11. Developing the Rice Genome in China
Huanming Yang BGI (Beijing Genomics Institute), China
In this presentation, I will share with you information about the development of the rice genome as well as other genomes in China, through international collaboration and international sharing of scientific data.
The field of genomics was cultivated by the Human Genome Project (HGP). It was important to China that its scientists made a contribution, even a small one, to the HGP.
China was a latecomer. One of my colleagues wrote: “The work was already underway in other countries, and China was way behind the curve. But Huanming convinced me that China’s involvement would represent a major advance for his country and for the Beijing Genome Center.” Our idea was that with just a simple contribution from us, together with all other countries, we could benefit from this project as could all the people in the world. It was so expensive that China alone could not afford it. That was the reason for us to support international scientific collaboration.
As a member of the International Bioethics Committee of UNESCO, I strongly proposed that the most important and urgent issue in bioethics at present is the immediate release and free sharing of the human genome data. In a period of 5 months, I submitted four proposals for free sharing of human genome data. Then on May 9, 2000, the director-general of UNESCO issued a statement on the free availability of human genome data. This led to the Group of 8 (G8) communiqué of the Okinawa meeting in July 2000, as well as the United Nations Millennium Declaration on September 19, 2000, to ensure free access to information on the human genome sequence.
As a unique member state from the developing countries in the HGP consortium, China’s contribution is not only a technical accomplishment, but also a recognized effort in the free sharing of human genome data. As Michael Morgan, who was responsible for the Wellcome Trust projects on genomics, said, “China’s unswerving support of open data release was an important factor in ensuring that the human genome sequence is the property of the whole world.” John Sulston, the leader of the HGP of the United Kingdom, said, “I especially salute the Chinese colleagues, who have affirmed the Human Genome Project’s common ownership by all mankind.”
The Human Genome Project has established a brilliant example for international collaboration and data sharing. With no participation in the HGP or no international data sharing, genomics in China today would not be so advanced.
With regard to the rice genome data, we freely shared and published the first draft sequence of the rice genome in April 2002. It was a big event in the history of natural science in China. That is the reason that the editorial department of Science magazine went to Beijing for the news release: “Science magazine honoring China’s sequencing of the rice genome.” I would like to quote from Science: “This team deserves enormous credit for their outstanding world-class accomplishment in a remarkably short time.”
It is true that the whole job was done in 74 days. Then we released all the data. Our database on the rice genome has been one of the most popular databases in global genomics. If regarding citations as one indicator of the impact of that paper, we are proud to see that our paper has been cited about 2,000 times up to now, and the citations are still increasing. The number of specific publications in one field could be another indication of the impact of the work in that field. Since the release and the sharing of the rice genome data, the research on rice has dramatically increased and has outnumbered that of wheat, though they were more or less the same at the starting line.
For our students, it would be difficult to imagine how research could be done on rice without referring to its genome sequences. This is also the case for all the researchers in the field of genomics. I will give you two examples showing how significant the impact of the free sharing of the rice genome sequence can be.
We are now collaborating with various institutes and universities within and outside China to sequence 10,000 rice accessions or strains. We believe that is an important stimulus for rice genome research and breeding. We also have analyzed the genomes of other important crops, such as maize and potatoes. A paper about this research will be published in a prestigious journal very soon. We also have analyzed the cucumber, and sequenced the genes related to sex expression, disease resistance, and so on. As soybean curd is an important food for the Chinese, we have also completed the genome analysis on soybeans.
We have analyzed the genome for chickens. One of the most important scientific discoveries is that the genome diversity of chickens originated before their domestication.
Together with other institutions, we have analyzed the genome for silkworms. The most thorough genome sequence data so far for the silkworm was released by BGI and other Chinese researchers many years ago. Now the job is continued by analyzing about 50 genomes, with the aim to reveal the domestication events and genes for this important insect.
We have analyzed the panda genome. Technically it is the first successful de novo assembly of a mammal, without the help from a genetic or physical map of this animal. We have also analyzed, in collaboration with our colleagues in the United States, the genome of two ant species to reveal how they organize their social activities.
We are also concerned about global climate change, so we have finished the sequencing part of some animals living in extreme environments, like the polar bear, penguin, the Tibetan antelope, and the camel.
We have initiated, together with our colleagues all around the world, the International 10K Genomes Project. About 25 percent of all the vertebrates are listed in this project. We have already begun studying the first batch of more than 100 species.
Now I would like to turn back to the human genome research. As I have already indicated, the Human Genome Project has shaped the field of human genomics, which is characterized by international collaboration and international data sharing. Our contribution to the HGP has been small, probably 1 percent, but we made about a 10 percent contribution with the International HapMap Project. We completed the first Asian human genome by means of the new-generation sequencers and published the results in Nature. The publication of the Asian genome revealed that the methods or sequencing technology available now are extremely powerful, as stated in a review paper in Science.
Perhaps you have seen that my institute is committed to sequencing more than 10,000 individuals before the end of this year. We are also an essential part of the Human Variome Project, as well as the International Cancer Genome Project. For the International Cancer Genome Project, we are committed to gastric cancer now, and we will expand our contribution to other cancers related to digestive systems. We have initiated the 1,000 Monogenic Diseases Project, based on our own experiences working on the Mendelian disorders. One of the most important discoveries in 2009 was the “human pan-genome”. We have identified that 0.6 ~ 1.5 percent of human genome sequences actually are population-specific.
We also analyzed people living in two different environments to reveal how their genomes have adapted to the environment. The population study of Tibetans and Han Chinese uncovered that regulation of the hypoxia response is central to high-altitude adaptation. That paper was published in Science.
We captured more than 80 percent of the human genome sequence from the hairs of a sample which was at least 4,000 years old. Then, we published the first catalogue of the human gut metagenomes, in which we identified at least 2,000 species of bacteria. Finally, we also have initiated a 10,000 Microbial Genome Project, as well as the Earth Microbiome Project, also through international collaboration.
As scientists, we acknowledge that we have a responsibility to the world. In 1994, I told my colleagues what we should bring back is not only the technology, but also the ethical principles. As co-chairman of the European Actions for Global Life Sciences, we developed this slogan: “To raise the banner of science and humanity.” I am president of BGI (formerly known as Beijing Genomics Institute), and the institute’s slogan is: “To raise the banner of innovation and ethics.”
BGI’s mission is to share what we have with others to promote global genomics, to work together with those who are unable to access what they need, and to work together with others to do something bigger, faster, cheaper, and better. It is, after all, so important to build the capacity in developing countries.
I think that international sharing of scientific data is not only an issue of science, nor merely an issue of economics, but also an issue of humanity and global harmony. In genomics, we Chinese already have benefited so much from it. We are told by our ancestors that nobody can be a hero without three partners. Genomics cannot be done alone.
BGI’s strategy for sustainable development requires international collaboration and data sharing. None of our achievements would have been gained without international collaboration. We are confident of our passionate young staff, but of course, enthusiasm is not enough for science. Innovation is the key for scientific development. Bioinformatics software is our major innovation. That is how we have made rice genome assembly possible, as well as most, if not all, of the software that we are using for next-generation sequencers.
Nonetheless, humbleness is rooted in our Chinese culture. We began with nothing, though now my institute is poised to become the biggest DNA sequencing lab in the world. When we are called the sequencing factory for the world, we are happy to have this nickname. According to Science, “BGI-Shenzhen enhances its reputation as the world’s largest sequencing center, deciphering an ant, the Asian human genome, the human methylome, and a gene catalog of the human gut microbes.” At the same time, we appreciate again all those who have helped us. Just like the Chinese proverb says: “When you drink sweet water from the well, do not forget who helped dig it.”
Željko Ivezić University of Washington, United States
I am going to summarize a few experiences we collected in astronomy with data sharing. I will first list a few questions that we deal with in astronomy just to set the stage. Then I will review what we learned from the Sloan Digital Sky Survey (SDSS) and similar surveys in the context of data sharing. My main point is that we have to submit ourselves to cost-benefit analysis and see whether or not it pays back.
What we are trying to do in astronomy is fairly well summarized in these three big questions:
1. How and when did the universe begin?
2. How did the structure (planets, stars, galaxies) in the universe form and evolve?
3. Is our planetary system unique (or, is there life anywhere else)?
You can rephrase this by asking if the physics we learned on earth is applicable to the rest of the universe, and if we can use the observations of the universe and of the heavens to learn more physics. These are, so to say, business questions, but we should never forget that, going from families visiting the Smithsonian Institution in Washington, D.C., to a small village in some corner of the world where grandparents tell their children the stories about stars, we are all fascinated by our place in the cosmos.
Over the last decade or so, there has been an explosion in new tools and methods. Due to the fast development of computer and information technologies, we now have new tools, methods, and cutting edge sky surveys that allow us to observe the whole sky in great detail. There are three frontiers in optical astronomy.
The first is to build ever-larger telescopes. These are, for example, the twin Keck telescopes in Hawaii. We build large telescopes not to get more detail, but to see fainter objects. The second frontier is to launch our telescopes in space, above the Earth’s atmosphere, which blurs the images of ground-based telescopes and absorbs all the light except visible and radio. The beautiful images from the Hubble Space Telescope are so detailed not because Hubble is exceptionally large, but because the images are not blurred by the atmosphere.
Both the Keck telescopes and Hubble more or less study one object at a time, but they cannot see the whole sky. In fact, all of the area in the sky that Hubble imaged to date is less than one-thousandth of the whole sky. With new technology, we can begin to cover the entire sky and get diverse and precise data on hundreds of millions of objects. That progress brought in the third frontier in astronomy: gathering digital data for these large numbers of sources and then using statistical analysis and data-mining methods to study them.
One of the key developments related to this symposium is that about a decade ago astronomers started sharing all these giant databases freely with the public. What do these data contain? First, they have images. Observers are interested in the position of the object in the sky, its brightness, the size and shape of its image. Objects are observed with different filters to get information about the spectral energy distribution. Images are processed to measure objects, such as stars and galaxies, and to construct catalogues, which are the most useful data products that we put in the public domain—of course, together with images.
Why are these catalogues important? We can use them to discover new objects, classify those new objects, study them statistically, and search for unusual objects. The larger the sample is, the more unusual stars or galaxies will be found. Then we can study cosmology and try to answer these questions:
When did the universe begin? How did it develop?
When astronomy databases are open to the public, more people can participate. One of the foremost examples of this new generation of surveys is the Sloan Digital Sky Survey (SDSS). It was the first time that we could get digital data, a color map of the sky, for a substantial fraction of the sky. There have been a number of projects in astronomy in the last decade that have built digital databases and made them available to the world’s public.
SDSS used a camera that has 120 megapixels. It used to be the largest camera in the world. For more than half a decade we collected measurements—positions, colors, shapes, and so on—for about 400 million objects. When the database containing these measurements was released to the public, even before a substantial fraction of science analysis was completed by the project team, that was a revolution in astronomy. This approach is still not accepted in all the fields, but I would argue that there are clear benefits of doing so.
The SDSS public portal provides astronomical data to anyone, anywhere. Two years ago, I was vacationing in Croatia, my country of origin, and I met a friend from Serbia who is an astronomer who had a house on the same island. We got all the data for a paper in four days by accessing this database through our laptops while drinking beer in a beach coffee bar.
The portal also has a special section with many exercises for teachers and supplemental material that shows K–12 teachers how to use these data in the classroom. There are literally tens of thousands of examples where the data were incorporated in school curricula. I would like to remind you that astronomy is very effective in attracting students to science, technology, engineering, and mathematics professions.
As a result of this public data release, again even before a substantial fraction of science was done by the project team itself, several thousand papers were published by scientists not associated with the SDSS—more papers than by the people who were members of the SDSS collaboration. The total data volume that was delivered through this portal was more than 100 times the size of the full SDSS database. There were more than 300 million Web hits over 6 years. The number of unique users was about 1 million. Compare this to about 10,000 professional astronomers. It is a huge impact. As a result of SDSS and previous work, the intellectual father of SDSS, Prof. Jim Gunn from Princeton University, was awarded the National Medal of Science by President Obama.
Over the last few years, even more portals have been developed. One was developed by Microsoft and called WorldWide Telescope. You can use it to get not only SDSS data, but just about every large astronomical dataset. Google Sky did a similar thing. A colleague who developed Google Sky with people from Google told me that as soon as they put it online, there were several million downloads. Google thought that someone was giving them a denial-of-service attack. That shows many people were interested.
What did we learn from this exercise of releasing data even before the project team did its own science? First, it requires higher standards. When you put something out to the public, you have to have documentation. You cannot put out documentation that is not spell-checked, for example. It is a very serious job to put something in the public domain. Then there are costs to keep it public, such as servers, help desks, and so on. These are the two main issues: higher standards and the cost of curation, and because there are costs, we cannot just say, let’s do it. We have to subject this idea of publicly releasing data to a cost-benefit analysis.
I will go through a list of the top 10 benefits that we think we extracted from our experience in astronomy. The caveat is, not all of them may apply to your field. Also, they may vary with time. For
example, 20 years ago in astronomy, not many astronomers would buy the arguments that I am going to present to you, but today most of them would. Things change with time.
The primary benefit, in the view of many, is that while you are taking data, and you have a finite lifetime for your project, if your dataset is complex, you need to subject it to rigorous analysis to be certain that everything is fine with your data-taking strategy, with your instrument, and so on. Some easy things, of course, you can see immediately. In astronomy, if you get a blank image, then you know that something is wrong. But there are effects at the 1 percent level. With these statistical surveys that have hundreds of millions of objects, we study such percent-level effects. Often they can be hidden systematically in the data that you cannot see with the naked eye; you cannot even see with simple analysis. You need to do complex analysis and cutting-edge research to discover something special with your data. If the data collecting is already finished, then it is too late. That is the main reason why you should do it early, before the end of your project.
Then, especially in astronomy, sometimes you want to take data with different facilities at the same time. In astronomy there are objects in the sky that do not exist forever. Supernova explosions last for a month or so; similarly, there are some asteroids that pass close to Earth, and then we do not see them for decades. There are events in the sky that last a short time and we want to deploy many facilities. To enable this, you have to release your data early.
If you release your data to the whole world, you will have many more users doing your science. With SDSS in particular, outsiders wrote more papers. Many of these papers had great ideas that were not even listed in the project book that was given to funding agencies to justify this investment of close to $200 million. More people mean more ideas.
If you release your data for everyone to work on, you ensure that all the scientific results will be reproducible. They can be verified and you will preserve your data for posterity. That is, again, very important in astronomy. Astronomers strongly believe that code should be released with the data, not just the final product. In particular, in astronomy, because of the atmospheric effects, we see from the ground only optical and radio wavelengths, but we learn a lot about the heavens by observing in X-ray and infrared bandwidths, among others, and using telescopes on satellites. To take this same argument further, cross-disciplinary science is enabled.
There are some fantastic examples from the astronomy–statistics–computer science boundary, where computer scientists use astronomical data to develop new ideas, which are then used once again in astronomy to do better science than astronomers could have done on our own. One can extend this also to other fields, because most disciplines have these giant datasets today. Many of the problems are the same: How do you do data mining in highly multidimensional space? How do you visualize massive datasets? How do you store and query 100 petabytes? These are all common problems. It is not efficient to try to solve them N times when we could solve them together.
Sometimes collaborating and promising to release the data are the only way to secure scarce resources, especially in astronomy where tools are becoming very expensive. Most of the easy things were done in the last century—things you could do in a basement, with one professor and three graduate students. Today’s more difficult problems require major resources. Also if you look at the gross domestic product (GDP) of a big country like the United States, in the 1950s, it was 50 percent of the world’s economy. Today, it is only one-quarter of the world GDP: therefore, it is now much more profitable for the United States to enter international partnerships than it was 50 years ago. In astronomy, very often when you have a major undertaking, you have to get everyone on board. That is why it is good to share your data.
Going back to the team that produced the data, we are all concerned about our careers, especially when
we are young postdoctoral researchers (postdocs) with small children to feed. We want to protect these young people. We want them in successful positions. Indeed, we heard many counterarguments against early release of data from SDSS, claiming that all of us who were postdocs at that time would not get jobs because other people would do the science. What happened in practice was that all of us who were postdocs had know-how about the dataset. We did all the early science, even though the data was public. There was a delay of a year to two for the world community to get on board. Furthermore, our know-how became a marketable commodity, so all the young postdocs who worked on SDSS—literally, all of them, a few dozen—are faculty today.
Then, especially in astronomy, but in other fields as well, education and public outreach are important aspects of our science endeavors. You may have heard about the Galaxy Zoo project, where members of the public visually classify galaxies from the SDSS. The project has already recruited 200,000 volunteers, who went through images of a million galaxies. Evidently, the general public is greatly fascinated by astronomy.
Finally, there are issues of ethics and broader impact. First, sharing is nice, as we all know from our kindergarten days. Furthermore, taxpayers paid for most of our projects. They have the right to see the results at every level of understanding, looking at astronomical images through Google Sky and WorldWide Telescope, all the way to seeing scientific results and translating these to understand better how the universe began, and on the other big questions.
A big aspect of sharing data worldwide is that we are enabling democratization of scientific research. In the context of developing countries, this public release of giant datasets allows small teams to do big science. That colleague of mine and I on the beach in Croatia were a pretty small team, just two of us, and we managed to do cutting-edge science because SDSS data were made public worldwide.
These are the top benefits, but then there are some other issues that we should consider when thinking about releasing data. For example, if you spend some resources to release the data, is anyone going to use them? How many customers do you expect?
Of course, there are different kinds of data. Sometimes they are of great use, like astronomical data. We saw millions of hits to public data releases of people looking at images of the sky. Sometimes data are so specific that even if you release them, not many people would use them—for example, giant datastreams from particle accelerators. That is another extreme.
Then there are security and proprietary issues. For example, a big astronomical project, Pan-STARRS, was done in collaboration with the Air Force. They have to mask some parts of images and not release them to the public. There are also issues of commercial gains and foreign competition.
Jim Gray, who was a Principal Researcher of Microsoft Research Lab, developed parts of those tools that SDSS used to manage and release data. He liked to joke that he loved to work with astronomical data because “they are worthless.” What he meant was that they do not have commercial value. With other databases, he had to be very careful about what was released.
If you are doing a major data release that can cost tens of millions of dollars, you have to justify the cost. Once, I listed these top benefits on one page. Being a scientist, I immediately asked myself if there is any predictive value in that list. Can we go through other projects and see if they are consistent with this reasoning? Should they publicly release their data? I will share with you two examples, one from astronomy and one from physics. The first one, an astronomical example, will be a new telescope, a successor to SDSS called the Large Synoptic Survey Telescope (LSST), for which I am the project scientist. Located in Chile, the collaboration includes more than 30 U.S. institutions and about a half
dozen European institutions. The shortest way to summarize the difference is that the SDSS gave us the first digital color map of its kind, a snapshot. With LSST, we will observe the sky in a similar fashion as SDSS, but with about 1,000 times greater resolution. Essentially, you can think of it as a digital movie—the greatest movie ever made. If you watched that movie, it would take you a full year without sleep, just staring at the screen. This telescope will have an 8-meter aperture, not a 2.5-meter one, like SDSS. Because its sensitivity will be greater, LSST will detect many more objects. Instead of 400 million objects detected with SDSS, we will collect 20 billion objects with this telescope. It will be the first time that astronomers will have cataloged more objects than living people on Earth. Everyone can get their own galaxy.
When you look at the data volume, SDSS was about 40 terabytes. This is roughly the data volume of the books in the U.S. Library of Congress. Now, when you want to compare LSST, about 100 petabytes, 100,000 terabytes, it is about the same as the volume of all the words ever printed in the world since Gutenberg. Of course, so much data can be problematic. We have 20 billion objects, and we have to track them in real time.
On my list of 10 benefits, indeed, all 10 of them apply to the LSST dataset. It is not a great surprise, because it is so similar to SDSS. But because of this clear win in the cost-benefit analysis, all of the data will be made public to the world. Phenomena that change in the sky will be released within 60 seconds. Then every year there will be a cumulative data release as well (about 10 petabytes of new data every year).
Let me tell you just a few words about another big project in physics, the Laser Interferometer Gravitational Wave Observatory (LIGO). There are two sites in Louisiana and Washington. The project is trying to detect gravitational waves. If it is successful, that is a Nobel Prize-class discovery that would have a great impact on our understanding of the fundamental physics. Should those data be released immediately, or not? It turns out that most of the top 10 benefits apply to LIGO, too, but it is a more difficult question.
To summarize, the issue of public data sharing is a matter of cost-benefit analysis. Often when you do a detailed analysis, you are led to the conclusion that you just have to do it. Given all the parameters of the problem, all the benefits, and all other forces that act upon you, you simply have to share the data.
Daniel I. Cheney Federal Aviation Administration, United States
What I would like to address today is an information system that the Federal Aviation Administration has developed regarding transport airplane accidents. It is an initiative to gather information and lessons learned from these accidents, because they definitely have been repeated. The origin of this effort goes back to some problems that we worked on in the late 1990s and the early 2000s. There were several accidents, some of them very large that were heavily covered in the media. When we looked at them carefully, they exposed deficiencies about the way the aircrafts were operated, shortcomings in the maintenance programs, and in the fundamentals of the design of the aircraft. The processes that linked them together were inconsistent. The closer we looked, the starker the inconsistencies were.
Four accidents drew particular attention. These were the TWA Flight 800, Swissair Flight 111, Alaska Airlines Flight 261, and the American Eagle Flight 4184.
From an investigator’s standpoint, the TWA Flight 800 accident investigation was a very long, complicated, and technical one. It took years to assemble the wreckage, and much research was done to understand the science behind the cause. At the end, we certainly knew more about fuel system flammability than we did before this accident. Much has been done to reduce the risk of this kind of accident on airplanes today. The Swissair Flight 111 accident was also a very complicated one. Much research was done on the subject of flammability of materials, particularly the materials used in the cabin for thermal acoustic protection. Flawed assumptions were a dominant characteristic of this accident. The Alaska Airlines Flight 261 accident involved a stabilizer control system that malfunctioned due to poor lubrication. Again, there were inconsistencies in the methods by which the aircraft was being maintained. The fourth accident that caused us to initiate this accident library was the American Eagle Flight 4184 accident in Indiana. There were flawed assumptions in the way that atmospheric icing would affect the airplane. The ice would actually accumulate on parts of the airfoil that had never been observed before. It was very unusual and, before this accident, it was a part of atmospheric icing that was not understood. The result was loss of flight control.
All these accidents and investigations resulted in a great deal of scientific research, testing, retesting, and evaluation. That research was documented, but was largely languishing in various archives and places not readily available to the general public.
Knowledge of these and other major accidents was basically being lost with the passage of time. The more years that passed, the more people forgot the causes, and the more new folks coming into this industry had no knowledge of these deficiencies and shortcomings at all, and we were seeing repeated accidents, which is unacceptable.
Awareness of information about previous accidents became key to understanding more recent accidents. The challenge that we undertook was to craft an information system that could take what could be well over a decade of work in doing the research and fixing things and make it freely and easily accessible to those who can benefit from it so we do not need to make these mistakes again.
More than a century ago, George Santayana of Harvard University wrote a paper titled, Life of Reason: Reason in Common Sense, saying that “those that cannot remember the past are condemned to repeat it.”1
_____________________
1 Available at http://iat.iupui.edu/Santayana
It really is true. We will continue to repeat things unless we are mindful about what we need to be careful. A large transport airplane accident is an enormous human tragedy, but a second tragedy would be not to learn from it and then cause similar accidents.
Let me now talk about the barriers. Some other presenters talked about barriers to science and barriers to information flow. Aviation, particularly accidents, is driven by these very real and powerful barriers:
• Fear of negative publicity. There is probably nothing more negative than a large transport airplane accident. It is very sad for all involved.
• Lengthy investigation. Some investigations take more than a decade in order to get all the science and research together, and get a go-forward path that is solid.
• Continual workforce turnover. Many of the people that come into these accident investigations will be involved for a decade or so and then move on, and we replace them with new people and fail to build corporate memory of the problems.
• The information technology (IT) tools. If you go back 30 or 40 years, before the Internet and the computational systems, it was very hard to capture all these data.
We have developed a “Lessons Learned from Transport Airplane Accidents” library2 that is organized, threat-based, and has search-and-sort capability. Its intent is to stop and reverse the loss of lessons so that others can benefit from what has taken 40 or 50 years to accumulate. Many of the speakers at this meeting, including myself, that came from out of town flew here on one of these airplanes. We take air travel almost for granted. The biggest risk is the car, not the airplane. We want to keep it that way and maintain a very safe aviation system.
Now let me talk about the portal itself. Right now there are 57 major accident modules on the site. We are working on more all the time. We have another five being crafted. It is relatively time-consuming to create this material and involves different stakeholders. Boeing, Airbus, General Electric, Pratt and Whitney, and the airline companies all have been very helpful. They realize that their work can benefit from getting this right.
The only information on the portal is information that is already publicly available. We are looking at maybe 10 to 15 years of hard work captured in a 15-minute read. Anybody who wants to know about accidents like these can get the big issues in about 15 minutes.
Regarding the organization of the library, when we first started this, it was very tempting to only look at issues from technical and scientific aspects, such as fire, structural issues, flight control issues, and things like that. Then we looked at several major accidents and asked ourselves: Are we really getting the maximum value of the material? If not, what are we missing? After looking at half a dozen big accidents that we had prototyped, we realized that we were missing the bigger issues beyond the technical and scientific ones; we were missing what may be an organizational problem or a human error aspect.
After the iterative process of developing the library, we agreed upon three perspectives of looking at accidents. First, we look at the accident from a perspective of what we call the airplane life cycle. This is the beginning, the operational, and the maintenance and repair perspective. Second, we examine the accident threat categories. In looking at accidents since the jet era began, it turns out that we can put them into 18 technical categories that are important. The third perspective is what we call the common themes. Every accident has a strong link to one of five common themes: flawed assumptions, human error,
_____________________
2 Available at http://accidents-ll.faa.gov/.
organizational lapses, preexisting failures, and unintended effects. Every miscue that has happened in aviation can be strongly linked to at least one of these. In some accidents we have found strong links to all five—not often, but there are some.
We have the site arranged so that you can search any one of these categories or themes. You can say, “I want to know about structural failures,” and you can pull out the accidents in that category. You can also look at specific examples of accidents from an organizational standpoint. These are important not because they provide the most lessons, but they are certainly the most deadly accidents that have ever happened. For each accident there is a quick 60-second read of the general accident summary. The overview section gives you the nuts and bolts of the accident, the science, and the research that was done.
What we also feel is very important in this kind of material is the value of the Internet and IT tools—streaming video, animation, interviews, and things like that. Where animation has not been done by the companies involved, we have created animation with the help of staff at the National Aeronautics and Space Administration. It has been a very good partnership.
The portal has been in the public arena for two and a half years now. We are still building it. We add 5 to 10 accidents per year, and we will do that for at least 2 more years. Then we will transition into a sustaining mode where, as accidents happen, we will add them, but we will be finished with doing the historical mining and catching up.
14. Integrated Disaster Research: Issues Around Data
Jane E. Rovins Integrated Research on Disaster Risk Programme, China
I am going to introduce a new program as well as talk about the data issues surrounding disasters, because this is a topic that the Integrated Research on Disaster Risk (IRDR) Programme has taken to heart. It is one of our three main projects. It comes up on a regular basis, and I spend as much time addressing disaster data questions as I do with risk-reduction questions.
So that you understand where I am coming from, I am a practitioner who happens to have academic credentials. I am a certified emergency manager. I spent about 10 years in the field getting mud on my boots before I came into research.
The main issues that are faced by the IRDR Programme in the disaster arena are globalization, population growth, widespread poverty, and climate change. Urban areas face unique challenges in these issues, so we have initiated several city-at-risk projects. The Coastal Cities at Risk project is a partnership between various countries and is funded by Canada. Another project is the Cities at Risk project, which is a partnership between IRDR, the Global Change System for Analysis, Research, and Training, and the East-West Center in Hawaii. These projects are geared toward the specialized needs of densely populated, rapidly developing cities.
In response to these issues, the International Council for Science (ICSU) decided to ask the following question: Why, despite the advances in science on hazards and disasters, do our losses continue to increase? They created the IRDR Science Plan to try to answer that question.3
In addressing natural and human-induced environmental hazards, we are taking an integrated approach to research through an international, multidisciplinary (natural, health, engineering, and social sciences, including socioeconomic analysis), collaborative research program. This, however, gets challenging. As you know, trying to bring even two or three of the natural science disciplines under one program can be difficult. IRDR is bringing all those disciplines in with the social sciences as well.
What exactly is our scope at IRDR? This is fairly straightforward, for those of you that have looked at natural hazards or disasters:
• Geophysical and hydrometeorological trigger events.
• Earthquakes, tsunamis, volcanoes, floods, storms (hurricanes, cyclones, typhoons), heat waves, droughts, wildfires, landslides, coastal erosion, climate change.
• Space weather and impact by near-Earth objects.
• Effects of human activities on creating or enhancing disasters, including land-use practices.
What we are not looking at are technological disasters and warfare. What do I mean by technological disasters? Take the Japanese earthquake and tsunami, for example. There were two natural disasters that created a technological problem with nuclear fallout. We would look at that in relationship to the earthquake and the tsunami. Had it been just a nuclear release caused by a failure at the plant, we probably would not have looked at it, but because it was created by natural hazards, we are incorporating it into our studies of the event.
_____________________
3 The Science Plan can be accessed at http://www.irdrinternational.org/.
The IRDR Science Plan, has three key objectives. All of these are very data intensive, as you will see. The first is the characterization of hazards, vulnerability, and risk. This includes the following:
• Identifying hazards and vulnerabilities leading to risks.
• Forecasting hazards and assessing risks.
• Dynamic modeling of risk.
This is mainly focused on identifying hazards and vulnerabilities, especially in developing countries. They do not necessarily have the means to identify and understand what their risk levels are (e.g., the forecasting of these hazards and assessing risk). The second objective is effective decision making in the context of complex and changing risk. This includes the following:
• Identifying relevant decision-making systems and their interactions.
• Understanding decision making in the context of environmental hazards.
• Improving the quality of decision-making practice.
The third objective has to do with how we ultimately reduce the risk and curb the losses through knowledge-based actions. In the disaster arena, there is much anecdotal information. That is historically on what the disaster field has based its decisions. Science is not good at getting information down to the practitioners. The practitioners do not understand a lot of the science, because it is presented in highly complicated expositions, using very statistics-heavy methods. It needs to be translated in such a way that they can actually understand it.
In this process, some of the things that we are trying to develop are long-term databases. Disasters have very short memories. Within a year or two of a major event, it is forgotten. Even in Florida, where they get hit all the time, they just forget.
How are we going to actually do this? The three crosscutting themes that we are implementing are as follows:
• Capacity building. Research is great, but if you do not train people how to use it, it does not do any good. We are partnering with several capacity-building organizations, in the United States and globally, to develop this.
• Looking at case studies and demonstration projects. People learn by seeing what has already happened.
• Addressing assessment, data management, and monitoring.
To illustrate the difficulties in achieving this kind of integration, we can think of the parable of the blind men and an elephant, where six blind men touch different parts of an elephant in a collective attempt to discern its true nature. The one who touched the elephant’s ears decided that an elephant must be like a fan. The one who touched its trunk determined that an elephant is like a snake, and similar conclusions were reached by those who felt horns like spears, skin like a wall, legs like tree trunks, and a tail like a rope. Similarly, with so many scientists from diverse fields “touching” disasters, it is a monumental task to integrate their respective works into a comprehensive, cohesive understanding of the nature of hazards and risk. The researchers will examine wind speed, wave action, rainfall, ground motion, and the like. They all have their own unique data issues and their own unique data links to social science aspects, such as religion, ethnicity, age, and gender, for example.
Ultimately, though, what we need to be doing is talking about risk reduction in the collective sense, to get an accurate picture of what an elephant looks like. When you try to integrate census data from the social
science side, which is collected differently and measured differently in every country around the world, with the natural sciences and how these issues are all measured, you come up with very different results. What IRDR is trying to do in the disaster arena is get researchers to look at all of the topics in a common platform and a common way. It is also very important that the private sector be brought into what we are doing.
Then there are problems with the quality and quantity of data on the impacts of disasters. This comes from several factors. Natural hazard management, right or wrong philosophically, is tied very closely to homeland security. When you start looking at disaster data and public infrastructure (e.g., police stations, fire stations, hospitals, power grids, water systems), the homeland security experts do not want to talk about it, because they are afraid that it will show vulnerabilities in the system. They would say, “We cannot talk about the water system, because then somebody would know how to poison the water system in a major city.” I can understand that logic, but when you look at a city like Beijing, with 22 million people, whether or not that water system continues to function in an earthquake is really important.
There is also the issue of the quantity of data. There are massive amounts of satellite data coming in from all over the world for a disaster, but if we look at information on disaster losses related to injury and death, there is actually very little information because there is no common agreement within the medical field of what defines death by disaster. Disaster is not a cause of death on a death certificate. In the recent tsunami in Japan, for example, people were washed out to sea—is that an immediate impact? Or what about people in Florida who died of carbon monoxide poisoning several days after the disaster because they ran generators in their garage? Is that still death by disaster, or is that just accidental? We have not figured that out yet, and this creates questions related to the quality of data and how they are being analyzed.
There are many other biases affecting data quality, such as those in the databases regarding various losses. Most data related to disasters look at post-disaster responses and the losses that have accumulated. We also have hazard biases, since hazards are defined differently in different parts of the world. Further still, there are temporal biases. How are hazards compared over time? Can we compare a disaster from the 1960s and 1970s to a disaster that just occurred today? Realistically, it is much more difficult, partially because of technology. The technology was not available then to measure things the way we do now. Other biases include the following:
Threshold biases. This becomes a bigger issue, because many the databases will, for example, not count an event as a disaster unless a certain number of people were affected. For example, a tornado this morning ripped through Greenburg, PA. That community had less than 10,000 people, but the event was not any less significant. For the 11 families affected, it was as devastating as Hurricane Katrina was to the Gulf Coast. This becomes a threshold bias, because every database has a different cutoff.
Accounting biases. This is really straightforward. How are they measuring the economic effects? What losses are included and how is that done? Even within the U.S. Federal Emergency Management Agency, they are not consistent. There is no consistency across the field or within individual countries.
Geographic bias. This becomes really interesting when you start talking about changing borders. An example that occurred recently was an earthquake on the China-Myanmar border. How China counts the damages and how Myanmar counts them can be very different. How do you look at the earthquake collectively when you only get half the information?
If you are interested in these issues, I encourage you to look at a recent article titled “When do losses
count? Six fallacies of natural hazard loss data”4 that looks directly at natural hazard loss data.
So what is needed? For one, users need to be educated about data biases and issues of social loss data. This is especially important at the policy-makers’ level. Within the social sciences sphere, this becomes an even bigger issue when you start looking at thresholds for age, income levels, and so on.
Making things comparable and accessible for human disaster loss data to support research and policy is another requirement. For example, the floods in Pakistan and China that occurred last year look very similar on many levels. The problem is, we cannot go in and scientifically compare them because the data were collected so differently for the two events.
Furthermore, existing databases need to be identified. We know what is out there globally, but what about the national and regional datasets? In Latin America, for example, they deal a lot on a regional level. What do the databases look like there versus here in the United States, in Canada, or in Europe? A lot of the data have global and regional trends. How do we make the sub-national databases accessible so that policy makers can look at them? We are working very closely with the United Nations International Strategy for Disaster Reduction (UNISDR) to try to make a lot of this information readily available to the policy makers so they can apply it in their areas.
The following are brief introductions to two activities already underway.
• The Risk Interpretation and Action project is looking at why people think they are at risk (or are not) and how they react. It is also taking into account the likelihood of the magnitude of events. Moreover, it is bringing together the physical sciences with the social sciences to look at the resiliency of physical infrastructure.
• Our job in the Forensic Investigations of Disasters (FORIN) project is to trace back through all the articles, data, and other research to figure out what happened leading up to and ultimately causing the disaster. What was the underlying cause for the event? It goes far beyond looking at why the communities are socially vulnerable. In Japan, for example, why was a nuclear reactor put in a tsunami zone? There must be an underlying reason somewhere back in history. This will be a series of case studies using a common template and methodology. The Japanese have already picked this up and will be looking at it in relationship to a previous earthquake, as well as the latest series of events.
How did IRDR get to this point, and who are our partners? We are cosponsored by ICSU, the UNISDR, and the International Social Science Council. Our partners are important, because this is about implementation. They are the national and international science institutes, national and international development agencies, and funding bodies. We are working with organizations such as the Department for International Development, which is the United Kingdom’s overseas development agency, and the U.S. Agency for International Development, looking at the majority of disasters that happen in developing countries. We have six national committees around the world. These take the IRDR principles and apply them in a national context. We also have the IRDR Centres of Excellence, the first being in Taipei. They will be doing research (national and international) as well as capacity-building programs, and they have funding for young scientists.
One of the things that we are not doing is trying to duplicate existing initiatives. We are trying to identify the gaps, fill the gaps, and partner with agencies that are already out there doing it. We provide them with expertise and solid scientific research.
_____________________
4 M. Gall, K. A. Borden, S. L. Cutter, 2009. When do losses count? Six fallacies of natural hazard loss data. BAMS 90 (6): 799–809.
We strive to enhance the capacity of the world to address these issues and make informed decisions. We are trying to get stakeholders to shift their focus from response and recovery to mitigation and prevention. How remarkable would it be if there were a major earthquake and we did not have to spend the next 10 years rebuilding? What we are trying to do also is to learn from the data—not only the mistakes and the lessons learned, but also the best practices and innovative approaches.
15. Understanding Brazilian Biodiversity: Examples Where More Data Sharing Makes the Difference
Vanderlei Canhos Reference Center on Environmental Information (CRIA), Brazil
Increasing consumption of natural resources is leading to a high rate of environmental degradation and biodiversity loss. In the last decade, large forest areas of the Brazilian Amazon Basin have been destroyed, including national park areas classified as high-priority areas for biodiversity conservation. To halt this destruction, real-time monitoring systems to support law enforcement are needed. Prompt and easy access to scientific and socioenvironmental data are critical for the development of real-time monitoring systems, law enforcement measures, and informed decision making. To promote conservation and sustainable use of biological resources, we need to improve access to quality and fit-for-use data, from molecules to ecosystems, deploying new tools that allow the real-time integration of different types of information, including legal regulations, biological data, and socioeconomic and environmental data.
Biological collections are repositories of reference materials and, as data centers, they provide the information necessary to understand the biocomplex interactions of organisms in ecosystems. Biological collections worldwide house an estimated 2.5 to 3 billion biodiversity objects. Of those, about 40 million samples are housed in Brazilian collections. Therefore, to improve knowledge of Brazilian biodiversity, we need to unlock and share the data associated with these Brazilian samples. The most efficient way to address this challenge is to develop a long-term, large-scale cooperative program focused on data sharing and data repatriation, with the deployment of state of-the-art information and communication technology.
The interoperability of e-infrastructures needs to be improved to facilitate the easy, open, and free access to biological data, tools, and Web services. This is the key to innovation in the life sciences, and is a critical factor in the design of support systems for informed decision making. Both the emergence of state-of-the-art computational tools and workflows to check and improve data quality and with the availability of Web services and tools that allow the visualization of models and data on a spatial scale are opening new avenues to improve our knowledge on biodiversity.
The 10-year existence of the speciesLink network5 demonstrated that the key success factors for network consolidation were the adoption of practical solutions to address the lack of adequate institutional infrastructure and the change in culture of sharing data. The network was developed as an inclusive and collaborative bottom-up effort, based on low-cost equipment and free or shareware software. The project implementation started in November 2001, and the network was launched in October 2002, releasing 5,000 records online. Three years after the network launch, approximately 700,000 records were available online.
In the early days of the project, many organizations had serious concerns about releasing and sharing data, but this problem was overcome as the benefits of collaborative networking became clear. At the end of 2010, the network reached 4 million records from nearly 200 collections, double the 2 million records expected to be reached in the initial growth rate.
The key factor in the consolidation of the speciesLink network was the adoption of an architecture (Figure 15-1) that allows the harvesting of data from all types of collections, from small laboratories to large institutional repositories, including collections with substandard computing facilities and poor Internet connectivity. The development of mechanisms to mirror the data in regional cache nodes, associated with mechanisms to transfer and store the data in a centralized database, allows the deployment of tools and
_____________________
5 Available at http://splink.cria.org.br.
services to flag potential errors (such as misspelled names and other outliers) and the provision of tools to improve data quality to the data providers and ensuring full control of the data served.
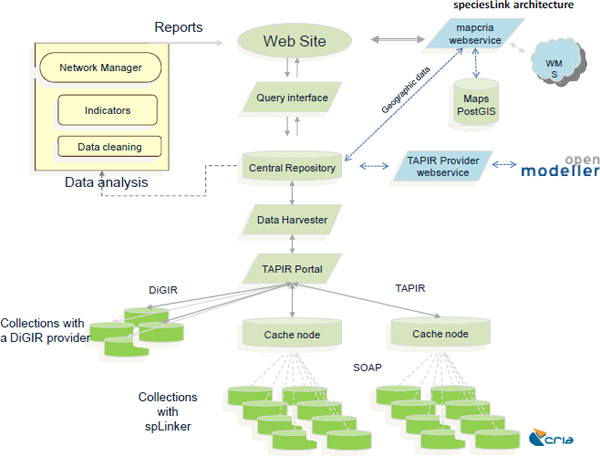
FIGURE 15-1 speciesLink network architecture
Source: From the speaker’s presentation at the symposium.
Speakers in this symposium from Tanzania and South Africa emphasized that a major problem to be addressed is the lack of confidence of data providers to release and share data openly. This problem has been addressed by the speciesLink network through the development of mechanisms that allow the mapping and filtering of sensitive data, and facilitating the blocking of records that are not to be shared. When the user carries out the search, the system indicates that collection has the data, but data can be blocked. If needed, users can contact the database curator to check the restrictions to get the data under special circumstances.
The system harvests the data from cache nodes and stores the data in the central depository. This dataset is used for the network management and the development of indicators of the network evolution. The daily data-cleaning reports are available online not only to data providers but also to the users. This provides an opportunity for the users to check if the data have the required quality and are fit for use. A set of tools and algorithms are available to develop ecological niche models within the openModeller framework.
The 10-year development of the Reference Center on Environmental Information’s (CRIA) biodiversity e-infrastructure is the basis for the effort to integrate the Brazilian and European e-infrastructures through the implementation of the “EU-Brazil Open Data and Cloud Computing e-Infrastructure6”. This is a scoping project cofunded by the European Commission and the National Council for Scientific and Technological Development in Brazil.
_____________________
6 Available at http://www.eubrazilopenbio.eu/.
One important feature of the CRIA’s system is the deployment of a statistics package to monitor usage. Figure 15-2 shows the continuous increase throughout the years, in which the green bar indicates the consolidated usage of all systems available at CRIA. The record is 4.2 million visits in 2010.
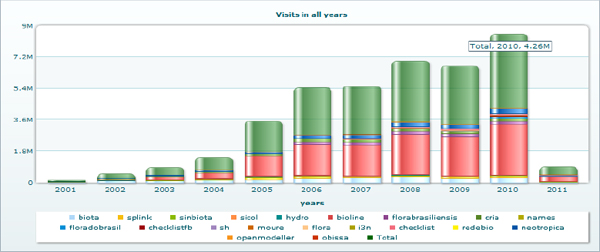
FIGURE 15-2 Visits to CRIA’s online systems in 2010
Source: From the speaker’s presentation at the symposium.
Considering that the bulk of information available at CRIA’s systems is primary data and taxonomic information, the figures are impressive, mainly because more than 80 percent of users are from Brazil. This is the result of investments made by the Brazilian government in improving Internet access. The numbers show the increasing demand for access to scientific biodiversity data and information.
A challenge that is currently being addressed by the CRIA team is the development of mechanisms to share biodiversity information with different user communities, meeting the needs of scientists, policy makers, and common citizens. The way that the data from images, maps, species, and specimen is transparently integrated and shown to the users is key to meet their needs. New Web services are being developed to show high-resolution images integrated with taxonomic information. This is the core of the implementation of the ongoing, long-term, large-scale repatriation program of data from international collections.
In a cooperative effort involving the National Museum of Natural History (MNHN) in Paris and the São Paulo Botanical Garden, CRIA developed the “August de Saint-Hilaire Virtual Herbarium-HVSH7”. This site integrates bibliographic data with higher-resolution images of maps, field notebooks, and samples of herbarium specimens collected during Saint-Hilaire’s travels in Brazil between 1816 and 1822. This material is stored at the MNHN in Paris and the Clermont-Ferrand Herbarium in France. The information system is the prototype that will be adopted to integrate high-resolution images of samples collected in Brazil. This is relevant, as the MNHN Herbarium is finalizing the processing of more than 6 million images of specimens collected worldwide. After processing the label information and improving the data quality by adding coordinates and revising the taxonomic information, the digital data will be integrated to the MNHN Sonnerat Information System.
Another successful example is the revision of the Brazilian List of Plant Species. To meet the first target of the Global Strategy for Plant Conservation approved at the Conference of Parties of the Convention on Biological Diversity (CBD) in 2002, all CBD member countries were supposed to produce an updated
_____________________
7 Available at http://hvsh.cria.org.br/.
working list of plants by 2010. In 2008, after defining the strategy to revise the Brazilian List of Plant Species, the Brazilian government contracted with CRIA to develop the information system to support the collaborative network of experts. The coordination task was delegated to the Rio de Janeiro Botanical Garden. After merging more than 40 databases in different formats into a single one, the 400 specialists in the network revised the list. It included more than 40,000 valid names of plant species and synonyms, was published in May 2010, and was presented at the CBD Nagoya meeting in October 20108. After the development of new functions, the system was reopened for the continuous review by experts.9
To conclude, we are living in a new information era where small nonprofit organizations like CRIA, in collaboration with local and international partners, can play an important role in the creation of virtual environments and e-infrastructures that integrate distributed data, information, and knowledge. These developments are opening new avenues for innovation in science and technology. In this respect the sharing of data, tools, and experiences is making the difference, and a big difference it is!
_____________________
8 Digital version available at http://floradobrasil.jbrj.gov.br/2010.
9 The official 2011 updated list is available at http://floradobrasil.jbrj.gov.br/2011/.
Victoria A. Bakhtina10 International Finance Corporation, United States
The focus of my presentation will be on social statistics as an instrument of strategic management for sustainable development processes. This topic is important because of the stronger interlinkages between social, economic, and ecological domains coming to the forefront of sustainable development progress, and a need to assess the impact of this progress on society.
The twenty-first century brings radical changes to determining the direction of economic progress, shifting the focus to solving the problems of innovative development, and transition to economies of knowledge based on intellectual resources, intellectual capital, science, and education. The ultimate goal is the improvement of people’s lives and the expansion of people’s choices and opportunities, together with sustainable, balanced, and harmonious development of society.
The modern vision of gradual sustainable development unifies three main components: economic, ecological, and social. The quality of the economic component is increasingly linked in people’s minds with the concept of human development. Human or social development becomes the main purpose, and material means become a condition of this development.
Enormous data capabilities in conjunction with scientific and technological progress allow for a systemic approach to management and a comprehensive review and vision of complex information, leading to a better understanding of the major challenges facing humanity. It is key, therefore, to create an information-analytic base for innovative development using pioneering models and high-quality data. We can benchmark and look at the world as a whole, and we can add to these descriptions of reality and infuse more precision to the understanding of real situations.
Another approach is extracting data on risks, both general and specific, and drawing conclusions about management and prevention of risk situations. Statistical analysis is a necessary tool and condition for justifying the taking of strategic decisions.
In the 1990s the Human Development Report (HDR) of the United Nations demonstrated how to change the approach to development, focusing on people, and opening an era of new opportunities for the policy and research agenda. History has shown that a sole focus on economic growth does not necessarily result in the highest achievement in human development, and countries with the highest economic growth can lag in health, education, and other key areas. Having people as the source and the purpose of development makes progress equitable and creates an enabling environment for each member of the community to be a participant in change.
Initially the Human Development Index (HDI) encompassed only longevity, adult literacy, and income. In 2010, HDI was adjusted to reflect inequality and was complemented by the Multidimensional Poverty Index (MPI) and the Gender Inequality Index (GII), which illustrate countries’ trends over the years and highlight important lessons for the future.
Assessments of “well-being” vary dramatically based on many aspects not covered by HDI: lack of self-respect and dignity, lack of access to assets and access to information, lack of time, insecurity, lack of
_____________________
10 The views expressed herein are those of the individual contributor and do not necessarily reflect the views of the IFC or its management.
freedom of choice, helplessness, social exclusion, and isolation. This list is far from complete and can be expanded based on any specific context. These aspects are often interconnected and can lead to the disempowerment of people. Further research is needed to shed more light on these areas.
Social statistics complement and enrich economic and environmental measures, and allow us to analyze the efficiency of policies. It is equally important on a macro level when conceptually assessing global phenomena, and on a micro level, for example, utilizing household surveys, when assessing the qualitative nature of information and tailoring the policies to a certain context.
The Human Development Index is important for developed and developing countries, allowing us to track cumulative progress year after year. Measurements should be standardized as much as possible around the globe. Complemented by MPI and GII data, HDI adds various nuances to country performance relative to similar countries. The HDI shows that inequality adjustment can drastically shift the perception of the countries. For example, you can see from Figure 16-1 that there are significant losses in HDI due to inequality in various components.
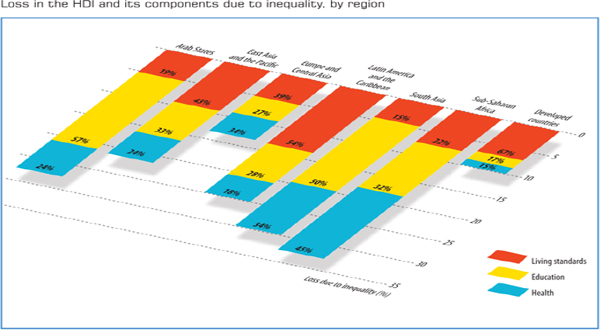
FIGURE 16-1 HDI index adjustments due to inequality.
Source: From the speaker’s presentation at the symposium.
Figure 16-2 shows six African countries: Ghana, Botswana, Gabon, Malawi, Rwanda, and Kenya. These countries are dispersed around the continent, differ in access to natural resources, and have different political situations. There is a strong desire in these countries for reforms.
FIGURE 16-2 Six African countries: Overall Satisfaction with Life
Source: From the speaker’s presentation at the symposium.
Rwanda has strong leadership, far-reaching reforms, and a roadmap outlining the transformation to a middle-income country by 2017. According to an International Monetary Fund report, reforms are on track and quantitative targets are being met. Malawi is a landlocked country and an agriculture-driven, smallholder-based economy. Ghana is one of the most robust democracies in Sub-Saharan Africa. Botswana is one of the most successful countries in Sub-Saharan Africa. Gabon is rated highest in human development in Sub-Saharan Africa. Kenya is one of the largest economies in East and Central Africa.
Figure 16-3 displays four of the six countries that are in the low human development quartile, according to the HDR. The main reasons are poverty, poor infrastructure, and limited access to health services and education.
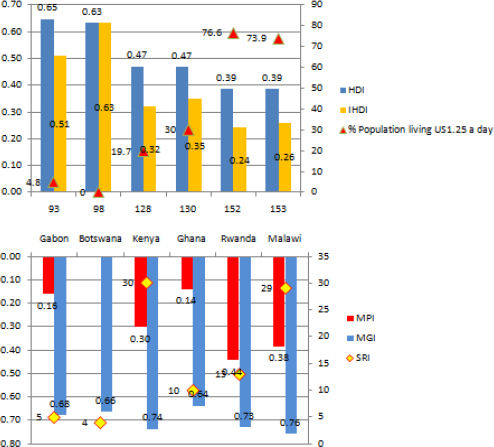
FIGURE 16-3 Multidimensional Poverty Index of Six African Countries
Data Source: HDR 2010; Figure is from the speaker’s presentation at the symposium.
Gabon and Botswana have a medium level of human development, and they are almost identical, standing at 0.65 and 0.63 HDI, respectively. Kenya and Ghana are close, with 0.47 HDI. Rwanda and Malawi both have 0.39 HDI.
Let us expand the picture inferred from the Human Development Index further, and take into consideration inequalities in income, longevity, and education. According to Figure 16-3, Gabon moves to 0.5 HDI when we review inequality-adjusted HDI. After adjustment for inequality, Ghana comes first in the set of four countries from the low human development cluster. Malawi is a little higher than Rwanda.
Now, add an additional measure—a Multidimensional Poverty Index (see Figure 16-3). Based on this index, Ghana supersedes Gabon, appearing as the country with the lowest MPI of the six countries. In gender inequality, Rwanda exhibits the most equality, followed by Botswana and Gabon. Kenya has the same GII as Ghana, and Malawi shows the largest inequality.
An additional measure to be considered in the analysis is developmental, or the sustainability, risk index based on region-specific features. The sustainability risk index shows how vulnerable the countries are to the targeted key risks. According to the latest research (Bakhtina, 201011), Botswana (rank 4) and Gabon (rank 5) falls into the cluster of least vulnerable countries, followed by Ghana (rank 10). Malawi (rank 29) falls into the medium-risk cluster, followed by Kenya (rank 30). Rwanda (rank 13) is the country least vulnerable to regional risks in the medium-risk cluster.
Now let us see how happy the people feel in the six countries under consideration. Overall satisfaction with life is highest in Malawi, followed by Ghana and Botswana, and those least satisfied with life are Kenyans. Females appear to be less happy. Is it a result of inequality? This presumption is not clear, as
_____________________
11 V. A. Bakhtina, 2010, Sub-Saharan Africa: Sustainability risk discussion (presented at CODATA 22 International Conference, Cape Town, October 24–27).
Malawi has the largest inequality among the six countries, but the highest life satisfaction (see Figure 16-4).
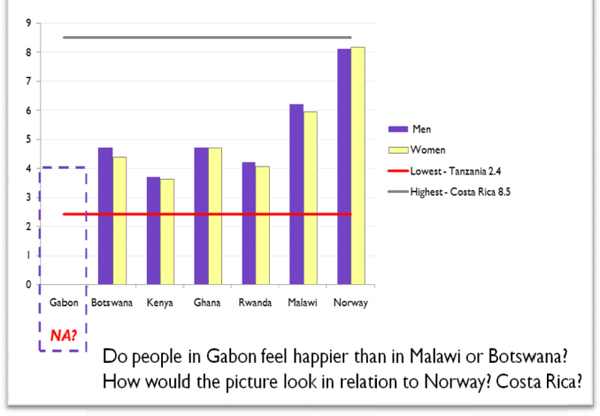
FIGURE 16-4 Six African Countries–Overall Satisfaction with Life.
Source: From the speaker’s presentation at the symposium. Credit: HDR 2010. Overall Satisfaction with Life is based on responses to a question about satisfaction with life in a Gallup World Poll
What conclusions can be made from these data? Would the data help us identify and flag key issues to address on the global scale? As we see from these statistics, life satisfaction has no strong relation to poverty and inequality.
When Malawi is compared with Norway, the country with highest HDI, the satisfaction-with-life index is 6 versus 8. (The lowest life-satisfaction level is observed in Tanzania at 2.4 and Togo at 2.6, the highest is in Costa Rica at 8.5.) We can also expand the research agenda: What policies would make people happier and inspired to act towards change and life-quality improvement?
Here, more information is needed: for example, measures of empowerment, unpaid women’s work, and domestic violence. What factors increase happiness of the population?
According to the HDR, “human development is the expansion of people’s freedoms to live long, healthy, and creative lives, to advance other goals they have reason to value, and to engage actively in shaping development equitably and sustainably on a shared planet. People are both the beneficiaries and drivers of human development as individuals and groups.”
Let us turn now to some compelling examples showing how social statistics were used to drive policy decisions. In 2009 in Mexico, the Constitution and the General Law of Social Development were amended based on a multidimensional poverty measure reflecting various deprivations the households face (National Council for Evaluation of Social Policy). The country is mapped based on the level of deprivation in at least one of six dimensions: education, health care, social security, housing quality, basic
household utilities, and access to food. The Mexican government uses the data to monitor the effectiveness of national social assistance programs.
The World Bank utilizes the Participatory Poverty Assessment (PPA) approach. The PPA allows the World Bank to incorporate views of the poor into the analysis of poverty, combine the results with other types of data, and communicate the findings to the policy makers, thus allowing the poor to influence policy. In numerous cases, PPA results contributed to a shift in World Bank lending programs. For example, in Ghana, PPAs contributed to a shift in the focus of reforms to rural infrastructure, quality and accessibility of health care, and education. In the 1990s, the World Bank focus in Nigeria was shifted to water and roads.
At the same time, PPAs were used in Thailand as a part of the Social Investment Project to increase the understanding of shifting patterns of vulnerability as the impact of the Asian crisis deepened, and to inform policy makers, strengthening the capacity of the country by consolidating various types of information. The Asian Development Bank also conducted similar assessments in Laos and the Philippines.
Social aspects are also key in disaster response and mitigation. Along with physical aspects, such as the probability of loss and risk, social aspects such as vulnerability and resilience should be considered. Fear, depression, despair, and post-traumatic stress can cause long-term consequences for the nation after a natural disaster. Apart from the assessment of hazards, probability, magnitude, and impact, a people-centered perspective is required to evaluate the susceptibility of the community to natural hazards. Interaction between hazard conditions and vulnerability conditions should be tracked dynamically (e.g., climate change impacts and economy impacts on migration of people to places susceptible to hazards like floods).
The World Health Organization points out that the concept of risk is associated with the perception of risk, and is a characteristic of society and culture. More effort is needed in community involvement in risk mapping and analysis, enhancing vulnerability assessments, understanding of risk perception, and the capacity to adjust.
The example of Mozambique shows how to link early warning of disaster and early action. After the flood in early 2000 left devastating consequences for the country, the social aspects of disaster mitigation were assessed and used to train the community to understand the risk and use warning information. The consequences of the 2007 and 2008 floods were significantly less severe.
Social factors contributing to cyclones’ severity in Bangladesh are cultural. Male heads of the household historically did not want to move to shelters “unsuitable for females,” with lack of privacy and poor sanitation. Families mostly adopted a wait-and-see strategy. Vulnerability was increased by late responses. Currently the issue is being addressed by educating the population.
The examples above show us how social statistics can serve as an instrument and basis for decision making and research on sustainable development processes. We are looking at the management process of decision making and utilizing this powerful instrument. We are showing how this instrument works on the global scale in the computation of indexes, which help us to understand objective trends. The focus is on the level at which the statistics are used. We reviewed three levels of application of social statistics: global, national, and local decision making. Many challenges remain, such as the sharing of data, the need for global statistical standards, data communication, intellectual property rights to the research results, and utilization of the data and research in policy decision making.
The benefits of sharing social data are many, and only a selection of them are given in this presentation. We should not forget that with benefits come responsibilities: To whom do we distribute the data? How do we protect the information against misinterpretation? How do we educate communities on how to use the data? How do we protect the data against terrorism? Who is accountable for quality, timeliness, and accessibility of data?
Let me conclude by reinforcing what I said earlier. We have tremendous opportunities. With the help of powerful technologies and new innovative models, we can promote and reinforce sustainable development. We can illustrate socioeconomic and ecological aspects of the development and address key risks. To validate, calibrate, and use the models, we need accurate and timely data and collective cooperation among statistical agencies and policy makers around the world. The assessment of the contribution of policies and financing mechanisms in the improvement of people’s lives and expansion of their freedoms, cannot possibly be done without extensive shared information on social statistics.
References
Human Development Report (UN), 2010.
Robb, Caroline M., Can the Poor Influence Policy? Participatory Poverty Assessment in the Developing World, 2nd ed. (International Bank for Reconstruction and Development–The World Bank, 2002).
World Disasters Report 2009, Focus on Early Warning, Early Action (International Federation of Red Cross and Red Crescent Societies, 2009).
Bakhtina, Victoria A., Sub-Saharan Africa: Sustainability Risk Discussion (paper presented at the CODATA 22 International Conference, Cape Town, October 24–27, 2010).
17. Remote Sensing and In Situ Measurements in the Global Earth Observation System of Systems
Curtis Woodcock Boston University, United States
What I would like to present today are my experiences related to the Group on Earth Observations (GEO), specifically the Forest Carbon Tracking (FCT) task. There are examples that relate to what we are interested in here, so I would like to share those.
The whole point of the FCT task has been to try to stay ahead of the problem of determining if and how developing countries could start to report on rates of deforestation and degradation. This is important so they could be eligible for compensation for reducing those rates, all in support of trying to reduce greenhouse gas emissions to the atmosphere to try to mitigate increasing carbon dioxide in the atmosphere. Then the main question is, can those countries report in a reasonable way? To do this accurately, there is a need for data.
The two primary kinds of data that are needed for this task are satellite observations and in situ forest data. I am mostly going to talk about the satellite observations. On the one hand, that is because if you think about data policy and data sharing, in situ observations are typically collected by people in their own backyard. On the other hand, satellite data are generally collected by somebody else. Certainly we ought to make the first step, that if we are going to collect data in other people’s backyards, we ought to at least share the data with them, rather than necessarily force them to share the data they collect. If we could at least take that step, it would be a good step forward.
GEO has identified coordination among space agencies for image acquisitions as its first and top priority as it moves forward. Different satellite programs have collected data to be contributed to this task, including those in Table 17-1:
Table 17-1 Image Acquisitions by Country
| ND Scene counts (vs submitted), 2010 | ||||||||||||
| Borneo | Brazil | Cameroon | Colombia | DRC | Guyana | Mexico | Peru | Sumatra | Tanzania | Tasmania | TOTAL | |
| CBERS | 2850 (14000) | 1013 | 495 | 1864 | 6004 (14339) | |||||||
| LANDSAT | 353 | 284 | 1097 | 731 | 225 | 1063 | 263 | 423 | 134 | 4354 (4366) | ||
| LANDSAT-INPE | 685 (2130) | 144 | 113 | 287 | 1189(2191) | |||||||
| PALSAR | 725 | 790(3356) | 150 | 803 | 1346 | 180 | 1004 (1286) | 891 | 380 | 580 | 20 | 6798 (9101) |
| RADARSAT | 35 | 38 (39) | 37 | 27 | 123 | 70 | 80 | 410 | ||||
| RESOLRCESAT | 756(1862) | 120 | 117 | 282 | 1149 (1888) | |||||||
Why I call it a first priority is that there has actually been some coordination of image acquisitions across different countries’ space agencies. Those data were collected for this task and has been contributed to countries where we are trying to demonstrate these technologies. Nonetheless, there are still questions about data access, even though the data were collected specifically for this task. Free and open sharing of data remains very much an issue.
Why is this so hard? It is hard because satellite missions are expensive. They range from the hundreds of millions to billions of dollars. What makes it frustrating is that the revenues that countries get from selling data that come from these missions rarely offset anything close to a significant fraction of the cost of those missions. Just logically, it does not match up very well. I am going to try to convince you that charging for the data significantly hinders the use of the data.
The example I am going to use is the Landsat mission, which is the oldest of the land remote-sensing missions in the United States. There have been seven satellites in the Landsat program. Two are barely functioning still. The eighth is scheduled to be launched in December 2012. What we have is a dataset from 38 years of satellite observations that is a little more than 2.5 million images. An image covers 185 by 185 kilometers of the surface of the Earth. If you think about all the costs associated with building and launching the satellites, the data cost somewhere between $5 billion and $10 billion. It is an expensive dataset. We spent a lot of money on generating this dataset, and we would like to see it get used effectively in the future.
The good news is that starting in October 2008, the U.S. Geological Survey (USGS), the agency that distributes the Landsat data, stopped charging for the data and provided access on a no-cost basis to anyone in the world. The data usage has gone up by a factor of 100 since it became freely available. As a taxpayer, think about this. As shown in Figure 17-2, before the data became freely available, in the biggest year ever for sales of Landsat data, they sold about 25,000 images. The income that was coming in from selling images, on an annual basis, was somewhere between about $5 million and, at the very best, $10 million. That is one-tenth of 1 percent of the cost of the dataset, on an annual basis, that they were getting back from selling the data.
After all these years, we are finally getting our money’s worth out of this dataset. This is the kind of effect that data policy can have on the ability to do research and applications.
The other thing that is interesting to note here is that I have been using Landsat data for my research over 35 years. All of a sudden, we are figuring out new and better ways to do a lot of things we never thought we could do in the past. It not only allows us to know what has happened in the past, but there are whole new categories of activities that are starting to show up, and it has been only 2 years since the distribution policy was changed. I think it is a transformative kind of experience for the use of satellite data.
Let me raise one sensitive issue about this data policy. Landsat is a U.S. satellite, but the United States has had international collaborators. The government sold licenses to receiving stations to get data over the years. There are about 2.5 million images archived in the United States and about 3 million images now in the international ground stations around the world. Although those other countries paid for the licenses to
collect the data, the United States now is giving the data away freely. At the same time, the history of the surface of the Earth is embedded in these datasets. We cannot go back and recollect the data. It is a really valuable dataset showing part of the history of the planet’s surface.
Here is where that effort stands. The USGS made an offer to the long list of countries that have data in their archives that if they would give us the data, we would reprocess them to the highest standards and return the data to them, and we would freely make available the data, if they want them. The United States has made that offer to these countries, but I think you can appreciate the delicate nature of this sort of discussion, when, after having asked people to pay for the data all these years ago, you are now saying that you want the data returned at no cost.
It really is an effort to bring into a standard state of affairs a very valuable dataset. A lot of this imagery is sitting around on media that is degrading. If we are going to do this right in the future, because we have not really done this correctly in the past, we need to think about the dimensions of international coordination and collaboration on earth observations. We now have dozens of countries running satellite observation missions, but in a very coordinated manner.
The first step is mission planning, so that there could actually be some compatibility between the datasets produced by these different organizations and countries. Then you have acquisition strategies. Can we get some coordination of acquisition strategies? For the first time, countries are actually beginning to collect data in concert with each other.
These satellite missions are expensive; there is no question about that. A big question then is risk mitigation. If multiple countries are going to put up comparable satellite missions, which is what is happening now, can we at least share risk with each other, so that if somebody’s mission fails, the other will provide the data that would have otherwise been collected by the other satellite mission? This is starting to happen to some extent between the European Space Agency and the U.S. Geological Survey, with the Sentinel satellites and the Landsat program.
If we are really going to take advantage of having multiple satellite systems collecting similar data in space, it is equally important to get the data processed and distributed in consistent fashion, so that we can actually start to make use of them. It is difficult for individual investigators to try to take data from multiple satellite systems and combine them if they do not have any coordination at the outset. This is something that, at an organizational level, would really help.
My concluding thoughts are not too surprising. One is that we need satellite observations for many societal benefit areas, such as effective research and monitoring on climate and deforestation. We are in the infancy of trying to do any significant international coordination of these missions and collaboration, and there are many benefits to doing it that way. Free and open access to Earth observation, in some ways, is most important for developing countries, because they can least afford to pay for the data.
I also work in an organization called the Global Observation of Forest Cover and Global Observation of Land Dynamics. We have two sets of thematically oriented groups—the forest group and a land cover group. I cochair the latter. We also support capacity building through a set of regional networks. These are groups of regional scientists with common interests in Earth observation as used for forest monitoring and other such research and applications.
We have been running data-sharing workshops for the last couple of years, sponsoring people from regional networks to come to the USGS Earth Resources Observation Systems Data Center in South Dakota, and letting them take away all the data they want. This removes many of the technology barriers.
They go home with hard disks full of satellite images that they can use and share in the region. It becomes a focused network.
We are going to continue to do this for at least the next 2 or 3 years. There was one workshop per year for each of the last 2 years, and we are going to do two more this year and in the next 2 years. If anybody wants to try to get people into one of these workshops, or is interested in a particular region or getting access to the data that has already been taken back to some of these regions, I would be happy to help connect people and try to coordinate those activities.
18. DISCUSSION BY THE WORKSHOP PARTICIPANTS
PARTICIPANT: This question is to Dan Cheney. Have you and the Federal Aviation Administration taken the next step? Now that you have all this wonderful scientific and technological experience data, have you started thinking about how you can exploit those data to make the entire air system both safer and more efficient?
MR. CHENEY: That really is the ultimate plan. The first step was to accumulate the knowledge. We are at the front end of taking the final step, and that is integrating the knowledge into our processes for maintaining and improving aviation safety. This initiative is only two and a half years old, so we are still in the middle stages of catching up on the history of aviation. We are laying the groundwork now for what to do with it. We know we have some gaps in knowledge in various aspects of aviation. So the question is, how do we use this to fill those gaps? We are not there yet. There is more work to do.
PARTICIPANT: I am thinking particularly about the accidents that did not happen. There is a huge amount of learning there that goes on inside the airline industry all the time, when these things are adequately reported. Sometimes they are not. There are malpractices with loading, with crew behavior in the cockpit, and so on. There are seriously overweight takeoffs and landings. We, as the general public, do not hear about those at all. There is an enormous amount of learning embedded in that.
MR. CHENEY: The aviation industry has benefited from a level of safety margins that exist in every aspect of safety. We have achieved internationally an unthinkable safety record when you look back at where we were 40 years ago and 30 years ago, and even 20 years ago. But it is because of the safety margins that are there. When we do have the overweight takeoff, when we do have the pilot that forgets to deploy a system, or a fire occurs, there are margins upon margins that result in that airplane not having an accident.
What we believe is the value in understanding the causes of yesterday’s accidents is to recognize that when you do load an airplane over gross or you do have a tired crew that forgets things, you effectively take away one of the five levels of margin or one of the three levels of margin for that flight. That in itself may not be catastrophic, but now you have put that plane in a level of vulnerability in which it would not otherwise have been in. The accident is nearly always because you have eaten up four or five levels, and you have nothing left. The close calls, the accidents that did not happen—and they are certainly going on every day—are mirror images of what has happened in the last 50 years since the jet era. Whether it is an A380 or a 787 of tomorrow, those margins are the margins.
I think our big return will be to have tomorrow’s decision makers and operators understand the importance of checklists, the importance of getting it right from a maintenance perspective, because getting it wrong takes the margin away on one level. Now we have a two-legged stool. One more mistake, and eventually we have an accident. The reason we are enjoying the safety we have is that it is a robust, margin-full industry. We are going to fight any threats of giving up any of the margins.
PARTICIPANT: One of the themes I got from several presentations is that a traditional problem for these kinds of data efforts is that data was locked up in disciplines, in silos. A lot of the presentations talked about how, especially for applied problems, you were able to overcome those traditional silos and put data together in new ways. The thing I am wondering about is, now that we have new problem-oriented silos, it seems there is always a danger of repeating history and having these new problem areas become silos on their own. I am just curious if you have thoughts on how to avoid that. To what extent in each of your areas are you really paying attention to other people’s standards and looking at interoperability between biodiversity and disasters, which may not have an obvious connection now, but in fact, because of deforestation and forest cover loss, may have a connection to land use and disaster loss? To what extent
do you think people are thinking, especially in developing countries, about not creating new barriers after having overcome the old barriers?
DR. CANHOS: I think that you raise a very critical point about standards and protocols for interoperability of databanks and data systems. When you look back at biological data, there were no standards and protocols for biodiversity data, although several organizations have worked in this area in recent years, such as the Global Biodiversity Information Facility (GBIF) in close collaboration with a Committee on Data for Science and Technology (CODATA) committee for developments and improvements in this area.
We do have big issues with silos. You talk here about different people working in different data areas such as biological data, species-oriented biological data and integration at a molecular level, genome data, and then integration with the information derived from satellite data, like land coverage. I think for local development, it is extremely important from the beginning to look at how the internationally agreed standards and protocols are developed. For the speciesLink network, when we started 10 years ago, our first approach was to see what was happening in the taxonomic databases working group. Also, after looking at what they were doing regarding biodiversity information, we decided to work in close collaboration with the Taxonomic Data Working Group (TDWG) and GBIF.
In conclusion, I think here we need the international organizations. We need CODATA and the International Council for Science (ICSU). All those efforts, like TDWG, are voluntary efforts. It is extremely difficult to develop standards and protocols on a voluntary basis. I think the major development was the support of GBIF to TDWG. This happened with $1.5 million that came from the Gordon and Betty Moore Foundation. Again, we need more involvement of international organizations. I think ICSU has an important role to play in the definition and further development of the data-related standards and protocols.
PARTICIPANT: Dr. Canhos, are you concerned about the sustainability of your biodiversity database?
DR. CANHOS: I am very concerned about the sustainability of those efforts. For me, it is difficult to get funds from the Brazilian government to maintain the speciesLink network. As to the content, from the beginning of the development of the network, we told the collections, “You can come in or leave the network anytime you want.” It is just like pressing a button to take all the data from your database from the network, but that did not happen. Today we have more than 200 collections. The cost is well distributed and includes the cost of development of the software and the tools. This was a well-funded project for 4 years. Now the cost is to upgrade all those tools and also to develop more tools. The more data use we get, the more new requests for new applications we receive. I think that is a challenge, but now we are in an age of integrating distributed intelligence and distributed data. That lowers the cost. That is public infrastructure. When you think about the industrialized countries, we are talking about huge legacy collections. Think about the Smithsonian Institution. They have more than 130 million biodiversity objects in the Smithsonian collections. The cost to digitize all this legacy information is very high. Finally, I think we need support from international organizations. We need ICSU and CODATA to continue advocating the importance of not only sharing the data but also finding means to gather this data and to treat the data so that it can be easily accessible by the whole community all over the world.
DR. BAKHTINA: Regarding involvement of international organizations, I want to add that the World Bank promotes democratization of development via open data access. Last week, a new initiative, Mapping for Results, was launched. This initiative allows civil societies, various organizations, and citizens to access large World Bank databases, to contribute to the database and to use the data. It also opens a direct dialogue with governments and citizens, in terms of transparency of government policy and delivery of public services.
Furthermore, it will promote transparency related to the development, and hopefully, create incentives for various types of organizations to improve data quality and increase data sharing.
PARTICIPANT: I am with the International Environmental Data Rescue Organization. We are a nonprofit organization whose mandate is to locate, rescue, and digitize every piece of historic environmental data we can find throughout the world. We have projects in 15 developing countries, and we have rescued probably 2 million to 30 million historic weather observations. One of the problems we have is that, for example, we have located about 30 million weather observations on microfiche. They were taken in about 1,000 observation sites throughout Africa. We are negotiating with the African Center for Meteorological Applications for Development in Niger to at least get the microfiche before they deteriorate to nothing. I am wondering, do any of your organizations have data in a format where you cannot share them? Right now 95 percent of our data are either on microfiche or on paper, and that is a very huge problem for us. I am wondering if anybody knows of any other organizations, other than our own, that actually seek out data on perishable media to rescue them before they are gone forever.
DR. WOODCOCK: The U.S. Geological Survey is doing that for Landsat data. They do get data on all kinds of customized software and media, where they have to go back and reconstruct and reconcile them. It is hard. I do not do that myself, but I have been convinced that it is a pretty big obstacle in many countries.
PARTICIPANT: I am not sure if I am the only librarian in the audience, but libraries are definitely aware of this. They have been looking at this for straight text documents, and now they are starting to look at it for both born-digital, which is also being lost at an alarming rate, and print objects. So there are institutions working on these problems, and the National Science Foundation is supporting some of that activity.
PARTICIPANT: There is also a new CODATA task group called Data at Risk, which is working to at least identify a broader range of scientific data, not just environmental. Another group is the Minnesota Population Center. There is a lot of recovery of old census data, going back and migrating data on old media and similar activities. There are different groups in different disciplines that are doing this.
PARTICIPANT: The National Oceanic and Atmospheric Administration also has a data-rescue activity. I imagine there are quite a few others. And the United Nations Educational, Scientific and Cultural Organization, I believe, has a digital heritage program.
PARTICIPANT: I have a question for Dan Cheney about the difficulty in getting access to the data that you have been talking about because of the sensitive nature of them. For the FAA data, it could be potentially sensitive for the companies that either built the planes or operate them, in legal exposure. I was wondering if that is mitigated by some kind of legislation that caps the exposure to lawsuits or if there is some kind of waiver of liability associated with the disclosure of the data.
MR. CHENEY: I do not think I mentioned this during the talk. There are four criteria that have to be met before we even begin to look at an accident: the official accident report is issued; the corrective action and accident results are finished; there has not been another accident or incident that would call into question the official accident findings; and litigation is finished. Only then, when all four of those are met, do we begin to work, and we only use publicly available information. Normally it is the information that was gathered during the course of the investigative process, and in the United States it is part of the public domain. We work with other countries’ accident-investigating bodies to secure access to their information that was gathered in the course of the investigation. We do not do any additional investigation. It is only what is already lying around in archives in pieces that is being lost. The task is to put it together in one cohesive place, look at it, and put it in a structure that makes sense. As far as litigation, the opportunity to
litigate has already come and gone. Could someone come back and say, “Well, we did not think about that. Let us review it again.” They could. Our greater-good concern, however, is that the information is so important that it cannot be lost, and we have to take this step and make sure that erosion is stopped.
PARTICIPANT: Do you also get data from other countries?
MR. CHENEY: We do. Not all are equally sharing, but there is outreach. As we have done networking and have improved our networking process, the efficiency is increasing, with recognition that this is a product that will benefit world aviation, not just U.S. aviation. It is getting much better.
PARTICIPANT: Victoria Bakhtina, how difficult is it to get access to the data that support the statistics for the different countries? Are the data collected by the countries themselves and made available to the World Bank, or does the World Bank pay people to collect the data and work with the different ministries? It is not clear how the data are compiled, and whether it is comparable kind of coverage.
DR. BAKHTINA: In terms of access, with the launch of open data policy, it becomes very easy. Now the set of all development reports that include underlying data will be available. Just visit the World Bank website, and you can easily download and research the statistics from different countries. Moreover, you can visualize the data and map the country statistics to the World Bank projects information. I am also using the UN data which are also publicly available. There are various methods and strategies of collecting the data and multiple approaches can be applied depending on what is measured. The World Bank Data Group partners with other organizations on data collection and statistical capacity-building. I would recommend that you consult the World Bank web site for details. When using any publicly available data, it is important to understand what is behind the numbers, and depending on the end goal of your research, determine what coverage would be acceptable, and most importantly, always conduct the analysis within a specific context.