Influence of Diet and Dietary Components on the Microbiome
As the workshop progressed, speakers explored in greater depth the impact of diet on the microbiome, how dietary influences on the microbiome contribute to human health and disease, and ways to modulate the microbiome to build and maintain health through the use of prebiotics and probiotics in food products. This chapter summarizes that discussion.
Through evolutionary experimentation, mammals have spent the last 120 million years successfully developing “the most efficient, effective and adaptable means of postnatal nutrient provision that has ever arisen among vertebrates: lactation.” —Blackburn (1993)
That a majority of people suffer from diet-dependent diseases raises the question, Is it possible to prevent disease through diet? In his exploration of the preventive potential of the human diet, Bruce German focuses his research on the one food that evolved to be preventive: human breast milk. The cost–benefit trade-off associated with human milk is key to understanding milk’s preventive potential, German explained. Everything in human milk costs the mother. “The mother is literally dissolving her tissues to make milk,” he said. Yet the third most abundant component in milk, the oligosaccharides, are undigestible by the infant. How can this be? How
__________________________
1 This section summarizes the presentation of Bruce German.
can such an abundant material of such a costly phenotype be undigestible by the individual for whom it is intended, that is, the infant?
What Are Human Milk Oligosaccharides (HMOs), and What Health Benefit Do They Provide the Infant?
Glycobiology2 is “disastrously, catastrophically complex,” German said. According to German, the number of possible glycans, or oligosaccharides, in a biological system is in the billions, based on the number of ways that sugars (the basic structural units of glycans) and linkages (the bonds between sugars) can be combined. This makes sense given that oligosaccharides on cell surfaces are the basis of a recognition system across all life forms. Carlito Lebrilla developed a methodology for analyzing glycan complexity in human milk, based on innovative separation technologies and very high-efficiency, high-accuracy mass spectrometry (Ninonuevo et al., 2006). His research team has constructed an annotated database of the nearly 200 highly variable structural compositions of HMO (Wu et al., 2010, 2011).
David Mills was among the first to address the question, What do HMOs do? He hypothesized that HMOs serve as a food source for the infant microbiome. However when he and his colleagues tested bacterial growth on HMO as a sole food source, none but Bifidobacterium infantis grew (Ward et al., 2007). “Perhaps we shouldn’t be surprised,” German said, given that B. infantis is a dominant member of the breast-fed-infant microbiome. Moreover, Mills and his group have discovered that only very specific strains of B. infantis grow on HMO medium. Even bacteria that grow very well on a variety of other sugar media are unable to grow on HMO medium (Marcobal et al., 2010; Ward et al., 2006). (The only other genus that appears to be able to grow on HMO medium is Bacteroides. However, as both German and, later during the question-and-answer period, Mills explained, B. infantis readily outcompetes Bacteroides when the two are grown together.) “What we are discovering about this remarkable interaction between milk oligosaccharides and this particular bacterium is remarkable,” German said. “Mothers are literally recruiting another life form to babysit their babies and using the oligosaccharides to direct the microbiome.”
How does the system work? For example, if oligosaccharides are serving as a food source for B. infantis, which oligosaccharides are being consumed? Mills and his group have discovered that unlike other bifidobacteria, B. infantis selectively cleaves and eliminates sialic acid–containing
__________________________
2 Glycobiology is the study of the structure, biosynthesis, and biology of glycans, also called oligosaccharides (i.e., sugar chains).
oligosaccharides (LoCascio et al., 2007; Ward et al., 2007). Only an estimated 4 to 38 percent of HMOs are sialyated; a higher proportion are fucosylated (40 to 70 percent) (Ninonuevo et al., 2006). Moreover, Mills’s team has also identified which B. infantis genes cleave what HMO linkages (Sela et al., 2011). Interestingly, in German’s opinion, the expression of the bacterial enzyme that actually cleaves the sialyated oligosaccharides is regulated by the abundance of HMOs in the growth medium. There is other evidence indicating that human milk sugars interact with the microbiome in ways that increase the value of the microbiome to the infant. For example, German mentioned the research of Helen Raybould’s group on B. infantis and its role in endocrine signaling in the infant intestine (Chichlowski et al., 2012).
The real question, in German’s opinion, is whether the association between HMOs and B. infantis persists as a phenotype in “real life.” That is, “does is really influence the bacteria in living babies?” Data on microbiome development through the first 12 weeks of an infant’s life show that initially Bifidobacterium is not present in the microbiome (manuscript in preparation), but by week 12 it emerges as a dominant member of the microbiome. Evidence from fecal sampling indicates that HMOs are not being digested during the first weeks of life, presumably because there are no bifidobacteria to digest them, but they begin to disappear from the infant feces at the same time Bifidobacterium begins to dominate the microbiome (manuscript in preparation). Thus, the association between HMOs and B. infantis is a “true symbiotic relationship,” German said. “It’s as important to feed the bacteria in the baby as the baby.”
German suggested that knowledge of human milk–microbiome symbiosis could be translated into practice in several ways. For example, he mentioned Mark Underwood’s research on the effects of administering a combination of B. infantis and HMOs to premature infants (manuscript in preparation).
HOST-MICROBE INTERACTIONS IN THE PERINATAL PERIOD3
There are very few data on the development of the gut microbiota in healthy infants, let alone how diet impacts that microbiota. Yet, there is a plethora of clinical and epidemiological data suggesting that breast-feeding promotes mucosal immune development and protects against many diseases. These data, combined with the fact that human milk contains a variety of bioactive proteins, carbohydrates, and lipids not present in infant formula, raise questions about whether and how the infant gut microbiota differs between breast-fed and formula-fed infants. Sharon Donovan’s long-term
__________________________
3 This section summarizes the presentation of Sharon Donovan.
research goal is to use noninvasive approaches to define how early nutrition shapes host-microbe interactions and influences intestinal development in breast-fed versus formula-fed infants. She hopes that the knowledge gained can be used to identify selective additives, such as bioactive proteins, prebiotics (including HMO), and probiotics that can be added to infant formula to provide some of the health benefits afforded by breast-feeding.
Differential Expression of Microbial Genes in Breast-Fed Versus Formula-Fed Infants
In what Donovan described as a “proof-of-concept” study, she and colleagues used a method developed by Robert Chapkin (Davidson et al., 1995) for isolating exfoliated epithelial cells from stool to identify genes differentially expressed in breast-fed versus formula-fed infants (Chapkin et al., 2010). Specifically, they analyzed stool samples collected at 3 months of age from vaginally delivered term infants who were medically certified as healthy and who were either exclusively breast-fed (N = 12) or formula-fed (N = 10) (Chapkin et al., 2010). The researchers gained institutional review board (IRB) approval to train the mothers themselves to collect the samples at home. The initial messenger RNA (mRNA) expression microarray analysis yielded 4,250 genes that were expressed in all infants. Of those, about 1,200 were significantly differentially expressed between breast-fed and formula-fed infants. Due to the small sample size and thus greater potential for false discovery, the scientists compared these 1,200 genes to a list of 546 that they had predicted could be differentially expressed based on their known roles in intestinal biology. This yielded 146 differentially expressed genes, to which researchers applied a linear discriminant analysis and coefficient of determination analyses developed by Edward Dougherty and colleagues (Dougherty et al., 2009; Kim et al., 2000) to identify the genes that best classified breast-fed versus formula-fed infants and those that were master regulators, respectively.
The strongest classifier was EPAS1, which encodes a protein involved in cellular response to hypoxia. Given that necrotizing enterocolitis (NEC) is associated with tissue hypoxia and that human milk has been shown to protect preterm infants from NEC, Donovan speculated that upregulation of EPAS1 in breast-fed infants might be helping those babies’ guts to tolerate hypoxic episodes.
Other genes that qualified as good classifiers are summarized in Table 5-1. The linear discriminant analysis methodology used allowed investigators to identify not just single genes that could be considered good classifiers of breast-fed versus formula-fed infants, but also two- and three-gene combinations (Chapkin et al., 2010).
Donovan speculated that these gene expression differences might ex-
TABLE 5-1 Exfoliated Epithelial Cell Genes Identified as Good Classifiers of Breast-Fed (BF) Versus Formula-Fed (FF) Infants, Based on a Linear Discriminant Analysis of Genetic Material Collected from Stool Samples
| Gene Name | Function | Fold change (BF/FF) | ||
| EPAS1 | Transcription factor (TF); cellular response to hypoxia | 3.3 | ||
| NR5A2 | TF, encodes liver receptor homologue-1 (LRH-1); development | 2.8 | ||
| NR3C1 | Encodes glucocorticoid receptor | 5.5 | ||
| PCDH7 | Encodes protocadherin-7; membrane protein | 3.9 | ||
| ITGB2 | Encodes integrin beta-2 (CD18); ICAM-1 receptor | 2.5 | ||
| FGF5 | Encodes fibroblast growth factor 5; mitogenesis and cell survival | 2.0 | ||
| TJP1 | Encodes ZO-1; intercellular tight junctions | 2.2 | ||
| MYB | TF, transcriptional transactivation; proto-oncogene | 2.8 | ||
| EPIM | Syntaxin 2/epimorphin; epithelial cell morphogenesis | 2.5 | ||
| BAD | BCL2-associated agonist of apoptosis | 4.0 | ||
| SOURCE: Chapkin et al., 2010. | ||||
plain some of the clinical and epidemiological evidence that has accumulated over the years showing that breast-fed babies’ guts develop differently and are less leaky than those of formula-fed babies. For example, the expression of TJPI, which encodes ZO-1, an intercellular protein that plays an important role in regulating tight junctions, was also upregulated in the cells from breast-fed babies. Additionally, expression of NR3C1, which encodes a glucocorticoid receptor that plays a role in gut differentiation, was fivefold higher in breast-fed than in formula-fed infants.
Using MetaCore, a bioinformatics tool that provides information about function, researchers found that some of the strongest signals were with combinations of genes that encode signaling pathways involved in fundamental pathways of intestinal stem cell proliferation and differentiation, such as WNT and NOTCH.
Donovan suggested that these various gene and gene network classifiers could serve as potential biomarkers for differentiating between breast-fed and formula-fed infants. Also, it would be interesting to see if addition of any of these to infant formula, in the form of a prebiotic or probiotic, would shift the gut microbiota toward the direction of breast-fed infants.
Next, Donovan and her team tested the hypothesis that the integration of infant (host) epithelial cell transcriptome and functionally profiled microbiome can be used to suggest important regulatory pathways of the microbiome affecting intestinal development in the first few months of life (Schwartz et al., 2012). Community-wide microbial gene expression in stool
from the same breast-fed (N = 6) and formula-fed (N = 6) infants from their earlier work (Chapkin et al., 2010) using established protocols (Poroyko et al., 2010) was evaluated using 454 pyrosequencing of DNA libraries created from stool samples. Taxonomic composition of the metagenome was analyzed with the metagenomics analysis server MG-RAST using similarity to a large nonredundant protein database. Using the same database, the sequence alignments for known microbial metabolic functions were tested against the SEED subsystems.4 At the phyla level, all formula-fed infants shared the same distinct signature, whereas breast-fed infants were more variable. Three of the breast-fed infants had similar profiles, but the other three “were sort of going to the beat of their own drummer,” said Donovan. She noted that all of the formula-fed infants were receiving the same formula but that breast milk composition can be highly variable.
Using established protocols for evaluating community-wide microbial gene expression in stool samples, the team observed a greater total percentage (i.e., percentage of total 16S ribosomal RNA [rRNA]) of Actinobacteria, Proteobacteria, and Bacteroidetes in breast-fed piglets as a group and a greater total percentage of Firmicutes and no Bacteroidetes in formula-fed infants as a group (see Figure 5-1) (Donovan et al., 2012). Donovan remarked that these findings warrant follow-up, given Peter Turnbaugh and colleagues’ observation that obesity in mice is associated with a higher Firmicutes:Bacteroidetes ratio. In addition, epidemiological studies have demonstrated that breast-feeding protects against the development of childhood obesity.
Which Human Genes Respond to Bacterial Signals?
An overarching theme of the workshop discussion was the importance and growing interest in understanding not just what bacteria are present in the microbiome, but how those bacteria are signaling in a way that impacts the human host biology. For example, Donovan expressed curiosity about whether differences in microbial gene expression between breast-fed and formula-fed infants impact host gene expression. A comparison of the functional SEED categories of the stool metagenome of breast-fed and formula-fed infants demonstrated a significantly higher proportion of virulence genes in the breast-fed infants. Next, the scientists applied a systematic and statistically rigorous analytic framework for the simultaneous examination of both host and microbial responses to dietary or environmental components in the early neonatal period. Specifically, using canonical correlation analysis,
__________________________
4 SEED is an open-source software platform that seeks to curate microbial genomic data into subsystems-based functional annotation (e.g., amino acid metabolism). More information is available online: www.theseed.org (accessed August 28, 2012).
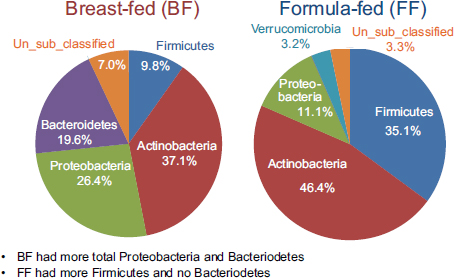
FIGURE 5-1 Results from a study on breast-fed versus formula-fed human infants showing variation in gut microbial composition.
SOURCE: Donovan et al., 2012.
Donovan and her team identified associations between the microbiome virulence genes and 11 host immunity defense genes (TACR1, VAV2, ALOX5, NDST, REL, BPILI, AOC3, KLRF1, DUOX2, IL1A, and SP2) (Schwartz et al., 2012). Donovan speculated on the potential biomarker usefulness of microbial sequencing, in this case as a way to predict host defense mechanisms. These findings suggest that simultaneously examining the multivariate structure underlying the microbiome and gut transcriptome leverages richer and fuller information content compared to analyses focusing on single datasets (e.g., only host transcriptome data or only microbiome data) and only single variables (e.g., gene-by-gene differential expression testing). The use of canonical correlation analysis can support the formulation of hypothesis-based studies by accurately identifying those genes active in commensal microbiome and host activities (Schwartz et al., 2012).
What Components in the Infant Diet Affect the Intestinal Microbiota?
Nutrients and bioactive components in human milk directly influence the development of the infant’s immune system, actively protect the infant from pathogenic infection, and facilitate establishment of the microbiota, the last of which is required to activate the mucosal immune system. Recent data suggest that HMOs contribute to many of these activities (Donovan et al., 2012). Oligosaccharides are the third most predominant component
of milk, after lactose and fat, and up to 200 different structural forms have been identified in human milk (Wu et al., 2010, 2011). Since HMOs are resistant to digestion by the infant and pass into the colon, Donovan described them as the fiber of human milk, a fact that she said hasn’t really been appreciated until recently. In the colon, they potentially function in a variety of ways, including as substrates for fermentation and the production of short-chain fatty acids. They can also serve as prebiotics for beneficial bacteria. Donovan referred to David Mills’s very elegant data showing that Bifidobacterium infantis metabolizes specific HMOs (see the previous section for a more detailed description of work conducted in the Mills laboratory and by Bruce German).
HMO composition is influenced partly by secretor status of the mother and whether she has the 2-fucosyltransferase gene; non-secretor mothers do not produce 2’-fucosyllactose (2’-FL), which is one of the primary HMOs in the milk of secretor mothers. Therefore, Donovan and others are exploring potential predictive associations between HMO composition and infant gut microbiota. Systematic evaluation of the impact of HMO on infant development, however, has been limited by the lack of sufficient quantities of pure HMOs to conduct animal or human feeding studies. However, in the near future, this limitation will be overcome through improved synthetic approaches, opening avenues of investigation into the biology of HMOs. Additionally, the availability of noninvasive methods of assessing outcomes in human infants (Chapkin et al., 2010; Schwartz et al., 2012) and high-throughput methods for measuring HMOs (Wu et al., 2010, 2011) and the infant microbiome (Schwartz et al., 2012) will facilitate our understanding of the role of HMOs in host-microbe interactions in the developing infant (Donovan et al., 2012).
THE RESISTOME AS A DRIVER OF THE MICROBIOME5
Food is not the only major driver of the microbiome. So too is the way we raise food, Ellen Silbergeld stated. Most food animals are grown very intensively, including through the use of animal feeds that contain antibiotics. Food and Drug Administration (FDA) data indicate that 80 percent of total antimicrobial production in the United States in 2009 was for use in animal feed.6 Silbergeld stressed that the use of antibiotics in animal feed is not for veterinary medical purposes; rather, antibiotics are added to feed
__________________________
5 This section summarizes the presentation of Ellen Silbergeld.
6 These values were calculated by the Center for a Livable Future based on data provided by the Food and Drug Administration. For more information, read the posting on its website: http://www.livablefutureblog.com/2010/12/new-fda-numbers-reveal-food-animals-consume-lion%E2%80%99s-share-of-antibiotics (accessed September 19, 2012).
as growth promotants. This is a new phenomenon in the history of antimicrobials and one with significant implications for what Silbergeld referred to as the “resistome.”
The Resistome
The term “resistome” was introduced by Wright (2007), who defined it as the collection of all the antibiotic resistance genes and their precursors in the entire microbial community of both pathogenic and nonpathogenic bacteria. Genes within the resistome encode molecular changes that confer phenotypic resistance to both general and specific antibiotic molecules. They are often clustered in cassettes and are transferable by plasmids, creating a pleiotropic efficacy. Multigene cassettes can encode other phenotypes, not just resistance.
An important feature of the resistome is that resistance genes are readily transferred from one bacterial cell to another via horizontal, or lateral, gene transfer (usually via plasmid-mediated transfers but also by conjugation). The classic model of antibiotic resistance describes a population of diverse organisms encountering antibiotic pressure, with some organisms being susceptible and some resistant and with the susceptible organisms dying and the resistant organisms persisting (Sommer and Dantas, 2011). However, that model does not account for the dynamic nature of horizontal gene transfer and the fact that a population of initially susceptible bacteria can accumulate expressible resistance genes over time. Even after a stressor is withdrawn, this system can be permanently altered by the experience. In a study of humans exposed to ciprofloxacin, Dethlefsen and Relman (2011) showed that resistant phenotypes persisted even after exposure ended.
Increasingly, horizontal gene transfer involves not just the sharing of single genes, but also the sharing of cassettes of multiple genes. Silbergeld explained how the extensive use of antimicrobials exerts multiple and repeated pressures on bacterial populations, resulting in sequential acquisition of resistance genes and buildup of multigene cassettes (Canton and Ruiz-Garbajosa, 2011). As empirical evidence of the buildup of transferable multigene cassettes, Silbergeld mentioned U.S. Department of Agriculture (USDA) data showing growth over time of extended multidrug resistance phenotypes of Escherichia coli in domestic animals (i.e., chicken, swine, cat, dog, dairy cattle) (Lindsey et al., 2011).
Not only is the resistome accruing more resistance genes, either singly or bundled in multigene cassettes, it also appears to be accumulating networks of preferential horizontal gene transfers, or “cliques” (Skippington and Ragan, 2011). Again, extensive antimicrobial use is exerting selective pressure, in this case for more active networks of horizontal gene transfer.
The bacteria involved in any given network, or clique, are not necessarily in close proximity and may not even share the same ecology.
Silbergeld described the resistome as being analogous to cloud computing, because it is a resource that can be externalized and accessed by various groups of bacteria and because the transfer of resistance genes via horizontal gene transfer is like transferring downloaded bytes of data. She stressed the importance of recognizing that the resistome encompasses both pathogenic and nonpathogenic bacteria and may include most of the bacteria in a specific microbiome.
From the Modern Livestock Farm to Humans: Implications for the Resistome
Selective pressures exerted by extensive antibiotic use abound in the modern livestock farm, which Silbergeld described as an “impressive laboratory for driving microbial evolution.” Danzeisen et al. (2011) sampled the microbiomes of chicken ceca after feeding chickens either control feed, feed with monensin (an antibiotic), or feed with virginiamycin (another antibiotic) and detected significant differences in microbiome content (e.g., percentage of total microbes represented by Lachnospiraceae versus Ruminococaceae). However, it is not just the flora that is changing in the face of increased antimicrobial pressure. Researchers are also reporting an increased prevalence of antibiotic-resistant phenotypes in food animals fed antibiotic-containing diets (Looft et al., 2012) (see Figure 5-2).
Importantly, the increased prevalence of resistant phenotypes being observed in the guts of farm animals persists not just in their microbiome but in the resistome at large, mainly because of the practice of land disposal of animal wastes without required pretreatment. For example, Nandi et al. (2004) traced the movement of resistance genes from poultry litter into the soil environment where poultry waste was deposited. Eventually, humans can potentially become exposed to those same resistance genes via several routes.
One way to represent the resistome and the way it transcends, or extends across, all of these different microbiomes, from farm animals to soil bacteria to the human gastrointestinal (GI) tract, is as a nested system of increasing complexity, where components occupy different spaces in the ecosystem and events that occur can eventually impact the human microbiome (Davis et al., 2011). Silbergeld’s research group is conducting an ongoing study of the historical ecology of the Chesapeake Bay to see if the appearance of resistance genes in sediment correlates with the introduction of intensive poultry production and the use of antibiotics in poultry feed.
Some experts consider antibiotic-resistant genes to be environmental pollutants that bioaccumulate over time (Martinez, 2009). Although antibiotics are generally not very persistent in the environment, if humans
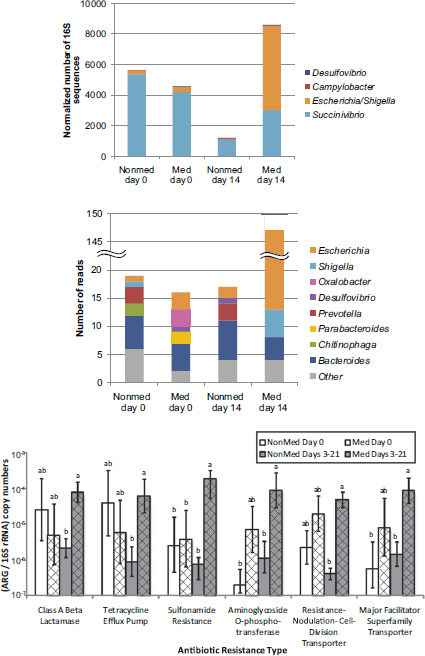
FIGURE 5-2 Results from a study showing a shift in gut microbiota (top figures) and an increased prevalence of antibiotic-resistant phenotypes (bottom figure) in food animals fed antibiotic-containing diets (“GPA feeds”).
NOTES: ARG = antibiotic-resistant gene; med = antibiotic-containing diet; nonmed = control. Columns with the same letter are not statistically significant (p > 0.05) within each resistance type.
SOURCE: Looft et al., 2012.
continuously add them to the environment—for example, by adding farm animal waste to soil—they can become persistently present, not just in any given microbiome but in the environment at large. Bioaccumulation of antibiotic resistance also occurs through the expansion of microbial populations, and this makes sense evolutionarily, according to Silbergeld. Scientists used to think of antimicrobial resistance as being costly for a microbe to maintain in the absence of antimicrobial pressure, but work by Levin et al. (2000) has shown that in many cases, it is less costly and more efficient for a bacterium to accumulate additional mutations that reduce these costs. This fact, Silbergeld said, may partly explain why resistant bacteria are so persistent even after the antimicrobial stressor is removed. Because of the large number of antimicrobials now in the environment, this may be an efficient evolutionary strategy, as suggested by Martinez (2009).
Silbergeld agreed with Vincent Young, Richard Darveau, and other speakers that the bad-bugs–good-drugs paradigm is too simplistic. Resistant genes can readily travel from one “bug” to another via horizontal gene transfer, either as naked DNA or on cassettes. Even studying the microbiome may not be enough. The challenge is to locate the resistome in space (e.g., Where is it within the microbiome? Where is it in the ecosystem?) and identify where gene transfer and “cross-talk” among microbes occur.
PROBIOTIC MECHANISMS OF ACTION7
A reader … may be surprised by my recommendation to absorb large quantities of microbes, as a general belief that microbes are harmful. This belief is erroneous. There are many useful microbes, amongst which the lactic bacilli have an honorable place. —Elie Metchnikov (1907)
While the fundamental conceptual framework for probiotics was laid out in the early 20th century by Elie Metchnikov (1845-1916), many of today’s scientists use the Food and Agriculture Organization-World Health Organization (FAO-WHO) (2002) definition as their working definition. FAO-WHO (2002) defines probiotics as living microorganisms that when administered in adequate amounts confer a health benefit on their host. James Versalovic called attention to three features of the FAO-WHO definition. First, probiotics are living, viable microorganisms. Second, for a microorganism to be considered a probiotic, it has to be administered in adequate amounts. Third, a probiotic must confer some kind of health benefit. For Versalovic, the question is, How do they confer that health benefit? Or, as he put it, how do they “optimize the functioning of our physiology”? Versalovic listed several potential mechanisms of action: stimulation of
__________________________
7 This section summarizes the presentation of James Versalovic.
immunity, suppression of immunity (both innate and adaptive immunity), promotion of intestinal epithelial cell development and migration, alteration of microbiome composition and function, enhanced recovery from infection, and antimicrobial functions.
Stimulation of Immunity
Probiotics can directly stimulate both adaptive and innate immunity (Thomas and Versalovic, 2010). They can also indirectly impact host immunity by enhancing host ability to digest and absorb nutrients that have an impact on the immune system. Both the direct and the indirect effects have been described in detail in a number of studies published over the past couple of decades. For example, Yamanaka et al. (2003) reported that gut bacteria drive Peyer’s patch8 development in rats, with germ-free rats having defective and immature lymphoid follicles and conventionalized rats having very mature gut lymphoid tissue. Indeed, it has been proposed that a key function of the gut microbiome might be to serve as a “treadmill” for the host immune system (Madara, 2004). By “tickling” Toll-like receptors and other receptors and signaling pathways that build host immunity, gut microbes might be keeping the host immune system “finely tuned and fit” and preparing the immune system for new challenges (e.g., antibiotic exposure, changes in diet). Evidence supports this hypothesis. As just one example, Prescott et al. (2008) reported that pregnant women who took either a Bifidobacterium or Lactobacillus probiotic had significantly elevated levels of interferon-gamma levels in their cord blood and that maternal probiotics may enhance interferon-gamma production in neonates. Interferon-gamma production has been associated with protection against allergic disease early in life (Macaubas et al., 2003). Prescott et al. (2008) also showed a significant effect of maternal probiotics on antibody production in breast milk. Breast milk immunoglobulin A (IgA) levels were significantly elevated at both 1 week and 3 months of age in the children of women who took either a B. lactis or L. rhamnosus probiotic. Although the difference in IgA levels between the children of the experimental and control women disappeared by 6 months of age, infants fed breast milk with elevated IgA levels early on may be receiving a critical head start in gaining passive immunity.
Suppression of Immunity
Evidence suggests that probiotics don’t just stimulate immunity, they also suppress it. Versalovic’s research group has conducted mouse model
__________________________
8 Peyer’s patches are aggregates of lymphoid follicles located in the epithelium of the small intestine.
studies showing that the probiotic Lactobacillus reuteri can suppress pro-inflammatory cytokines by secreting very small soluble factor(s) (Thomas and Versalovic, 2010).9 Using nuclear magnetic resonance (NMR) and other advanced metabolomic technologies, Thomas et al. (2012) identified one of these soluble factors as histamine and reported a correlation between elevated levels of bacterial-derived histamine and potent tumor necrosis factor (TNF) suppression in human monocytoid cells. According to Versalovic, the correlation has been found in a variety of other cell lines as well. As far as mechanism, Thomas et al. (2012) showed in vitro that bacterial-derived histamine suppresses TNF production by binding to histamine type 2 (H2) receptors and blocking mitogen-activated protein (MAP) kinase signaling pathways. “This was clearly surprising that we would have histamine as an immunoregulatory molecule,” Versalovic said. “But the real punch line is not the histamine. It’s histidine.”
Histidine is a dietary component that serves as the chemical precursor to histamine. Several research groups have identified three genes involved in histidine-to-histamine biosynthesis: hdcA, hdcB, and hdcP (Copeland et al., 1989; Martin et al., 2005; Trip et al., 2011). Versalovic’s team generated single-gene knockouts of the probiotic strain L. reuteri ATCC 6475 for all three genes and showed in vitro that knocking out any one of the three genes eliminates a large chunk—40 percent or more—of bacterial TNF inhibitory activity (Thomas et al., 2012). The research group is testing the single-gene knockouts in vivo. Unpublished data indicate that L. reuteri 6475 hdcA knockouts administered to mice via orogastric gavage cause a diminished ability to attenuate colitis following a TNBS (2,4,6-trinitrobenzenesulfonic acid) challenge.10 By contrast, when mice are administered wild-type probiotic, their disease state ameliorates. The next step, Versalovic noted, is to add more histidine to the diet via modified mouse chow and see if that enhances the anti-inflammatory effect of the wild-type probiotic.
Histamine is “a large part of the story,” Versalovic said, but it’s not the whole story. The “other part of the punch line” is that histamine operates as an immunomodulatory compound only in the presence of the H2 receptor. The H1 receptor, by contrast, triggers a proinflammatory response. So the effect of histamine—and histidine—in the diet is dependent on the relative distribution of H2 versus H1 receptors. Versalovic’s team is exploring the biogeography of H2 receptors and ways in which they can be enhanced.
__________________________
9 Versalovic noted that most of his research on probiotics is with L. reuteri. Not only is it a widely used probiotic, it is also an indigenous member of the human microbiome (Reuter, 2001).
10 The TNBS challenge is standard protocol for inducing murine colitis, that is, the mouse equivalent of human inflammatory bowel disease.
Other research groups are thinking along the same line, that is, that a dietary amino acid, in this case histidine (specifically, L-histidine), can be converted by gut bacteria into another compound with immunomodulatory effects (Andou et al., 2009; Blumberg and Strober, 2001).
Promotion of the Intestinal Epithelium
In addition to their impact on host immunity, probiotics also impact development of the intestinal epithelium. For example, Preidis et al. (2012a,b) demonstrated that gut bacteria can prevent or ameliorate Rotavirus- associated acute gastroenteritis in a neonatal mouse model. Administering two different strains of Lactobacillus reuteri led to about a 1-day reduction in disease duration, which is a significant amount of time in the case of acute infection. One L. reuteri strain in particular had a dramatic effect on maturation of epithelial cell walls. It is not clear what the signals are, that is, how these microbes are enhancing the ability of the epithelium to differentiate. Data from expression profiling studies suggest that L. reuteri may be promoting the sloughing of Rotavirus-infected cells by altering the actin cytoskeleton and weakening attachments of the basement membrane, thereby increasing epithelial cell migration and turnover.
Other Mechanisms of Action
In addition to host immunity and intestinal epithelium development, probiotics can also influence human health and disease by enhancing microbiome diversity or, more compellingly, by changing microbiome gene expression. They can also impact antimicrobial production by the microbiome (e.g., L. reuteri produces reuterin). Versalovic suggested that a probiotic antimicrobial strategy could replace the widespread use of antibiotics in animal farms.
The Future
Rapidly advancing knowledge of the microbiome could be used to create “designer strains” of probiotics that enhance health or prevent disease (Preidis and Versalovic, 2009). Alternatively, it might be possible to select natural strains of probiotics that make foods even more effective and functional than they already are in terms of health maintenance and disease prevention.
PREBIOTIC MECHANISMS OF ACTION11
Food ingredients and novel compounds are increasingly being examined for their ability to do more than provide nutrition, according to George Fahey. Most of this expanding research activity is focused on health promotion or disease reduction. At the top of the list of food ingredients being studied for nonnutrition activity are the nondigestible oligosaccharides (NDOs). Nutritionally, NDOs are known mostly for their low caloric value and ability to enhance mineral absorption, but they are also becoming known for their potential to lower the risk of infections and diarrhea, modulate the immune system, and modulate the microbiota. At the beginning of the prebiotic era, in the mid-1990s (Gibson and Roberfroid, 1995), scientists spent a great deal of time in particular thinking about how NDOs might be used to increase the presence of beneficial bifidobacteria (members of the genus Bifidobacterium) and lactobacilli (members of the genus Lactobacillus) in the microbiota while decreasing the presence of pathogenic bacteria. Many NDOs have the ability to alter the composition of the colonic microbiota in a positive manner, thus satisfying a key criterion for what defines a prebiotic—the selective stimulation of growth and/or activity of those bacteria that contribute to colonic and host health.
What Are the Major Dietary Sources of Prebiotics?
There are several well-established major dietary sources of prebiotics, primarily fructins (including chicory root extract, inulin, oligofructose, and short-chain fructooligosaccharides). Two other major dietary sources of prebiotics are the galactooligosaccharides and the stool softener lactulose.
There is a long list of potential prebiotic candidates, including soybean oligosaccharides, glucooligosaccharides, cyclodextrins, gentiooligosaccharides, oligodextrans, glucorinic acid, pectic oligosaccharides, isomaltooligo-saccharides, lactosucrose, xylooligosaccharides, human milk oligosaccharides, mannanoligosaccharides (yeast cell wall), lactose, resistant starch and derivatives, oligosaccharides from melobiose, N-acetylchitooligosaccharides, poly-dextrose, sugar alcohols, and konjac glucomannan. These are widely variable types of compounds, Fahey noted. Several are natural ingredients (e.g., soybean oligosaccharides), and several are widely used in both human and animal diets (e.g., yeast cell wall, which is very rich in mannanoligosaccharides). Some are very simple from the point of view of chemical composition (e.g., glucooligosaccharides); others are “really strange,” according to Fahey (e.g., N-acetylchitooligosaccharide). These and other candidates are considered “potential” because research on their prebiotic characteristics is incomplete,
__________________________
11 This section summarizes the presentation of George Fahey.
and they have not been shown to meet all of the specific requirements of the current working definition of a prebiotic.
Prebiotics and prebiotic candidates are produced from a variety of raw materials, such as chicory, artichoke, beet, cow’s milk, starch, and soybean (Mussatto and Mancilha, 2007). Production typically involves extracting an intermediate product (e.g., inulin from chicory and artichoke, sucrose from beet, lactose from cow’s milk, soluble starch from starch, soybean whey and xylan from soybean) and then using one of several processes (i.e., hydrolysis, transglycosylation, isomerization, extraction) to isolate the actual prebiotic.
How Do Prebiotics Modify the Composition of the Microbiota?
The effect of a prebiotic (or potential prebiotic) on bacterial growth depends on the type of prebiotic (or potential prebiotic) ingested. In a study on the effect of resistant starch on fecal microbiota in 10 healthy human volunteers, Martinez et al. (2010) observed significant changes in the relative proportions of various bacterial taxa depending on the type of resistant starch ingested. Researchers fed the volunteers three types of crackers in a 17-week double-blind crossover study: RS2 (crackers made with Hi-Maize 260, a resistant starch 2), RS4 (crackers made with a chemically modified, phosphorylated, cross-linked type 4 resistant starch, Fibersym RW), and native wheat starch (the control). All subjects consumed 33 grams of resistant starch per day. Consumption of RS4 increased the proportion of phylum Firmicutes and decreased the proportions of Bacteroidetes and Actinobacteria relative to the control and, in the case of Firmicutes and Actinobacteria, relative to the consumption of resistant starch 2 as well. At the family level, researchers observed increased proportions of Bifidobacteriaceae and Porphyromonadaceae in the RS4 treatment and decreased proportions of Ruminococcaceae and Erysipelotrichaceae relative to the control. At the genus level, they observed increased proportions of Parabacteroides and Bifidobacterium and decreased proportions of Faecalibacterium and Dorea in the RS4 treatment compared to the control.
As another example of the variable effects of different types of prebiotics, in a randomized, double-blind, placebo-controlled, crossover study of 20 healthy men between 21 and 28 years of age, Hooda et al. (2012) observed significant differences in the proportion of bacterial genera detected in feces depending on which of three types of fiber were consumed. Researchers fed the volunteers three fiber bars per day, with each bar containing either no supplemental fiber or 7 grams of either polydextrose (PDX) or soluble corn fiber (SCF). They observed large and significant increases in the
proportions of Faecalibacterium, Phascolarctobacterium, and Dialister in feces following consumption of both PDX and SCF and a small but statistically significant increase in Lactobacillus following consumption of SCF.
The extent to which a prebiotic (or potential prebiotic) stimulates microbial growth depends not just on the type of substance ingested, but also on its dietary concentration. In a 16-week study of galactooligosaccharides (GalOS) in 18 healthy human volunteers between 19 and 50 years of age, Davis et al. (2010) reported two major findings. First, based on culture enumeration, the concentration of Bifidobacterium in feces increased significantly among volunteers who were fed 5 or 10 grams of GalOS per day, compared to baseline. There was no significant increase in Bifidobacterium in the feces of individuals fed either 0 or 2.5 grams of GalOS per day. Second, individuals fed 10 grams of GalOS per day also had significantly more total anaerobes in their feces compared to baseline. None of the other treatment groups showed a change in total anaerobe concentration. Using quantitative real-time PCR (qRT-PCR) to measure the bifidogenic effects of GalOS, again the researchers found a significant increase in bifidogenic activity among individuals fed 5 or 10 grams of GalOS per day, but not among individuals fed 0 or 2.5 grams per day. With respect to which bacteria are affected by GalOS consumption, Davis and colleagues (2010) pyrosequenced the V1-V3 region of 16S ribosomal DNA (rDNA) and found that GalOS consumption did not impact the diversity of fecal microbes but did impact the relative proportions of bacterial taxa at the phylum, family, genus, and species levels. For example, consumption of 10 grams of GalOS per day increased the proportion of the family Bifidobacteriaceae from 1.56 percent at baseline to 6.14 percent. Within that family, consumption of 10 grams of GalOS per day increased the proportion of Bifidobacterium from 1.28 percent at baseline to 5.20 percent.
Fahey referred to previous speakers’ comments on the considerable variability that exists among individuals with respect to how their microbiota respond to dietary intervention. Not surprisingly, Davis et al. (2010) observed highly variable responses among individuals at the phylum, family, genus, and species levels. Fahey recognized the limitations that individual variability sets up for a study based on a sample size of 18, but asserted that 18 is a manageable number for such an intensive study.
Sometimes prebiotics are used as supplements at very low levels, even though much of the murine research on inulin, for example, involves daily administration of the human equivalent of 30 to 35 grams of fermentable substrate, which is within the range of the dietary reference intake (DRI)
for men.12 Human studies show variable effects depending on dietary concentration.
In addition to the type of prebiotic or potential prebiotic ingested and dose, a multitude of other factors influence the way a prebiotic impacts the GI microbiota, including gastric emptying time, intestinal transit time, nutrient digestibility, fecal bulk and frequency of defecation, short-chain fatty acid (SCFA) production, intestinal morphology, gut immune modulation, and the GI microbiota itself.
Are Prebiotics Effective in Achieving Host Health Benefits?
Much of the research on prebiotics is in healthy individuals. Many studies have shown significant increases in the so-called beneficial microbes (i.e., bifidobacteria and lactobacilli) following consumption of GalOS, inulin, and other prebiotics—in healthy individuals. Researchers have also reported increases in butyrate producers (e.g., Eubacterium, Faecalibacterium, Roseburia) following consumption of resistant starch, polydextrose, soluble corn fiber, and other prebiotics—again, in healthy individuals. Fahey urged more studies on diseased populations, given that many microbes are associated with disease. For example, inflammatory bowel disease conditions are known to be associated with a decreased proportion of Faecalibacterium. Studies have shown that SCF, PDX, inulin, fructooligosaccharides, pea fiber, and other prebiotics or potential prebiotics can impact Faecalibacterium. The question remains, Do those same prebiotics or potential prebiotics alleviate inflammatory bowel disease conditions via their impact on Faecalibacterium?
Fahey also urged more consideration of how prebiotics impact microbial metabolites, especially butyrate and other SFCAs. Studies have shown that inulin, fructooligosaccharides, and GalOS can increase SFCA levels, but what impact do prebiotics have on the toxic end products of fermentation such as ammonia, phenols, and indoles? Hooda and colleagues (2012) measured some of those toxic end products as part of a larger microbe-health index principal component analysis and found that Lachnospiraceae, Lactobacillaceae, and Veillonellaceae were all negatively correlated with ammonia, phenols, and indoles. Additionally, Lachnospiraceae and Lactobacillaceae were positively correlated with total SCFA, and Veillonellaceae was positively correlated with fiber intake. Veillonellaceae was also negatively correlated with total branched-chain fatty acids (BCFAs).
As an example of a prebiotic study that both used a disease model and
__________________________
12 The DRI for total fiber (combination of dietary fiber and functional fiber) for men is 31 grams per day for the 9- to 13-year age group, 38 grams per day for the 14- to 50-year age group, and 30 grams per day for men aged 51 and older (IOM, 2002).
examined metabolic outcome, Everard et al. (2011) fed ob/ob mice either a control diet or a diet supplemented with oligofructose for 5 weeks. At the end of the 5 weeks, they sequenced the V1-V3 region of 16S rDNA and conducted various glucose metabolism tests. The results of the 16S rDNA analysis showed phylum-level increases in Bacteroidetes, Actinobacteria, and Proteobacteria and a decrease in Firmicutes. At the family level, they detected Bifidobacteriaceae in the prebiotic-fed group, but not in the controls. At the genus level, again they detected Bifidobacterium only in the prebiotic-fed mice. The glucose tolerance testing showed several positive metabolic outcomes associated with prebiotic consumption: lower fasting glycemia level, improved glucose tolerance, decreased fat-to-muscle mass ratio, decreased plasma triglycerides, improved gut barrier function, lower plasma lipopolysaccharide (LPS) concentrations, and reduced expression of oxidative stress and inflammatory markers.
Potential Ways to Advance the Field of Prebiotics
In addition to more research on diseased populations and on microbial metabolites, Fahey suggested several other ways to advance the field of prebiotics. First, conduct more compositional analyses of potential prebiotics. “We do too little of that,” he said. Knowing the monomeric composition, chain length, linkages, branching, side chains, and other features of the structural composition of a prebiotic can help to interpret the biological data. Second, examine prebiotic activities in natural foods, such as soybean products, beet fiber, and whole grains and their co-products. Third, continue to look beyond the bifidobacteria. Fourth, study microbiome–health index relationships, à la Hooda et al. (2012).
TRANSLATION OF PROBIOTIC SCIENCE INTO PROBIOTIC FOODS13
How can knowledge about the microbiome influence the design of healthy food, including probiotic foods? Scientists know that probiotics can impact the microbiome, both directly and indirectly, as James Versalovic described during his presentation (O’Toole and Cooney, 2008). They also know that probiotics can impact health. What they don’t know, according to Mary Ellen Sanders, is whether probiotic impacts on the microbiome are directly responsible for the observed human health benefits. Most studies that correlate microbiome changes and human health benefit do not reveal anything about causality. So the question remains, Do probiotics have a beneficial effect on health through their direct or indirect actions on the
__________________________
13 This section summarizes the presentation of Mary Ellen Sanders.
microbiome? “I would say the answer to that is likely yes,” Sanders said, but such causality needs to be confirmed. Sanders provided an overview of demonstrated effects of probiotics on the microbiome and on health and scientific challenges to translating this knowledge into probiotic foods.
Impact of Probiotics on the Microbiome
There is plentiful evidence of the effects of probiotics on the microbiome, especially intestinal microbiota (Sanders, 2011). The most common impact of probiotics on the intestinal microbiota, or more accurately the fecal microbiota, is an increase in the particular strain that the test individuals have been fed. Probiotics expand across a wide taxonomic range and even include yeast (i.e., Saccharomyces). However, researchers usually feed their test subjects probiotics that they know will survive intestinal transit. Another common observation is changes in metabolic parameters that can be either local or pan-organismal. These changes can be observed not just in the feces or colon, but also in the urine and in other tissues. Probiotics have also been observed to impact pathogens, as evidenced by changes in the infectivity and toxicity of pathogens. Researchers have also observed changes in the community structure of indigenous microbiota, although results vary among probiotics and among studies. For example, different probiotics have been shown to increase community evenness, functional redundancy, and specific types of potentially beneficial bacteria. Finally, and one of the more interesting effects in Sanders’s opinion, is that probiotics have been shown to encourage homeostasis, or stability, of the microbiota. It has been hypothesized that maintaining the microbiota in an “even state” has beneficial physiological effects; microbiota that maintain a certain evenness, or stability, may be able to rebound more quickly when perturbed by an antibiotic or other stressor (Sanders, 2011).
When considering beneficial effects of probiotics on the microbiome, it must be remembered that experts have yet to reach consensus on what a healthy microbiome looks like. This fact makes it difficult to know which probiotic effects on the microbiome are likely to translate into health benefits for the host.
Demonstrated Health Benefits of Probiotics
The demonstrated health benefits of probiotics go beyond the gut. Researchers have investigated a wide range of end points, including oral microbiology (e.g., dental caries), allergies (e.g., atopic dermatitis, asthma), vaginal infections, mental function, skin microbiology, acute upper respiratory tract infections, and various global end points (e.g., growth parameters of undernourished children, reduced absences from work or day care,
quality-of-life indicators). Even within the gut, a wide range of end points have been tested. These include acute diarrhea, antibiotic-associated diarrhea, travelers’ diarrhea, C. difficile infection, lactose digestion, irritable bowel syndrome (IBS) symptoms, colic, inflammatory bowel conditions, and gut pain sensation.
In Sanders’s opinion, the field is embracing evidence-based approaches to conclusions on the health effects of probiotics, as demonstrated by the many systematic reviews and meta-analyses that have been published. As of November 2011 (the time of this IOM workshop), Sanders had identified 66 such reviews in the scientific literature. The end points cover a very broad range of body sites and conditions, including NEC; infant growth; persistent diarrhea; radiation-induced diarrhea; antibiotic-associated diarrhea; travelers’ diarrhea; H. pylori; Crohn’s disease, ulcerative colitis, and pouchitis; IBS; digestive symptoms; allergy; critical care or hospital infections; bacterial vaginosis; acute respiratory tract infections; and safety. Sanders noted that most of these end points are drug (i.e., can cure, treat, mitigate, or prevent disease) and not food end points. A common misperception is that a probiotic dose needs to be at least 109 in order to be effective, Sanders noted, but there is no single best minimum dose. Rather, whatever dose was used in the human study that showed a significant positive effect should be the minimum dose for that probiotic (Savino et al., 2007; Whorwell et al., 2006).
Significance of Strain Specificity
For the past 20 years, researchers have been emphasizing the importance of strain specificity. Plentiful evidence from animal models shows this to be the case, according to Sanders. The effectiveness of one strain of a species does not necessarily mean that other strains are equally effective. In a comparison of five different commercial probiotic preparations, Canani et al. (2007) observed variable effects on the duration of diarrhea in children, with only two of the commercial preparations demonstrating effectiveness (see Table 5-2). Sanders suggested that part of the reason that commercial products labeled as probiotic do not necessarily have similar effects could be that variable combinations of strains are used.
The challenge of strain specificity raises the question, Are there shared effects among phylogenetically related strains? For example, almost all studies on the two yogurt-containing probiotics Streptococcus thermophilus and Lactobacillus bulgaricus have demonstrated reduced lactose maldigestion in people who are lactose-intolerant. Regardless of strain, all yogurts containing at least 108 live starter S. thermophilus and L. bulgaricus per gram can bear the claim that the product will improve lactose digestion in individuals with lactose maldigestion (EFSA, 2010). Sanders asked, Could
TABLE 5-2 Comparison of Five Commercial Probiotic Preparations on Duration of Diarrhea in Children
| Treatment | Median (IQR) Duration (hours) | Estimated Difference (95% CI) | PValue |
| Oral rehydration solution alone | 115.5 (95.2-127) | — | — |
| Lactobacillus casei subp rhamnosus GG | 78.5 (56.5-104.5) | –32 (–41 to –23) | <0.001 |
| Saccharomyces boulardil | 105.0 (90-104.5) | –5 (–13 to 5) | 0.38 |
| Bacillus clausii | 118.0 (95.2-128.7) | 1 (–7 to 8) | 0.76 |
| L. delbrueckii var. bulgaricus, L. acidophilus, Streptococcus thermophilus, B. bifidum | 70.0 (49-101) | –37 (–47 to –25) | <0.001 |
| Enterococcus faecium SF 68 | 115.0 (89-144) | 2 (–5 to 11) | 0.61 |
NOTE: N = 571 children aged 3-36 months presenting with acute diarrhea; 5-day treatment period. CI = confidence interval; IQR = interquartile range.
SOURCE: Canani et al., 2007.
other general claims be made about groups of phylogenetically related strains? For example, can multiple strains of Bifidobacterium improve digestive comfort, or might multiple strains of Lactobacillus increase short-chain fatty acids in the colon? To be convincing, these more general claims will require demonstrating that multiple strains of the same taxonomic group have the same effect and either that a common mechanism of action among the strains mediates this effect or that different mechanisms of action among the strains result in the same effect. Sanders was hopeful that “there may be a time in this field when enough accumulated data are present that we are able to say that although certain activities are definitely linked to certain strains, others do seem to be more broadly attributable to broader microbiological categories or phylogenetic types.”
Challenges to Translating Probiotic Science into Probiotic Foods
While the plethora of probiotic products on the market seems to suggest a lack of any barriers to the development of probiotic foods, in fact there are many scientific, regulatory, technological, and marketing challenges. Sanders elaborated on a couple of key scientific and regulatory challenges.
In addition to strain specificity, another major scientific challenge is that the magnitude of the demonstrated effect must be meaningful. Sanders
remarked that if experimental study does not demonstrate a meaningful effect, nothing of interest has really been demonstrated. This raises the question, What is a meaningful magnitude of effect? She asked members of the workshop audience whether they considered an intervention that decreases absences from school by half a day per year enough of a magnitude of effect to consider the intervention worthwhile. Few audience members nodded yes. Yet, that is exactly the magnitude of effect demonstrated in a study on hand-washing by children—and it was enough of an effect to justify a national hand-washing campaign by the Centers for Disease Control and Prevention. Using the same end point—missed school days—Leyer et al. (2009) showed that a 6-month course of L. acidophilus NCFM/B. animalis Bi-07 probiotic resulted in more than 1 fewer missed school days per child. That is double the effect of hand-washing, Sanders noted. “Sometimes in the probiotic field we kind of beat ourselves up because we don’t have these overwhelmingly huge magnitudes of effect,” she said. “But maybe we don’t need them.”
Yet another major scientific challenge is that not all studies demonstrate the same effects. Mixed results reflect the considerable individual-level variation in microbiota that has been demonstrated many times (Candela et al., 2010). They also reflect the prevalence of underpowered, small-N probiotic studies. An underpowered study providing no evidence of an effect is very different from a sufficiently powered study providing evidence of no effect. Sanders said, “We need to be able to distinguish that and possibly quit running underpowered studies.”
In addition to these scientific challenges, Sanders suggested that new regulatory challenges may end up discouraging future probiotic research. In October 2010, FDA issued a guidance on determining when human research studies require Investigational New Drug (IND) applications (FDA, 2010). Sanders interpreted the guidance to mean that any human study on the cure, treatment, mitigation, or prevention of disease or on the structure or function of the body is a drug study and therefore can only be conducted with an IND. “If it’s ever finalized in this form,” she said, “it will have a chilling effect on research” in probiotics.
In conclusion, Sanders touched on another regulatory challenge that would be explored in depth by subsequent workshop speakers (see Chapter 6), that is, the current regulatory framework for product claims. Probiotic products sitting side by side on the store shelf can be very different regarding both content and scientific evidence for safety and efficacy, but allowable information on claims doesn’t enable consumers and health care professionals to differentiate among products. Sanders mentioned Proctor and Gamble’s Align. The claim is that Align builds and supports a healthy digestive system, but the scientific evidence is for improved symptoms associated with irritable bowel syndrome (O’Mahony et al., 2005; Whorwell
et al., 2006). The range of allowed claims, even with documented evidence, is too narrow. Sanders commented that consumers and health care providers should be provided with truthful information so that they can make informed choices about probiotics.
Experiments within a well-controlled laboratory or clinical setting may indicate that a particular bacterium is a highly effective probiotic, but in reality the probiotic may not be effective if its bioactivity is lost before it is able to confer any health benefit. Many studies have shown that probiotic bacteria lose their activity over time if they are placed in foods that have not been correctly designed to accommodate those bacteria, according to David Julian McClements. There has been a dramatic increase in probiotic viability studies over the past decade, with many studies showing appreciable reductions in probiotic viability during food storage or during transit through the human GI tract. For example, Priya et al. (2011) reported a 108- to 109-fold decrease in the number of viable probiotic organisms by the time they reached the small intestine.
Delivery System Design
McClements’s research program revolves around the design of encapsulation systems, that is, structured delivery systems that encapsulate, protect, and deliver bioactive compounds to an appropriate site of action within the GI tract. While most of his work is with nutraceuticals, he asserted that the same systems are amenable to utilization for the encapsulation and delivery of live bacteria. In the previously mentioned Priya et al. (2011) study, in addition to researchers observing a dramatic decrease in the number of viable bacteria reaching the colon, they also observed substantial improvement upon encapsulating the bacteria in a multilayer polymer coating (see Figure 5-3).
McClements noted two key considerations to keep in mind when designing delivery systems for probiotics. First, whether a probiotic delivery system will work or not depends on the strain of bacteria, the nature of the delivery system, and the kind of food in which the bacteria is being delivered. He remarked that the tremendous variability observed in the results of probiotic viability studies reflects variation in these factors. Second, foods are low-profit-margin materials. The food industry wants the simplest, cheapest, and most robust solutions to any given problem. Yet, because they are difficult to make and require complicated processing operations,
__________________________
14 This section summarizes the presentation of David Julian McClements.
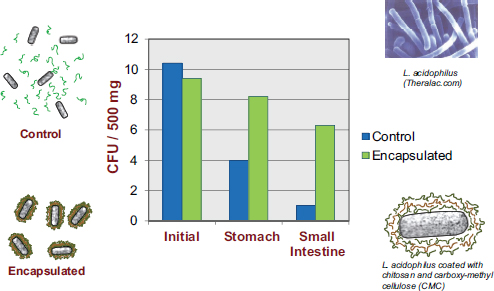
FIGURE 5-3 Results showing increased viability of encapsulated probiotics, relative to non-encapsulated probiotics, as they pass through the human GI tract.
NOTE: CFU = colony-forming unit.
SOURCE: Priya et al., 2011.
delivery systems are often expensive. There are other options for protection, such as controlling food matrix properties (e.g., nutrients, pH, ionic strength, temperature, oxygen, other properties that affect bacterial survival) and selecting resistant microbial strains (e.g., bile- and acid-resistant strains, strains resistant to some of the other stressors to which bacteria are likely to be exposed). Third, any potential delivery system used should not adversely affect the desirable quality attributes of a food, such as appearance, taste, texture, and shelf life, because this will affect the likelihood that an individual will continue to purchase and consume the food.
Encapsulation Technology
In many respects, McClements noted, developing delivery systems for probiotics is similar to what drug manufacturers do when they develop delivery systems for drugs. However, the challenge is even greater because of greater constraints on the types of components that can be used with foods and the complexity of the food matrix. A drug can be placed in a capsule, pill, or syrup, but a probiotic, if it is going to be consumed regularly, needs to be placed in a food matrix in such a way that it does not adversely affect the appearance, taste, texture, or stability (shelf life) of the food, according to McClements. Plus, food products encounter a series of
different stressors during manufacturing, storage, transport, and preparation (e.g., thermal processing, chilling, freezing, dehydration), any of which could affect probiotic viability.
Food delivery system design has been driven in part by the nano-technology revolution, which has provided new tools that can be used to create molecular and colloidal structures to encapsulate bacteria, protect them from the various challenges they are likely to encounter in food and as they pass through the GI tract, and release the encapsulated bacteria at specific sites in the GI tract. The three most common structural designs are embedding, coating, and a hybrid embedding-coating approach. Embedding involves trapping the probiotic within some sort of solid or liquid matrix made of proteins, polysaccharides, or other components. Coating involves covering the probiotic with one or more layers of dietary fibers, proteins, or other substances. A hybrid embedding-coating approach involves trapping the bacteria within a matrix and then coating the matrix.
Encapsulation technologies are different for dry foods (e.g., cereals, powders, breads) versus wet foods (e.g., beverages, yogurts). Dried products are typically encapsulated using spray drying, yielding 50 micrometer dried powder microencapsulated probiotics. The particles dissolve when exposed to water, releasing the probiotic. Spray drying protects probiotics during food storage but not upon exposure to the human body. A variety of different technologies can be used for wet product encapsulation, including coacervation, bead formation, emulsion formation, and coating. Bead formation involves mixing the probiotics with a polymer solution (such as alginate) and then dripping the mixture into a gelling solution (such as calcium chloride) to form beads with probiotics encapsulated inside. Co-acervation involves mixing the probiotics with a mixture of positive and negative polymers to form a hydrogel bead with probiotics trapped inside. Emulsion formation involves using a water-and-oil emulsion to make filled biopolymer particles with the probiotic contained inside. Finally, coating methods involve coating a negatively charged bacterium with a positively charged polymer (monolayer) or a series of positive and negatively charged polymers (multilayer). The advantage of these three technologies is that they maintain their structure when diluted. Unlike dried encapsulation systems, they do not fall apart upon exposure to the human body and can be designed to maintain their viability until they reach a specific region of the GI tract.
Controlling probiotic viability is a challenge. The delivery system has to be designed to withstand the variable challenges it encounters along the length of the GI tract, such as high acidity, lipase activity, antimicrobial activity, and oxygen levels. The stomach is arguably the harshest environment, with a pH typically between 1 and 3, and with enzymes that can break down the delivery system or attack the probiotic (e.g., lipases, proteases),
bile salts, and other stressors. To overcome these challenges, researchers have devised many ways to manipulate delivery system properties to ensure that a system remains viable along the length of the GI tract. Viability is influenced by different characteristics of encapsulation systems: particle size—larger particles are usually more stable during transit through the GI tract; composition—the digestibility of the matrix components determines their response; nutritional profile—the type of nutrients present within a matrix may influence probiotic viability; physical state—solid particles are often more stable than liquid ones; permeability—the pore size of matrices can be changed so that digestive enzymes, bile salts, and other stressors cannot access the probiotic; and environmental responsiveness (e.g., microencapsulated probiotics can be designed to swell or shrink under different pH conditions or ionic strengths). Changing the electrical charge on the delivery system particles can impact where in the GI tract the encapsulated probiotic is likely to attach (e.g., on the mucin layer in a certain region of the GI tract).
Once a system has been designed and developed, it is tested in vitro, which usually involves simulating the mouth, stomach, small intestine, and colon by controlling pH and the types of enzymes and minerals present. In vitro testing can be used to screen different types of delivery systems or various alterations in a delivery system (e.g., Chavarri et al., 2010; Priya et al., 2011). Eventually the system needs to be tested in vivo by using animal and human studies, according to McClements.
Challenges and Opportunities
In conclusion, McClements stated that there is great potential for delivery systems to protect probiotics in foods and within the body. Probiotics that otherwise might not survive in foods under normal conditions or in the human body might need the protection that encapsulation affords. However, in addition to being effective, delivery systems must be economical (i.e., many of the technologies that have been tested in vitro are too expensive for commercial application), practical (e.g., they have to be constructed from food-grade materials), and without any potentially adverse effects.
While several encapsulation systems have been shown to be effective in vitro, a major challenge to translating these results into commercial products is the non–food-grade nature of some of the materials used. The encapsulated probiotics tested in both Priya et al. (2011) and Chavarri et al. (2010) were engineered with a chitosan coating. Not only is chitosan not food-grade, it is also antimicrobial, which could have adverse effects. Sensory quality poses another challenge. The particles tested in Chavarri et al. (2010) were in the millimeter size range, which is too large to incorporate
into a food matrix. The mouth detects as undesirable (in texture) anything larger than about 50 micrometers.
HOW THE MICROBIOME REVOLUTION FUELS FUNCTIONAL FOOD RESEARCH15
The main mission of Danone is to help people build and preserve their health capital through food, according to Johan van Hylckama Vlieg. Danone offers consumers products that serve all life stages, from babies (i.e., foods focused on infant nutrition) to older adults (including foods for people with specific nutritional requirements, also referred to as “medical nutrition”).
However, Danone—and the food industry at large—is up against some new challenges, not the least of which is a changing demographic context. In 2013, five countries will represent 47 percent of the global population and 45 percent of the global gross domestic product (GDP): China, India, the United States, Indonesia, and Brazil. “These are mostly new target populations,” van Hylckama Vlieg said. Much of the research on the microbiome, diet, and health to date has focused on the “classical First World context,” that is, Western Europe and the United States. Added to that is the trend in global aging. The age pyramid in 2015 is expected to be drastically different than it was in 2000, with large geographic differences in aging trends.
These demographic challenges are compounded by the fact that the food industry is often held responsible for the trend in obesity, although many other lifestyle factors contribute to obesity. Consequently, consumer associations are requesting more transparency on food composition, origin, and value for money. Yet there is an important role for the food industry to play in improving health for the global population. Science—in particular the science of the microbiome—is providing new tools and knowledge to manage these challenges. With respect to the microbiome, researchers are identifying a growing number of microbiota signatures and activities associated with health and disease (e.g., energy metabolism, production and availability of nutrients, cardiovascular health, cell proliferation and cancer, gut-brain axis and emotion, immune maturation and functioning, gut comfort, pathogen protection). More interestingly, in van Hylckama Vlieg’s opinion, is that researchers are beginning to identify not just associations, but causal relationships between microbiome signatures and activities associated with specific health benefits. The food we eat is also the major source of growth for our gut microbiota and thereby may be an effective way to steer its composition and activity. The question is, How can we
__________________________
15 This section summarizes the presentation of Johan van Hylckama Vlieg.
leverage this science on the microbiome to develop products that maintain or improve health? Establishing causal relationship allows for microbes to be targeted, and the next step is to identify specific active ingredients and components that target these microbes and their impact on host health. Or, as van Hylckama Vlieg expressed, “Science provides increased rationale for functional food concepts using pre- and probiotics that bring a clear health benefit to consumers.”
Leveraging the Microbiome for Health
In fact, humans have been leveraging the microbiome for a long time, initially through animal husbandry and consumption of fermented foods and moving toward science-based evidence for dietary intake. Fermented foods are important constituents of the human diet worldwide, and use of these foods dates back approximately 10,000 years (Evershed et al., 2008). These fermentations are often carried out with lactic acid bacteria. As van Hylckama Vlieg explained, lactic acid bacteria are also natural inhabitants of the human GI tract, so foods fermented by lactic acid bacteria are effectively supplementing the indigenous microbiota. In fact, lactic acid bacteria could be considered “domesticated” microbes. By substituting “microbe” for “animal” or “plant,” they fit the Webster’s dictionary definition of domesticated: “to adapt (an animal or plant) to life in intimate association with and to the advantage of humans.”16
As an example of how Danone has leveraged the microbiome for health, van Hylckama Vlieg highlighted work on a fermented food product containing Bifidobacterium animalis subsp. lactis strain CNCM I-2494. The impact of FMP on the gut microbiome and host health was explored using a specific mouse model (T-bet-/- and Rag2-/- knockout mice, also known as TRUC mice) that spontaneously develops gut inflammation resembling human ulcerative colitis (Garrett et al., 2007). Garrett et al. (2007) reported that antibiotics reverse inflammation in TRUC mice, indicating a microbial etiology. Plus, when they co-housed TRUC and wild-type mice, they observed colitis in the wild-type mice as well, indicating the communicability of whatever gut microbes were associated with inflammation in the TRUC mice.
While a population of individuals with ulcerative colitis is far removed from Danone’s target population, using that sort of extreme model can provide cleaner data and reveal mechanisms more readily than would be possible using other methods. Using the TRUC mouse model, Veiga et al. (2010) analyzed the TRUC gut microbiota by 16S rDNA sequencing and observed low levels of Bifidobacterium in the colon compared to non-inflamed (wild-
__________________________
16 See http://www.merriam-webster.com/dictionary/domesticate.
type) mice. They observed that feeding the TRUC mice 100 milligrams of FMP per day decreased intestinal inflammation (<0.0001 compared to TRUC mice fed the same amount of nonfermented food product) and increased levels of short-chain fatty acids (acetic acid, propionic acid, butyric acid). They also observed increased levels of lactate-consuming bacteria in the FMP-fed TRUC mice. The researchers showed “very elegantly,” according to van Hylckama Vlieg, that the newly altered intestinal environment in FMP-fed mice inhibits growth of colitogenic bacteria. Research on the impact of FMPs on TRUC mouse microbiome and host health is ongoing. Meanwhile, another study with the same FMP product on the gut microbiomes of healthy twin pairs and gnotobiotic mice demonstrated that FMPs do not cause any major perturbation of the dominant microbiota of healthy human subjects. However, this product does trigger distinct responses in the activity of the microbiome detected through transcriptomics (McNulty et al., 2011). Also, in both the mouse model and human samples, the metabolic activity of gut Bifidobacterium animalis subsp. lactis is dramatically altered. B. animalis subsp. lactis harvests and grows on the xylooligosaccharides derived from dietary components, which demonstrates that strain is becoming an active member in the gut microbial community.
Microbiome Biomarkers: Implications for Personalized Nutrition
In addition to research on mouse models and human samples, Danone is a partner of the MetaHIT Consortium. Acknowledging controversy about the biological relevance as expressed during the meeting regarding the existence of enterotypes, van Hylckama Vlieg remarked that regardless, “the observation is there” (Arumugam et al., 2011). The question is, What are the implications for the food industry? Do observations of enterotypes or any other clusters of markers in the microbiome indicate that people have specific nutritional needs, perhaps specific probiotic needs, depending on their microbiota compositions? It may be possible that these issues will lead to the emergence of personalized, or categorized, nutrition in coming years.
In preparation for these coming years, Danone is building and maintaining a culture collection of more than 3,000 microbes, mainly lactic acid bacteria. The collection includes strains isolated from multiple geographic areas (i.e., from various countries), ecoystems (i.e., from dairy products, cereals, plants, and stools), and time periods (i.e., from the 1960s through the present) and contains more than 80 species. They have sequenced nearly 100 genomes in the collection. “It is a very powerful platform for discovery,” van Hylckama Vlieg said. Much of its power stems from its focus on strain diversity. In a comparative genome hybridization study of 42 strains of Lactobacillus plantarum, Siezen and colleagues (2010) found
that while about 70 percent of the genome of L. plantarum is shared among all strains, there are many genes not shared by all strains and many strain-specific genes.
Strain individuality raises the key question: What is the correlation with functional diversity? Currently, Danone investigators have been exploring the “pangenome” of multiple strains of two species, L. rhamnosus and L. paracasei, for gene-function correlations. Researchers are building a genome diversity database and coupling that with an extensive phenotyping program. Such studies will help to identify both common activities shared by many strains and specific features that can only be delivered by a few strains.
According to van Hylckama Vlieg, Danone scientists are looking at phenotypes related to carbohydrate utilization and short-chain fatty acid production, antimicrobial activity, and immune modulation. As an example of the type of results they are collecting, a sequencing analysis of 12 strains of L. rhamnosus and 30 strains of L. paracasei indicated that about 30 to 40 percent of the genome is “core,” that is, shared among all strains. However, up to 25 percent of genes are strain-specific, with different strains having different immune function effects (i.e., based on an NF-kappa B-type screen on HT29 cells).
Most of the discussion during the question-and-answer period at the end of this session revolved around three major issues: (1) functional consequences of modulating the microbiome through food, (2) whether there are any known adverse effects of prebiotic and probiotic interventions, and (3) experimental design and study size.
Focus on Function
A recurring theme of the workshop was the importance of functional, not just compositional, changes to the microbiome as a result of dietary (or antibiotic) intervention. For example, as summarized in this chapter, Johan van Hylckama Vlieg mentioned research results demonstrating that Activia does not cause any major perturbation of the microbiota but does trigger distinct transcriptomic responses related to the metabolic activity of Bifidobacterium animalis. Also, George Fahey commented on the results of a study showing several positive metabolic outcomes associated with oligofructose consumption in mice. Finally, James Versalovic elaborated on the many ways that probiotics can impact host immunity. An audience member asked whether, given that food products appear to impact the microbiome not by recolonizing (so not by changing the taxonomic makeup of the mi-
crobiome), by rather through signaling (changing the way the microbiome is functioning), does it matter whether the food product in question is a live organism (i.e., a probiotic) or an inert substance (i.e., a prebiotic)? In other words, is the differentiation between probiotic and prebiotic a false dichotomy? Are the conceptual barriers between probiotic and prebiotic artificial?
Versalovic agreed that as the field moves forward, hopefully the dialogue will move beyond definitions and focus more on how food is impacting health via its effect on the microbiome. The value of the discovery of histamine as a microbially produced molecule that impacts host immunity isn’t the histamine itself. Rather, its value is that “it has pointed us in a whole new direction—to look at amino acid conversions.” Are other amino acids being converted by microbes? What other enzymatic machinery is the microbiome providing for the biochemical conversion of dietary information into biological signals? Versalovic said, “We’re on entirely new trails in the wilderness.” His research group is also studying another microbially catalyzed bioconversion of a host dietary substance into a biological signal, that is, glutamate into gamma-aminobutyric acid (GABA). He speculated on the potential regulatory role of the gut microbiome with respect to how much dietary information is actually being converted in vivo. “What’s really intriguing here,” he said, “is that that may be how the microbiome is really contributing to health and physiology … providing the enzymatic machinery, the metabolic pathways, at a particular location. It could be the GI tract. It could be the airways. It could be the oral cavity … providing that machinery that then allows [for the conversion of] the dietary content into [a] signal for the body.”
Adverse Effects?
An audience member asked whether there were any known adverse effects of prebiotic or probiotic interventions. Mary Ellen Sanders replied that the National Institutes of Health (NIH) recently commissioned a review on probiotic safety that covered hundreds of prevention and treatment-of-disease studies of a variety of microbes (Hempel et al., 2011). The review concluded that most studies have not been adequately designed to address safety. Sanders speculated that the lack of safety data is likely due to the fact that probiotics have always been viewed as a food. They have not been viewed as drugs or as something that potentially could cause problems, at least not when administered to healthy people. So in the past, most researchers jumped right into efficacy studies, bypassing what would be the equivalent of a phase I drug study. Today, more researchers are designing their studies with safety factors in mind. Meanwhile, to Sanders’s knowledge, at least for the most commonly researched microbes there is no or
very little association of use in a normal population with any adverse effects of real concern. There are some notable exceptions, she said. For example, Besselink et al. (2008) reported increased mortality after administration of probiotics to patients with severe pancreatitis. Sanders cautioned that not only do different microorganisms have different safety patterns, but that the health status of the host also influences safety.
Dan Levy emphasized the need to examine strain-specific safety effects. He said, “The tendency has been to say, well, we’re exposed to all of these lactic acid bacteria with no adverse effects.” But not all lactic acid bacteria are the same. While some strains may cause no adverse effects, that is not necessarily the case for all strains. This is especially worrisome when a strain is chosen for a unique property, that very uniqueness suggesting that the strain does not behave like others, particularly when selecting for strains intended to have specific and perhaps novel physiological effects on the consumer. “You have to stop lumping all lactic acid bacteria together,” Levy said. “We’re looking at specific physiological mechanisms associated with specific strains, and we have to study the good, the bad, and the ugly.”
Experimental Design and Study Size
The discussion of microbiome markers of health and disease prompted a couple of comments on experimental design. Ellen Silbergeld expressed concern that too many studies on the microbiome are “almost entirely underpowered,” raising serious questions about the value of the information being provided by those studies. If a study is too underpowered to provide any evidence of an effect, then what is the value of that study? “If you really want to move this field forward,” she said, “you really have to start considering your study design.” In response, Johan van Hylckama Vlieg remarked that many small studies, such as the twin study that he mentioned, are intended to be exploratory and hypothesis-generating. They are not intended to yield the same type of results that large-scale clinical trials provide. Silbergeld then wondered whether any of the larger studies actually meet the criteria of a clinical trial. It is not clear that any do. Versalovic questioned whether large-scale clinical trials are even appropriate for microbiome studies. Given such extreme individual-level variation in microbiome composition and function, he said, “I don’t know that we can do trials the same way anymore,” indicating the need for further discussion on issues of study design.
Andou, A., T. Hisamatsu, S. Okamoto, H. Chinen, N. Kamada, T. Kobayashi, M. Hashimoto, T. Okutsu, K. Shimbo, T. Takeda, H. Matsumoto, A. Sato, H. Ohtsu, M. Suzuki, and T. Hibi. 2009. Dietary histidine ameliorates murine colitis by inhibition of proinflammatory cytokine production from macrophages. Gastroenterology 136(2):564-574.
Arumugam, M., J. Raes, E. Pelletier, D. Le Paslier, T. Yamada, D. R. Mende, G. R. Fernandes, J. Tap, T. Bruls, J. M. Batto, M. Bertalan, N. Borruel, F. Casellas, L. Fernandez, L. Gautier, T. Hansen, M. Hattori, T. Hayashi, M. Kleerebezem, K. Kurokawa, M. Leclerc, F. Levenez, C. Manichanh, H. B. Nielsen, T. Nielsen, N. Pons, J. Poulain, J. Qin, T. Sicheritz-Ponten, S. Tims, D. Torrents, E. Ugarte, E. G. Zoetendal, J. Wang, F. Guarner, O. Pedersen, W. M. de Vos, S. Brunak, J. Dore, H.I.T.C. Meta, M. Antolin, F. Artiguenave, H. M. Blottiere, M. Almeida, C. Brechot, C. Cara, C. Chervaux, A. Cultrone, C. Delorme, G. Denariaz, R. Dervyn, K. U. Foerstner, C. Friss, M. van de Guchte, E. Guedon, F. Haimet, W. Huber, J. van Hylckama Vlieg, A. Jamet, C. Juste, G. Kaci, J. Knol, O. Lakhdari, S. Layec, K. Le Roux, E. Maguin, A. Merieux, R. Melo Minardi, C. M’Rini, J. Muller, R. Oozeer, J. Parkhill, P. Renault, M. Rescigno, N. Sanchez, S. Sunagawa, A. Torrejon, K. Turner, G. Vandemeulebrouck, E. Varela, Y. Winogradsky, G. Zeller, J. Weissenbach, S. D. Ehrlich, and P. Bork. 2011. Enterotypes of the human gut microbiome. Nature 473(7346):174-180.
Besselink, M. G., H. C. van Santvoort, E. Buskens, M. A. Boermeester, H. van Goor, H. M. Timmerman, V. B. Nieuwenhuijs, T. L. Bollen, B. van Ramshorst, B. J. Witteman, C. Rosman, R. J. Ploeg, M. A. Brink, A. F. Schaapherder, C. H. Dejong, P. J. Wahab, C. J. van Laarhoven, E. van der Harst, C. H. van Eijck, M. A. Cuesta, L. M. Akkermans, H. G. Gooszen, and Dutch Acute Pancreatitis Study. 2008. Probiotic prophylaxis in predicted severe acute pancreatitis: A randomised, double-blind, placebo-controlled trial. Lancet 371(9613):651-659.
Blackburn, D. G. 1993. Lactation: Historical patterns and potential for manipulation. Journal of Dairy Science 76(10):3195-3212.
Blumberg, R. S., and W. Strober. 2001. Prospects for research in inflammatory bowel disease. JAMA 285(5):643-647.
Canani, R. B., P. Cirillo, G. Terrin, L. Cesarano, M. I. Spagnuolo, A. De Vincenzo, F. Albano, A. Passariello, G. De Marco, F. Manguso, and A. Guarino. 2007. Probiotics for treatment of acute diarrhoea in children: Randomised clinical trial of five different preparations. BMJ 335(7615):340.
Candela, M., C. Consolandi, M. Severgnini, E. Biagi, B. Castiglioni, B. Vitali, G. De Bellis, and P. Brigidi. 2010. High taxonomic level fingerprint of the human intestinal microbiota by ligase detection reaction—universal array approach. BMC Microbiology 10:116.
Canton, R., and P. Ruiz-Garbajosa. 2011. Co-resistance: An opportunity for the bacteria and resistance genes. Current Opinion in Pharmacology 11(5):477-485.
Chapkin, R. S., C. Zhao, I. Ivanov, L. A. Davidson, J. S. Goldsby, J. R. Lupton, R. A. Mathai, M. H. Monaco, D. Rai, W. M. Russell, S. M. Donovan, and E. R. Dougherty. 2010. Noninvasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells. American Journal of Physiology: Gastrointestinal and Liver Physiology 298(5):G582-G589.
Chavarri, M., I. Maranon, R. Ares, F. C. Ibanez, F. Marzo, and C. Villaran Mdel. 2010. Microencapsulation of a probiotic and prebiotic in alginate-chitosan capsules improves survival in simulated gastro-intestinal conditions. International Journal of Food Microbiology 142(1-2):185-189.
Chichlowski, M., G. De Lartigue, J. B. German, H. E. Raybould, and D. A. Mills. 2012. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. Journal of Pediatric Gastroenterology and Nutrition 55(3):321-327.
Copeland, W. C., J. D. Domena, and J. D. Robertus. 1989. The molecular cloning, sequence and expression of the hdcB gene from Lactobacillus 30a. Gene 85(1):259-265.
Danzeisen, J. L., H. B. Kim, R. E. Isaacson, Z. J. Tu, and T. J. Johnson. 2011. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS ONE 6(11):e27949.
Davidson, L. A., Y. H. Jiang, J. R. Lupton, and R. S. Chapkin. 1995. Noninvasive detection of putative biomarkers for colon cancer using fecal messenger RNA. Cancer Epidemiology, Biomarkers & Prevention 4(6):643-647.
Davis, L. M., I. Martinez, J. Walter, and R. Hutkins. 2010. A dose dependent impact of prebiotic galactooligosaccharides on the intestinal microbiota of healthy adults. International Journal of Food Microbiology 144(2):285-292.
Davis, M. F., L. B. Price, C. M. Liu, and E. K. Silbergeld. 2011. An ecological perspective on U.S. industrial poultry production: The role of anthropogenic ecosystems on the emergence of drug-resistant bacteria from agricultural environments. Current Opinion in Microbiology 14(3):244-250.
Dethlefsen, L., and D. A. Relman. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. PNAS 108(Suppl 1):4554-4561.
Donovan, S. M., M. Wang, M. Li, I. Friedberg, S. L. Schwartz, and R. S. Chapkin. 2012. Host-microbe interactions in the neonatal intestine: Role of human milk oligosaccharides. Advances in Nutrition 3(3):450S-455S.
Dougherty, E. R., M. Brun, J. M. Trent, and M. L. Bittner. 2009. Conditioning-based modeling of contextual genomic regulation. IEEE/ACM Transactions on Computational Biology and Bioinformatics 6(2):310-320.
EFSA (European Food Safety Authority). 2010. Scientific opinion on the substantiation of health claims related to live yoghurt cultures and improved lactose digestion (ID 1143, 2976) pursuant to article 13(1) of regulation (EC) No. 1924/2006. European Food Safety Authority Journal 8(10):1763. http://www.efsa.europa.eu/en/efsajournal/doc/1763.pdf.
Everard, A., V. Lazarevic, M. Derrien, M. Girard, G. G. Muccioli, A. M. Neyrinck, S. Possemiers, A. Van Holle, P. Francois, W. M. de Vos, N. M. Delzenne, J. Schrenzel, and P. D. Cani. 2011. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60(11):2775-2786.
Evershed, R. P., S. Payne, A. G. Sherratt, M. S. Copley, J. Coolidge, D. Urem-Kotsu, K. Kotsakis, M. Ozdogan, A. E. Ozdogan, O. Nieuwenhuyse, P. M. Akkermans, D. Bailey, R. R. Andeescu, S. Campbell, S. Farid, I. Hodder, N. Yalman, M. Ozbasaran, E. Bicakci, Y. Garfinkel, T. Levy, and M. M. Burton. 2008. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature 455(7212):528-531.
FAO-WHO (Food and Agriculture Organization-World Health Organization). 2002. Guidelines for the evaluation of probiotics in food. ftp://ftp.fao.org/es/esn/food/wgreport2.pdf.
FDA (Food and Drug Administration). 2010. Guidance for industry: Investigational New Drug applications (INDs)—determining whether human research studies can be conducted without an IND. Draft guidance. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM229175.pdf.
Garrett, W. S., G. M. Lord, S. Punit, G. Lugo-Villarino, S. K. Mazmanian, S. Ito, J. N. Glickman, and L. H. Glimcher. 2007. Communicable ulcerative colitis induced by t-bet deficiency in the innate immune system. Cell 131(1):33-45.
Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. Journal of Nutrition 125(6):1401-1412.
Hempel, S., S. Newberry, A. Ruelaz, Z. Wang, J. N. Miles, M. J. Suttorp, B. Johnsen, R. Shanman, W. Slusser, N. Fu, A. Smith, B. Roth, J. Polak, A. Motala, T. Perry, and P. G. Shekelle. 2011. Safety of probiotics to reduce risk and prevent or treat disease. Evidence report/technology assessment No. 200. (Prepared by the Southern California Evidence-Based Practice Center under Contract No. 290-2007-10062-1.) AHRQ Publication No. 11-e007. Rockville, MD: Agency for Healthcare Research and Quality.
Hooda, S., B. M. Boler, M. C. Serao, J. M. Brulc, M. A. Staeger, T. W. Boileau, S. E. Dowd, G. C. Fahey, Jr., and K. S. Swanson. 2012. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. Journal of Nutrition 142(7):1259-1265.
IOM (Institute of Medicine). 2002. Dietary reference intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein, and amino acids. Washington, DC: The National Academies Press.
Kim, S., E. R. Dougherty, M. L. Bittner, Y. Chen, K. Sivakumar, P. Meltzer, and J. M. Trent. 2000. General nonlinear framework for the analysis of gene interaction via multivariate expression arrays. Journal of Biomedical Optics 5(4):411-424.
Levin, B. R., V. Perrot, and N. Walker. 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154(3):985-997.
Leyer, G. J., S. Li, M. E. Mubasher, C. Reifer, and A. C. Ouwehand. 2009. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics 124(2):e172-e179.
Lindsey, R. L., J. G. Frye, S. N. Thitaram, R. J. Meinersmann, P. J. Fedorka-Cray, and M. D. Englen. 2011. Characterization of multidrug-resistant Escherichia coli by antimicrobial resistance profiles, plasmid replicon typing, and pulsed-field gel electrophoresis. Microbial Drug Resistance 17(2):157-163.
LoCascio, R. G., M. R. Ninonuevo, S. L. Freeman, D. A. Sela, R. Grimm, C. B. Lebrilla, D. A. Mills, and J. B. German. 2007. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. Journal of Agricultural and Food Chemistry 55(22):8914-8919.
Looft, T., T. A. Johnson, H. K. Allen, D. O. Bayles, D. P. Alt, R. D. Stedtfeld, W. J. Sul, T. M. Stedtfeld, B. Chai, J. R. Cole, S. A. Hashsham, J. M. Tiedje, and T. B. Stanton. 2012. In-feed antibiotic effects on the swine intestinal microbiome. PNAS 109(5):1691-1696.
Macaubas, C., N. H. de Klerk, B. J. Holt, C. Wee, G. Kendall, M. Firth, P. D. Sly, and P. G. Holt. 2003. Association between antenatal cytokine production and the development of atopy and asthma at age 6 years. Lancet 362(9391):1192-1197.
Madara, J. 2004. Building an intestine—architectural contributions of commensal bacteria. New England Journal of Medicine 351(16):1685-1686.
Marcobal, A., M. Barboza, J. W. Froehlich, D. E. Block, J. B. German, C. B. Lebrilla, and D. A. Mills. 2010. Consumption of human milk oligosaccharides by gut-related microbes. Journal of Agricultural and Food Chemistry 58(9):5334-5340.
Martin, M. C., M. Fernandez, D. M. Linares, and M. A. Alvarez. 2005. Sequencing, characterization and transcriptional analysis of the histidine decarboxylase operon of Lactobacillus buchneri. Microbiology 151(Pt 4):1219-1228.
Martinez, I., J. Kim, P. R. Duffy, V. L. Schlegel, and J. Walter. 2010. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE 5(11):e15046.
Martinez, J. L. 2009. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environmental Pollution 157(11):2893-2902.
McNulty, N. P., T. Yatsunenko, A. Hsiao, J. J. Faith, B. D. Muegge, A. L. Goodman, B. Henrissat, R. Oozeer, S. Cools-Portier, G. Gobert, C. Chervaux, D. Knights, C. A. Lozupone, R. Knight, A. E. Duncan, J. R. Bain, M. J. Muehlbauer, C. B. Newgard, A. C. Heath, and J. I. Gordon. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science Translational Medicine 3(106):106ra106.
Metchnikov, E. 1907. The prolongation of life. London: William Heinemann.
Mussatto, S. I., and I. M. Mancilha. 2007. Non-digestible oligosaccharides: A review. Carbohydrate Polymers 68(3):587-597.
Nandi, S., J. J. Maurer, C. Hofacre, and A. O. Summers. 2004. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. PNAS 101(18):7118-7122.
Ninonuevo, M. R., Y. Park, H. Yin, J. Zhang, R. E. Ward, B. H. Clowers, J. B. German, S. L. Freeman, K. Killeen, R. Grimm, and C. B. Lebrilla. 2006. A strategy for annotating the human milk glycome. Journal of Agricultural and Food Chemistry 54(20):7471-7480.
O’Mahony, L., J. McCarthy, P. Kelly, G. Hurley, F. Luo, K. Chen, G. C. O’Sullivan, B. Kiely, J. K. Collins, F. Shanahan, and E. M. Quigley. 2005. Lactobacillus and Bifidobacterium in irritable bowel syndrome: Symptom responses and relationship to cytokine profiles. Gastroenterology 128(3):541-551.
O’Toole, P. W., and J. C. Cooney. 2008. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdisciplinary Perspectives on Infectious Diseases 2008:175285.
Poroyko, V., J. R. White, M. Wang, S. Donovan, J. Alverdy, D. C. Liu, and M. J. Morowitz. 2010. Gut microbial gene expression in mother-fed and formula-fed piglets. PLoS ONE 5(8):e12459.
Preidis, G. A., and J. Versalovic. 2009. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: Gastroenterology enters the metagenomics era. Gastroenterology 136(6):2015-2031.
Preidis, G. A., D. M. Saulnier, S. E. Blutt, T. A. Mistretta, K. P. Riehle, A. M. Major, S. F. Venable, J. P. Barrish, M. J. Finegold, J. F. Petrosino, R. L. Guerrant, M. E. Conner, and J. Versalovic. 2012a. Host response to probiotics determined by nutritional status of rotavirus-infected neonatal mice. Journal of Pediatric Gastroenterology and Nutrition 55(3):299-307.
Preidis, G. A., D. M. Saulnier, S. E. Blutt, T. A. Mistretta, K. P. Riehle, A. M. Major, S. F. Venable, M. J. Finegold, J. F. Petrosino, M. E. Conner, and J. Versalovic. 2012b. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB Journal 26(5):1960-1969.
Prescott, S. L., K. Wickens, L. Westcott, W. Jung, H. Currie, P. N. Black, T. V. Stanley, E. A. Mitchell, P. Fitzharris, R. Siebers, L. Wu, J. Crane, and Probiotic Study Group. 2008. Supplementation with Lactobacillus rhamnosus or Bifidobacterium lactis probiotics in pregnancy increases cord blood interferon-gamma and breast milk transforming growth factor-beta and immunoglobin A detection. Clinical and Experimental Allergy 38(10):1606-1614.
Priya, A. J., S. P. Vijayalakshmi, and A. M. Raichur. 2011. Enhanced survival of probiotic Lactobacillus acidophilus by encapsulation with nanostructured polyelectrolyte layers through layer-by-layer approach. Journal of Agricultural and Food Chemistry 59(21):11838-11845.
Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: Composition and succession. Current Issues in Intestinal Microbiology 2(2):43-53.
Sanders, M. E. 2011. Impact of probiotics on colonizing microbiota of the gut. Journal of Clinical Gastroenterology 45(Suppl):S115-S119.
Savino, F., E. Pelle, E. Palumeri, R. Oggero, and R. Miniero. 2007. Lactobacillus reuteri (American type culture collection strain 55730) versus simethicone in the treatment of infantile colic: A prospective randomized study. Pediatrics 119(1):e124-e130.
Schwartz, S., I. Friedberg, I. V. Ivanov, L. A. Davidson, J. S. Goldsby, D. B. Dahl, D. Herman, M. Wang, S. M. Donovan, and R. S. Chapkin. 2012. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biology 13(4):r32.
Sela, D. A., Y. Li, L. Lerno, S. Wu, A. M. Marcobal, J. B. German, X. Chen, C. B. Lebrilla, and D. A. Mills. 2011. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. Journal of Biological Chemistry 286(14):11909-11918.
Siezen, R. J., V. A. Tzeneva, A. Castioni, M. Wels, H. T. Phan, J. L. Rademaker, M. J. Starrenburg, M. Kleerebezem, D. Molenaar, and J. E. van Hylckama Vlieg. 2010. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environmental Microbiology 12(3):758-773.
Skippington, E., and M. A. Ragan. 2011. Lateral genetic transfer and the construction of genetic exchange communities. Federation of European Microbiological Societies Microbiology Reviews 35(5):707-735.
Sommer, M. O., and G. Dantas. 2011. Antibiotics and the resistant microbiome. Current Opinion in Microbiology 14(5):556-563.
Thomas, C. M., and J. Versalovic. 2010. Probiotics-host communication: Modulation of signaling pathways in the intestine. Gut Microbes 1(3):148-163.
Thomas, C. M., T. Hong, J. P. van Pijkeren, P. Hemarajata, D. V. Trinh, W. Hu, R. A. Britton, M. Kalkum, and J. Versalovic. 2012. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE 7(2):e31951.
Trip, H., N. L. Mulder, F. P. Rattray, and J. S. Lolkema. 2011. HdcB, a novel enzyme catalysing maturation of pyruvoyl-dependent histidine decarboxylase. Molecular Microbiology 79(4):861-871.
Veiga, P., C. A. Gallini, C. Beal, M. Michaud, M. L. Delaney, A. DuBois, A. Khlebnikov, J. E. van Hylckama Vlieg, S. Punit, J. N. Glickman, A. Onderdonk, L. H. Glimcher, and W. S. Garrett. 2010. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. PNAS 107(42):18132-18137.
Ward, R. E., M. Ninonuevo, D. A. Mills, C. B. Lebrilla, and J. B. German. 2006. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Applied and Environmental Microbiology 72(6):4497-4499.
———. 2007. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Molecular Nutrition and Food Research 51(11):1398-1405.
Whorwell, P. J., L. Altringer, J. Morel, Y. Bond, D. Charbonneau, L. O’Mahony, B. Kiely, F. Shanahan, and E. M. Quigley. 2006. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. American Journal of Gastroenterology 101(7):1581-1590.
Wright, G. D. 2007. The antibiotic resistome: The nexus of chemical and genetic diversity. Nature Reviews Microbiology 5(3):175-186.
Wu, S., N. Tao, J. B. German, R. Grimm, and C. B. Lebrilla. 2010. Development of an annotated library of neutral human milk oligosaccharides. Journal of Proteome Research 9(8):4138-4151.
Wu, S., R. Grimm, J. B. German, and C. B. Lebrilla. 2011. Annotation and structural analysis of sialylated human milk oligosaccharides. Journal of Proteome Research 10(2):856-868.
Yamanaka, T., L. Helgeland, I. N. Farstad, H. Fukushima, T. Midtvedt, and P. Brandtzaeg. 2003. Microbial colonization drives lymphocyte accumulation and differentiation in the follicle-associated epithelium of Peyer’s patches. Journal of Immunology 170(2):816-822.








































