Interaction Between the Microbiome and Health and Environment
Two major overarching themes that emerged early during the course of the workshop were the intimate role the microbiome plays at the interface between humans and their environment and the key role the microbiome plays in human health. This chapter summarizes the presentations that explored in detail the environment-human-microbiome dynamic in the early years of life (i.e., how vaginal versus cesarean [C-section] deliveries impact the fetal microbiome and are associated with fetal health), in the oral environment (i.e., how periopathogens cause oral disease by interfering with the community in a subversive way), and in the adult gastrointestinal (GI) tract (i.e., how the indigenous microbiome influences the effect of a pathogen).
OVERVIEW OF PEDIATRIC CLINICAL IMPLICATIONS AND INTERVENTIONS1
One of the first studies conducted on early development of the human intestinal microbiome revealed some interesting findings, including that a baby’s first stool contains microbes and that a baby’s first antibiotic treatment has a marked effect on microbes in the GI tract (Palmer et al., 2007). More recently, Koenig et al. (2011) demonstrated in a case study of one infant how microbial diversity in the GI tract increases over time during the first year of life, with the introduction of specific types of foods causing phylum-level changes in microbial composition. Koenig et al. (2011) also
__________________________
1 This section summarizes the presentation of Josef Neu.
reported having found microbial DNA in the meconium (i.e., first stool). Josef Neu speculated on the implications of these and other recent microbiome research in pediatric populations. For example, that the first stool contains microbes suggests not only that a fetal microbiome exists, but also that its existence could relate to prematurity.
Fetal Microbiome: Clinical Implications
Evidence of microbes in the meconium refutes the popular notion that the mammalian fetal intestine is sterile and that the first exposure to maternal microbiota occurs during passage through the birth canal, according to Neu. Goldenberg and colleagues (2000) suggested that first exposure could occur during the last trimester of pregnancy, that is, babies are bathed in amniotic fluid that may contain microbes that have ascended from the vagina and translocated through the choriodecidual membrane. DiGiulio and colleagues (2008) examined the possibility by analyzing stored amniotic fluids of babies born at various gestational times. Using both culture-based and polymerase chain reaction (PCR) techniques, they found that gestational age was inversely correlated with microbial presence and quantity. In other words, babies born prematurely had more microbes in their amniotic fluids. The researchers also reported a positive correlation between both their culture and PCR results and amniotic fluid concentrations of white blood cells and interleukin-6 (IL-6), suggesting that microbial presence and quantity are associated with intestinal inflammation. Nanthakumar and colleagues (2000) had previously reported an inverse relationship between maturity and IL-8 expression, also suggesting that prematurity could be associated with an intestine-derived inflammatory response to microbes swallowed by the fetus through the amniotic fluid.
Together, these data suggest that when microbes are swallowed by the fetus, the ensuing infection increases the output of inflammatory mediators (e.g., IL-6, IL-8) and thereby potentially triggers premature labor as well as other problems (e.g., necrotizing enterocolitis, chronic lung disease, neurodevelopmental delays). Because amniotic fluid is difficult to sample, the next best evidence available for testing this hypothetical scenario comes from the baby’s first stool. “If you think of it from the perspective that the baby’s meconium is actually a reflection of what has been going on in utero in terms of the swallowing of the microbes,” Neu said, “meconium could potentially be a very valuable source of information.” Using data from meconium samples, he and his colleagues reported lower microbial diversity among less mature babies (Mshvildadze et al., 2010). More recent, unpublished data also show correlations between gestational age and phylum-level diversity—for example, a fairly strong negative correlation between gestational age and Actinobacteria (Triplett and Neu, unpublished
data). Neu noted that the Gardnerella genus (which is in the Actinobacteria phylum), when it is associated with bacterial vaginosis, has been associated with premature delivery.
Diseases in Preterm Babies: Role of the Microbiome
About 7 percent of babies in neonatal intensive care units (ICUs) who weigh less than 1,500 grams develop necrotizing enterocolitis (NEC). About 30 percent of babies who develop NEC do not survive. Of those who do survive, about 50 percent suffer significant neurodevelopmental delays. Symptoms include abdominal distention, redness around the belly button, and specific X-ray findings (Neu and Walker, 2011). Surgical treatment for NEC often results in a shortened gut, which requires about $1.5 million in medical care during the first 5 years of life.
In an ongoing microbiota study of babies with NEC, Neu and colleagues have been collecting weekly stool samples from NEC babies and carefully matched non-NEC babies (i.e., matched with respect to gestational age, size, time in the neonatal ICU). Their first results revealed differences in demography (i.e., babies with NEC were more likely to be formula-fed than breast milk–fed), antibiotic administration (i.e., babies with NEC were administered more antibiotics than control babies were), and fetal microbiota (Mai et al., 2011). With respect to fetal microbiota, the NEC and control babies demonstrated a marked difference in Firmicutes prevalence one week before diagnosis (60.68 percent in NEC babies, compared to 31.49 percent in controls) and a marked difference in Proteobacteria composition within 72 hours of diagnosis (70.9 percent in NEC babies, compared to 31.49 percent in controls). At the species, or operational taxonomic unit (OTU), level, there appear to be significant differences in Klebsiella spp. and Cronobacter spp.
Another recent study reported similar findings with respect to the relationship between antibiotic administration and NEC, with greater antibiotic use increasing the risk of NEC (Alexander et al., 2011). According to Clark and colleagues (2006), antibiotics are among the top 10 drugs administered to babies in neonatal ICUs, with about 95 percent of all babies being administered at least 48-72 hours of either ampicillin or gentamicin. As reviewed by Preidis and Versalovic (2009), an association between antibiotic administration and lower microbial species diversity has been observed in infants.
Late-onset sepsis is another prevalent disease among premature babies, affecting about 37 percent of babies born at less than 28 weeks’ gestation, with fetal microbiota associations. According to Neu, coagulase-negative Staphylococcus spp., Escherichia coli, Klebsiella spp., Pseudomonas spp., and Enterococcus spp. are the most common microorganisms in blood
cultures of babies with late-onset sepsis. Unpublished data from Neu’s research group show lower overall microbial diversity in sepsis babies 2 weeks before diagnosis, with few Proteobacteria detected. At the onset of sepsis, Proteobacteria bloom, which Neu observed is similar to what happens with NEC.
C-Section Versus Vaginal Delivery: Microbiome Differences
Neu explored in more detail a subject that Lita Proctor touched on, that is, microbiome differences between babies born vaginally versus those born via C-section. It is an important topic, Neu argued, because of the impact of the microbiome on development of the immune system during the first year of life and because of the growing number of C-section deliveries worldwide. In the United States, C-sections have increased from 24 to 34 percent over the past 15 years; in large cities in China, C-section delivery rates reach 60 percent; and in some South American countries, for example, Argentina and Brazil, C-section deliveries in private hospitals approximate 100 percent. Neu and Rushing (2011) listed a range of health outcomes associated with C-section deliveries, including allergic rhinitis, asthma, celiac disease, diabetes mellitus, and gastroenteritis.
With C-section delivery, lack of exposure to the vaginal microbiota results in “abnormal” microbial seeding of the GI tract and “abnormal” development of immunity, according to Neu. Dominguez-Bello and colleagues (2010) reported that with vaginal delivery, the baby’s first stool microbiota closely resembled the mother’s vaginal microbiota, whereas with C-section delivery, the baby’s first stool microbiota closely resembles the mother’s skin microbiota. Neu mentioned some unpublished data that show not only differences in microbial presence between C-section and vaginal deliveries, but also changes in those differences over time. Major phylum-level differences that exist at week 1 (e.g., greater Proteobacteria abundance in C-section babies, greater Bacteroides abundance in vaginal babies) disappear by week 4, while certain genus-level differences that are not present at week 1 emerge at week 4 (e.g., relative abundance of Enterococcus).
Pediatric Microbiome: Implications for Long-Term Health and Disease
In conclusion, Neu briefly described yet another early-life disease, type 1 diabetes, that has been associated with the pediatric microbiome. In addition to genetic predisposition and other factors, researchers have found significant differences in microbial ecology between children who develop type 1 diabetes and children who do not (Brown et al., 2011; Vaarala et al., 2008). Butyrate-producing bacteria appear to be especially important for maintaining a healthy gut and preventing type 1 diabetes (see Figure 3-1).
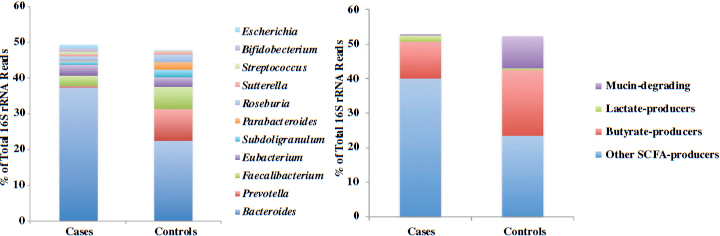
FIGURE 3-1 Differences in genus-level microbiome content between children who develop type 1 diabetes and children who do not.
NOTE: rRNA = ribosomal RNA.
SOURCE: Brown et al., 2011.
IMPACT OF MICROBIOME ON ORAL HEALTH AND DISEASE2
The oral environment operates under a “different paradigm” from other parts of the GI tract, according to Richard Darveau. A key difference between the oral and intestinal environments, one with significant implications for differentiating health and disease, is that two-way communication between the inside and outside environments is a regular feature of even a healthy oral cavity. Unlike the intestinal epithelium, which is characterized by tight junctions, junctional epithelium in the oral cavity is very loosely organized. The looseness allows for constant neutrophil movement from the vasculature to the gingival crevice. Elsewhere in the GI tract, neutrophil movement is a sign of inflammation or disease. In the oral cavity, it is “normal.” Similarly, inflammatory cytokines are widely present in healthy mouths, where they play a key role in healthy tissue development and function. There are just “a lot more of them” in diseased mouths, Darveau explained. So the innate immune defense system is highly active even in healthy tissue. For example, Yoshioka and colleagues (2008) showed that plaque from both clinically healthy and diseased sites can stimulate both Toll-like receptor-2 (TLR-2)-mediated and TLR-4-mediated inflammatory responses. Darveau described disease as a “disruption in homeostasis,” that is, a disruption in the healthy relationship between oral microbes and the host tissue—one that causes increased inflammation and, eventually, bone and teeth loss (Darveau, 2010).
Constant movement across the junctional epithelium in the oral cavity, combined with the fact that the periodontium is a highly vascularized tissue, implicates periodontitis as a contributing factor to systemic disease. Darveau remarked that while the mechanisms are still unclear, researchers have reported clinical associations between dental and systemic diseases (Zelkha et al., 2010).
Another important difference between the oral and gut microbiomes is the ease of sampling the former. Scientists have conducted “thousands and thousands of analyses” of the oral microbiome, according to Darveau, providing the data to paint a good picture of healthy versus diseased oral bacterial consortia. A healthy oral bacterial consortium is characterized by mostly Gram-positive bacteria, whereas a periopathogenic bacterial consortium is characterized by mostly Gram-negative bacteria. Years ago, Darveau was involved in work that led to the identification of three of these Gram-negative bacteria collectively known as the “red complex bacteria”: Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. The three species are associated both with each other and with periodontitis (Socransky et al., 1998). Much of Darveau’s research is on P. gingivalis, a
__________________________
2 This section summarizes the presentation of Richard Darveau.
late colonizer in ecological succession of the oral microbial biofilm that lands on the top layer of already formed biofilms (Zijnge et al., 2010).
Health as a Homeostatic Relationship Between Commensal Bacteria and Their Host
Initially, Darveau hypothesized that P. gingivalis produces a potent inflammation-inducing lipopolysaccharide (LPS). However, as described in Al-Qutub et al. (2006), he and his research team found that P. gingivalis produces a very complex LPS, one with a lot of structural heterogeneity and specific structural alterations that actually result in reduced inflammation under certain conditions (e.g., hemin concentration can influence LPS structure). So contrary to initial suspicion, Darveau said that P. gingivalis LPS “is very weak at activating inflammation.” In some cases, it actually inhibits TLR-4-mediated inflammation (Coats et al., 2005, 2007). Other researchers have confirmed Darveau’s findings, showing in a similar fashion that P. gingivalis is not only not a strong inducer of inflammation, but also an excellent modulator of host inflammatory response (Darveau, 2010; Hajishengallis et al., 2011).
Refutation of his initial hypothesis led Darveau to propose a new hypothesis: that P. gingivalis is a keystone species in the oral microbiota and that it impacts the host immune system not directly, but by subverting innate immunity in a way that prevents the host from detecting and clearing not just P. gingivalis but other oral microbes as well (Darveau, 2009, 2010). For example, by inhibiting TLR-4-mediated inflammation, P. gingivalis might be inhibiting the ability of TLR-4 to sense not only its own presence (and clear P. gingivalis) but also the presence of other microbes. Darveau noted that P. gingivalis can disrupt host tissue homeostasis via other mechanisms as well, for example, by inhibiting host cell secretion of the chemokine IL-8 in response to other oral microbes, not just in response to P. gingivalis (Darveau et al., 1998). Keystone species do not need to be present in large numbers, Darveau said, in order to exert global effects on the community.
To test the concept of P. gingivalis as a keystone species, Darveau and colleagues colonized both wild-type and germ-free mice with P. gingivalis and detected alveolar bone loss and an increase in total oral bacterial load in the wild-type but not the germ-free mice after 6 weeks (Hajishengallis et al., 2011). The results suggest that commensals must be present in order to induce a diseased state. Wondering if commensals alone could cause bone loss, Hajishengallis et al. (2011) also co-caged germ-free with wild-type mice (i.e., uninfected with P. gingivalis) and measured bone loss after 16 weeks. They reported that, yes, the germ-free mice showed bone loss after 16 weeks of having been co-caged with wild-type mice. Thus, naturally
existing commensals can cause eventual bone loss even in the absence of infection with P. gingivalis, but P. gingivalis does accelerate bone loss (i.e., when P. gingivalis is present, bone loss occurs at 6 weeks versus 16 weeks). Additionally, Hajishengallis et al. (2011) showed that both P. gingivalis-induced and natural bone loss require complement.3 Complement receptor knockout mice showed no signs of bone loss even in the presence of P. gingivalis. Together, these results suggest that P. gingivalis accelerates natural bone loss by exploiting and modulating naturally existing commensal interaction with complement.
Recent, unpublished data in mice underscore the important role that commensals play in periodontal disease (Zenobia et al., manuscript in preparation). The data indicate that CXCL1 (a mouse analog of human IL-8) is expressed in both conventionally reared and germ-free mice, but that expression of CXCL2 (another mouse analogue of human IL-8) requires the presence of commensals. In mice, both CXCL1 and CXCL2 are needed for the “normal” neutrophil migration that characterizes a healthy oral environment. In humans, IL-8 is believed to be a key mediator in tissue production. Darveau concluded that we “need to know more concerning oral commensal bacteria contribution to health.” For example, which oral commensal bacteria contribute to neutrophil migration in health? Can modulation of commensal bacteria improve health in certain individuals?
IMPACT OF MICROBIOME ON GASTROINTESTINAL HEALTH4
Medical students today are learning how to think about microbes in a different way from when Vincent Young was a student. “Find the bug, find the drug, because the only good bug is a dead bug” was the mantra, a way of thinking that originated with Koch’s postulates (1882).5 However, microbes play a much more complex role in human health and disease than previously thought. Today, Young is teaching his students to think not about “bad” and “good” bugs, but rather good and bad communities of microbial organisms.
__________________________
3 The complement system comprises about 25 proteins that work together to assist, or complement, the action of antibodies in destroying bacteria.
4 This section summarizes the presentation of Vincent Young.
5 Young’s rendition of Koch’s postulates was that the pathogen must be found in all cases of disease, the pathogen must be isolated from the host and grown in culture, the pathogen must re-create disease when given to a susceptible host, and the pathogen must be re-isolated from the experimental host.
A New Way of Thinking About Microbes: Clostridium difficile as a Case Study
Young presented a “case study” of Clostridium difficile infection to illustrate this change in paradigm. Clostridium difficile was associated with disease in the 1970s, when researchers fulfilled Koch’s postulates to identify C. difficile as the causative agent of clindamycin-associated colitis (Bartlett et al., 1977). The case involved a 56-year-old man with chronic obstructive pulmonary disease (COPD) due to long-term cigarette use. The man was admitted with probable pneumonia and, as is standard of care for patients with suspected pneumonia, he was treated with broad-spectrum antibiotics. Although his pulmonary disease improved with antibiotics, on hospital day 3, the patient developed abdominal pain, diarrhea, and hypotension and was transferred to the intensive care unit, all as a result of a C. difficile infection. This is a “typical case,” of C. difficile infection, Young said, where antibiotic treatment for one infection results in infection with the intestinal pathogen.
The “dogma” regarding C. difficile that Young was taught as a medical student was that the indigenous microbiota somehow prevents colonization by C. difficile. Accordingly, C. difficile erupts when antibiotics disturb the indigenous microbiota; colonization resistance against C. difficile is lost; and the patient is susceptible to spores of the pathogen, which are present in the hospital environment. When patients start showing signs of C. difficile infection, they are typically prescribed yet another antibiotic, usually metronidazole or vancomycin. Although this antibiotic treatment directed against C. difficile generally results in improvement, there can be problems with recurrence, with about 25 percent of patients redeveloping symptoms after ending antibiotic treatment. Importantly, recurrence can develop even in the absence of any further original antibiotic treatment and is thought to reflect continued imbalance in the microbiota that does not correct after stopping antibiotics. Although these hypotheses regarding the relationship between C. difficile and the indigenous microbiota were proposed shortly after it was proven that the pathogen caused antibiotic-associated colitis, they have only recently been examined experimentally.
Young challenges his students to consider other ways to think about C. difficile, reminding them that the indigenous gut microbiome not only has massive metabolic capacity, but also serves many vital functions. Importantly, it has been proposed that one of those functions is a protective one and that indigenous microbiota confer on the gut what Young called “colonization resistance.” Without any additional insult to the microbiota, an estimated 25 percent of treated C. difficile patients do not have enough colonization resistance to withstand continued exposure to C. difficile
(Maroo and Lamont, 2006). “So there is something wrong,” Young said. The question is, What?
Young and his research team have collected data demonstrating that individuals who have recurrent disease have lower diversity of their indigenous gut microbiota compared to individuals who do not and to healthy individuals. Specifically, Chang and colleagues (2008) examined the diversity of the gut microbiota using culture-independent methods that involve retrieving 16S ribosomal RNA (rRNA)-encoding gene data to distinguish different bacteria. The analysis of these data was accomplished by constructing what are known as rarefaction curves6 for gut microbiota in three groups of patients: (1) individuals successfully treated for C. difficile with a single round of metronidazole or vancomycin, (2) individuals with recurrent disease, and (3) controls. The rarefaction curves showed that individuals with recurrent disease had the least amount of gut microbial diversity. Although the gut microbiome diversity in individuals who were successfully treated for C. difficile was not markedly different from that of the controls, it was at the lower end of what would be considered “normal.” However, intestinal microbial diversity in patients with recurrence was much lower than in the other two groups.
This new knowledge that refractory C. difficile disease is associated with lower gut microbiome diversity helps explain the efficacy of an “alternative” treatment for C. difficile, which has been known of for years but has had a recent resurgence given the increasing burden of C. difficile infection. Instead of administering repeated courses of antibiotics in an attempt to kill the “bad” bug that keeps reappearing, physicians try to treat recurrent C. difficile with what is known as microbiota transplantation. By administering a new microbiota (in the form of the administration of fecal material from a healthy individual), the intention is to restore microbiota diversity and therefore colonization resistance. Despite the obvious “ick factor” of this treatment, it has become an option for patients with multiple recurrences with a greater than 95 percent success rate, according to Young (Gough et al., 2011).
__________________________
6 Young explained that rarefaction analysis is a tool from classical ecology that provides a general sense of the abundance of different species or, in the case of 16S microbiome data, operational taxonomic units (i.e., bacterial types defined by similar 16S-encoding gene sequences). Rarefaction curves are created by repeatedly sampling the data and plotting the number of unique observations as a function of sampling effort. As the number of samples increases, the number of unique observations decreases. An exhaustive sampling of a community yields a flat curve, indicating that no new species should be identified no matter how many additional samples are taken. When comparing two rarefaction curves derived by sampling two communities, the curve that lies below the other at a given level of sampling indicates that the community from which the curve was derived is less diverse.
Resilience of Gut Microbial Community Structure
How does colonization resistance become compromised in the first place? Evidence suggests that insults to the microbiome, such as antibiotic treatment, can have long-lasting effects on the indigenous microbial community. Young and colleagues treated mice with antibiotics, let the animals recover from the antibiotic stress in a sterile environment, and then observed what happened when they were either left alone in a sterile environment or co-housed with a donor mouse (Antonopoulos et al., 2009). They found that mice left alone, with no donor mouse present to repopulate their guts, had microbiota that looked very similar to each other but very different from microbiota in mice that had been co-housed with donors. Even 6 weeks after stopping antibiotic treatment, mice left alone had much lower microbiota diversity than the other mice. However, as with humans, if fecal transplantation is done, diversity can return to normal.
Subsequent mouse research showed that with respect to C. difficile infection, colonization resistance can be overcome by the administration of specific (but not all) antibiotics or combinations of antibiotics. Although early work with antibiotic-treated mice was unsuccessful in modeling human C. difficile infection, Chen and colleagues (2008) were able to establish disease by pretreating the mice with a cocktail of five antibiotics before treating them with clindamycin and challenging them with C. difficile. Young’s team recreated the Chen et al. (2008) model and found that a pretreatment of five antibiotics without clindamycin did not cause disease, that clindamycin alone without pretreatment allowed transient colonization without disease (i.e., the infected mice shed bacteria briefly but showed no signs of inflammation), and that the combination of the pretreatment antibiotic cocktail followed by clindamycin allowed C. difficile colonization and the development of disease (Reeves et al., 2011). The severity of disease in the cocktail-plus-clindamycin treatment group varied. About half of the animals became very ill clinically, while the other half were able to maintain their health even though their gut epithelia became inflamed. The sicker animals also had more C. difficile and bacterial toxin present in their intestinal tissue.
With respect to microbial taxonomic composition, researchers observed high levels of the Firmicutes families, especially Lachnospiraceae genera (i.e., important short-chain fatty acid producers), and some Bacteroidetes families in untreated mice, but mice in the cocktail-plus-clindamycin treatment group bloomed Proteobacteria (e.g., E. coli, which Young described as only a “minority player” in a healthy gut). The microbiota in mice that developed clinical illness remained dominated by Proteobacteria over time, while the microbiota of mice that suffered some inflammation but did not become clinically ill eventually reverted to “healthy” Lachnospiraceaedominated communities (Reeves et al., 2011, 2012).
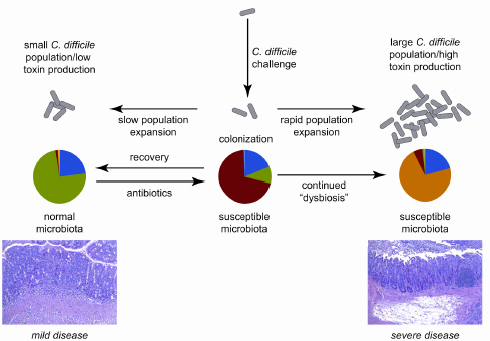
FIGURE 3-2 Model of the interaction between dynamics of the gut microbiota and C. difficile in antibiotic-treated mice, with clinical outcome being determined by the balance between recovery of the indigenous gut microbiota following antibiotic withdrawal and growth of the C. difficile population.
SOURCE: Reeves et al., 2011.
Young’s team repeated the experiment using cefoperazone instead of clindamycin and observed that all mice administered cefoperazone died as soon as they were infected with C. difficile. Moreover, colonization resistance was lowered so much that their microbiomes became pure cultures of C. difficile, and the microbiota were unable to restore colonization resistance even after some recovery time.
Young’s interpretation of the results is that colonization resistance recovery following an antibiotic assault seems to depend on which is happening faster—growth of the indigenous microbiota or growth of C. difficile (see Figure 3-2). Restoring balance in the community, or preventing imbalance, could be the basis for yet another new therapeutic approach to managing C. difficile. For example, Young mentioned the dissertation research of one of his students demonstrating that Lachnospiraceae bacteria are associated with greater colonization resistance. He wondered whether restoring balance might be simply a matter of adding “more bugs” in the “right combination.”
Alexander, V. N., V. Northrup, and M. J. Bizzarro. 2011. Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. Journal of Pediatrics 159(3):392-397.
Al-Qutub, M. N., P. H. Braham, L. M. Karimi-Naser, X. Liu, C. A. Genco, and R. P. Darveau. 2006. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infection and Immunity 74(8):4474-4485.
Antonopoulos, D. A., S. M. Huse, H. G. Morrison, T. M. Schmidt, M. L. Sogin, and V. B. Young. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infection and Immunology 77(6):2367-2375.
Bartlett, J. G., A. B. Onderdonk, R. L. Cisneros, and D. L. Kasper. 1977. Clindamycin-associated colitis due to a toxin-producing species of clostridium in hamsters. Journal of Infectious Diseases 136(5):701-705.
Brown, C. T., A. G. Davis-Richardson, A. Giongo, K. A. Gano, D. B. Crabb, N. Mukherjee, G. Casella, J. C. Drew, J. Ilonen, M. Knip, H. Hyoty, R. Veijola, T. Simell, O. Simell, J. Neu, C. H. Wasserfall, D. Schatz, M. A. Atkinson, and E. W. Triplett. 2011. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE 6(10):e25792.
Chang, J. Y., D. A. Antonopoulos, A. Kalra, A. Tonelli, W. T. Khalife, T. M. Schmidt, and V. B. Young. 2008. Decreased diversity of the fecal microbiome in recurrent clostridium difficile-associated diarrhea. Journal of Infectious Diseases 197(3):435-438.
Chen, X., K. Katchar, J. D. Goldsmith, N. Nanthakumar, A. Cheknis, D. N. Gerding, and C. P. Kelly. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135(6):1984-1992.
Clark, R. H., B. T. Bloom, A. R. Spitzer, and D. R. Gerstmann. 2006. Reported medication use in the neonatal intensive care unit: Data from a large national data set. Pediatrics 117(6):1979-1987.
Coats, S. R., T. T. Pham, B. W. Bainbridge, R. A. Reife, and R. P. Darveau. 2005. Md-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the tlr4 signaling complex. Journal of Immunology 175(7):4490-4498.
Coats, S. R., C. T. Do, L. M. Karimi-Naser, P. H. Braham, and R. P. Darveau. 2007. Antagonistic lipopolysaccharides block E. coli lipopolysaccharide function at human tlr4 via interaction with the human md-2 lipopolysaccharide binding site. Cell Microbiology 9(5):1191-1202.
Darveau, R. P. 2009. The oral microbial consortium’s interaction with the periodontal innate defense system. DNA and Cell Biology 28(8):389-395.
———. 2010. Periodontitis: A polymicrobial disruption of host homeostasis. Nature Reviews Microbiology 8(7):481-490.
Darveau, R. P., C. M. Belton, R. A. Reife, and R. J. Lamont. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infection and Immunology 66(4):1660-1665.
DiGiulio, D. B., R. Romero, H. P. Amogan, J. P. Kusanovic, E. M. Bik, F. Gotsch, C. J. Kim, O. Erez, S. Edwin, and D. A. Relman. 2008. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: A molecular and culture-based investigation. PLoS ONE 3(8):e3056.
Dominguez-Bello, M. G., E. K. Costello, M. Contreras, M. Magris, G. Hidalgo, N. Fierer, and R. Knight. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. PNAS 107(26):11971-11975.
Goldenberg, R. L., J. C. Hauth, and W. W. Andrews. 2000. Intrauterine infection and preterm delivery. New England Journal of Medicine 342(20):1500-1507.
Gough, E., H. Shaikh, and A. R. Manges. 2011. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clinical Infectious Diseases 53(10):994-1002.
Hajishengallis, G., S. Liang, M. A. Payne, A. Hashim, R. Jotwani, M. A. Eskan, M. L. McIntosh, A. Alsam, K. L. Kirkwood, J. D. Lambris, R. P. Darveau, and M. A. Curtis. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10(5):497-506.
Koenig, J. E., A. Spor, N. Scalfone, A. D. Fricker, J. Stombaugh, R. Knight, L. T. Angenent, and R. E. Ley. 2011. Succession of microbial consortia in the developing infant gut microbiome. PNAS 108(Suppl 1):4578-4585.
Mai, V., C. M. Young, M. Ukhanova, X. Wang, Y. Sun, G. Casella, D. Theriaque, N. Li, R. Sharma, M. Hudak, and J. Neu. 2011. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS ONE 6(6):e20647.
Maroo, S., and J. T. Lamont. 2006. Recurrent Clostridium difficile. Gastroenterology 130(4): 1311-1316.
Mshvildadze, M., J. Neu, J. Shuster, D. Theriaque, N. Li, and V. Mai. 2010. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. Journal of Pediatrics 156(1):20-25.
Nanthakumar, N. N., R. D. Fusunyan, I. Sanderson, and W. A. Walker. 2000. Inflammation in the developing human intestine: A possible pathophysiologic contribution to necrotizing enterocolitis. PNAS 97(11):6043-6048.
Neu, J., and J. Rushing. 2011. Cesarean versus vaginal delivery: Long-term infant outcomes and the hygiene hypothesis. Clinics in Perinatology 38(2):321-331.
Neu, J., and W. A. Walker. 2011. Medical progress: Necrotizing enterocolitis. New England Journal of Medicine 364(3):255-264.
Palmer, C., E. M. Bik, D. B. DiGiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biology 5(7):e177.
Preidis, G. A., and J. Versalovic. 2009. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: Gastroenterology enters the metagenomics era. Gastroenterology 136(6):2015-2031.
Reeves, A. E., C. M. Theriot, I. L. Bergin, G. B. Huffnagle, P. D. Schloss, and V. B. Young. 2011. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2(3):145-158.
Reeves, A. E., M. J. Koenigsknecht, I. L. Bergin, and V. B. Young. 2012. Suppression of Clostridium difficile in the gastrointestinal tract of germ-free mice inoculated with a murine lachnospiraceae isolate. Infection and Immunity 80(11):3786-3794.
Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. Journal of Clinical Periodontology 25(2):134-144.
Vaarala, O., M. A. Atkinson, and J. Neu. 2008. The “perfect storm” for type 1 diabetes: The complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 57(10):2555-2562.
Yoshioka, H., A. Yoshimura, T. Kaneko, D. T. Golenbock, and Y. Hara. 2008. Analysis of the activity to induce toll-like receptor (tlr)2- and tlr4-mediated stimulation of supragingival plaque. Journal of Periodontology 79(5):920-928.
Zelkha, S. A., R. W. Freilich, and S. Amar. 2010. Periodontal innate immune mechanisms relevant to atherosclerosis and obesity. Periodontology 2000 54(1):207-221.
Zijnge, V., M. B. van Leeuwen, J. E. Degener, F. Abbas, T. Thurnheer, R. Gmur, and H. J. Harmsen. 2010. Oral biofilm architecture on natural teeth. PLoS ONE 5(2):e9321.














