24- Linking Data to Publications: Towards the Execution of Papers
Anita De Waard1
Elsevier Labs and the University of Utrecht, The Netherlands
First, I would like to say that I am not representing all commercial publishers and that I have not even coordinated this talk with my colleagues at Elsevier, so this is my personal perspective on the issues being discussed here.
I think it is useful when we are talking about integrating data with publications to look at where data fit within the scientific process. The KEfED model developed by Gully Burns2 can help in this regard.
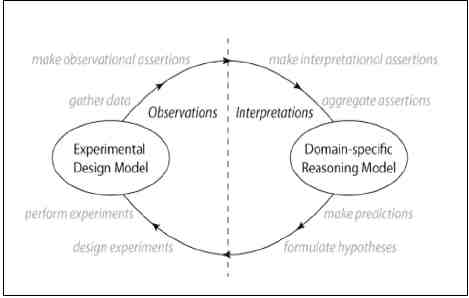
FIGURE 24-1 KEfED model “Cycle of Scientific Investigation.”
Essentially, in doing research we start thinking about the background and making some hypotheses. This is basically experimental science. You do an experimental design, you manipulate some external objects, and then you have observations. From those observations, you gather what is called data. Then you do some statistical analysis, and come up with some findings. In general, the data support your claims and findings. What happens in a publication is
______________________
1 Presentation slides are available at http://www.sites.nationalacademies.org/PGA/brdi/PGA_064019.
2 Gully APC Burns and Thomas A. Russ. 2009. Biomedical knowledge engineering tools based on experimental design: a case study based on neuroanatomical tract-tracing experiments. In Proceedings of the fifth international conference on Knowledge capture (K-CAP ‘09). ACM, New York, NY, USA, 173-174. DOI=10.1145/1597735.1597768 http://www.doi.acm.org/10.1145/1597735.1597768.
that you make a representation of your thoughts through language. These are the bases with which I would like to start.
Currently, the scientific community is storing data in repositories. We link to publications and vice-versa. The example that is commonly used is that people add PDFs and spreadsheets to their papers. This is pretty useless because we are not doing anything with these documents. Having them does not mean we can find the dataset.
In general, I believe that datasets should all be available for server search and that sets and subsets of that data should be made freely accessible, whenever possible. Overall, commercial publishers are not interested in owning or charging for research data or running those repositories. There might be exceptions, but in general, this is the case.
In my view, most publishers are very interested in working with data repositories and believe that it would be very useful if there were one place where we can find data items. It would be useful if an identifier is persistent and unique and that if the content changes, the identifier changes as well. Also, it would be very useful if the data would link back to the publication. It would be more interesting if we have data in a repository and can link them to some content from within a publication. Not only from the top level, but from within the publication. There are some examples of this. What my lab has been doing currently is tagging entities and linking them to databases. This involves some manual as well as some automated work.
More interesting, I think, is the fact that we can now create claim evidence networks that span across documents, so we can have a statement that can be backed up in a table or a reference in another publication or in another data center. At least at Elsevier, we are very invested in the idea of linked data. We have developed something that we call a satellite, which is essentially a way to describe a Linked Data annotation, in RDF. We are using Dublin Core and SWAN’s provenance and authoring/version ontology to identify the provenance.
We are very happy to develop this with people like Paul Groth and Herbert van de Sompel and others to have an ontology that connects to their work. The idea is that we can have some files that link to our XML at any level of granularity. There are files that sit outside the publication or the data center but we can still link one to the other. I think this is a very promising way to move forward.
What would be really interesting is if we had the opportunity to completely re-think science publishing. Why only change where the data is located: why not change the whole process? In my opinion, what is key is that scientists should be allowed to do their research process the way they want. We do not want to put more obstacles in front of the busy scientists who are already struggling to do their work. In fact, I think that the publishers would like to help them. So, if they have an experimental design, perhaps they can put a copy of it in the repository and put a link to it in their paper. Similarly, there are reports of observations. Perhaps there can be some way to deposit these reports in a repository and to pull them into their paper, code their statistics in a same way, and then draw the conclusions.
For the publisher and probably for the reader, it is incredibly important to maintain the context that the data have (e.g., the experimental context, the reason you did the experiment, the time
involved, and the like). There is a narrative context and we are using it to prove a point, so the data act as a key point for life scientists to communicate with other scientists. There are big questions that we are tackling and it is very important that the data are maintained and preserved.
Now let me ask this question: why do not the scientists themselves keep track of their own experimental design, their observed results and their code of statistics? They can share part of this with the publisher. Similarly, they can share with the data repositories. They can share the experimental design, the data and the code of statistics, using cloud computing. Imagine scientists using the cloud to store their research, find their results, experiments, and observations. I think it is truly important that as research keeps building, there are good systems in which researchers can keep track of their own data, store them, and add appropriate metadata.
The assignment of unique identifiers plays a central role in the advertisement of these materials. Data centers are able to connect datasets and promote them. They can also advertise them. The role of data centers in terms of quality control and access is very critical and, as we saw earlier in this meeting, this differs from one field to another.
So, if we are publishing a paper with data, all we need to do is to deposit our document in a repository and allow access to an editor or somebody who we think can evaluate our work. Then, we would have access to the collective thoughts as well as to links to the data, to the workflow, to the other science components, and to a publisher or somebody in the role of validating quality.
I think these and similar practices will connect more in the future and publishers, data repositories, and perhaps software developers (e.g., Microsoft, Google, Skype, Twitter, or Dropbox) will be involved in these processes. We all use commercial software all the time. These programs are very good at building tools that help us communicate. Therefore, it is very useful to have such companies working with us on improving communication between scientists by encouraging them to build better software and applications.
Citizen science was mentioned earlier as well. Citizens can also play a key role in these processes and we should be keen to involve them. Again, some technological components and applications are now in place and can facilitate these processes.
Let me conclude by emphasizing that, in my view, publishers are not interested in owning or charging for data. We believe in identifiers and embrace open standards and I think that scientists should keep track of their own work. We certainly believe in a future where science is shared and stored in a better and productive way, as well as in working together with all stakeholders to make it happen.
This page intentionally left blank.




