and samples accessible to future researchers are central to ensuring that the understanding of environmental phenomenon continues to grow and evolve with the science. Ensuring that all data collected with federal funds are archived and accessible is critically important, although ideally that would be the norm for all environmental data collected with public or private funds. It is also important to develop sample archives in which materials are appropriately stored for analysis or reanalysis later. New measurement techniques are constantly emerging and providing useful insights. When it is feasible
to do the new analyses with stored samples, and thereby create a long-term record of exposure or change, the resulting insights can be invaluable for understanding the implications of new observations (see Rothamsted 2012 for an example of the information that this type of long-term data potentially could provide).
USING NEW SCIENCE TO DRIVE SAFER TECHNOLOGIES AND PRODUCTS
In addition to using new tools and technologies to address the major challenges identified in Chapter 2, it will be important for EPA to continue to look for new ways of preventing environmental problems before they arise. The tools and technologies for measuring and managing scientific data outlined in this chapter have generally been thought of in the context of refined risk-assessment processes. The use of scientific information for the purposes of risk assessment is focused in large part on detailed and nuanced problem identification—that is, a holistic understanding of causes and mechanisms. Such work is important and valuable in understanding how toxicants and other stressors affect environmental health and ecosystems, and at times is required by statute. However, the focus on problem identification often occurs at the expense of efforts to use scientific tools to develop safer technologies and solutions. Consideration of whether functional, cost-effective, and safer alternative manufacturing processes or materials exist that could reduce or eliminate risks while still stimulating innovation is not often part of the risk-assessment processes undertaken by EPA. Given the changing nature of chemical exposures in the United States, from large point sources to disperse, non-point exposures, the traditional tools of exposure assessment and control will likely be insufficient to prevent exposure to chemicals and it may be more effective to place a greater focus on preventing exposure through design changes. NRC (2009) outlined a framework for risk assessment in which the assessment process is tied to evaluating risk-management options rather than the safety of single hazards.
Defining problems without a comparable effort to find solutions greatly diminishes the value of the agency’s applied research efforts and may impede its mission to protect human health and the environment. Furthermore, if EPA’s actions lead to a change in technology, chemical, or practice, there is a responsibility to understand alternatives and to support a path forward that is environmentally sound, technically feasible, and economically viable (Tickner 2011). Sarewitz et al. (2010) have proposed the Sustainable Solutions Agenda as an alternative approach to think about sustainability problems in the context of complex systems. As noted in other parts of this report, uncertainty is an inevitable part of decision-making processes surrounding complex risks. The Sustainable Solutions Agenda asks a different set of questions about such problems, from asking whether “x causes y” or leads to an “unacceptable” risk to “given current knowledge of the possibility that x causes y, is there a way to move to-
ward more sustainable practice by reducing or replacing x while preserving some or most of its benefits?” Such a focus on solutions through alternatives assessment processes can support the agency’s dual science and engineering goals: protection and innovation.
The Pollution Prevention Act of 1990 established the principle that all EPA environmental protection efforts should be based on the prevention or reduction of pollution at the source. Pollution prevention was viewed as such an important program for the agency that its coordination initially occurred through the administrator’s office (EPA 2008b). On the basis of the Pollution Prevention Act’s mandates, EPA’s Office of Research and Development (ORD) and the Office of Pollution Prevention and Toxics (OPPT) embarked on a wide array of initiatives to develop tools, information sources, technologies, approaches, and initiatives to advance pollution prevention. Those have resulted in making EPA a global leader for the application of science and engineering for prevention. Examples of those initiatives are the Cleaner Technologies Substitutes Assessments, the Green Suppliers Network, and a suite of tools to integrate consideration of pollution prevention into chemical design. With more resources and increased coordination at the highest levels of leadership in the agency, EPA has the ability to enhance its support for safer technologies and products. Some mechanisms through which enhanced support can be accomplished include funding research on safer chemistry; building tools so that designers outside the agency can create safer chemicals, products, and processes; providing simple data integration dashboards that will help companies identify and evaluate safer alternatives to chemicals and materials of concern; and setting up consistent guidelines, frameworks, and metrics for evaluating safer chemicals and products.
EPA has taken global leadership in three fields of innovative solutions-oriented science: pollution prevention, Design for the Environment, and green chemistry and engineering. This suite of programs compromises non-regulatory approaches that protect the environment and human health by designing or redesigning processes and products to reduce the use and release of toxic materials. Green chemistry and engineering focuses on molecular design, Design for the Environment focuses on evaluating the safest chemistries and designs for a particular functional use, and pollution prevention focuses on reducing or eliminating emissions and waste in the manufacturing process. The three programs have evolved and changed over time and are overlapping in many ways, but they address different parts of the production process, from chemical design to the use of chemicals in product design to the application in manufacturing. Despite the overlapping connections, the three programs have not been fully integrated in EPA’s administrative structures within or between ORD and OPPT, which may ultimately limit the impact and effectiveness of the programs.
Pollution prevention, Design for the Environment, and green chemistry and engineering share a number of common features. First, they have a strong emphasis on education and assistance. To support the change in mindset from “controlling exposure to hazardous materials” to “preventing generation of haz-
ardous materials” or “reducing the hazards of the materials of commerce”, each program has developed educational materials. Technical assistance and tools, methods, and expertise are also provided, and research efforts are initiated through ORD and OPPT. Second, they align environmental protection with economic development. A strong incentive for participation in the programs derives from the potential for economic advantages that result from alternative approaches. Using less material or less toxic materials can reduce costs. Innovative solutions driven by environmental concerns can open new markets. Third, they promote strong partnerships between agencies, industry, nongovernment organizations, and academic institutions. The programs recognize participants from outside the agency, including all the stakeholders in the chemical enterprise, as partners that are needed to implement the changes that the programs promote. The partners bring content expertise, research and development resources, and commercialization pipelines that are essential for implementing change and bringing improved products or processes into the marketplace. Fourth, they provide a mechanism for nimbleness. In emphasizing the search for innovative solutions to specific problems, the programs are nimble. In each case, small supporting efforts within the agency support a framework that harnesses and leverages the efforts of innovators in industry, academe, and nongovernment organizations. And fifth, they are a form of voluntary action. Each program promotes participation through incentives as opposed to regulatory approaches. Self-interest of the participants rewards and reinforces participation. These programs are described in greater detail below.
Pollution Prevention
Launched in 1990 through the Pollution Prevention Act (EPA 2011c), EPA’s pollution-prevention program was a paradigm shift for the agency in its focus on preventing the generation of waste (source reduction) as opposed to the previous command-and-control, “end-of-pipe” solutions (Browner 1993). The agency recognized that the new approach could be more cost-effective and provide competitive advantages for companies that adopted it. EPA’s pollution-prevention efforts have focused on partnerships for prevention in various sectors, such as the automobile, electronics, and health-care sectors; technical support through a network of federal and state technical-assistance providers; technology-development projects; demonstration projects to evaluate technologies; sustainable procurement; and the development of tools for evaluating pollution-prevention options. EPA has established a 2011–2014 strategic plan for pollution prevention that outlines directions for the program (EPA 2010). It has a responsibility to ensure that the market is moving in the right direction, and this can be accomplished through some of the mechanisms described above. Box 3-2 shows an example of how the private sector has influenced the market without the use of regulatory mandates.
BOX 3-2 Example of Private Industry’s Influence on the
Supply Chain without Regulatory Mandates
The increasingly global nature of production, coupled with the expanding number of chemicals used in commerce, presents a daunting challenge for protection of human health and ecosystem quality. It is also challenging for either regulations or the underlying science to keep pace. When the complex interactions between chemicals and their vast production networks are considered, the problems become even more daunting inasmuch as they span organizational and national boundaries.
Partly as a response to those challenges, corporations in several industries have begun to issue supply-chain mandates in which they demand changes in production processes and material inputs from suppliers over which they exert economic influence. Under those mandates, firms in a corporation’s supply chain are obliged to meet customer expectations and adopt specific requirements with the promise of future contracts or under the threat of discontinuation of business. The private-sector policies are rapidly emerging as part of a new generation of quasiregulatory policy tools whereby private organizations use economic leverage to effect changes in pursuit of the public good. The efficacy of such mandates is still an open question, but their emergence signals an important evolution and opportunity in the development of strategies aimed at improving consumer protections against exposure to harmful substances. Although such private-sector actions are not a substitute for effective chemical regulations, if developed and executed correctly, they can augment public policies substantially.
An example of the potential influence of managing supply chains is Wal-Mart’s commitment to selling high-efficiency light bulbs and the effects that policy had on energy use in the United States (Barbaro 2007). By working with its suppliers on product quality and price, they fulfilled a commitment to quadruple sales of efficient light bulbs. These sales were equal to the maximum sustainable sales—that is, the number of light bulbs per household and the technology available. This effort radically reduced the electricity used by their customers, reducing demand for electricity that was equal to the output of three or four 700 MW power plants.
Another example that demonstrates the power of the marketplace for chemical substitution is bisphenol-A in polycarbonate water bottles (Tickner 2011). As a result of consumer campaigns, emerging science, and state regulations, major retailers of water bottles, such as REI, rapidly switched from polycarbonate to alternative materials. However, in the switch, little research was undertaken on the alternative materials, which has the potential to lead to health and environmental concerns at a later time.
Design for the Environment
Established in 1992, EPA’s Design for Environment program is a model of stakeholder-engaged product design to reduce the environmental effects of consumer products. The Design for Environment concept “encourages busi-
nesses to incorporate environmental and health considerations in the design and redesign of products and processes” (EPA 2001). It merges several non-regulatory, voluntary initiatives related to the synthesis of chemicals that are safer, an analysis of the risks related to these chemicals, and the development of alternative chemicals and technologies (EPA 2012e). It promotes a collaborative process to improve product design, provides information and tools on design strategies and alternative ingredients, and uses technical assistance, design methods, and a labeling program to create incentives for participation. To achieve its goals, Design for Environment has undertaken cutting-edge research on tools and approaches for advancing safer product design, undertaken a number of supply chain partnerships on more sustainable materials, and engaged in significant outreach with industry and other partners (EPA 1999). Design for the Environment partnerships consider human health and environmental implications, the performance of products, and the economic effects of traditional and alternative chemicals, materials, technologies, and processes (EPA 2006b). In recent years, a primary goal of the Design for the Environment program has been to achieve “informed substitution”, that is, moving from a chemical that raises health or environmental concerns to chemicals that are known to be safer or to nonchemical alternatives (EPA 2009). According to EPA (2009), “the goals of informed substitution are to minimize the likelihood of unintended consequences, which can result from a precautionary switch away from a chemical of concern without fully understanding the profile of potential alternatives, and to enable a course of action based on the best information—on the environment and human health—that is available or can be estimated.” Design for the Environment achieves its goals through both alternatives-assessment processes and recognition programs. Through its alternatives-assessment processes, Design for the Environment has evaluated alternatives to polybrominated diphenylethers in furniture flame retardants, tetrabromobisphenol-A in printed-circuit boards, and bisphenol-A in thermal cash-register tape, and is examining alternatives to phthalates in wire and cable. The Design for the Environment Safer Product Labeling Program evaluates products and labels the ones that meet the program’s safety standards (EPA 2012f). EPA establishes minimum toxicologic criteria for individual cleaning-formulation constituents, and manufacturers submit alternatives that meet the criteria for third-party certification, creating a marketplace for alternative formulations. EPA has developed detailed transparent criteria for evaluation for both programs.
Green Chemistry and Engineering
Green chemistry is another innovative approach to environmental protection that emerged from the Pollution Prevention Act of 1990 (EPA 2011d). Green chemistry and engineering incorporates hazard reduction and waste minimization into design at the molecular level to reduce hazards throughout the life cycle. Green chemistry has been defined as “the utilization of a set of principles that reduces or eliminates the use or generation of hazardous substances in
the design, manufacture and application of chemical products” and is guided by 12 design principles set forth by Anastas and Warner (2000). The approach goes well beyond reduction of waste and process optimization—it drives the redesign of production and processes at the molecular level. The scope of green chemistry extends beyond alternative-chemicals assessment; when alternatives are not available or suitable, green chemistry principles can be used to design new substances that have environmental performance incorporated at the design stage. The principles address effects throughout the life cycle of a material or product and spur new solutions that represent system-wide reduction in effects as opposed to reduction in effects in only one facet of the life cycle.
Case studies have been used to show how green chemistry and engineering reduce costs and spur innovation by exploring entirely new approaches that are driven by life-cycle thinking and systems thinking. EPA’s green chemistry efforts center around research on innovative technologies, provision of tools to evaluate and design green chemistries and processes, and recognition of leadership in green chemistry innovation. For example, the Presidential Green Chemistry Challenge Awards Program (established in 1995) has been used to reward success in green chemistry and to communicate the value of the approach for reducing effects and advancing commercial interests. According to EPA (EPA 2011e),
During the program’s life, EPA has received more than 1,400 nominations and presented awards to 82 winners. Winning technologies alone are responsible for reducing the use or generation of more than 199 million pounds of hazardous chemicals, saving 21 billion gallons of water, and eliminating 57 million pounds of carbon dioxide releases to the air. These benefits are in addition to significant energy and cost savings by the winners and their customers.
Future Implications for Innovation
The three programs described above demonstrate the potential for innovative approaches to advance and use scientific knowledge to protect health and the environment through the redesign of chemicals, materials, and products. They also show the role that EPA can play in driving decisions by providing high-quality scientific information. Since their inception, the three programs have had important data needs and strong links to data generation and tools development. For example, pollution-prevention efforts have been measured using data on chemical release, waste, and use generated by the Toxics Release Inventory and in accordance with several state-level pollution-prevention bills. EPA’s Sustainable Futures Initiative has relied on data from EPA’s New Chemicals Program and EPA’s expertise in chemical assessment and structure—activity relationships to develop tools to assist chemical designers in developing safer products (EPA 2012g). Similarly, efforts are underway to utilize data developed
through EPA’s ToxCast and ExpoCast programs for the development and evaluation of options for green chemistry and Design for the Environment. A 2011 workshop hosted by the NRC Committee on Emerging Science for Environmental Health Decisions specifically explored applying 21st century toxicology to green chemical and material design (NRC 2011).
The science and engineering tools and technologies for measuring, monitoring, and managing environmental and health data outlined in Chapter 3 can provide essential information to drive sustainable solutions through prevention programs. For example, data on chemicals used in media and sensing can be important in setting priorities among chemicals, processes, and products for prevention actions and for measuring results of such actions; toxicogenomics and exposure data are critical for supporting design and evaluation of new technologies, comparing alternatives throughout their life cycles, and helping to avoid unintended consequences; and crowdsourcing and social-media tools provide a mechanism for sharing information about successful innovations and enhancing existing technical support and demonstration efforts. In addition to traditional environmental sciences, there is a critical need for behavioral and social sciences in advancing the development and adoption of safer chemicals, materials, and products. The data that these scientific disciplines provide are important inputs for characterizing and making the economic case for new technologies, for understanding business and consumer behavior, and for effecting the behavioral changes necessary to ensure such innovations take root in such a way that consumer preferences recognize safer materials.
In today’s information age, explosive amounts of data are generated through all kinds of media, for different purposes, and by commercial or research organizations in both the private and public sectors. It will be a cornerstone of the future of science in general, and EPA’s future science in particular, that EPA be able to harvest and synthesize the large amounts of data that transcend geopolitical and scientific disciplinary boundaries. Taking advantage of these data requires a variety of techniques, led by careful problem formulation to ensure that the appropriate data are being collected or analyzed. It also requires state-of-the-art capability in data integration and synthesis, particularly in the areas of data-mining, and in modeling of biologic systems with biostatistics, computer simulation, and other emerging methods. Although the committee notes that it is imperative that EPA be conversant in the latest tools and technologies, a subset of which are discussed in this chapter, it also recognizes that there are substantial constraints on resources. In many cases, building capacity in new and emerging technologies can be achieved through strategic collaborations and should not come at the expense of core disciplines relevant to its mission. These core disciplines include, but are not limited to, statistics, chemistry, economics, environmental engineering, ecology, toxicology, epidemiology, ex-
posure science, and risk assessment. Regardless, leveraging insights from new tools and technologies will be necessary to address some of the emerging problems of the 21st century.
Aboytes, R., G.D. Di Giovanni, F.A. Abrams, C. Rheinecker, W. Mcelroy, N. Shaw, and M.W. Lechevallier. 2004. Detection of infectious Cryptosporidium in filtered drinking water. J. Am. Water Works Assoc. 96(9):88-98. Appel, K.W., S.J. Ro-selle, R.C. Gilliam, and J.E. Pleim. 2010. Sensitivity of the Community Multiscale Air Quality (CMAQ) model v.4.7 results for the eastern United States to MM5 and WRF meteorological drivers. Geosci. Model Dev. 3(1):169-188.
Ahsan, H., Y. Chen, F. Parvez, M. Argos, A.I. Hussain, H. Momotaj, D. Levy, A. van Geen, G. Howe, and J. Graziano. 2006. Health Effects of Arsenic Longitudinal Study (HEALS): Description of a multidisciplinary epidemiologic investigation. J. Expo. Sci. Environ. Epidemiol. 16:191-205.
Anastas, P.T., and J.C. Warner. 2000. Green Chemistry: Theory and Practice. New York: Oxford University Press.
Appel, K.W., S.J. Roselle, R.C. Gilliam, and J.E. Pleim. 2010. Sensitivity of the Community Multiscale Air Quality (CMAQ) model v4.7 results for the eastern United States to MM5 and WRF meteorological drivers. Geoscientific Model Development. 3(1):169-188.
ASCE (American Society of Civil Engineers). 2004. Interim Voluntary Guidelines for Designing an Online Contaminant Monitoring System. American Society of Civil Engineers, Reston, VA [online]. Available: http://www.michigan.gov/documents/deq/deq-wb-wws-asceocms_265136_7.pdf [accessed Apr. 5, 2012].
Aw, T.G., and J.B. Rose. 2012. Detection of pathogens in water: From phylochips to qPCR to pyrosequencing. Curr. Opin. Biotechnol. 23(3):422-430.
Baker, D., and M.J. Nieuwenhuijsen. 2008. Environmental Epidemiology: Study Methods and Application. Oxford: Oxford University Press.
Barbaro, M. 2007. Wal-Mart puts some muscle behind power-sipping bulbs. January 2, 2007. New York Times [online]. Available: http://www.nytimes.com/2007/01/02/business/02bulb.html?pagewanted=all [accessed Mary 17, 2012].
Bergen, K.M., S.J. Goetz, R.O. Dubayah, G.M. Henebry, C.T. Hunsaker, M.L. Imhoff, R.F. Nelson, G.G. Parker, and V.C. Radeloff. 2009. Remote sensing of vegetation 3-D structure for biodiversity and habitat: Review and implications for LiDAR and radar spaceborne missions. J. Geophys. Res. Biogeosci. 114:G00E06, doi:10.1029/ 2008JG000883.
Bibby, K., E. Viau, and J. Peccia. 2010. Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Res. 44(14):4252-4260.
Bibby, K., E. Viau, and J. Peccia. 2011. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 52(4):386-392.
Bierman, P., M. Lewis, B. Ostendorf, and J. Tanner. 2011. A review of methods for analysing spatial and temporal patterns in coastal water quality. Ecol. Indic. 11(1): 103-114.
Blaauboer, B.J. 2010. Biokinetic modeling and in vitro-in vivo extrapolations. J. Toxicol. Environ. Health B Crit. Rev. 13(2-4):242-252.
Browner, C.M. 1993. Pollution Prevention Takes Center State. EPA Journal, July/September 1993 [online]. Available: http://www.epa.gov/aboutepa/history/topics/ppa/01.html [accessed Apr. 9, 2012].
Byappanahalli, M.N., R.L. Whitman, D.A. Shively, and M.B. Nevers. 2010. Linking non-culturable (qPCR) and culturable enterococci densities with hydrometeorological conditions. Sci. Total Environ. 408(16):3096-3101.
CAMRA. 2012. Welcome to the CAMRA Microbial Risk Assessment Wiki (CAM-RAwiki) [online]. Available: http://wiki.camra.msu.edu/index.php?title=Main_Page [accessed Apr. 10, 2012].
Cangelosi, G.A., K.M. Weigel, C. Lefthand-Begay, and J.S. Meschke. 2010. Molecular detection of viable bacterial pathogens in water by ratiometric pre-rRNA analysis. Appl. Environ. Microbiol. 76(3):960-962.
CDC (Centers for Disease Control and Prevention). 2012. National Report on Human Exposure to Environmental Chemicals. Centers for Disease Control and Prevention [online]. Available: http://www.cdc.gov/exposurereport/ [accessed Mar. 30, 2012].
Christensen, V.G., P.P. Rasmussen, and A.C. Ziegler. 2002. Real-time water quality monitoring and regression analysis to estimate nutrient and bacteria concentrations in Kansas streams. Water Sci. Technol. 45(9):205-211.
Cooper, S., F. Khatib, A. Treuille, J. Barbero, J. Lee, M. Beenen, A. Leaver-Fay, D. Baker, Z. Popović, and Foldit Players. 2010a. Predicting protein structures with a multiplayer online game. Nature 466(7307):756-760.
Cooper, S., A. Treuille, J. Barbero, A. Leaver-Fay, K. Tuite, F. Khatib, A.C. Snyder, M. Beenen, D. Salesin, D. Baker, Z. Popović, and Foldit Players. 2010b. The challenge of designing scientific discovery games. Pp. 40-47 in Proceedings of the Fifth International Conference on the Foundations of Digital Games (FDG 2010), June 19-21, Monterey, CA. New York: ACM [online]. Available: http://www.cs.washington.edu/homes/zoran/foldit-fdg10.pdf [accessed Mar. 30, 2012].
Cooper, S., F. Khatib, I. Makedon, H. Lu, J. Barbero, D. Baker, J. Fogarty, Z. Popović, and Foldit Players. 2011. Analysis of social gameplay macros in the Foldit cookbook. Pp. 9-14 in Proceedings of the Sixth International Conference on the Foundations of Digital Games (FDG 2011), June 28-July 1, 2011, Bordeaux, France. New York: ACM [online]. Available: http://grail.cs.washington.edu/projects/protein-game/foldit-fdg11.pdf [accessed Mar. 30, 2012].
Coppus, R., and A.C. Imeson. 2002. Extreme events controlling erosion and sediment transport in a semi-arid sub-Andean valley. Earth Surf. Proc. Land. 27(13):1365-1375.
Crouse, D.L., P.A. Peters, A. van Donkelaar, M.S. Goldberg, P.J. Villeneuve, O. Bioron, S. Khan, D.O. Atari, J. Jerrett, C.A. Pope, M. Brauer, J.R. Brook, R.V. Martin, D. Stieb, and R.T. Burnett. 2012. Risk of nonaccidental and cardiovascular mortality in relation to long-term exposure to low concentrations of fine particular matter: A Canadian national-level cohort study. Environ. Health Perspect. 120(5):708-714.
CUAHSI (Consortium of Universities for the Advancement of Hydrologic Science, Inc.). 2012a Hydrologic Information System [online]. Available: http://his.cuahsi.org [accessed Apr. 23, 2012].
CUAHSI (Consortium of Universities for the Advancement of Hydrologic Science, Inc.). 2012b. CUAHSI — Data Access, Publication and Analysis [online]. Available: http://www.cuahsi.org/his.html [accessed Apr. 23, 2012].
Diez Roux, A.V. 2011. Complex systems thinking and current impasses in health disparities research. Am. J. Public Health 101(9):1627-1634.
Djikeng, A., R. Kuzmickas, N.G. Anderson, and D.J. Spiro. 2009. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS ONE 4(9):e7264.
Dominici, F., R.D. Peng, C.D. Barr, and M.L. Bell. 2010. Protecting human health from air pollution: Shifting from a single-pollutant to a multipollutant approach. Epidemiology 21(2):187-194.
Eder, B., D. Kang, R. Mathur, J. Pleim, S. Yu, T.A. Otte, and G. Pouliot. 2009. Performance evaluation of the National Air Quality Forecast Capability for the summer of 2007. Atmos. Environ. 43(14): 2312-2320.
Egeghy, P.P., R. Judson, S. Gangwal, S. Mosher, D. Smith, J. Vail, and E.A. Cohen Hubal. 2012. The exposure data landscape for manufactured chemicals. Sci. Total Environ. 414(1):159-166.
Elliott, P., and T.C. Peakman. 2008. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int. J. Epidemiol. 37(2):234-244.
Environmental Information Exchange Network. 2011. Exchange Network Leadership Council [online]. Available: http://www.exchangenetwork.net/about/network-management/exchange-network-leadership-council/ [accessed Apr. 2, 2012].
EPA (US Environmental Protection Agency). 1984. Risk Assessment and Management: Framework for Decision Making. EPA 600/9-85-002. US Environmental Protection Agency, Washington, DC.
EPA (US Environmental Protection Agency). 1986. Ambient Water Quality Criteria for Bacteria-1986: Bacteriological Water Quality Criteria for Marine and Fresh Recreational Waters. EPA-440/5-84-002. Office of Water Regulations and Standards, US Environmental Protection Agency, Washington, DC. January 1986 [online]. Available: http://water.epa.gov/action/advisories/drinking/upload/2009_04_13_beaches_1986crit.pdf [accessed Apr. 3, 2012].
EPA (US Environmental Protection Agency). 1997. Approval of EPA Method 1613 for Analysis of Dioxins and Furans in Wastewater. Fact Sheet, July 1997. Office of Water, US Environmental Protection Agency [online]. Available: http://water.epa.gov/scitech/methods/cwa/organics/dioxins/1613-fs.cfm [accessed Apr. 2, 2012].
EPA (US Environmental Protection Agency). 1999. Design for the Environment: Building Partnerships for Environmental Improvement. EPA744-R-99-003. Office of Pollution Prevention and Toxics, US Environmental Protection Agency, Washington, DC.
EPA (US Environmental Protection Agency). 2000. Risk Characterization Handbook. EPA 100-B-00-002. Office of Science Policy, Office of Research and Development, US Environmental Protection Agency, Washington, DC [online]. Available: http://www.epa.gov/stpc/pdfs/rchandbk.pdf [accessed Aug. 1, 2012].
EPA (US Environmental Protection Agency). 2001. The US Environmental Protection Agency’s Design for Environment Program. EPA744-F-00-020. Office of Pollution Prevention and Toxics, US Environmental Protection Agency, Washington, DC. May 2001 [online]. Available: http://www.epa.gov/dfe/pubs/tools/DfEBrochure.pdf [accessed Apr. 11, 2012].
EPA (US Environmental Protection Agency). 2002. Method 1600: Enterococci in Water by Membrane Filtration Using membrane-Enterococcus Indoxyl-β-D-Glucoside Agar (mEI). EPA-821-R-02-022. Office of Water, US Environmental Protection Agency, Washington DC [online]. Available: http://www.caslab.com/EPA-Methods/PDF/EPA-Method-1600.pdf [accessed Apr. 3, 2012].
EPA (US Environmental Protection Agency). 2005. Microbial Source Tracking Guide Document. EPA-600/R-05/064. Office of Research and Development, US Envi-
ronmental Protection Agency, Washington DC. June 2005 [online]. Available: http://www.ces.purdue.edu/waterquality/resources/MSTGuide.pdf [accessed Apr.3, 2012].
EPA (US Environmental Protection Agency). 2006a. Implementing the BEACH Act of 2000. Report to Congress. US Environmental Protection Agency [online]. Available: http://water.epa.gov/type/oceb/beaches/report_index.cfm [accessed Aug. 13, 2012].
EPA (US Environmental Protection Agency). 2006b. Design for the Environment Partnership Highlights. US Environmental Protection Agency, March 2006 [online]. Available: http://www.epa.gov/dfe/pubs/about/dfe-highlights06b.pdf [accessed Apr. 11, 2012].
EPA (US Environmental Protection Agency). 2008a. US EPA Office of Research and Development Computational Toxicology Research Program Implementation Plan for Fiscal Years 2009 to 2012: Providing High-Throughput Decision Support Tools for Screening and Assessing Chemical Exposure, Hazard, and Risk. US Environmental Protection Agency [online]. Available: http://epa.gov/ncct/download_files/basic_information/CTRP2_Implementation_Plan_FY09_12.pdf [accessed Apr. 2, 2012].
EPA (US Environmental Protection Agency). 2008b. Evaluation of EPA Efforts to Integrate Pollution Prevention Policy throughout EPA and at Other Federal Agencies. Office of Pollution, Prevention and Toxics, US Environmental Protection Agency. October 2008 [online]. Available: http://www.epa.gov/p2/pubs/docs/p2integration.pdf [accessed Apr. 20, 2012].
EPA (US Environmental Protection Agency). 2009. EPA’s DfE Standard for Safer Cleaning Products (SSCP). US Environmental Protection Agency. June 2009 [online]. Available: http://www.epa.gov/opptintr/dfe/pubs/projects/formulat/dfe_criteria_for_cleaning_products_10_09.pdf [accessed Apr. 10, 2012].
EPA (US Environmental Protection Agency). 2010. FY 2011-2015 EPA Strategic Plan: Achieving Our Vision. US Environmental Protection Agency, September 30, 2010 [online]. Available: http://nepis.epa.gov/Adobe/PDF/P1008YOS.pdf [accessed Apr. 2, 2012].
EPA (US Environmental Protection Agency). 2011a. National-Scale Mercury Risk Assessment Supporting the Appropriate and Necessary Finding for Coal- and Oil-Fired Electric Generating Units, Draft. Technical Support Document. EPA-452/D-11-002. Office of Air Quality Planning and Standards, US Environmental Protection Agency, Research Triangle Park, NC. March 2011 [online]. Available: http://yosemite.epa.gov/sab/sabproduct.nsf/02ad90b136fc21ef85256eba00436459/9F048172004D93BB8525783900503486/$File/hg_risk_tsd_3-17-11.pdf [accessed Apr. 4, 2012].
EPA (US Environmental Protection Agency). 2011b. Inventory of US Greenhouse Gas Emissions and Sinks: 1990–2009. EPA 430-R-11-005. US Environmental Protection Agency, Washington, DC. April 15, 2011 [online]. Available: http://www.epa.gov/climatechange/Downloads/ghgemissions/US-GHG-Inventory-2012-Main-Text.pdf [accessed Aug. 2, 2012].
EPA (US Environmental Protection Agency). 2011c. Pollution Prevention (P2): Basic Information. US Environmental Protection Agency [online]. Available: http://www.epa.gov/p2/pubs/basic.htm [accessed Apr. 9, 2012].
EPA (US Environmental Protection Agency). 2011d. Green Chemistry Program at EPA. US Environmental Protection Agency [online]. Available: http://www.epa.gov/greenchemistry/pubs/epa_gc.html [accessed Apr. 9, 2012].
EPA (US Environmental Protection Agency). 2011e. EPA Honors Winners of 2011 Presidential Greem Chemistry Challenge Awards. US Environmental Protection Agency, News: June 20, 2011 [online]. Available: http://yosemite.epa.gov/opa/admpress.nsf/0/93C78AFC58096165852578B5004B1E99 [accessed Apr. 9, 2012].
EPA (US Environmental Protection Agency). 2012a. Computational Toxicology Research, Research Publications. US Environmental Protection Agency [online]. Available: http://www.epa.gov/ncct/publications.html [accessed July 10, 2012].
EPA (US Environmental Protection Agency). 2012b. Quantifying Methane Abatement Efficiency at Three Municipal Solid Waste Landfills. Final Report. EPA/600/R-12/003. Prepared by ARCADIS US, Inc., Durham, NC, for Office of Research and Development, US Environmental Protection Agency, Research Triangle Park, NC. January 2012.
EPA (US Environmental Protection Agency). 2012c. Solid Waste and Emergency Response Discussion Forum. US Environmental Protection Agency [online]. Available: http://blog.epa.gov/oswerforum/ [accessed Apr. 4, 2012].
EPA (US Environmental Protection Agency). 2012d. Watershed Central. Office of Water, US Environmental Protection Agency [online]. Available: http://water.epa.gov/type/watersheds/datait/watershedcentral/index.cfm [accessed Apr. 4, 2012].
EPA (US Environmental Protection Agency). 2012e. Program History. Design for the Environment. US Environmental Protection Agency [online]. Available: http://www.epa.gov/dfe/pubs/about/history.htm [accessed Apr. 9, 2012].
EPA (US Environmental protection Agency). 2012f. Safe Product Labeling. Design for Environment, US Environmental Protection Agency [online]. Available: http://www.epa.gov/opptintr/dfe/pubs/projects/formulat/saferproductlabeling.htm [accessed Apr. 10, 2012].
EPA (US Environmental Protection Agency). 2012g. Sustainable Future. US Environmental Protection Agency [online]. Available: http://www.epa.gov/oppt/sf/ [accessed Apr. 9, 2012].
Ferebee, M., Jr. 2011. New Tools, New Rules, New Year. Inside. Business News, December 2, 2011 [online]. Available: http://insidebiz.com/news/new-tools-new-rules-new-year-whether-its-through-new-technology-processes-behavior-modeling-les [accessed Apr. 5, 2012].
Fishman, J., K.W. Bowman, J.P. Burrows, A. Richter, K.V. Chance, D.P. Edwards, R.V. Martin, G.A. Morris, R. B. Pierce, J.R. Ziemke, J.A. Al-Saadi, J.K. Creilson, T.K. Schaack, and A.M. Thompson. 2008. Remote sensing of tropospheric pollution from space. Bull. Amer. Meteor. Soc. 89(6):805-821.
Foldit. 2012. The Science behind Foldit. Solve the Puzzles for Science [online]. Available: http://fold.it/portal/info/science [accessed Apr. 4, 2012].
Fong, T.T., and E.K. Lipp. 2005. Enteric viruses of humans and animals in aquatic environments: Health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 69(2):357-371.
Gilski, M., M. Kazmierczyk, S. Krzywda, H. Zábranská, S. Cooper, Z. Popović, F. Khatib, F. DiMaio, J. Thompson, D. Baker, I. Pichová, and M. Jaskolski. 2011. High-resolution structure of a retroviral protease folded as a monomer. Acta Cryst. D67:907-914.
Goetz, S.J., and R.O. Dubayah. 2011. Advances in remote sensing technology and implications for measuring and monitoring forest carbon stocks and change. Carbon Manag. 2(3):231-244.
Greenbaum, D., and R. Shaikh. 2010. First steps toward multipollutant science for air quality decisions. Epidemiology 21(2):195-197.
Gurney, K.R., R.M. Law, A.S Denning, P.J. Rayner, D. Baker, P. Bousquet, L. Bruhwiler, Y.H. Chen, P. Ciais, S. Fan, I.Y. Fung, M. Gloor, M. Heimann, K. Hi-guchi, J. John, T. Maki, S. Maksyutov, K. Masarie, P. Peylin, M. Prather, B.C. Pak, J. Randerson, J. Sarmiento, S. Taguchi, T. Takahashi, and C.W. Yuen. 2002. Towards robust regional estimates of CO2 sources and sinks using atmospheric transport models. Nature 415(6872):626-630.
Haas, C.N. 1983. Estimation of risk due to low doses of microorganisms: A comparison of alternative methodologies. Am. J. Epidemiol. 118(4):573-582.
Haas, C.H., J.B. Rose, and C.P. Gerba, eds. 1999. Quantitative Microbial Risk Assessment. New York: John Wiley and Sons.
Hall, F.G., K. Bergen, J.B. Blair, R. Dubayah, R. Houghton, G. Hurtt, J. Kellndorfer, M. Lefsky, J. Ranson, S. Saatchi, H.H. Shugart, and D. Wickland. 2011. Characterizing 3D vegetation structure from space: Mission requirements. Remote Sens. Environ. 115(11):2753-2775.
Hall, J., A.D. Zaffiro, R.B. Marx,, P. C. Kefauver, E.R. Krishnan, R. Haught, and J.G. Herrmann. 2007. On-line water quality parameters as indicators of distribution system contamination. J. Am. Water Works Assoc. 99(1):66-77.
Hayes, D.J., D.P. Turner, G. Stinson, A.D. McGuire, Y. Wei, T.O. West, L.S. Heath, B. de Jong, B.G. McConkey, R.A. Birdsey, W.A. Kurz, A.R. Jacobson, D.N. Huntzinger, Y. Pan, W. Mac Post, and R.B. Cook. 2012. Reconciling estimates of contemporary North American carbon balance among terrestrial biosphere models, atmospheric inversions, and a new approach for estimating net ecosystem exchange from inventory-based data. Global Change Biol. 18(4):1282-1299.
He, S. 2003. Informatics: A brief survey. Electron. Libr. 21(2):117-122.
Heald, C.L., D.J. Jacob, R.J. Park, B. Alexander, T.D. Fairlie, R.M. Yantosca, and D.A. Chu. 2006. Transpacific transport of Asian anthropogenic aerosols and its impact on surface air quality in the United States. J. Geophys. Res. 111:D14310, doi:10.1029/2005JD006847.
Hill, V.R., A.L. Polaczyk, D. Hahn, J. Narayanan, T.L. Cromeans, J.M. Roberts, and J.E. Amburgey. 2005. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 71(11):6878-6884.
Hystad, P., E. Setton, A. Cervantes, K. Poplawski, S. Deschenes, M. Brauer, A. van Donkelaar, L. Lamsal, R. Martin, M. Jerrett, and P. Demers. 2011. Creating national air pollution models for population exposure assessment in Canada. Environ. Health Perspect. 119(8):1123-1129.
Jean, J., Y. Perrodin, C. Pivot, D. Trep, M. Perraud, J. Droguet, F. Tissot-Guerraz, F. Locher. 2012. Identification and prioritization of bioaccumulable pharmaceutical substances discharged in hospital effluents. J. Environ. Manage. 103:113-121.
Jeong, Y., B.F. Sanders, and S.B. Grant. 2006. The information content of high-frequency environmental monitoring data signals pollution events in the coastal ocean. Environ. Sci. Technol. 40(20):6215-6220.
Johns, D.O., L.W. Stanek, K. Walker, S. Benromdhane, B. Hubbell, M. Ross, R.B. Devlin, D.L. Costa, and D. S. Greenbaum. 2012. Practical advancement of multipollutant scientific and risk assessment approaches for ambient air pollution. Environ. Health Perspect. 102(9):1238-1242.
Jones, L., J.D. Parker, and P. Mendola. 2010. Blood Lead and Mercury Levels in Pregnant Women in the United States, 2003-2008. NCHS Data Brief No. 52. US Department of Health & Human Services, Centers for Disease Control and Prevention, National
Center for Health Statistics, Hyattsville, MD. December 2010 [online]. Available: http://www.cdc.gov/nchs/data/databriefs/db52.pdf [accessed Apr. 2, 2012].
Judson, R.S., K.A. Houck, R.J. Kavlock, T.B. Knudsen, M.T. Martin, H.M. Mortensen, D.M. Reif, D.M. Rotroff, I. Shah, A.M. Richard, and D.J. Dix. 2010. In vitro screening of environmental chemicals for targeted testing prioritization: The Tox-Cast project. Environ Health Perspect. 118(4):485-492.
Judson, R.S., R.J. Kavlock, R.W. Setzer, E.A. Cohen-Hubal, M.T. Martin, T.B. Knudsen, K.A. Houck, R.S. Thomas, B.A. Wetmore, and D.J. Dix. 2011. Estimating toxic-ity-related biological pathway altering doses for high-throughput chemical risk assessment. Chem. Res. Toxicol. 24(4):451-462.
Kaiser, J. 2012. Overhaul of the US Child Health Study Concerns Investigators. Science. 335:1032.
Khatib, F., S. Cooper, M.D. Tyka, K. Xu, I. Makedon, Z. Popović, D. Baker, and Foldit Players. 2011a. Algorithm discovery by protein folding game players. Proc. Natl. Acad. Sci. 108(47):18949-18953.
Khatib, F., F. DiMaio, Foldit Contenders Group, Foldit Void Crushers Group, S. Cooper, M. Kazmierczyk, M. Gilski, S. Krzywda, H. Zábranská, I. Pichová, J. Thompson, Z. Popović, M. Jaskolski, and D. Baker. 2011b. Crystal structure of a monomeric retroviral protease solved by protein folding game players. Nat. Struct. Mol. Biol. 18(10):1175-1177.
Koetz, B., G.Q. Sun, F. Morsdorf, K.J. Ranson, M. Kneubuhler, K. Itten, and B. Allgower. 2007. Fusion of imaging spectrometer and LiDAR data over combined radiative transfer models for forest canopy characterization. Remote Sens. Environ. 106(4):449-459.
Kristiansson, E., J. Fick, A. Janzon, R. Grabic, C. Rutgersson, B. Weijdegård, H. Söderström, and D.G.J. Larsson. 2011. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE 6(2):e17038.
Lioy, P.J., and S.M. Rappaport. 2011. Exposure science and the exposome: An opportunity for coherence in the environmental health sciences. Environ. Health Perspect. 119(11):A466-A467.
Lioy, P.J., S.S. Isukapalli, L. Trasande, L. Thorpe, M. Dellarco, C. Weisel, P.G. Georgopoulos, C. Yung, S. Alimokhtari, M. Brown, and P.J. Landrigan. 2009. Using national and local extant data to characterize environmental exposures in the national children’s study: Queens County, New York. Environ. Health Perspect. 117(10):1494-1504.
Long, S.C., and J.D. Plummer. 2004. Assessing land use impacts on water quality using microbial source tracking. J. Am. Water Resour. Assoc. 40(6):1433-1448.
Longley, D., and M. Shain. 1985. Dictionary of Information Technology, 2 Ed. London: Macmillan Press.
Loperfido, J.V., C.L. Just, and J.L. Schnoor. 2009. High-frequency diel dissolved oxygen stream data modeled for variable temperature and scale. J. Environ. Eng.-ASCE 135(12):1250-1256.
Loperfido, J.V., P. Beyer, C.L. Just, and J.L. Schnoor. 2010a. Uses and biases of volunteer water quality data. Environ. Sci. Technol. 44(19):7193-7199.
Loperfido, J.V., C.L. Just, A.N. Papanicolaou, and J.J. Schnoor. 2010b. In situ sensing to understand diel turbidity cycles, suspended solids, and nutrient transport in Clear Creek, Iowa. Water Resour. Res. 46:W06525, doi: 10.1029/2009WR008293.
Lovett, G.M., D.A. Burns, C.T. Driscoll, J.C. Jenkins, M.J. Mitchell, L. Rustad, J.B. Shanley, G.E. Likens, and R. Haeuber. 2007. Who needs environmental monitoring? Front. Ecol. Environ. 5(5):253-260.
Martin, R.V. 2008. Satellite remote sensing of surface air quality. Atmos. Environ. 42(34):7823-7843.
Matthews, M.W. 2011. A current review of empirical procedures of remote sensing in inland and near-coastal transitional waters. Int. J. Remote Sens. 32(21):6855-6899.
McHale, C.M., L. Zhang, A.E. Hubbard, and M.T. Smith. 2010. Toxicogenomic profiling of chemically exposed humans in risk assessment. Mutat. Res. 705(3):172-183.
Medema, G.J., W. Hoogenboezem, A.J. van der Veer, H.A. Ketelaars, W.A. Hijnen, and P.J. Nobel. 2003. Quantitative risk assessment of Cryptosporidium in surface water treatment. Water Sci. Technol. 47(3):241-247.
Mertes, L.A.K. 2002. Remote sensing of riverine landscapes. Freshwater Biol. 47(4):799-816.
Messer, J.W., and A.P. Dufour. 1998. A rapid, specific membrane filtration procedure for enumeration of enterococci in recreational water. Appl. Environ. Microbiol. 64(2):678-680.
Messner, M., S. Shaw, S. Regli, K. Rotert, V. Blank, and J. Soller. 2006. An approach for developing a national estimate of waterborne disease due to drinking water and a national estimate model application. J. Wat. Health 4(suppl. 2):201-240.
Morris, G.A., S. Hersey, A.M. Thompson, S. Pawson, J.E. Nielsen, P.R. Colarco, W.W. McMillan, A. Stohl, S. Turquety, J. Warner, B.J. Johnson, T.L. Kucsera, D.E. Larko, J. Oltmans, and J.C. Witte. 2006. Alaskan and Canadian forest fires exacerbate ozone pollution over Houston, Texas, on 19 and 20 July 2004. J. Geophys. Res. 111:D24S03, doi:10.1029/2006JD007090.
Napelenok, S.L., R.W. Pinder, A.B. Gilliland, and R.V. Martin. 2008. A method for evaluating spatially-resolved NOx emissions using Kalman filter inversion, direct sensivitities, and space-based NO2 observations. Atmos. Chem. Phys. 8(18):5603-5614.
Neng, N.R., and J.M. Nogueira. 2012. Development of a bar adsorptive micro-extraction-large-volume injection-gas chromatography-mass spectrometric method for pharmaceuticals and personal care products in environmental water matrices. Anal. Bioanal. Chem. 402(3):1355-1364.
NHLBI (National Heart, Lung and Blood Institute). 2011. NHLBI Population Studies Database [online]. Available: http://apps.nhlbi.nih.gov/popstudies/ [accessed Oct. 11, 2011].
NIH (National Institutes of Health). 2012. Agricultural Health Study [online]. Available: http://aghealth.nci.nih.gov/ [accessed May 3, 2012].
Noble, R.T., A.D. Blackwood, J.F. Griffith, C.D. McGee, and S.B. Weisberg. 2010. Comparison of rapid quantitative PCR-based and conventional culture-based methods for enumeration of Enterococcus spp. and Escherichia coli in recreational waters. Appl. Environ. Microbiol. 76(22):7437-7443.
Nowak, P., S. Bowen, and P. Cabot. 2006. Disproportionality as a framework for linking social and biophysical systems. Soc. Natur. Resour. 19(2):153-173.
NRC (National Research Council). 1983. Risk Assessment in Federal Government: Managing the Process. Washington, DC: National Academy Science.
NRC (National Research Council). 2007a. Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press.
NRC (National Research Council). 2007b. Models in Environmental Regulatory Decision Making. Washington, DC: National Academies Press.
NRC (National Research Council). 2009. Science and Decisions: Advancing Risk Assessment. Washington, DC: National Academies Press.
NRC (National Research Council). 2011. Emerging Science for Environmental Health, Decisions: Workshop: Applying 21st Century Toxicology to Green Chemical and Material Design [online]. Available: http://nas-sites.org/emergingscience/workshops/green-chemistry/workshop-presentations-green-chemistry/ [accessed Apr. 11, 2012].
NRC (National Research Council). 2012. A Research Strategy for Environmental, Health and Safety Aspects of Engineered Nanomaterials. Washington, DC: National Academy Press.
NRC/IOM (National Research Council and Institute of Medicine). 2008. The National Children’s Study Research Plan: A Review. Washington, DC: The National Academies Press.
NRDC (National Resource Defense Council). 2011. Testing the Waters: A Guide to Water Quality at Vacation Beaches, 21st Annual Report. National Resource Defense Council [online]. Available: http://www.nrdc.org/water/oceans/ttw/titinx.asp [accessed Apr. 6, 2012].
Ostby, F.P. 1999. Improved accuracy in severe storm forecasting by the Severe Local Storms Unit during the last 25 years: Then versus now. Weather Forecast. 14(4): 526-543.
Paules, R. 2003. Phenotypic anchoring: Linking cause and effect. Environ. Health Perspect. 111(6): A338-A339.
Pepper, I.L., J.P. Brooks, R.G. Sinclair, P.L. Gurian, and C.P. Gerba. 2010. Pathogens and indicators in United States Class B Biosolids: National and historic distributions. J. Environ. Qual. 39(6): 2185-2190.
Pillmann, W., W. Geiger, and K. Voigt. 2006. Survey of environmental informatics in Europe. Environ. Modell. Softw. 21(11):1519-1527.
Preuss, P. 2011. ORD Innovation. Presentation to the Science Advisory Board and Board of Scientific Counselors. June 29-30, 2011 [online]. Available: http://yosemite.epa.gov/sab/sabproduct.nsf/962C02E5D5A1587B852578BC005BD263/$File/CORRECTED+PWP+PPT+SAB-BOSC+for+posting.pdf [accessed April 10, 2012].
PSP (Puget Sound Partnership). 2011. The Action Agenda in South Central Puget Sound, Appendix C. Puget Sound National Estuary Program Management Conference Overview. Draft, December 9, 2011 [online]. Available: http://www.psp.wa.gov/downloads/AA2011/120911/AA-draft-120911-appendixC.pdf [accessed Apr. 3, 2011].
Rappaport, S.M., and M.T. Smith. 2010. Environment and disease risks. Science 330 (6003):460-461.
Riboli, E., K.J. Hunt, N. Slimani, N.T. Ferrari, T. Norat, M. Fahey, U.R. Charrondière, B. Hémon, C. Casagrande, J. Vignat, K. Overvad, A. Tjønneland, F. Clavel-Chapelon, A. Thiébaut, J. Wahrendorf, H. Boeing, D. Trichopoulos, A. Trichopou-lou, P. Vineis, D. Palli, H.B. Bueno-De-Mesquita, P.H. Peeters, E. Lund, D. Engeset, C.A. González, A. Barricarte, G. Berglund, G. Hallmans, N.E. Day, T.J. Key, R. Kaaks, and R. Saracci. 2002. European Prospective Investigation into Cancer and Nutrition (EPIC): Study populations and data collection. Public Health Nutr. 5(6B):1113-1124.
Ridolfi, L., P. D’Odorico, A. Porporato, and I. Rodriguez-Iturbe. 2003. Stochastic soil moisture dynamics along a hillslope. J. Hydrol. 272(1-4):264-275.
Rosario, K., C. Nilsson, Y.W. Lim, Y.J. Ruan, and M. Breitbart. 2009. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 11(11):2806-2820.
Rose, J.B. 1988. Occurrence and significance of Cryptosporidium in water. J. Am. Water Works Assoc. 80(2):53-58.
Rothamsted. 2012. The Rothamsted Archive. Rothamsted Research, Harpenden, U.K. [online]. Available: http://www.rothamsted.ac.uk/Content.php?Section=Resources&Page=RothamstedArchive [accessed Apr. 3, 2012].
Rotroff, D.M., B.A. Wetmore, D.J. Dix, S.S. Ferguson, H.J. Clewell, K.A. Houck, E.L. Lecluyse, M.E. Andersen, R.S. Judson, C.M. Smith, M.A. Sochaski, R.J. Kavlock, F. Boellmann, M.T. Martin, D.M. Reif, J.F. Wambaugh, and R.S. Thomas. 2010. Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicol Sci. 117(2):348-358.
Sarewitz, D., D. Kriebel, R. Clapp, C. Crumbley, P. Hoppin, M. Jacobs, and J. Tickner. 2010. The Sustainable Solutions Agenda. The Consortium for Science, Policy and Outcomes, Arizona State University, Lowell Center for Sustainable Production, University of Massachusetts Lowell [online]. Available: http://www.sustainableproduction.org/downlods/SSABooklet.pdf [accessed Apr. 10, 2012].
Sauch, J.F. 1985. Use of immunofluorescence and phase-contract microscopy for detection and identification of Gardia cysts in water samples. Appl. Environ. Microbiol. 50(6):1434-1438.
Scavia, D., N.N. Rabalais, R.E. Turner, D. Justic, and W.J. Wiseman. 2003. Predicting the response of Gulf of Mexico hypoxia to variations in Mississippi River nitrogen load. Limnol. Oceanogr. 48(3):951-956.
Schaepman, M.E., S.L. Ustin, A.J. Plaza, T.H. Painter, J. Verrelst, and S.L. Liang. 2009. Earth system science related imaging spectroscopy: An assessment. Remote Sens. Environ. 113(1):S123-S137.
Schober, S.E., T.H. Sinks, R.L. Jones, P.M. Bolger, M. McDowell, J. Osterloh, E.S. Garrett, R.A. Canady, C.F. Dillon, Y. Sun, C.B. Joseph, and K.R. Mahaffey. 2003. Blood mercury levels in US children and women of childbearing age, 1999-2000. JAMA 289(13):1667-1674.
Schwalm, C.R., C.A. Williams, K. Schaefer, R. Anderson, M.A. Arain, I. Baker, A. Barr, T.A. Black, G. Chen, J.M. Chen, P. Ciais, K.J. Davis, A. Desai, M. Dietze, D. Dragoni, M.L. Fischer, L.B. Flanagan, R. Grant, L. Gu, D. Hollinger, R.C. Izaur-ralde, C. Kucharik, P. Lafleur, B.E. Law, L. Li, Z. Li, S. Liu, E. Lokupitiya, Y. Luo, S. Ma, H. Margolis, R. Matamala, H. McCoughey, R.K. Monson, W.C. Oechel, C. Peng, B. Poulter, D.T. Price, D.M. Riciutto, W. Riley, A.K. Sahoo, M. Sprintsin, J. Sun, H. Tian, C. Tonitto, H. Verbeeck, and S.B. Verma. 2010. A model-data intercomparison of CO2 exchange across North America: Results from the North American carbon program site synthesis. J. Geophys. Res. 115:G00H05, doi:10.1029/2009JG001229.
Seminara, D., M.J. Khoury, T.R. O’Brien, T. Manolio, M.L. Gwinn, J. Little, J.P. Hig-gins, J.L. Bernstein, P. Boffetta, M. Bondy, M.S. Bray, P.E. Brenchley, P.A. Buffler, J.P. Casas, A.P. Chokkalingam, J. Danesh, G. Davey Smith, S. Dolan, R. Duncan, N.A. Gruis, M. Hashibe, D. Hunter, M.R. Jarvelin, B. Malmer, D.M. Maraganore, J.A. Newton-Bishop, E. Riboli, G. Salanti, E. Taioli, N. Timpson, A.G. Uitterlinden, P. Vineis, N. Wareham, D.M. Winn, R. Zimmern, and J.P. Io-annidis. 2007. The emergence of networks in human genome epidemiology: Challenges and opportunities. Epidemiology 18(1):1-8.
Shanks, O. 2011. Global Inter-lab Fecal Source Tracking Methods Comparison Study. Presentation at 2011 National Beach Conference, March 16, 2011, Miami, FL.
Sheldon, L.S., and E.A. Cohen Hubal. 2009. Exposure as part of a systems approach for assessing risk. Environ. Health Perspect. 117(8):1181-1184.
Shukla, S., C.Y. Yu, J.D. Hardin, and F.H. Jaber. 2006. Wireless data acquisition and control systems for agricultural water management projects. HortTechnology 16(4):595-604.
Sivapalan, M., K. Takeuchi, S.W. Franks, V.K. Gupta, H. Karambiri, V. Lakshmi, X. Liang, J.J. McDonnell, E.M. Mendiondo, P.E. O’Connell, T. Oki, J.W. Pomeroy, D. Schertzer, S. Uhlenbrook, and E. Zehe. 2003. IAHS decade on Predictions in Ungauged Basins (PUB), 2003–2012: Shaping an exciting future for the hydro-logical sciences. Hydrol. Sci. 48(6):857-880.
Slifko, T.R., D. Friedman, J.B. Rose, and W. Jakubowski. 1997. An in vitro method for detecting infectious Cryptosporidium oocysts with cell culture. Appl. Environ. Microbiol. 63(9):3669-3675.
Slifko, T.R., D.E. Huffman, and J.B. Rose. 1999. A most probable assay for enumeration of infectious Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 65(9): 3936-3941.
Srinivasan, S., A. Aslan, I. Xagoraraki, E. Alocilja, and J.B. Rose. 2011. Escherichia coli, enterococci, and Bacteroides thetaiotaomicron qPCR signals through wastewater and septage treatment. Water. Res. 45(8):2561-2572.
Tapscot, D., and A.D. Willams. 2010. MacroWikinomics - Rebooting Business and the World. New York: Penguin.
Tian, D., D.S. Cohan, S. Napelenok, M. Bergin, Y. Hu, M. Chang, and A.G. Russell. 2010. Uncertainty analysis of ozone formation and response to emission controls using higher-order sensitivities. J. Air Waste Manage. Assoc. 60(7):797-804.
Tickner, J.A. 2011. Science of problems, science of solutions or both? A case example of bisphenol. Am. J. Epidemiol. Community Health 65(8):649-650.
Tornero-Velez, R., P.P. Egeghy, and E.A. Cohen Hubal. 2012. Biogeographical analysis of chemical co-occurrence data to identify priorities for mixtures research. Risk Anal. 32(2):224-236.
Tsow, F., E. Forzani, A. Rai, R. Wang, R. Tsui, S. Mastrioianni, C. Knobbe, A.J. Gandolfi, and N.J. Tao. 2009. A wearable and wireless sensor system for real-time monitoring of toxic environmental volatile organic compounds. 9(12):1734-1740.
University of Washington. 2011. MESA Air Pollution [online]. Available: http://depts.washington.edu/mesaair/ [accessed Apr. 3, 2012].
van Donkelaar, A., R.V. Martin, M. Brauer, R. Kahn, R. Levy, C. Verduzco, and P.J. Villeneuve. 2010. Global estimates of ambient fine particulate matter concentrations from satellite-based aerosol optical depth: Development and application. Environ. Health Perspect. 118(6):847-855.
Vandenberghe, V., P.L. Goethals, A. Van Griensven, J. Meirlaen, N. De Pauw, P. Vanrolleghem, and W. Bauwens. 2005. Application of automated measurement stations for continuous water quality monitoring of the Dender river in Flanders, Belgium. Environ. Monit. Assess. 108(1-3):85-98.
Varma, M., R. Field, M. Stinson, B. Rukovets, L. Wymer, and R. Hauglanda. 2009. Quantitative real-time PCR analysis of total and propidium monoazide-resistant fecal indicator bacteria in wastewater. Water Res. 43(19):4790-4801.
Wade, T.J., R.L. Calderon, E. Sams, M. Beach, K.P. Brenner, A.H. Williams, and A.P. Dufour. 2006. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestional illness. Environ. Health Perspect. 114(1):24-28.
Wang, K., S.E. Franklin, X. Guo, and M. Cattet. 2010. Remote sensing of ecology, biodiversity and conservation: A review from the perspective of remote sensing specialists. Sensors 10(11):9647-9667.
WATERS Network. 2009. Living in the Water Environment: The WATERS Network Science Plan, May 15, 2009 [online]. Available: http://www.watersnet.org/docs/WATERS_Network_SciencePlan_2009May15.pdf [accessed Aug. 8, 2012].
Weis, B.K., D. Balshaw, J.R. Barr, D. Brown, M. Ellisman, P. Lioy, G. Omenn, J.D. Potter, M.T. Smith, L. Sohn, W.A. Suk, S. Sumner, J. Swenberg, D.R. Walt, S. Watkins, C. Thompson, and S.H. Wilson. 2005. Personalized exposure assessment: Promising approaches for human environmental health research. Environ Health Perspect 113(7):840-848.
Wetmore, B.A., J.F. Wambaugh, S.S. Ferguson, M.A. Sochaski, D.M. Rotroff, K. Freeman, H.J. Clewell, III, D.J. Dix, M.E. Andersen, K.A. Houck, B. Allen, R.S. Judson, R. Singh, R.J. Kavlock, A.M. Richard, and R.S. Thomas. 2012. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol. Sci. 125(1):157-174.
WHO (World Health Organization). 2004. Guidelines for Drinking-Water Quality: Volume 1 Recommendations, 3rd Ed. Geneva: World Health Organization [online]. Available: http://www.who.int/water_sanitation_health/dwq/GDWQ2004web.pdf [accessed Aug. 1, 2012].
Wild, C.P. 2005. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 14(8):1847-1850
Willett, W.C., W.J. Blot, G.A. Colditz, A.R. Folsom, B.E. Henderson, and M.J. Stampfer. 2007. Merging and emerging cohorts: Not worth the wait. Nature 445(7125):257-258.
Yates, M.V., J. Malley, P. Rochelle, and R. Hoffman. 2006. Effect of adenovirus resistance on UV disinfection requirements: A report on the state of adenovirus science. J. Am. Water Works Assoc. 98(6):93-106.
Ye, L., and T. Zhang. 2011. Pathogenic bacteria in sewage treatment plants as revealed by 454 pyrosequencing. Environ. Sci. Technol. 45(17):7173-7179.
Zegura, B., E. Heath, A. Cernosa, and M. Filipic. 2009. Combination of in vitro bioassays for the determination of cytotoxic and genotoxic potential of wastewater, surface water and drinking water samples. Chemosphere. 75(11):1453-1460.





















































