Monica Duke1
The University of Bath
I am the project manager at “SageCite,” a project funded by JISC (the expert organization on information and digital information for education and research in the United Kingdom) through the Managing Research Data program in the United Kingdom. The project focuses on disease network modeling within Sage Bionetworks.
SageCite was funded between August 2010 and July 2011 to develop and test a citation framework linking data, methods, and publications. The domain of bio-informatics provided a case study, and the project builds on existing infrastructure and tools: myExperiment and the Sage Commons. Sage Commons is an initiative of Sage Bionetworks to build a platform to share data in bio-informatics. Citations of complex network models of disease and associated data will be embedded in leading publications, exploring issues concerning the citation of data including the compound nature of datasets, description standards, and identifiers. The project has international links with the Concept Web Alliance and Bio2RDF. The partners are UKOLN, the University of Manchester and the British Library (representing DataCite), with contributions from Nature Genetics and PLoS2
The project was structured through a number of work packages comprising:
• Review and evaluation of options and approaches for data citation.
• Understanding the requirements for citing large-scale network models of disease and compound research objects.
• Demonstration of a citation-enabled workflow using a linked data approach. http://blogs.ukoln.ac.uk/sagecite/demo/
• Benefits mapping using the KRDS2 (Keeping Research Data Safe) taxonomy. http://www.beagrie.com/SageCite-KRDS_BenefitsWorksheet.pdf
• Technical and policy implications of citation by leading publishers. http://blogs.ukoln.ac.uk/sagecite/publisher-interviews/
• Dissemination across communities (bio-informatics and research and information communities).
Sage Bionetworks is a non-profit organization located in Seattle, WA that is creating resources for community-based, data-intensive biological discovery. Their work is motivated by the belief that it is necessary to have community-based analysis to build accurate models. They are also driven by the fact that no one single body has all the data required to build accurate models, so different stakeholders come together and contribute. Sage Bionetworks provides the data infrastructure, the culture, and the norms to make this happen.
______________________
1 Presentation slides are available at http://sites.nationalacademies.org/PGA/brdi/PGA_064019.
2 See project description at: http://blogs.ukoln.ac.uk/sagecite/.
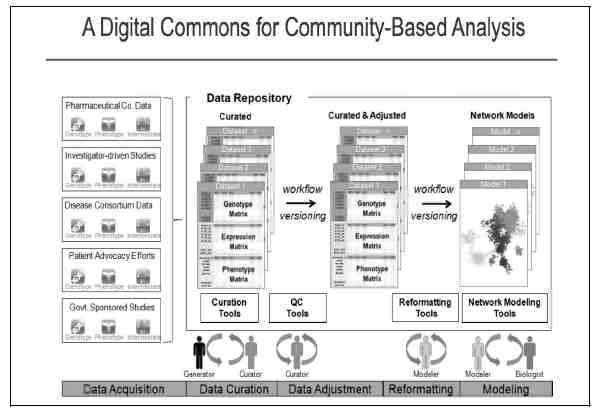
FIGURE 20-1 A digital commons for community-based analysis.
There are different types of data that the platform hosts and they come from different sources. For example, data can be obtained from pharmaceutical companies, disease consortia, investigators, patient advocacy organizations, and from government sponsored studies. There are seven stages in the data processing pipeline. The pipeline requires as input a combination of phenotypic, genetic, and expression data that need to be processed to determine a list of genes associated with diseases. The following figure shows an (idealized) description of these steps, each of which is likely to be performed by a different scientist who specializes in that area. One scientist acts as the project lead.
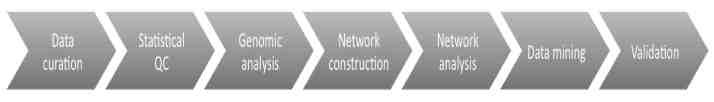
FIGURE 20-2 Stages in lifecycle.
Stage 1: Data Curation— This consists of basic data validation to ensure integrity and completeness of the data (although some files use common formats, others have considerable variety.) The datasets include microarray data and clinical data. This step ensures that the format of the data is understood and the required metadata is present.
Stage 2: Statistical QC— Actual values in data are validated for quality to check for experimental artifacts. The checks made are dependent on the type of data set and involves the
use of R scripts (for statistical computing) or specialized gene analysis tools (like “Plink.”). The output is a normalized dataset.
Stage 3: Genomic Analysis— This involves identifying regions in the genome associated with clinical phenotypes and other molecular traits. The Sage Genetic Analysis Pipeline, which consists of a set of R and C programs, is used. Statistical analysis is applied to identify interesting loci significantly associated with specific phenotypes (e.g., clinical phenotypes such as cQTL).
Stage 4: Network Construction— This stage focuses on building a network using a statistical technique to capture how biological entities, such as genes, are related to each other. Networks can contain up to 100 thousand nodes. In the network, nodes represent biological entities of some type (a gene, a protein, or even a physiological trait) and edges represent relationships between pairs of nodes. The output could be a correlation network (undirected graph) or a Bayesian network (directed, acyclic graph).
Stage 5: Network Analysis— This involves examining the network to determine how its function can be modulated by a specific subset of biological nodes. The output may be a list of genes or a sub-network. The networks from the previous steps are analyzed using techniques like Key Driver Analysis to determine a subset of interest.
Stage 6: Data Mining— A report validating claims from network analysis is produced by a domain specialist with knowledge of the study domain. This stage uses resources from the literature and public databases to assess the predictions. The information is used to annotate network models to build the case for the involvement of genes in the functioning of the network.
Stage 7: Experiment Validation— In the final stage, laboratory experiments are devised and performed to test the claims of the model. Validation is not carried out at Sage Bionetworks, but is completed in partnership with Sage Bionetworks collaborators.
Such a complex process presents challenges for reproducibility and citation. Data curation is required as a first step to do basic data validation to ensure integrity and completeness, and to ensure that the format of the data is understood and the required metadata is present. Agreed standards are also required for data sharing. We have to make sure that the data from different sources can be described, shared, used, and make the discovery process easier.
The project has employed the workflow tool, Taverna3, which helps to document the data processes and enables the workflow to be re-enacted. The workflow can also be registered with a Digital Object Identifier (DOI). Capturing the workflow and assigning an identifier supports better citation because the cited resource is more re-usable, and strengthens the reproducibility and validation of the research.
Finally, we can describe the challenges for using data citation with the purpose of giving attribution and supporting reproducibility within this specific context. The challenges for attribution include:
______________________
• Preserving a link with the original data.
— The data that enters the processing pipeline originates from several sources with different methods of identification of that data (or none). Some discipline-based repositories have their own identifiers that are culturally the norm within the discipline, but may not be well-known within other communities, or may not fit in technically with global identifier infrastructures.
— Creating Bi-directional links.
It is not sufficient to keep links which go in one direction only from processed data to the originating data. The originators of the data (e.g., discipline repositories) would also like to track usage. Therefore links need to be maintained in both directions. However, systems of notification of usage and tracking have not yet been developed.
• Attributing data creators.
— Identifying the party that created or contributed the data may not be straightforward and may have confidentiality issues (e.g., where medical data identifies specific populations). The situation is made more complex through developments of sites like PatientsLikeMe, where individuals are choosing to contribute their data. The range of entities and individuals who expect to be credited can be expected to grow and identifying new categories of data contributors (such as the individual patient) will create new challenges.
• Defining creation of new intellectual objects, e.g., a curated dataset.
With a complex process the community needs to agree what represents a new intellectual object that should be attributed. A curated dataset represents a significant input from the curator to make the object usable, but is the curated dataset a new distinct object that should be attributed and identified separately to the original data?
• Cultural challenge in recognizing non-standard contributions; micro-attribution.
Traditionally there has been emphasis on publications as a measure of contribution for the purposes of career advancement and peer recognition. A culture change is required if other categories of contribution (such as curation effort and data sharing) are to be recognised. Unless these contributions are recognised there will be little motivation to put in the effort to attribute them and create citation mechanisms around them. Microattribution is a developing idea in data citation to recognise smaller contributions and was used in the description of genetic variation in a paper in Nature Genetics in March 2011.4
• New metrics.
______________________
As new types of contribution are recognised complementary metrics and mechanisms for measurement will be required. Communities will need to decide what should be measured and services will need to be devised to track data citation and measurements.
• Identification of contributors.
With multi-step processes where individuals with different roles contribute, methods will be needed to describe the role of individuals and their contributions, particularly if non- traditional contributions such as data curation, data processing, data analysis, software, or process development are to be attributed.
Reproducibility:
• Identification and granularity.
— Discipline identifiers, global identifiers.
SageCite has taken a workflow capture approach to preserving the steps of the process to make it reproducible. When assigning identifiers for citation purposes decisions are required to decide at which level of granularity unique identifiers should be issued. Is it sufficient to identify the work flow or do individual steps need to be assigned their own identifiers? When should discipline identifiers be incorporated and how are these associated with identifiers assigned from a global system?
— How much value has been added since the data entered the workflow?
— One argument for deciding when a new identifier is required is to assess the value added to the data since it has been in the pipeline. To ensure reproducibility and provenance tracking, links need to be kept between value-added versions which have acquired a new identity in the pipeline and the original data.
• Identifying processes and software.
For the purposes of reproducibility it is not only the data that needs to be identified and cited. The tools and the workflow applied need to be referred to and accessed. The exact details are not always recorded and although some generic tools (such as R) are sometimes cited, the specific scripts used must be curated in order to become citable objects.
This page intentionally left blank.
DISCUSSION BY WORKSHOP PARTICIPANTS
Moderated by David Kochalko
PARTICIPANT: This session reminded me that the history of documentation is full of powerful systems that died because they were too much work to operate.
MR. KOCHALKO: I think it is difficult to draw meaningful distinctions between locators and identifiers. I also think it is important to understand that when some providers change a version number they will also change the identifier, which maintains parity so that each version of a dataset can both be located and uniquely cited. When providers refuse to issue new identifiers, they make it difficult to associate the version of work with its location and a unique citable identifier. I think that these factors have to be accommodated and that it is still possible to maintain version histories, which is a really fundamental.
MR. PARSONS: Quite honestly, most earth science data are not well versioned currently. What we have found is that an accurate citation is highly coupled with provenance and we, as a community, are just now beginning to fully address provenance. My data center recently got some money to develop a so-called climate data record, which is meant to be the gold standard of a long time series, in this case, brightness temperatures measured from passive microwave sensing satellites. What we discovered is that the dataset that can be perfectly reproduced was actually not the best dataset because scientists had made decisions over its 30-year history that they were not necessarily documented in a way that could be reproduced by a machine. My point is that the provenance is really key and it is a developing field.
PARTICIPANT: The major versions approach is good, but the other approach, which I believe the British Oceanographic Data Center (BODC) is using, is periodic snapshots. Either way, it is not an identifier.
DR. CALLAGHAN: At the BODC and by extension the rest of the UK National Environmental Research Centre data centers, when we post a DOI on a dataset, we are saying that it is frozen in time and will not change as far as we can possibly manage it. If a dataset is still being updated, it will not have a DOI. It will still be accessible and citable, using URIs and URLs, but it will not have a DOI association.
PARTICIPANT: When you say a “dataset”, do you mean, for example, the time series of the history of the Earth’s temperature?
DR. CALLAGHAN: For those situations where we do have an ongoing time series, we divide it into decades or years or even months, if appropriate. That kind of dataset, however, is picked because once you have recorded the data, they are not going to change. One will not go back after the fact to change what is in that particular time period, unless there is a major problem. In that case, you have to redo the dataset or revise the calibration and then you issue a new DOI with a new version.
PARTICIPANT: The data processed by DataCite there are freely accessible by anybody. What happens if the commercial data curator decides to get out of the business? What happens to the data?
DR. BRASE: First of all, we believe that the access to the data should be free of charge, but there is no strict rule that they always should be free of charge. We therefore work together with data centers that need to get some compensation for the data and we encourage them to make access as free as possible. The data centers do not seek to make profit, but there are some that do provide access to the data for a fee, or the data are only available to the members of some institution.
Now to your question about what happens to the data? That is always an issue and that is a situation for which we still have not found a perfect solution. One of the good things about assigning an identifier to a dataset is that you always can ensure that when somebody references the DOI name of a dataset that is no longer available, they will not get a 404-error, but they will receive a page describing that this dataset is no longer available and where the last known version can be accessed. This is always a possibility, but the idea of DataCite is that, ideally, if we would have a situation where one data center would cease to exist, we would try to find other data centers to take over the data and ensure that DOI name refers to the current version of the data. If that does not work, then we would direct the DOI to a page describing why the data set is no longer available.
PARTICIPANT: I wonder if anyone would like to reflect on the citation systems that have two parties who can be credited: the publishers and data providers. I just noticed that the people we have at this workshop are mostly from the provider side. Are we designing a system that is driven by the data centers and their interests, but not necessarily by the data providers?
DR. CALLAGHAN: Basically, data providers are interested in getting data citation working. We know this because we have asked them, at least in the meteorological sciences. We also have had a few cases of data providers coming to us and asking, “can we get the DOI for our dataset?” or “when will the data be citable?” So, there is interest in the scientific community. As data center managers, our job is to get data from the data providers, but if they do not show any interest in data citation then it is not in our interest to do anything about it.
MR. PARSONS: I will briefly add that if we have the identifier, we do not really need to include the role of the distributor or publisher in the citation. I would like people to think that they are getting a higher quality product out of the National Snow and Ice Data Center, but then I think we also have to be careful about citation being the credit mechanism. For example, I am getting push-back now from the funding agencies that want to have NASA or NOAA in the citation. We never did that with literary citations. Why do we have to do it now?
DR. CHAVAN: I think while it is clear that the basic motivation for data citation is mainly for the authors of the data, publishers of the data could get their work properly recognized as well. If you look at it from the usability perspective, there is equal responsibility on the part of the users of the data by making sure that they cite the data as adequately as possible. This brings in some complexity (e.g., when data are contributed by multiple parties) when the user actually uses records from each of the datasets or subsets of the data. This is exactly why we have been
promoting the practice of having user-driven citations on top of the publisher’s data citation. I think that it was technically vital previously to have author-driven citations or publisher-driven citations, but I think as the digital area progresses we will need to promote both. The user-driven citations will be the key to authenticate or verify the validity of the interpretations on the dataset that they have actually used.
PARTICIPANT: Is there a possibility that Microsoft can tell me my true worth? If we are already indexing 24 million documents and doing many other things that are not measured in terms of scholarship, we might be able to begin to get at that through the Microsoft Bing index. I could actually come up with a number that was measured across all of my scholarship. This would seem to be something that could change the way people think about how scholarship is measured. It seems we need that kind of metric and maybe it is within reach.
PARTICIPANT: This is the kind of metric that we would like to make available. We are not affiliated with the Bing index, but we have the opportunity to work with that team, combine indexes, and run those kinds of searches and present data in a meaningful way. We are not doing that right now. If the community comes together and indicates that they would be very interested, that would be a good step forward. Maybe there could be a large-scale aggregator service of data citations?
This page intentionally left blank.
This page intentionally left blank.












