This page intentionally left blank.
Interoperability, Standards, and Linked Data
James Hendler
-Rensselaer Polytechnic Institute-
I will focus on the technological challenge of online knowledge discovery. I will first address interoperability issues and then talk about some promising developments.
Scientists can learn a lot about data sharing just from observing what is happening in the outside world. There are many data initiatives and developments outside academia to which we should be paying more attention.
Expressive ontologies are not a scalable solution. They are a necessary solution in certain domains, but they do not solve the interoperability problem widely. They allow a scientist to build his or her silo better, and sometimes they even let the silo get a little wider, but they are not good at breaking down the silos unless the scientist can start linking ontologies, in which case he or she has to deal with the ontology interoperability issue, as well as the costs and difficulty of building them and their brittleness.
I am known for the slogan “A little semantics goes a long way.” I have said this so often that about 10 years ago people started calling it the Hendler hypothesis at the Defense Advanced Research Projects Agency (DARPA).
We are used to thinking that a major science problem is searching for information in papers, and we have forgotten that we also have to find and use the data underlying the science. A traditional scientific view of the world might be, “I get my data, I do my work, and then I want to share it.” But the sharing should be part of how we do science, and other issues such as visualization, archiving, and integration should be in that life cycle too. I will talk about these issues, and then I will discuss the kinds of technologies that are starting to mature in this area. These technologies have not yet solved the problems of science and, in fact, largely have not been applied to the problems of science.
Scientists do use extremely large databases, and many of these data are crucial to society. On the other hand, we scientists tend to be fairly pejorative about something like Facebook, because Facebook is not being used to solve the problems of the world. I wish science could get the number of minutes per day that Facebook gets, which is roughly equivalent to the entire workforce of a very large company for a year. We are also not used to thinking about Facebook as confronting a data challenge per se, but it collects, according to the published figures, 25 terabytes of logged data per day. That sounds like the kind of numbers for large science data collections. Facebook’s valuation is estimated to be well over $33 billion, which is the size of the entire National Institutes of Health budget. Not surprisingly, they are able to deal with some of these data issues that are discussed here. We need to look at what they are doing and determine if we could take advantage of some of their approaches.
I do not have similar numbers for Google, because they have not been published. The last good estimate I could find was in 2008, which was 20 petabytes per day, but that was 3 years ago. That also does not include the exchange of or the storage of YouTube data, which I cannot find good numbers about either. Google’s valuation in 2011 is about $190 billion,
which is roughly a third of the Department of Defense budget. Not the research budget—the entire budget.
Therefore, if we think that this kind of work is expensive, yes, it is, but there are other people doing it, and they are investing large amounts of money. It is not surprising that they have been focusing on big data problems in a way different from scientists, and they have been able to explore some areas that are very difficult for us to explore. If a researcher wanted to buy 10,000 computers for data mining purposes, it would difficult to do because of the lack of resources.
Several speakers talked about semantics in the context of annotation and finding related work in the research literature. It is an important problem, but I do not think it is the key unaddressed problem in science. In fact, I would contend that we have spent a huge amount of money on that problem, much of it on trying to reinvent things that already exist in a better form from open sources in the real world.
Most companies today that want to work with natural language processing start with open-source software. For example, there is a company that is taking everything from the Twitter stream, running it through a number of natural language tools, and doing some categorization, visualization, and other similar work. They did not build any of their language processing software. I am also told that Watson, the IBM Jeopardy computer, had a large team of programmers, but, in fact, that the basic language technology used was mostly off the shelf, and that it was mostly statistical.
Semantic MedLine is a project that the Department of Health and Human Services has invested in. It does a fairly good job but does not understand the literature sufficiently to find exactly what we want. But we are not able to do that in any kind of literature, and Google is working on that problem as well. Hence, I do not see the point of yet another program to do that for yet another subfield or against yet another domain. Instead, we need to start thinking about how to put these kinds of technologies together.
The Web is a hard technology to monitor and track because it moves very fast. It has been moving very fast for a long time, however, and, as scientists, we need to start taking advantage of it much more than reinventing it.
There are a few different tools and models on the Web that are worth thinking about. One is to move away from purely relational models; from assuming that the only way to share data is to have a single common model of the data. In other words, to put data in a database or to create a data warehouse, we need to have a model, and that is done for a particular kind of query efficiency.
Sometimes, however, that efficiency is not the most important factor. Google had to move away from traditional databases to deal with the 20 petabytes of data generated every day. Nobody has a data warehouse that does that, so Google has been inventing many useful techniques. “Big Data” was one of their names for their file system. We cannot easily get the details of how Google does it, but we can get published papers from Google people that will be useful in learning how to do similar work. The NoSQL community is a fairly large and growing movement of people who are saying that when you are dealing with large volumes of data there is a need for something different from the traditional data warehouse.
I had a discussion with some server experts who said, “Just give us the schemas, and we can do this work.” I pointed out that we cannot always get the schemas, that some of these datasets were not even built using database technologies with schemas, and in some cases someone else owns the schema and will not share it. We can still obtain the data, however, either through an Application Programming Interface (API) or through a data-sharing Web site.
Although Facebook’s and Google’s big data solutions are proprietary, the economics of applications, cell phones, and other new technologies are pushing toward much more interoperability and exchangeability for the data, which is mandating some sort of a semantics approach. Consider, for example, the “like” buttons on Facebook. Facebook basically wants their tools and content to be everywhere, not just in Facebook, which means it has to start thinking more about how to get those data, who will get that data, whether it wants data to be shared or not, and what formats and technologies to use. As a result, there are many technologies behind that “like” button. Similarly, there are a number of approaches that Google employs to find better meanings for their advertising technologies. What happens is that Google recognizes that at one point it will not be able do the work by itself, because there is a long-tailed distribution on the Web. This means that it has to move the work to where Webmasters and other people will be able to develop the semantic representations for their domains.
That is happening with all the search engines, and the big issue now in that area is simple metadata and lightweight semantics. The notion of the complex ontology is getting replaced by the notion of fairly simple descriptive terms—that is, I can probably describe the data in my dataset with 10 or 12 different terms that will be enough for me to put in a federated catalog for people to decide whether they want to read my data dictionary.
The idea here is that if the data are going to be in a 200-page, carefully constructed metadata format with the required field-specific types, and they have to be compliant with the standards of the Internet Engineering Task Force and ISO (International Standards Organization), the vocabularies get harder to work with. It is an engineering problem that is very similar to the integration problem, and what many people are realizing is that you can arrange the data hierarchically and get good results for small investments.
Here is an example. Many governments are putting raw data on the Web now. They are not just putting visualizations of the data online; they are putting the datasets themselves. From a political point of view, the two biggest motivations are enhanced transparency and the chance to inspire innovation. Promoting innovation can proceed in two different ways. First, the governments are hoping that some people will figure out how to make useful and innovative tools using the data. More important, especially for local governments, is that they pay many people to build Web sites and interactive applications that they cannot afford to build anymore.
For instance, a government agency may be able to pay one contractor to build a big system for a problem that is of high priority, but it may have 57 more priorities that it cannot afford to fund. The governments need someone to develop those applications, so if it makes the data available, at least some of those applications are done by other people. The development of such applications is out of the agency’s control, but if the work is getting done at no cost or for some small amount of money, it can start planning strategically for other priorities.
The United States and the United Kingdom are the two leading providers in terms of organizing their data. The United States has about 300,000 datasets, most of them geodata. If
we take out the geodata, there are probably 20,000 to 30,000 datasets that have meaningful raw data. The United Kingdom has less, but it probably has the best in the sense that, for example, you can get the street crime data for the entire country for a number of years. They are releasing very high-quality data down to a local level.
Many countries, including ours, are releasing scientifically interesting data, but you would have to work to find them. To use these data, combining them with other data, can be more important than just looking at them. Those entities releasing data include countries, nongovernmental organizations, cities, and other groups. For example, one of the Indian tribes in New York State has released much of its casino data.
There are groups all over the world working with these data. My group is one, and the Massachusetts Institute of Technology has a group working jointly with Southampton University, mostly on U.K. data. Many of these groups are in academia, but there are many nonacademic groups doing this kind of work. One of my suggestions is to think about data applications. You may have a large database, and if parts of it can be made available through an application, an interface, or through an API, people would be using the data in a sharable—and often an attributable—way.
Getting back to science issues such as attribution and credit, we have one of our government applications in the iTunes store. I know exactly how many people downloaded it yesterday, and how many people are still using an old version.
When some students and I were in China, we discovered that China was releasing much of its economic data, so we took China’s gross domestic product (GDP) data and the U.S. GDP data to do a comparison. To do that comparison, we needed to find the exchange rate. Luckily, there is a dataset from the Federal Reserve that has all of the exchange rates for the U.S. dollar weekly for the past 50 years or so. We got those data, we mashed them together, and we got a chart that looks like Figure 3-1:
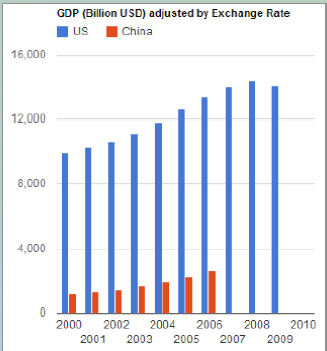
FIGURE 3-1 GDP Chart.
SOURCE: James Hendler
We also divided the GDP by the population (data we found in Wikipedia), so that we could click a button to go between total GDP and per capita GDP. The model was built in less than 8 hours, including the conversion of the data, the Web interface, and the visualization. That is a game-changer. It would completely change the way we work if we could get this down to a few minutes. When my group started working with this kind of technology several years ago, it would take us weeks to do this kind of work. Part of the improvement is because some of the technology was immature, part of it is because the tools are now available commercially, and another part is because there is now visualization technology freely available on the Web. Building visualizations is labor intensive, so by moving to simple visualizations, we can use visualizations much earlier. We are also able to link government data to social media.
One interesting question that has not really been part of the discussion that we have had in the scientific community is how to find data. For example, we noticed that most of the U.S. government data were about the states, and the metadata would represent the data as being about the 50 states, but very few of the sets actually covered all the states. Some states were missing. Some databases included American Samoa, Guam, Puerto Rico, et cetera, and the District of Columbia (which is not officially a state). In this case, there is a very loose definition of a state as opposed to a territory, and no one has much problem with that. But if a researcher wants to find datasets about Alaska, it can be erroneous to just assume that all of the datasets that say they cover the states will actually have Alaska data.
The other problem is that we cannot search for the keyword “Alaska” within the datasets. If there is a column that represents the states, it may be called Alaska, it may be called AK, it may be called state two, or it may be called S17:14B/X. The government has terabytes of data, so how do we find the data that are for Alaska, for example? We cannot just call for building a data warehouse and rationalizing the process, because these datasets are being released by different people in different agencies in different ways.
Thus, metadata becomes very important. Simple and easy-to-collect metadata can allow building faceted browsers and similar tools. It is a real research challenge, however, to determine the kind of metadata for real scientific data that are powerful enough to allow useful searches. With the government data, one problem is that all of the foreign databases are in their own languages, and it is hard for those of us who are English language speakers to figure out what is in the Chinese dataset, unless you hire a Chinese student. You cannot just use Google Translate and expect the result to be academically useful.
People are starting to consider integrating text search and data search. This is an application that one of my staff did, joined with Elsevier’s new SciVerse, which was featured on the U.S. Figure 3-2).
What this application does is that when we are doing a keyword search for scientific data, it is also looking for datasets that might be related to that same term. We are using very lightweight ontologies that mostly just use the keyword. We are working on making it better. We think it would be good when someone is looking for papers, the data in or about that paper were available, but also finding what is in the world’s data repositories that might be useful.
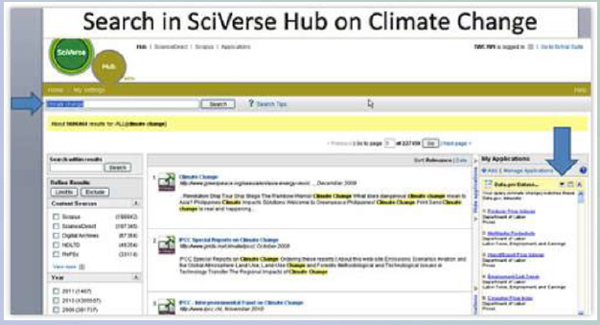
FIGURE 3-2 Search in SciVerse Hub on Climate Change.
Source: James Hendler
When we start thinking about data integration, using data, or searching for data, one thing that happens in the discovery space is that the data become part of the hypothesis formation. That means that looking at, visualizing, and exchanging the data cannot be an expensive add-on to a project. It has got to become a very key part of the standard scientific workflow, with appropriate tools to reduce the costs.
What is promising in this area? We have been looking at linked open data issues outside of science, and some of its promise has been explored (mostly within the ontology area). Genomics and astronomy are two fields where we have actually seen semantic Web technologies deployed, but many other science communities are still mostly thinking about their own data holdings, not about being part of a much larger data universe. It is interesting that when we talk to the Environmental Protection Agency, or to the National Oceanic and Atmospheric Administration, or similar agencies, they say that they are providing data to many communities, so they cannot easily use the ontologies of a particular one. Hence, we need to learn how to map between these approaches.
How do I know what is in a large data store? How do I know what is in a virtual observatory? I need services, metadata, and APIs. I also need to know the rules for using the data.
Other issues we need to think about are related to policies. If I take someone’s data from a paper, mix them with someone else’s data, and republish them, those people would probably like to get credit for the data being reused. Or if I do some work on your data that you do not like, you might want to rectify it. How do we do that?
National Technological Needs and Issues
Deborah Crawford
-Drexel University-
I am currently the vice provost for research at Drexel University, but I used to work at the National Science Foundation (NSF). I spent almost 20 years at NSF and was fairly significantly involved in the agency’s cyberinfrastructure activities. My experience within NSF and now at Drexel has provided me with an interesting perspective on what data-intensive science means in a research university that aspires to be more research intensive. The main message of my talk is that there have been many advances in data-intensive science in some fields, but we have massive amounts of work still to do if data-intensive science is to realize its full potential across all of the disciplines.
There have been many reports issued over the past decade that address the importance of data-intensive science and the role of information technology in advancing science. An important one is the Atkins report that Dr. Hey referred to earlier. In that report, Dan Atkins of the University of Michigan and his committee speak of revolutionizing the conduct of science and engineering research and education through cyberinfrastructure, and they examine democratizing science and engineering. Those were tremendously powerful statements in 2003, and they stimulated a great deal of excitement within the scientific community.
Since then, there have been a number of reports that have specifically addressed data-intensive science, several of which were released in the past couple of years. This is a quote from a joint workshop between the NSF and the U.K. Joint Information Systems Committee that was held in 2007: “The widespread availability of digital content creates opportunities for new kinds of research and scholarship that are qualitatively different from traditional ways of using academic publications and research data.”
What we have heard so far in this workshop was from those who I would describe as the visionaries and the trailblazers in science and engineering, representing those communities that were very motivated to take advantage of information technology in their work in order to advance their field of science.
I want to focus now on the long tail of science—the researchers in those fields where the immediate advantages of information technology and collaboration are not so readily apparent. There is tremendous opportunity in those communities, but we do not quite know how to realize those opportunities.
I would like to provide a snapshot of some surveys of researchers working in different communities. The main message is that computer-mediated knowledge discovery practices vary widely among scientific communities and among colleges and universities. In the United States and in the United Kingdom, for example, this is certainly true. There are some colleges and universities that know how to take advantage of their digital technologies and their digital capabilities, while there are others that simply are way behind the curve.
There are three fundamental issues that communities or universities must address: (1) What kinds of data and information are made open, at what stage in the research process, and
how? (2) To which groups of people are the data and information made available and on what terms or conditions? (3) Who develops and who has access to the tools and training to leverage the power of this discovery modality? These are fundamental questions and are key to the realization of data-intensive science.
I am going to talk about two case studies, one conducted in the United Kingdom by the Digital Curation Center, and one conducted within Yale University. Both point to some key features we need to address.
The study done by the Digital Curation Center in the United Kingdom was called “Open to All.” Its purpose was to understand how the principles of digitally enabled openness are translated into practice in a range of disciplines. These are the kinds of questions we need to be asking ourselves to determine the actions that we need to take to make sure that this modality of science is accessible and advantageous to everyone.
In this study, the authors characterized a research life-cycle model and asked different communities how they were using digital technologies within the context of that life cycle to further their science. They surveyed groups among six communities: chemistry, astronomy, image bioinformatics, clinical neuroimaging, language technology, and epidemiology. The range of responses within those different communities was fascinating to see. Surprisingly chemistry was the trailblazing community, at least among the individuals surveyed for this study. The chemists were using social networking tools, Open Notebook Science, wikis, and all the modalities of digital technologies to collaborate, and to collect, analyze, and publish their data. Everything was quite seamless from a community that I had not anticipated to be one of the trailblazing communities.
It was interesting to hear from the clinical neuroimaging group. They were so skeptical of the value of data-intensive science and open data sharing that in this study they refused to even disclose their identities as individuals. We therefore went from one extreme to the other, and we saw the range of practices and values within different scientific communities.
From their conversations with these communities, the group that conducted these case studies in the United Kingdom came up with a list of the perceived benefits of open data-intensive science. It included improving the efficiency of research and not duplicating previous efforts, sharing findings, sharing best practices, and increasing the quality of research and scholarly rigor. This last one was especially true for the members of the chemistry community who were surveyed. They saw a great opportunity in making available in blogs the day-to-day data that they collected—not just raw data, but derived data. They found tremendous benefits in making that open to the community to provide more scholarly rigor.
Among the other perceived benefits were enhancing the visibility of access to science, enabling researchers to ask new questions, and enhancing collaboration in community-building, which all of the groups surveyed agreed was a benefit. There was also a perceived benefit of increasing the economic and social impact of research about which the report was ambiguous. Although economic and social impacts each are treated separately as a perceived benefit, the sense was that the real value could not actually be measured. So, there was a question: Can we create economic value by making our data—essentially our intellectual property—much more openly accessible?
One of the perceived impediments was the lack of evidence of benefits. Researchers who felt this way were not motivated to make their data openly available. Other impediments were the lack of clear incentives to change and the values in academe not being consistent with open data sharing. For tenure and promotion and the drive to publish, the perceptions were that the only way to publish is in the open literature and that it is not in a researcher’s interest to share data, because someone else may use the data to advance farther than the one who shared the data. Competitiveness is a big issue.
The conflict with the culture of independence and competition is absolutely related to the lack of clear incentives to change. Other impediments were inadequate skills, a lack of time, and insufficient access to resources. Another big concern was how to train both the scientists who are practicing today and the scientists and engineers of the future. Researchers were also worried about how long it took them to prepare their data for open access, about quality, and about ethical, legal, and other restrictions to access. Those were big issues, especially in the life sciences community.
The report’s recommendations called for policies and practices for data managing and sharing. Communities are desperate for guidance on these issues. What have the trailblazers learned that can be shared and applied more broadly? Contributions to the research infrastructure should be valued more—that is something we have heard often. Training and professional development should be provided, and there should be an increased awareness of the potential of open business models. That is related to the attitude among researchers that their data are their intellectual property and they want to derive some value from that; thus, they feel that if they make their data openly available, they are giving that value away. Assessment and quality assurance mechanisms should be developed.
The study done by the Yale University Research Data Taskforce was conducted by an organization within Yale called the Office of Digital Assets and Infrastructure, which is an organization established to accrue the value over time of the digital assets that result from research and scholarly activity within the institution. The office conducted this study to determine the requirements and components of a coherent technical infrastructure, to provide service definitions, and to recommend comprehensive policies to support the life-cycle management of research data at Yale.
Given the discussion earlier about research libraries, this is interesting. Here is Yale University doing a survey that includes both the librarians and the information technology enterprise staff at the university to determine what their faculty base most needs for managing digital data. Very much like the other survey, this one found that data-sharing practices vary widely among the disciplines they surveyed. The researcher has the most at stake when determining what the data-sharing practices are. Yale is going to create an institutional repository to secure, store, and share large volumes of data.
There are certainly some institutional pioneers in this area, such as the Massachusetts Institute of Technology and Indiana University, and much of what I characterize as “slow followers.” A major concern is how an institution can afford to create and maintain infrastructure like this.
Yale University understands that it needs to develop and deliver research data curation services and tools to all of its interested parties, not only in science and engineering, but also in the humanities and in the arts. Recognizing the importance of persistent access and
preservation, Yale is trying to establish a digital-preservation program to determine what they need to preserve data in the long term and what value will be accrued from it.
Data ownership was a big issue in the report. Who actually owns data? Most faculty believed they owned their data. It will be important to develop clear policies regarding data ownership, especially in those communities where data sharing is not a standard practice or is not valued.
My main message, then, is that changes are in play at multiple scales: international, national, institutional, individual, and community. Most of the effort at this time has been focused on the community level, because communities have, for the most part, driven the transition to open data sharing in communities where clear value accrues from that sharing. Culture does not change because we desire it to change; it changes when our practices change. The bottom line is that all of this costs money. We will need to determine the appropriate balance between the cost of improved access to scientific opportunity and the benefits that accrue from it, and those are not easy questions to answer.
DISCUSSANT: Is it going to get to the point where IBM’s Watson knows the scientific literature and we do not ever need to do any work ourselves on the literature databases?
DR. HENDLER: No. For Jeopardy, Watson took many fairly high-end processors to beat Ken Jennings. For the medical version, they are trying to get that down to a more manageable number—maybe 20–50. They have already announced that the first issue they are pursuing with Watson is differential diagnosis, helping doctors access the medical literature. They designed the computer to be repurposable. This was the plan until 2009, but once they had the agreement that they were going to play Ken Jennings; they stopped doing anything except Jeopardy-related work. Now that they have won the game, they are back to the earlier focus. My guess is it will be 5 or 10 years until that becomes widely deployable.
DR. HEY: I am interested in the financial implications of sharing literature and data. Just as the music industry has to deal with what happens when someone makes a digital copy and distributes it for free, openly available data changes the business model. Similarly, with publications, we do not need printing presses anymore; we have the Web, and we can make digital copies and disseminate them. The big budgets that go to the publishers with their present business models perpetuate the system, but the system is being broken and it is up to universities and university libraries to respond. They should not be doing such deals with the publishers. They should be thinking about their role in the future. They should be thinking about how libraries can assist the research process of the institutions. I think the answer can be found in places like the iSchools. It is up to places like Drexel University and Rensselaer Polytechnic Institute to move forward, but how do we get that to take place? How do we catalyze the major research libraries and major research institutions to act together?
DR. CRAWFORD: There is nothing like a crisis to stimulate action. I know that at Drexel we are having conversations about the future of our libraries. We have an iSchool as well, so there is an opportunity there, because having shared interests in reinventing the model is essentially what it will take. There are trailblazer institutions, and it is a question of whether we can unite and define what the library of the future is in this digital world. But I absolutely agree that it is necessary. It is the future of the knowledge enterprise. I had a meeting with Drexel’s president last week in which we talked about the iSchool being the centerpiece of the twenty-first-century university and what that means in the context of research and scholarly activity.
DR. HENDLER: I would also mention projects like VIVO12, which is funded by the National Science Foundation. It has gotten a few schools involved, and it is growing. They have established the sharing of data and integrating that for the purpose of tracking researchers who agree to become a part of that system. Each library can maintain its own holdings. If I have a list of all my papers that I can get to my university in a way that they can curate, share, and archive, then it does not necessarily mean anymore that it must be in a particular book, in a particular building. We are seeing a lot of pressure in that direction, and it is promising that several of these big libraries are helping to lead that area.
__________________________
12 See: http://vivo.ufl.edu
I think the idea of a “catalog of catalogs,” the federated catalogs for not just the library holdings but also the people resources at a university—the individual papers and publications of the people, including who, what, when, where aspects—is a very powerful and interesting idea. Exactly how it will be funded remains to be seen, but once we get 100,000 or a million people accessing a page, the money can be found to maintain it. The problem is that until we reach that number, we cannot figure out how to monetize it. This is a situation in which scalable solutions that are experimented with and evolved may be better than trying to design a solution all in one step.
DR. ABELSON: When scientists say, “My data is my intellectual property,” I am not sure under what legal regime someone’s data is their intellectual property. Perhaps it is their trade secret or something similar. But there is tremendous confusion, and I think it is good to have some healthy criticism when people use that language. I would like to resist having people saying their data are their intellectual property.
DR. CRAWFORD: It is a misguided perception, but there is a lot of that.
DR. HENDLER: Dr. Abelson has been working on how we can make these policies more explicit through common-use licenses and the data more sharable, to actually tie the policies in a computable way onto the data and assets.
Sociocultural, Institutional, and Organizational Factors
Session Chair: Michael Lesk
-Rutgers University-
This page intentionally left blank.
Clifford Lynch
-Coalition for Networked Information-
Sociocultural factors in this area cover a very wide range of topics. In preparing these remarks, I deliberately wanted to avoid some of the very common themes that I expected to hear in this workshop and to focus on a few additional important, but less widely discussed, issues.
I will not thoroughly discuss, for example, the issue of incentives, which is obviously a major problem. The one point I will make in this regard, however, is that we have not talked much about the incentive to receive research funding in the first place, which is a fairly compelling one. If certain data-sharing behaviors are a condition of receiving research funds, and adherence to these behaviors is enforced as a condition of continuing eligibility for funding, this would be a real incentive.
It is interesting to see what is happening in some of the communities with foundations dedicated to the cure of specific, often rare, diseases. These foundations tend to be very focused on what they are doing, which is searching for treatments and cures. It is now common for them to insist that the scientists they fund should make their data openly available at least to other researchers within that community, on an immediate basis. This is the phenomenon of a funder who is focused on solving a problem and is not very interested in accommodating much long-term career-building by the participants. We will see more of this focus.
Let me move to the main issues I want to address. First, I want to talk about the scholarly literature and the realities of mining that literature, both as a research project in and of itself (i.e., to study what is going on in science) and as a pragmatic means of, for example, finding relevant earlier work. There is a longstanding history of researchers trying to extract hypotheses, insights, or facts by computational analysis of large bodies of scholarly literature, sometimes in conjunction with ancillary datasets of various kinds and sometimes not. What has happened with the way people compute on the Web is that we are coming to think about the scholarly record as an object of computation and to see that this computation can facilitate the discovery of new knowledge. It is important to understand that while the potential of this approach is great, the pragmatics of realizing it are phenomenally challenging.
It is clear that very large amounts of the scholarly literature are in one or another electronic format, some formats being far more hospitable to computation than others. They are often available in multiple formats, with the most computationally friendly ones only available internally on the publishers’ sites, since they are not designed to be directly read by human readers. But the licenses that universities typically enter into with publishers to use collections of online journals specifically forbid mass copying of the material from those journal collections, as someone would need to do to build up a corpus to compute upon. In fact, if a researcher tries to download a large number of journal articles from many of these databases, the librarian at that university will get a call from one of the publishers, because they have got mass-download detectors that notice a big spike of downloads. The publisher will demand that
the university librarian do something about this or it will shut off the university until it fixes this issue, at which point the researcher will get a phone call acquainting him or her with the terms and conditions under which the university has access.
Fixing this issue is not simply a matter of renegotiating one contract. A large university will have hundreds of such contracts, and thus far, publishers have not been very accommodating of this, with a few exceptions, mostly open-access publishers that do not require license agreements anyway. Of course, there are legitimate publisher concerns that need to be recognized in the renegotiations, but they are not simple to address. In short, there is a substantial pragmatic barrier to getting literature to mine. Note also that any comprehensive subject corpus, most of the time, needs to be drawn from a range of publishers, not just one.
Even if someone can get past the license issues, a second very real challenge to literature mining is the need to normalize data from a wide range of sources into a consistent format. One of the previous presenters talked about what it took to make sense out of one gene array dataset and the cleanup that was needed for that process. Imagine trying to deal with 50 or 60 different data sources, some of which vary depending on whether the journal issue is from 2003 or 1992, or is something from the 1960s that had to be converted into digital form. This is a very significant amount of work, particularly if it has to be done over and over again by different literature-mining groups.
A final subissue on barriers to mining the current scholarly literature base is related to copyright issues. Consider, for example, what happens if someone is performing a computational process on tens of thousands of copyrighted objects. Is the output a derivative work? If so, producing a derivative work from thousands of copyrights raises all sorts of questions. For example, if we did the same computation, but omitted 10 articles, does the fact that we included the 10 articles in our original computation make the computation a derivative of those 10 as well as of the other 9,990, even though they made no difference in the final product? There is not much legal certainty about anything in this area, as far as I know.
Let me address some practical barriers to mining the existing literature base. Today there is little consensus about the role of publishers in facilitating literature mining. We could imagine scenarios in which publishers, or some third party working with publishers, offer literature mining as a service for a database created just for that purpose. This might reduce some barriers, but it also raises concerns about the economics of text mining and the flexibility of the mining technology that it would be possible to apply to the literature base.
Focusing on the idea of computing on a corpus of scholarly literature and a body of experimental results, we also need to think about how to change the character and composition of this corpus to make it as valuable as possible for this kind of computational analysis. For example, it has often been observed that the published record, especially when it was read solely by human beings, has a strong bias toward positive results. People rarely get articles published in high-visibility venues saying, “I tried these six approaches, and none of them worked,” or “This compound seems to be good for nothing, it does not react with anything.” These are valuable contributions in a computational environment, and we need to think carefully, especially in a world full of robotic experimental equipment and large-scale screening systems, about how we get more of this negative information out there so that we can compute on it.
A closely related issue is the tremendous amount of data that industry produces and represents negative results or at least “prepositive results.” We should be able to disseminate and share these results. We need to sort through the notion of value in data, particularly (but far from exclusively) as it affects industry.
There is a school of thought that a database has value even if we invested in it and it proved to be a worthless investment, because if we keep it secret, we can force your competitors to waste time and money recreating that database themselves. Thus, it will consume resources that they could have spent elsewhere, and that will help us in competition. We need to reject that thinking, especially in environments such as the pharmaceutical arena, where we are struggling to contain costs and improve productive research. There is an enormous social cost to secrecy for negative advantage rather than for positive advantage. I do not know how we fix it other than by talking openly about it, but it does seem that we should look for ways to make as much negative data available as possible, in addition to positive data, to facilitate computation on the scholarly record.
One of the other challenges we face is how to make it easy for scientists and scholars to share data. We heard in some of the earlier presentations how difficult that can be. One key is simply giving scientists places to put materials that can be openly available to the world or that can be shared on a rather controlled basis. Anyone who has tried to do inter-institutional data sharing without making them public to the world—to share data just with groups of collaborators—knows how horrible that can be, because you do not have a common federated identity-management system across those institutions, so you have to resort to all kinds of clumsy ad hoc solutions.
One of the contributions that supporting organizations, such as information technology and libraries, can make is to help supply infrastructure that supports simple sharing. I see institutional repositories as key—but far from exclusive—players in that role. We see this from the discipline-related repositories that exist for specific genres of data. Those make sharing very easy, but we need to look for more mechanisms to facilitate that.
The last area I want to talk about is privacy. We face a big burden dealing with personally identifiable data of various kinds, the need in university settings to get involved with institutional review boards (IRBs), and the specific challenges of sharing and reusing data, which seems philosophically opposed to what most IRBs want to achieve. For personally identifiable data, they want to be as constraining and specific as possible. We need to recognize that we are getting into situations where we need to be able to reuse data more fluidly, and we are going to need some serious conversations about how this interacts with policies about privacy and informed consent.
There are several possible approaches in this regard. In many cases we want to anonymize the information, and we need to learn how to do a better job of that technically and to look for easy ways to share anonymized data with an agreement that the recipient will not attempt to de-anonymize it as a second line of defense. In other cases, personally identifiable information may be needed, particularly as we look at the interfaces between personal histories of various kinds and genomics or other kinds of population studies.
I wonder whether there are not some new ways to think about this issue. There is a tremendous interest in citizen science, as was mentioned earlier. There are many people who are interested in documenting their own behaviors, their own physical health. I was at a
meeting recently looking at developments in personal archiving, and it is clear there is a growing interest in collecting and sharing this kind of information among a sector of the population. We also have some very interesting developments in personal genotyping, for example, where communities of people are sharing the results that they get back from companies like 23andMe that do this sort of work.
I wonder if there is some way to connect these kinds of social changes into the process, or at least to think about whether we can do something better than the very constrained kinds of informed consent approaches that are in use today. We are significantly limited right now in doing the kinds of knowledge discovery that cross different kinds of databases, ranging from biological databases to databases about people and about behaviors and populations. I would suggest that is a major cultural and social issue that we need to be exploring in detail.
Paul Edwards
-University of Michigan-
The focus of my talk is on institutions and open data. I want to start with a success story. The meteorological data infrastructure is one of the oldest global infrastructures. It goes back to the 1850s. In the interwar period, a rudimentary, global data-sharing system emerged that used shortwave radio, telegraph, and many other technologies. At that time, meteorologists threw away almost all the data they received, because they had no way to use it. Once they got computers, they became able to process that data. Since the 1960s, the combination of a global observing system with many kinds of instruments linked to a global telecommunications system has been used. The system pours data from all over the world into computers that make forecasts and data products and then delivers those to national meteorological services, and it works well.
However, we do have a problem. Figure 3-3 is a graph of the famous global temperature curve as collected by different investigators since the nineteenth century.
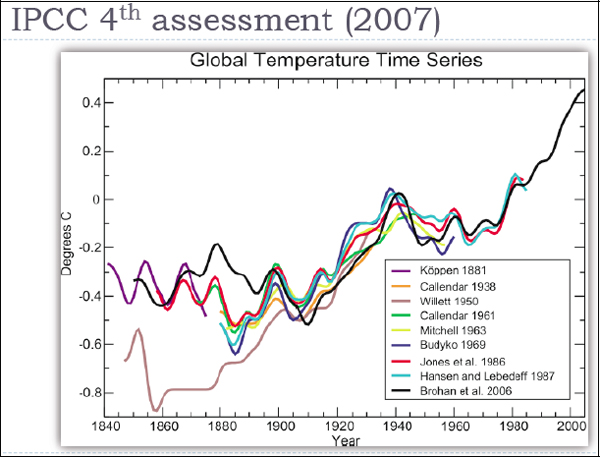
FIGURE 3-3 Global temperature time series.
SOURCE: Intergovernmental Panel on Climate Change
There are enormous differences in the temperatures recorded by the datasets from the nineteenth century and the early part of the twentieth century, especially before 1940. This is not much data by today’s standards, but the ability to crunch these numbers was not there before computers. Before 1963, almost all of these datasets included fewer than 450 stations. They did this because these were the stations known to be highly reliable. They were the stations that had been there over the entire period, and which had not made many changes to their locations, instrumentation, and so on. So their data were considered reliable.
The later temperature curves, put together in 1986, 2006, and more recently, use computer models to add in data from stations that were not trustworthy enough earlier—either because their records were incomplete or because they had gone through some changes, such as having been moved or engulfed by an urban environment, which made their record suspect. A recent project at the University of California, Berkeley is trawling the Web to find station records and add them into a global dataset containing more than 30,000 stations. The quality of that dataset is open to question, but it is one of these Web-sourcing ideas that we have heard a lot about today in other presentations.
This story offers an example of an extremely open data system that has existed for more than 150 years. One of the reasons the weather data system works so well is that in the early days of weather data reporting, data were freely exchanged, because they had no economic value. We could not do anything with them, because it was difficult to make much use of them in many ways, so the international telegraph system began to carry weather data at no cost. Since sharing these data was advantageous to everyone, they did it freely.
In the 1990s, that started to change. Some countries are charging for weather data products in an effort to recoup the costs of their observing systems. Figure 3-4 shows datasets that the European Center for Medium Range Weather Forecasts is trying to recuperate now, in order to add them into the global climate record and improve the quality of that record. There are many of these datasets.
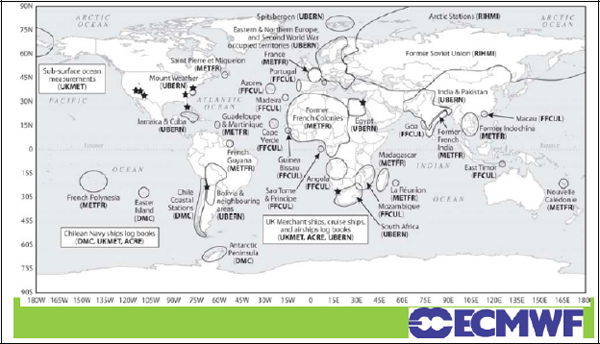
FIGURE 3-4 Focus on pre-1957 meteorological data in sensitive regions.
SOURCE: European Centre for Medium-Range Weather Forecasts
The point of this story is partly that the effort to build this digital climate data record has already been going on for almost 30 years, and it is still under way. This is difficult work, even though the relative size of these datasets is small compared to the size of some of the databases we have now. The results of the global collection effort are good, however. Figure 3-5 presents four independent analyses of the global temperature record. They converge much better—even in the nineteenth century—than the versions created earlier.
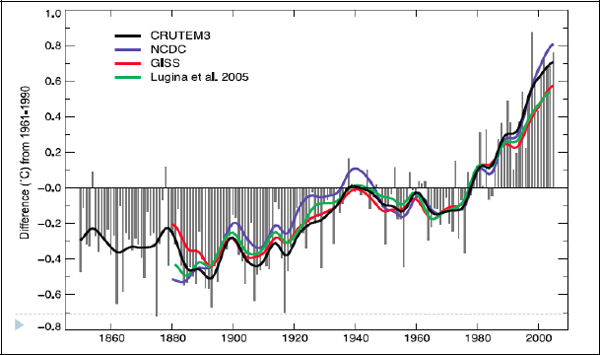
FIGURE 3-5 IPCC AR4 (2007).
SOURCE: Intergovernmental Panel on Climate Change
I want to focus now on data problems. Some of these are institutional and some of them are not. Most of us still work at a local workplace, and almost all of us work for some kind of hierarchical organization that pays our salary. This means that the issues that concern us locally about the institution that we work for will almost always trump concerns about things that are remote. This is true for various psychological and institutional reasons. Studies of face-to-face interaction show, for instance, that it generally works better and faster than interaction at a distance. There is also a trust aspect. The people you know are the people you are more likely to share with, and whose data you are more likely to trust.
The story about meteorology and climate science has some temporal issues that have not come up much in this discussion, but are nonetheless very important. It is easy to think that the data we are trying to federate and collect will be there forever, but of course they will not. The audience may be familiar with the story when Mars data were lost, because the software with which data tapes were made had been lost. Digital information is quite fragile, because the strings of bits cannot be interpreted without the code that created them—and that code is dependent on other code, such as operating systems. Losing data by losing the software and hardware context is the kind of problem that could happen again and that already does happen quite often.
Metadata is another issue that appears in the climate and weather data story. It is not obvious to us what people in the future will need to know about the data we collect now—and
we cannot know that with any certainty. We may not collect what they will need at all, or what we collect may not be sufficient.
Finally, there is a figurative dimension to distance, which is the distance between disciplines. The great promise of open data is that we can work on problems that have never been studied before, because they required collaborations among different fields. That is also a potential failing, however, because if an ecologist wants to work with data from, for example, an atmospheric physicist, the ecologist might be willing to accept that person’s data at face value. After all, what does the ecologist know about atmospheric physics? Yet those data may be defective in various ways. We may need to make an extra effort to know more about those data and the problems they may have.
This raises the problem of trust. The further away that some user is from the source of the data, the more possible it is for the members of that user community to find it completely unpersuasive. Hence, sharing data will not automatically facilitate communication and understanding.
The world we think we are in, and want to be in, is now thoroughly networked, with easy sharing of data and information among all scientists and disciplines. But the world that we are actually in is often much more hierarchical and stove-piped. In science, the hierarchy begins with the subdisciplines, rises to the disciplines, and rises again to larger traditions of scientific practice (Figures 3-6 and 3-7). Studying real-world problems, such as climate change, involves many disciplines, but the place where your scientific reputation is made is at the much lower level of your subdiscipline.
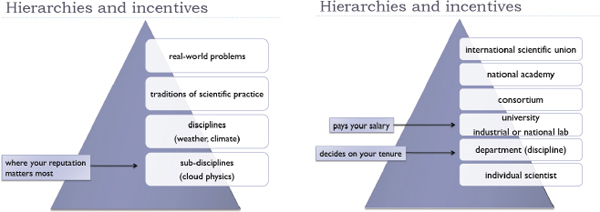
FIGURES 3-6 and 3-7 Hierarchies and incentives.
SOURCE: Paul Edwards
The same observation is true for our institutions. Academic scientists are individual, they work in a department, and the department is part of a university. There might be levels above that all the way up to international professional societies—but the place that pays the salary is the university, and the place where the scientists’ reputation is evaluated is their department. Therefore, these relatively local entities can often trump larger collaborations, networks, virtual organizations, and so on.
One of the phenomena we see in the formation of many infrastructures is a phase in which we build gateways—systems that allow us to move information, for example, from one place to another without having to worry about the particular features of every infrastructure involved in the transfer. The meteorological information infrastructure, the World Weather Watch of the U.N. World Meteorological Organization, is a great example. Its bottom level consists of more than 150 national meteorological services. Each one contributes its observational data to the global observing system. The global communication system sends them to central processing centers, where data from the entire planet are processed and then returned to the national weather services.
This is great for understanding and forecasting the weather, but once we start confronting larger problems, such as climate (which involves many Earth systems besides the atmosphere), the World Weather Watch is no longer enough. Therefore, we get another, higher level of institution. For example, the Global Earth Observing System is a collection of systems, and each of those systems is, in fact, a collection of other systems. What I see when I look at the world of scientific institutions is a constant need to climb up a level to try to get an understanding of the general area that is being studied.
I have done some work with people in the Global Organization of Earth System Science Portals (GO-ESSP). These are people who build tools like the Earth System Grid. They are concerned about adequate support and coherence. The problem they face is that their activities for GO-ESSP are, in general, unfunded. They feel strongly that it is necessary, because they want to do things that link the various portals and make them into a more coherent, interoperating system—but nobody is paying them to do that. Thus, again, the institutional level of concern and the one at which they actually want to work are different and conflicting.
My late colleague, Susan Leigh Star, coined this phrase: “Metadata are nobody’s job.” Everybody wants good metadata, but nobody knows who is supposed to create them. We would like to push it off onto the scientists, but they do not want to do it. They are moving on to the next project after they have finished their datasets, and they do not want to go back and document what they have done. Perhaps there is a class of data managers who can handle it, or perhaps it should be the “crowd.” Maybe somehow younger scientists will do it. What we often see in labs is that this work is pushed down onto the graduate students. Maybe social scientists have the magic answer. I am a social scientist, and, as far as I know, we do not.
My research group has been doing some ethnographic work with environmental scientists, and we have observed that there is an obsession with metadata as a product. In other words, metadata are conceived as a kind of library catalog that will describe all the data that someone has. The reality of data sharing, however, is quite often that metadata exist mainly as an ephemeral process. Many people say that the most important issue when dealing with data collected by someone else is to talk to that person. “I need to know more about what you did, how you collected this data, what it means, what the formats are”—there is an endless list of questions, and it is impossible that all of those will ever be answered by a catalog-style metadata product. One lesson, therefore, is that when someone is conceiving metadata, creating channels for communication among researchers is at least as important as creating the ultimate catalog that we would all like to have.
Finally, I am going to talk about data and software. Data cannot be read without software. An even more important point is that many things that we call data now are actually the product of software. Climate model output is an example, as are instrument data after they have been collected and processed through some sort of data model to be put into the final dataset that is used in a general publication. Which of those things are we talking about? If it is the latter, we need to know exactly how it was processed.
For the last 15 years or so, the journal Money, Credit, and Banking has required authors to submit both their code and their data so that their results, in principle, could be replicated. In 2006, researchers found that of 150 articles only 58 actually had some data or a code, and of those, for only 15 could the results be replicated with the code and data the authors provided. The journal editors concluded that there needs to be a stricter system for people who are submitting their code and data. But my question is, how much is it worth? What they did not do was attempt to communicate with the authors to find the missing information.
My last points are related to the problem of the academic reputation system. This is the way the system works now: Scientists have some data sources, they use them to do research, they publish the results, get citations, and that builds their reputations. Somewhere in there are services, and that is where data sharing and software, as well as the writing of scientific software, both sit.
My colleague, James Howison at Carnegie Mellon University, has been thinking about how we might design a reputation system that would work for scientific software writing. Many scientists write little bits of code, or sometimes much more elaborate code. That is part of their job, but it is not their main job. Yet once they have written the software, they get involved in the problem that if their code is going to stay active, they have to maintain it. There is an ecology of software, and as the operating systems and associated software libraries change, this piece of scientific software will eventually stop working. Therefore, the scientist has to keep updating it, to keep modernizing it, if people are going to keep using it. Sometimes this turns into a sideline, and the scientist actually becomes a software developer. More often, however, what happens is that the code eventually just dies, and then there is the problem of replication. How can the research that used it be replicated?
I collect a lot of data, and then what happens to it? Sharing the data requires work from me. Writing out all the metadata is work. One of the best examples we have of a well-functioning collection of large datasets is the collection for CMIP-5, the Coupled Model Intercomparison Project, that my colleague from Unidata spoke about earlier. It is working well because there is a major incentive, which is that if someone wants to have data in the Intergovernmental Panel on Climate Change (IPCC) report, that is the only way to do it. The scientist must fill out the metadata questionnaire. How long does filling out the metadata questionnaire take? Up to 30 days for each model, because there are many runs and the questionnaire has to be filled out separately for each run.
The point is that data sharing is work, and it always will be work. Thus, one of our big issues is to find ways to pay people for that and then also to include it in reputation systems so that they can get credit for it.
There is a conflict between the mandates of institutions like the IPCC, the National Science Foundation, and the National Institutes of Health that want us to publish and share data, but there are few career incentives for sharing data. The career incentives are for the
results that come from the data. That is true of building software. It is also true of sharing data. Moreover, the problems that software developers face are also there for people who want to share data, though perhaps to a lesser degree. Data have to be maintained, which is why the issue of curation is a critical one. Who will maintain the data?
There are many possible solutions, however, and some of them will work for some areas. It is important to realize that there is no one thing called data, and that the differences among scientific fields are enormous. What works in one area may not work in another. Maybe we are talking about a virtual organization, or maybe it is a crowd-sourcing project. Among the people I have been working with, we have often heard Wikipedia, Linux, Mozilla, and Apache held up as models for the future: “This is what we will do. Open-source software works. Open-access models work.” I do not think so. For many fields, there is going to be a problem, because there are not enough people with the right skills.
Metadata standards may help, but someone needs carrots or sticks to get that process to happen. I have heard many people say that the young people will do it. They assume the young people will know how to do it, but if we do not know how to do it, why is it that they will know? They need to be trained. Studies of young people using even simple things like Google Search show that most of them do not know how to use Google Search in an effective and efficient way, and that is equally true of young scientists. Some of them will be very good, but most of them will not, and it might be that we actually need new paradigms of education.
I love the idea of science informatics. Perhaps it will come from an iSchool, but the iSchools we have are not set up for this yet. Perhaps it will come from science departments, but most of the science departments I know are not set up to do that either. Science informatics is not computer science, and most computer science departments are not interested in this problem at all except for their own kinds of models and data. Therefore, it will probably be some combination of computer science, iSchools, and domain science. And the combination may need to be different for different domains and problems.
DISCUSSANT: There was a discussion about how difficult it is to get large amounts of text for literature analysis, but people have been trying to do that since the 1970s, long before copyrights became a big issue, and there are large bodies of text that are not available. Do you think the technology works even if someone could get the text, because I do not see that?
DR. LYNCH: Parts of the technology work, and in fact there are some successes to which we can point. The notion that someone has a computer that reads the literature on a certain subject and understands it in a deep way is fairly farfetched. One of the things that Google has suggested, however, is that when someone gets enough data, he or she can do some interesting work through statistical correlations.
DISCUSSANT: There is a general problem about where the credit lies, where the value lies, and who is responsible. But the main question is: What is the business model? Who is responsible among universities for ensuring that there is a cyberinfrastructure and that appropriate credit is given?
DR. LYNCH: I do not think there are simple answers. I do suspect that even in a period where resources are very tight, if we look carefully at most of the cyberinfrastructure investment, it is largely cost effective. This investment should be making the overall research enterprise more effective and providing some real leverage. I think those are the parts that will probably move ahead.
One of the issues in the area of data that we need to be very mindful of is that data preservation per se is almost a pure overhead activity, especially when we get past the first couple of years when people might want to reproduce new results. The activity with the payoff is reuse. That, not preservation, leads to new discovery, so it is important to be focused on facilitating reuse and not just looking for the longest possible list of data that we need to preserve. There are clearly materials we should preserve even if there are not obvious reuses for them right now, but we need to be judicious in that category and compete for the resources.
DISCUSSANT: It used to be that the reason an institution would have a repository was because there were physical objects that made sense to keep locally at an institution. Individuals did not have a chance to use an institutional repository unless they were at an institution.
Consider, for example, a person’s personal photography collection. Perhaps this person is in a bizarre hybrid situation, where he or she tries different services and ends up with a few hard drives, where many copies of this material are saved. There should be a different solution, however, where that person could take advantage of a big institution. Long-term trust is the main problem. He or she does not want those photographs to be lost. What if those companies go out of business? That person would still want everything on a personal hard drive.
If we move from digital photographs to scholarly output, the issues are the same. Why is Harvard University, for example, going to have a repository for everybody’s information, when it could just have some enterprise solution that gave everything to Microsoft? What is the balance between an institution and the individual?
DR. LYNCH: I am not sure that I parse the landscape the same way you do. I would say that the very critical issue about an institutional repository is that an institution is taking
responsibility for long-term preservation. Institutions and corporations, unlike people, may live forever, or at least indefinitely, and have processes that allow them to operate on much longer timescales than individuals.
DISCUSSANT: If a university is paying money to another institution, is that still an institutional repository?
DR. LYNCH: Yes. The way I view this kind of arrangement is that if the institution identifies the data and takes responsibility for their preservation, then it is making a much longer commitment than an individual could do, perhaps setting up a trust fund or some other mechanism that outlives the individual. Operationally, we may see many universities over time contracting the preservation and curation of specific kinds of data to other universities, or to other for-profit or not-for-profit archives. We may also see the emergence of more consortia, as well as straight-out contracting. The locus of ultimate responsibility is the key thing here.
DISCUSSANT: Does that mean that the universities can act like the individual researcher?
DR. LYNCH: Except that presumably the universities have thought through issues like multisourcing and what happens when a supplier goes broke, how to back up the data, how to audit contractors and move data from one contractor to the next, and other similar issues. The other point I want to make is that we often confuse distribution systems with preservations systems. Flickr is a great example. People put their pictures up on Flickr, and I think of that as a means of distributing them, making them available, and building some social interaction around them. However, there are people who believe this is a preservation venue, and I would urge them not to think that. If you really want your pictures, by all means put them up on Flickr or anywhere else, but keep a set for yourself offsite and make lots of copies.
DR. LESK: In terms of how we get somebody to do the preservation and the data curation, assuming that scientific data is indeed worth having—I wish we had more data on that point—the data are either a public good or a private good. If they are a private good, it means that the owner gets credit for having them, presumably by getting tenure. However, that means waiting for universities to give tenure for collecting data instead of for research results or for the papers interpreting the data. Coming from a university where departments complain that the administration is too slow, I am not optimistic about that.
The other option is that the data are a public good. Dr. Berman made a comment earlier about unfunded mandates. I have the feeling that if the National Science Foundation (NSF) imposed a data management mandate—we are not going to get any more pages in the proposal, but we are going to get 5 percent more money as the upper limit of what we can ask for—I think the world would be completely different. Obviously there would have to be a requirement that the 5 percent be spent on the data curation, but it would completely change the attitude about how the research community does this.
DR. SPENGLER: It is important to point out that the NSF asks for a data management plan. It does not ask a scientist to preserve the data. It does not ask that there always be copies.
Even more important, if we read all the details, we do say that we are willing to have the scientist put requests in the proposed budget for curation purposes. We say that we are willing to support it.
DISCUSSANT: If scientists decide that their data are worth preserving, is there some sort of mechanism for them to appeal that?
DR. SPENGLER: Some of my colleagues and some of the directorates had planned to include in the NSF data management policy that a grantee has to keep the data for 5 years beyond the award period. That got taken out.
DISCUSSANT: Is there a current plan for that?
DR. SPENGLER: No. Some disciplines have mechanisms set up for that, however. For example, if we look at some of the programs within the NSF, we will see that they do not normally say that the NSF wants data management plans. However, researchers will get in trouble if they do not have the data deposited in the designated database when it is time for their annual report or project renewal, because it will not be approved. Hence, some places have thought long term about it, but it requires a very large commitment by a given domain. The social sciences have stepped up in their resources, as has astronomy. Many disciplines and organizations have made more of a commitment, but questions about the long-term cost are still open.
DR. LYNCH: There are some interesting developments at the institutional level as well. For example, earlier this year Princeton University offered a storage service for their researchers that allows them to pay in advance for storage at some rate per terabyte, allegedly for eternity, or as long as Princeton will exist. The university made some projections about the cost of technology, the refresh rate, and the return on capital for the prefunding. We can argue about whether the numbers are right, but it is interesting to see someone step forward with a model like that, and I think we will probably see some more of it.
Policy and Legal Factors
Session Chair: Michael Carroll
-Washington College of Law, American University-
There was a comment made earlier about how researchers think of themselves as hunter-gatherers. This goes to the philosopher John Locke’s labor theory: I did it, and therefore I own it, and I have a property right in it.
Lawyers think of property in different ways, but I want to highlight the fact that there is a “there” there, even though the words “intellectual property” are being imprecisely used in this context. The way that an intellectual property lawyer might think about ownership and rights over the data is in terms of the rights over the first copy of the zeroes and ones. In a way, this is almost like tangible property in the sense that when there is only one copy, these zeroes and ones might be owned insofar as I have the medium on which these zeroes and ones sit, and you cannot have it unless I say so. This is a claim of ownership over a form of property.
Intellectual property rights are a layer above the zeroes and ones. They are entirely intangible, and these are rights that tell us what uses are acceptable. May we access the zeroes and ones? May we copy them? May we change the order of the zeroes and ones? These are use regulations.
Think about intellectual property law as access and use regulations that apply to the zeroes and ones rather than being embedded directly in the zeroes and ones. Once we make this conceptual separation, when we think about the policy issues, we are thinking about the regulations over data acquisition, data reuse, and the like.
This page intentionally left blank.
Michael Madison
-University of Pittsburgh School of Law-
I want to highlight some central legal questions in this area. In particular, I am going to focus on intellectual property (IP) law questions. There are some interesting and important privacy law issues as well, but I am not going to discuss those.
Figure 3-8 highlights in a very simplified, stylized way how I organize my thoughts on this topic.
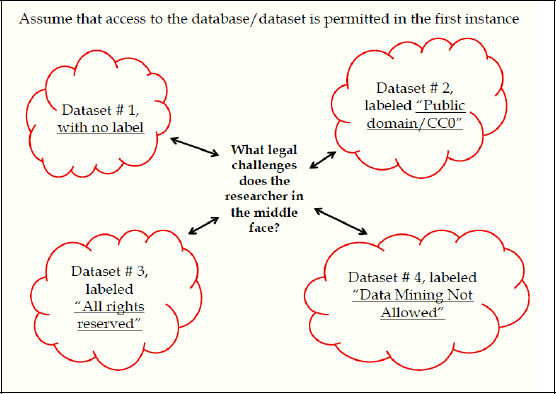
FIGURE 3-8 Intellectual property challenges to data access.
SOURCE: Michael Madison
There are four clouds of different datasets, and a researcher in the middle who is extracting—or recombining or using—data from each of these sources. There is a different kind of a label—or, in one case at least, no label whatsoever—attached to each of those datasets, specifying terms and conditions under which data from that source might be used, recombined, shared, and so forth. The question is, What does the person in the middle do, and how does the legal system frame that set of questions?
In the upper left corner, there is no label whatsoever, just a pot of data. The lower left is “all rights reserved,” or the standard copyright-style notice that would be attached to any kind of copyrighted work and that is often attached to materials that are not actually copyrighted, often out of ignorance or sometimes in an attempt to deliberately claim rights in things that cannot be claimed. It is a default copyright label.
On the lower right side is a context-specific restriction that is phrased “data mining not allowed.” This is to illustrate the idea that people who collect data or create copyrighted works increasingly have the power to custom-design labels or notices that they attach to their works before they send their works out into the world, and they expect both the world to respect that label and the legal system to enforce it somehow.
In the upper right corner is a label of a type that is increasingly common, used to specify or declare that the material inside the pot is in the public domain or, if it was not in the public domain in the first place but in fact was copyrighted to begin with, has now been dedicated to the public domain by some kind of affirmative notice or label. It is the CC0 label, which is a form of Creative Commons license that basically declares that the material covered by the license was copyrighted, but the author of the material, or the owner of the copyrighted material, is affirmatively dedicating that content to the public domain. One of the questions I will address is the extent to which that kind of labeling or notice device is actually effective.
There are three general categories I want to cover, with questions in each. One is the default legal status of data in these environments, both with respect to the underlying data and also with respect to the adjacent information, formats, coding, metadata, and literature that arise from these things. The second one pertains to contracts, licenses, notices, restrictive notices, and enabling notices. The third category is what the legal system has to say about the problem of managing a commons or managing a community of shared resources.
I will begin with some default copyright rules within the intellectual property system—just what the rules are. First, data are treated in copyright law as facts and therefore cannot be copyrighted. They are by design in the public domain. This is one of the few black-letter legal rules that is easy to state. This would include experimental results, observations, sensor readings, and the like. So for people who assert that they own their data, if we were to apply a formal legal model to this, they cannot own the individual items in the dataset. They might, however, be able to claim a copyright in the compilation of the data or the database as a whole.
Regarding collections of data and databases, copyright law gives us this phrase “selection, coordination, or arrangement.” If the selection, coordination, or arrangement of the data, or if the selection, coordination, or arrangement of practically anything, is original—that is, it has been touched by human judgment or creativity in some modest way—then there is at least the spark of a copyright that attaches to that collection. Not to the individual items, but to the collection.
If it is a collection of facts or a dataset that has been compiled in a way that has been organized by human judgment, then copyright may well apply, although the scope of the copyright may be relatively narrow. Copyright comes in different widths. There is narrower copyright for fact-oriented materials and broader copyright for classically entertainment-oriented things such as novels, films, and popular music.
When I say a “narrow” copyright, or what copyright lawyers would call a “thin” copyright, I mean that infringement of that kind of copyright will generally consist of verbatim or wholesale copying of the contents of the work. Copyright law also has this idea of substantially similar copying that can be infringement, but it generally would not apply to a thin copyright in a compilation of facts.
Codes, formats, metadata, data structures, or anything that involves any kind of interpretation or characterization of the data up to and including actual scholarly papers or scholarly output is going to be eligible for copyright protection. The more human judgment, human creativity, or human originality that goes into the production of that material, the stronger the copyright can be. Thus, a scientist’s scholarly papers, output, and the literature itself will be copyright-protected in a much more robust way than the underlying datasets. In short, there is a spectrum of copyright protection that starts at a relatively early stage, immediately after the creation or identification of the individual data point.
I will note two other points about default rules. First, if we are talking about a source in the European Union (E.U.), the European Union has the 1996 Database Directive, which operates in Europe in parallel with copyright rights. The Database Directive has some overlap with copyright and some distinct character. The most important feature of the Database Directive is that it prohibits extraction and reuse of a substantial part of the database. Therefore, picking out bits and pieces of an E.U. database may not violate the Database Directive, but if the Directive applies, then using the database wholesale may violate the Database Directive (in the manner that the Directive has been incorporated into national law) as well as local European copyright law, and perhaps other laws.
Copyright law in the United States is subject to fair use. Fair use refers to the use of particular portions of copyrighted works for critical purposes: teaching, research, scholarship, study, and so forth. Fair use in this area is not an especially reliable or useful tool, although it is potentially quite important in several other respects.
One possible application of fair use is to defend the copying of a copyrighted work at an intermediate stage on the way to producing something else. For example, U.S. case law in one instance focuses on intermediate copying for reverse engineering an underlying software product. If someone wants to build an interoperable or complementary software product—the case law generally deals with video games—then it may be permissible to make an intermediate copy of a copyrighted work to extract underlying public domain ideas and then use those public domain ideas to build a complementary work.
A related way in which fair use might help would be related to building and using tools in the database area. If someone wants to be able to connect to an Application Programming Interface to make use of a particular tool, fair use might be a way, using this case law, to exploit that particular possibility.
Second, turning to licenses and contracts, it is important to note that all the default rules, with some exceptions, can be changed by voluntary agreement, by a license or a contract, or a license bundled into a contract. There is some controversy about the extent to which fair use can be negotiated away in a license or a contract. The better view is that fair use is a mandatory legal requirement, but there is some dialogue about that in the world of law practice.
The most important point about contracts and licenses is that there is a distinction between a unilateral notice, or a license as a concept of property law, and a contract, which is something that you would voluntarily assent to. In practice, both in the software world generally and in this space of datasets, there is much ambiguity and confusion about the legal status of particular terms and conditions that come bundled with these collections of data.
A contract is something that typically requires that someone agrees to it, for example, by clicking on the “I Agree” or “I Assent” or “I Acknowledge” button. A license is a property term that grants a person permission to do things and that might require him or her to do some things with the copyrighted material, but a license does not necessarily require that the person agree to something in return. That is the first area that makes licenses tricky.
The second area that makes licenses unclear, besides the fact that a person is not necessarily required to agree to them, is that someone might not necessarily see them. They might come in machine-readable form, but a researcher might not have a human-readable equivalent. Or a researcher might have a human-readable version of a license, but might not have a machine-readable or automated discovery–readable version of the license. Therefore, it might not be apparent enough for a researcher actually to understand that there are some terms and conditions to deal with there.
A third complication is that particular terms and conditions, as illustrated on Figure 3-8, might be different, incompatible, or at least not perfectly aligned from one dataset to another. Thus, if a scientist is doing a large-scale project that entails extracting data or using data from many different sources, that scientist might end up with multiple legal obligations and rights for different chunks of the data. That has been a big problem.
Turning to open-source software, much of the discussion about such software focuses on the General Public License as the standard license for open-source software products. There are many licenses, however, that are open source in their style and their content, and those open-source licenses may not all say precisely the same thing. Therefore, we must be careful not only to read the particular open-source license that we are dealing with but also to be aware that if the source code comes from different sources, with different open-source licenses, we may end up with multiple obligations bundled in a single product.
The next issue about licenses is a bit more optimistic. In the IP world, people tend to think of licenses as a way to prevent people from doing certain things: I will share my content with you, so long as you do not steal it, cheat on it, share it, forward it, and the like. The underlying concept of license–as-permission can be enabling, however. Increasingly over the last decade, there has been innovation in the law in designing licenses that authorize people to do things that you want them to do. Creative Commons licenses are the paradigmatic example of this. Open-source licenses are often used in this way as well. The fact that lawyers have made innovations like Creative Commons and open-source licenses demonstrates the possibility that others could create all kinds of additional new mechanisms for enabling what people can do, alongside ways for restricting what people can do.
Both for the restrictive licenses and enabling licenses, the law is not fully clear regarding which, if any, of this is actually enforceable. The Creative Commons and open-source licenses are put out there and characterized and conceptualized within their respective communities as licenses rather than contracts. The idea behind them is that they are enforceable because the license is attached to the underlying copyrighted object and travels with the underlying copyrighted object. Anybody who encounters the underlying copyrighted object requires access to it along with the accompanying obligation, which is, I think, an interesting and useful concept.
That concept has not genuinely been tested on its merits in the courts, and the problem with it is that there is a dark underside that some people have identified. That is, it is one thing
to attach a happy public domain style or “please use this and share it” kind of style notice, but what is the difference, legally speaking, between attaching a “please go and share this” notice and attaching a “please do not do anything I do not like with this” notice? Suppose I were to share that dataset with you and attach a notice that said you may not do anything: You may not publish your results, you may not criticize my method, and you may not combine or collaborate with anybody else. Legally speaking, it is difficult to distinguish the restrictive notice from the enabling notice, and the legal system has not really come to terms with how to justify one and not the other.
The last point in this area has to do with the character of the enforcement of the license. So long as these licenses are designed by private parties and are attached to copyrighted works or databases by private parties, then enforcement lies with the copyright owner or with the holder of the database or whoever attached the notice. That creates obvious problems in inconsistency and conflicts. One solution has to do with the identity of the enforcer. Having either a government agent or some other third-party neutral in the middle, as custodian of the legal rights for purposes of monitoring and enforcement, is something to consider when designing an enforceable scheme that would accomplish the actual goals of the enterprise.
I am now going to move to the last section, which concerns management of data collected from a variety of sources. Assume that a researcher has identified and overcome various licensing hurdles and IP-rights hurdles associated with the underlying data. The researcher has a project that has data from various sources. Management of this database is often understood primarily by policy choices, but there is also an important role here for law. My research at the University of Pittsburgh focuses on commons communities, with commons understood as a structured or designed pool of resources or a sharing environment. Here is the question: If we have a choice between maintaining public domain status of the underlying content or understanding the mechanics of a particular license or contractual regime, are there potential upsides to a license or contractual regime that might make that more attractive than a public domain alternative?
Consider the questions in Box 3-1:
Managing the results of data collection
Forward-looking issues related to governing a data commons:
- Data/dataset integrity.
- Translation and interoperability of data from different sources, in different formats: designing and enforcing standards; managing and maintaining data consistency. (Ensuring PD status of data may be inadequate to deal with this challenge.)
- Who has access to the new collection of data, and for what purposes?
- What are participants’ duties and rights regarding standardization and data consistency, and re-sharing, re-combining, re-using data?
- How is compliance monitored and enforced?
- When do those duties and rights pass to downstream parties who did not obtain the data in the first place?
- Compare the costs and benefits of government-sponsored enforcement with those of private enforcement via licenses and contracts, and with those of informal/community enforcement.
BOX 3-1
SOURCE: Michael Madison
Box 3-1 identifies governance topics that relate to a community of researchers or to a particular researcher, using data from various resources that might be bundled in a license instrument and made enforceable as part of that license. It is important to identify both the “what”—that is, where things are coming from in terms of the underlying data—and the “who.” We want to decide who can participate in this particular enterprise and which researchers are eligible. We also want to know what their obligations will be, what their duties will be relative to each other and relative to the data, and also what the enforcement mechanism will be. Who has authority to enforce breaches of the underlying protocols? What are the carrots and sticks associated with ensuring compliance, monitoring performance, and making sure that the data is preserved in its integrity over time and combined in a way that is suitable to the underlying license obligations?
This last point is particularly important, and the license instrument can be especially useful for managing downstream obligations. When the questions are how these duties will pass to the next generation of researchers and who you may or may not know about in the beginning, then having a well-designed license instrument might be able to help you specify who those people are and what their obligations will be. That is the mechanics of an open-source software license, but the concept can be ported into this area as well.
In closing, I will quickly respond to the claim that the legal system might provide clarity and a deterministic foundation on which to build policy analysis or other analysis. That is not the case. There is no one-size-fits-all solution that the legal system can offer to these problems. What the Creative Commons example and the open-source software community have shown with its licensing experience is that the set of tools available to manage these collections is much more diverse and complex than it was 10 or 15 years ago.
Knowledge Discovery in Open Networked Environments: Some Policy Issues
Gregory A. Jackson
-EDUCAUSE-
The Coleman Report, released in 1966, was one of the first big studies of American elementary and secondary education, and especially of equality therein. Some years after the initial report, I was working in a group that was reanalyzing data from that report. We had one of what we were told were two remaining copies of a dataset that had been collected as part of that study but had never been used. It was a 12-inch reel of half-inch magnetic tape containing the Coleman “Principals” data, which derived from a survey of principals in high schools and elementary schools.
The first challenge was to decipher the tape itself, which meant trying every possible combination of labeling protocol, track count, and parity bit until one of them yielded data rather than gibberish. Once we did that—the tape turned out to be seven track, even parity, unlabeled—the next challenge was to make sure the codebooks (which gave the data layout—the schema, in modern terms—but not the original questions or choice sets) matched what was on the tape. By the time we did all that, we had decided that the Principals data were not that relevant to our work, and so we put the analysis and the tape aside, and eventually the tape was thrown away.
Unfortunately, apparently what I had had was actually the last remaining copy of the Principals data. The other so-called original copy had been discarded on the assumption that our research group would keep the tape. That illustrates what can happen with the LOCKSS strategy (lots of copies keeps stuff safe) for data preservation: If everybody thinks somebody else is keeping a copy, then LOCKSS does not work very well.
Many of us who work in the social sciences, particularly in the policy area, never gather our own data. The research we do is almost always based on data that were collected by someone else, but typically not analyzed by them. This notion that data collectors should be separate from data analysts is actually very well established and routine in my field of work.
The Coleman work came early in my doctoral studies. Most of my work on research projects back then (at the Huron Institute, the Center for the Study of Public Policy, and Harvard’s Administration, Planning, and Social Policy programs in education) involved secondary analysis. Later, when it came time to do my own research, I used a large secondary dataset from the National Longitudinal Study of the High School Class of 1972 (NLS72). This study went on for years and is now a huge longitudinal array of data.
My research question, based on the first NLS72 follow-up, was whether financial aid made any difference in students’ decisions whether to enter college. The answer is yes, but the more complex question is whether that effect is big enough to matter. NLS72 taught me how important the relationship was between those who gather data and those who use it, and so it is good to be here today to reflect on what I have learned since then.
My current employer, EDUCAUSE, is an association of most of the higher-education institutions in the United States and some elsewhere. Among other things, we collect data from our members on different questions: How much do you spend on personal computers? How many helpdesk staff do you have? To whom does the chief information officer report? We gather all of these data, and then our members and many other people use the Core Data Service for all sorts of purposes (Figure 3-9).
FIGURE 3-9 EDUCAUSE Core Data Service.
SOURCE: Gregory A. Jackson
One of the issues that struck us over time is that we get very few questions from people about what a data item actually means. Users apparently make their own assumptions about what each item of data means, and so although they are all producing research based ostensibly on the same data from the same source, because of this interpretation problem they sometimes get very different results, even if they proceed identically from a statistical point of view.
If issues like this go unexamined, then research based on secondary, “discovered” sources can be very misleading. It is critical, in doing such analysis, to be clear about some important attributes of data that are discovered rather than collected directly. I want to touch quickly on five attributes of discovered data that warrant attention: quality, location, format, access, and support.
Quality
The classic quality problem for secondary analysis is that people use data for a given purpose without understanding that the data collection may have been inappropriate for that purpose. There are two general issues here. One has to do with very traditional measures of data quality: whether the questions were valid and reliable, what the methodology was, and other such attributes. Since that dimension of quality is well understood, I will not discuss it further.
The other is something most people do not usually consider a quality issue, but any archivist would say it is absolutely critical: We have to think about where data came from and why they were gathered—their provenance—because why people do things makes a difference in how they do them, and how people do things makes a difference in whether data are reusable.
We hear arguments that this is not true in the hard sciences and is completely true in the social sciences, but the reverse is equally true much of the time. The question of why someone gathered data, therefore, is very important.
One key element of provenance I call primacy, which is whether we are getting data from the people who gathered them or there have been intermediaries along the way. People often do not consider that. They say, “I've found some relevant data,” and that is the end of it.
I was once assigned, as part of a huge Huron Institute review of federal programs for young children, to determine what we knew about latchkey kids. These are children who come home after school when their parents are not home, and let themselves in with a key (in the popular imagery of the time, with the key kept around their necks on a string). The question was, How many latchkey kids are there?
This was pre-Google, so I did a lot of library research and discovered that there were many studies attempting to answer this question. Curiously, though, all of them estimated about the same number of latchkey kids. I was intrigued by that, because I had done enough data work by then to find such consistency improbable.
I looked more deeply into the studies, determining where each researcher had gotten his or her data. The result was that every one of these studies traced to a single study, and that single study had been done in one town by someone who was arguing for a particular public policy and therefore was interested in showing that the number was relatively high. The original purpose of the data had been lost as they were reused by other researchers, and by the time I reviewed the finding, people thought that the latchkey-kid phenomenon was well and robustly understood based on multiple studies.
The same thing can happen with data mining. We can see multiple studies and think everyone has got separate data, but in reality everyone is using the same data. Therefore, provenance and primacy become very important issues.
Location
In many cases communicating data becomes a financial issue: How do I get data from there to here? If the amount of data is small, the problem can be solved without trade-offs. However, for enormous collections of data—x-ray data from a satellite, for example, or even financial transaction data from supermarkets—how data get from there to here and where and how they are stored become policy issues, because sometimes the only way to get data from source to user and to store them are by summarizing, filtering, or otherwise cleaning or compressing the data. Large datasets gathered elsewhere frequently are subject to such preprocessing, especially when they involve images or substantial detection noise, and this is important for the secondary analyst to know.
Constraints on data located elsewhere arise too. There may be copyright constraints: Someone can use the data, but cannot keep them, or can use them and keep them, but cannot publish anything unless the data collector gets to see—or, worse, approve—the results. All of these things typically have to do with where the data are located, because the conditions accompany the data from where they were. Unlike the original data collector, the secondary analyst has no ability to change the conditions.
Format
There are fewer and fewer libraries that actually have working lantern slide projectors such as the one shown in Figure 3-10. Yet there are many lantern slides that are the only records of certain events or images. In most cases, nobody has the funds to digitize or otherwise preserve those slides. As is the case for lantern slides, whether data can be used depends on their format, and so format affects what data are available. There are three separate kinds of issues related to format: robustness, degradation, and description.

FIGURE 3-10 Projector for obsolete format photographic slides.
SOURCE: Gregory A. Jackson
Robustness has to do with the chain of custody, and especially with accidental or intentional changes. Part of the reason the seven-track Coleman had even parity was so that I could check each 6 bits of data to make sure the ones and zeroes added up to an even number. If they did not, something had happened to those 6 bits in transit, and the associated byte of data could not be trusted. Hence, one question about secondary data is: Is the format robust? Does it resist change, or at least indicate it? That is, does data format stand up to the vagaries of time and of technology?
Degradation is about losing data over time, which happens to all datasets regardless of format. Error-correction mechanisms can sometimes restore data that have been lost, especially if multiple copies of the data exist, but none of those mechanisms is perfect. It is important to know how data might have degraded, and especially what measures have been employed to combat or reverse degradation.
Finally, most data are useless without a description of the items in this dataset: not just how the data are recorded on the medium or the database schema, but also how they were
measured, what the different values are, what was done when there were missing data, whether any data correction was done, and so on.
Therefore, as a matter of policy, the “codebook” becomes an important piece of format. Sometimes codebooks come with data, but quite often we get to the codebook by a path that is different from the one leading to the data, or we have to infer what the codebook was by reading someone’s prior research. Both are dangerous. That is what we had to do with the Coleman Principals data, for example, because all we had were summary tables. We had to deduce what the questions were and which questions had which values. It is probably just as well we never used the data for analysis.
Access
Two policy issues arise. The first access issue is promotion: Researchers trying to market their data. The risks in that should be obvious, and it is important that secondary analysts seek out the best data sources rather than vice versa.
As an example, as I was preparing this presentation, a furor erupted over a public-radio fundraiser who had been recorded apparently offering to tilt coverage in return for sponsorship. That is not the data issue—rather, it was the flood of “experts” offering themselves as commentators as the media frenzy erupted. Experts were available, apparently and ironically, to document almost any perspective on the relationship between media funding and reporting. The experts that the Chronicle of Philanthropy quoted were very different from the ones Forbes quoted.
The second issue is restriction. Some data are sensitive, and people may not want them to be seen. There are regulations and standard practices to handle this, but sometimes people go further and attempt to censor specific data values rather than restrict access. The most frequent problem is the desire on the part of data collectors to control analysis or publications based on their data.
Most cases, of course, lie somewhere between promotion and censorship. The key policy point is, all data flow through a process in which there may be some degree of promotion or censorship, and secondary analysts ignore that at their peril.
Support
Support has become a big issue for institutions. Suppose a researcher on Campus A uses data that originated with a researcher at Campus B. A whole set of issues arises. Some of them are technical issues. Some of them are coding issues. Many of these I have already mentioned under Location and Format, above.
A wonderful New Yorker cartoon13 captures the issue perfectly: A car is driving in circles in front of a garage, and one mechanic says to another, “At what point does this become our problem?”
Whatever the issues are, whom does the researcher at Campus A approach for help? For substantive questions, the answer is often doctoral students at A, but a better answer might come from the researcher at B. For technical things, A’s central Information Technology organization might be the better source of help, but some technical questions can only be solved with guidance from the originator at B. Is support for secondary analysis an institutional role, or the researcher’s responsibility? That is, do all costs of research flow to the principal investigator, or are they part of central research infrastructure? In either case, does the responsibility for support lie with the originator—B—or with the secondary researcher? These questions often get answered financially rather than substantively, to the detriment of data quality.
When data collection carries a requirement that access to the data be preserved for some period beyond the research project, a second support question arises. I spoke with someone earlier at this meeting, for example, about the problem of faculty moving from institution to institution. Suppose that a faculty member comes to Campus C and gets a National Science Foundation (NSF) grant. The grant is to the institution. The researcher gathers some data, does his or her own analysis, publishes, and becomes famous. Fame has its rewards: Campus D makes an offer too good to refuse, off the faculty member goes, and now Campus C is responsible for providing access to the data to another researcher, at Campus E. The original principal investigator is gone, and NSF probably has redirected the funds to Campus D, so C is now paying the costs of serving E’s researcher out of C’s own funds. There is no good answer to this issue, and most of the regulations that cause the problem pretend it does not exist.
A Caution about Cautions
Let me conclude by citing a favorite Frazz comic, which I do not have permission to reproduce here. Frazz is a great strip, a worthy successor to the legendary Calvin and Hobbes. Frazz is a renaissance man, an avid runner who works as a school janitor. In one strip, he starts reading directions: “Do not use in the bathtub.” Caulfield (a student, Frazz’s protégé, quite possibly Calvin grown up a bit) reads on: “Nor while operating a motor vehicle.” They continue reading: “And not to be used near a fire extinguisher, not recommended for unclogging plumbing, and you do not stick it in your ear and turn it on.”
Finally, Caulfield says, “Okay, I think we are good,” and he puts on his helmet. Then the principal, watching the kid, who is wearing skates and about to try rocketing himself down an iced-over sidewalk by pointing a leaf blower backwards and turning it on, says, “This
__________________________
13 Available at http://gregj.us/pFj6EN.
cannot be a good idea.” To which Frazz replies, “When the warnings are that comprehensive, the implication is that they are complete.”
If there is a warning about policy advice, it is that the list I just gave cannot possibly be complete. The kinds of things we have talked about today require constant thought!
DISCUSSANT: Is there any foreign country where you think the law works well? There are a variety of regimes around the world, ranging from Afghanistan, which has no intellectual property laws at all, up to us. Is there some place where you think it works well?
MR. CARROLL: When the United States invaded Iraq, shortly after we occupied that country we sent Hilary Rosen, the former head of the recording industry, to Iraq to help them write their copyright law. Generally speaking, the United States has been fairly aggressive over the past 12 or so years in negotiating bilateral trade agreements with a host of modest countries around the world that are explicitly designed to export U.S.-style intellectual property rights, including most all of copyright.
DISCUSSANT: At the moment, the countries that are not part of the Berne Convention include Afghanistan and Somalia and other places, such as East Timor.
MR. CARROLL: The U.S. weather data policies were among the first examples of an open approach to public information. Many of these countries, particularly in Europe, had the cost recovery model, but now Europe is starting to see that open data might be important to promote the innovation that can be built on open platforms and that the cost recovery model for sharing weather data might be inhibiting the pace of innovation. Therefore, one of the arguments in favor of open—rather than closed—systems is that with an open system we get innovation that cannot otherwise be predicted.
DISCUSSANT: I want to make sure that I do not come across as being overly negative or critical of U.S.-style intellectual property rights. One of the great strengths of the U.S.-style copyright system is that it has supported a very lively innovation space over the last decade or two. There is a fair amount of potential for using the existing copyright levers—including the public domain lever, enabling licensing, and so forth, all of which exist as part of the default landscape of American intellectual property law—to build legal institutions that enable the kind of collaboration that we have been talking about today.
MR. UHLIR: I would like to elaborate on the E.U. Database Directive. It is a block to the kinds of automated knowledge discovery and open networking environments that we are discussing here. One of the negative aspects is that an insubstantial part of the database gives rise to the right. So if we take it either qualitatively or quantitatively, as defined by the owner of the database, this leaves the user completely unaware of how much extraction of the data is really a cause of action or an infringement. Second, there is no economic harm needed to trigger the statute. Third, it applies to government data as well as to private-sector data. Fourth, it is potentially a perpetual right, because it is triggered for another 15 years every time the database is updated substantially. Fifth, it is an unprecedented exclusive property right in that it protects investment rather than intellectual creativity, which is a fundamental aspect of copyright law. And sixth, there are no mandatory exclusions or limitations. For example, even though this was supposed to harmonize the law in the European Union, there are at least three E. U. countries that have no exclusions whatsoever, no exceptions for scholarly use or education or any other purpose. This applies to factual information rather than creative content. It is thus highly pernicious in the sense that the controls over factual information and the reintegration of information absolutely forbid the type of reuse of information that scientists typically make.
MR. MADISON: In reaction to that, I want to note that none of this legal landscape is static. It is all moving at different rates of speed. The E.U. Database Directive was a major panic button in the intellectual property policy space for a long time. As Mr. Carroll said, to a certain degree it seems that the panic mode seems to have receded, although on the U.S. side of the Atlantic there seems to be renewed interest in the misappropriation doctrine relative to information from databases as a way to avoid some of the limitations that he described. There needs to be ongoing vigilance.
The other consideration in this area is that, as is often the case, the best defense is a good offense when trying to mitigate the harmful effects of some barriers like this. One way to offset some of the attractiveness of either the E.U.-style regulation or renewed interest in tort law, appropriations-style activity would be for universities, consortia, funders, and others to work proactively to use some of these legal levers, particularly in the area of commons, to build these commons proactively and then to have a proof of concept to show how they can actually succeed in specific settings. There are opportunities to build some of these legal questions into the design of the research enterprises from the beginning. Suppose, for example, that we take the National Science Foundation (NSF) data management plan and leverage off of that, from a legal standpoint, to see if that cannot be made to succeed in some other settings as a proof of concept.
DISCUSSANT: Could you address the problems that face chief information officers (CIOs) of universities, given that there are often residual complaints on these problems?
DR. JACKSON: CIOs are the people typically in charge of the boring part of the use of information technology on a campus. They make sure the training is run, for example. Scientific data discovery poses three major challenges for CIOs.
The first is, particularly for high-volume or time-sensitive scientific data, the question of whether the network will be there to move data in the right way at the right time and whether the storage is there to receive it. There is a set of infrastructure issues, all of which are episodic, meaning that most of the time the datasets sit there unused, then occasionally something happens that results in a peak use that demands all the capacity, and then back to the state of being relatively unused. This is a characteristic of most high-performance networks and computing. It is typically not true for storage. It is a very difficult argument for a CIO to make that the university should invest a lot of money in infrastructure that, on average, is not used. The argument that somebody needs it occasionally is a very difficult argument to make.
The second issue that CIOs face is the support question. This happens, for example, when a faculty member moves on, or a postdoc gets a faculty appointment elsewhere, or a doctoral student finishes a degree. Suddenly someone is looking around and saying, “Who will I rely on for support?” Of course, they will mention the overhead rate and say that the institution should provide the support.
The third issue, which is growing very fast, is the question of how we store data that the institution is required to hold and make available. This is an enormous technical challenge, not because it is hard technically, but because the technology is expensive and should be shared. The institutions are not good at sharing these data or putting them into the Google cloud, however. It sounds attractive, but it is hard to write the right contract. This means that someone at the university must take care of these problems, make sure that the data do not degrade, that error correction gets done, and so on.
One of my favorite questions to ask is, You have a body of data that you need to put in one place and have it available 10 years from now—what mechanism do you choose to do that? There is one good answer to that question, which is that you give it to a relatively expensive service to do it. And every so often—typically a matter of months, maybe every year—this service, which has two copies of the data, is going to read the two copies, compare them to one another, and do a certain amount of error correction and restoration. That is the only way to be sure that the data will be there 10 years from now.
Putting the data on CDs or DVDs does not work. It actually does work if you can manage to print it out on archival quality paper and then copy it, but a lot of data cannot be rendered that way.
MR. CARROLL: Think about the law as providing the default terms of use that apply to data. It was pointed out that those default terms of use are fairly uncertain for data, because it can be difficult to tell exactly where the line is between what is copyrighted and what is not, especially for certain types of data, such as image data.
If we do not do anything, we are still making a policy choice, we are choosing the default terms. Let the uncertainty be the rule.
To defend the NSF’s unfunded data management mandate, I think part of what NSF is saying is that researchers have to confront what the policy choices are, what the data management choices are. The researchers can no longer leave those unstated or underdetermined. This is a good idea, as we think about a network environment where people have to work together and we think about longitudinal issues, the demand for the intellectual resources, and the institutional resources, that we should think about such things as what happens to the data tomorrow or 5 or 10 years from now and who has rights to that data. These are the kinds of decisions that are worth making now, because as the flood of data continues, these questions will only become more pressing.
When we lawyers are doing our job right, we are problem solvers. And when we are doing our job really right, we are identifying problems before they have emerged and become serious, and are helping develop a solution. All of these data management issues are only going to become more pressing, and if we leave all these areas undetermined and not thought about, we will end up in a much worse situation in the future. We will find ourselves dependent on somebody else’s data, and suddenly they have some ability to claim rights and hold us up, because we did not think about the use and access issues in advance.
Therefore, one important point from this discussion is that these are issues that deserve attention. Attention is expensive, but if we do not attend to it now, it will only be more expensive later.
3. How Can We Tell? What Needs to Be Known and Studied to Improve Potential for Success?
Session Chair: Francine Berman
-Rensselaer Polytechnic Institute-
This page intentionally left blank.
Francine Berman
-Rensselaer Polytechnic Institute-
To set the stage for this session, I want to start by talking about how research itself is evolving. Modern researchers approach their work in multiple ways. There were two approaches that were mentioned in our discussions earlier.
One approach is analogous to looking for a needle in a haystack. The needle is a well-defined question, for example, Does P=NP?, which is a classic question in computer science. We know what to do to see if our proposed solution addresses the question, and often the question itself will suggest various approaches to a solution.
Another approach is analogous to identifying where the haystacks are. We may have huge amounts of data and be seeking patterns in the data that are likely to provide useful insights and information. For example, the original Seti@Home application combed through massive amounts of data to identify patterns that may indicate extraterrestrial intelligence of some sort.
To support modern research, today’s digital data are being analyzed, used in models and simulations, used to create additional derived data (e.g., visualizations and movies from computer simulations), distributed among a wide variety of participants for filtering, and so on. There is almost no area of modern research that is not being transformed by the availability and ubiquity of digital data.
Another emerging differentiator between today’s modes of research and the past is the broad spectrum of individuals involved. In traditional settings, research was done by professional experts—professors, individuals whose profession is research and development, and other experts. Research was their “day job,” and they spent a substantial amount of time doing it. Today we are increasingly seeing some research being done by the “crowd,” many individuals spending varying amounts of time contributing, who are not necessarily professional experts. The millions who have contributed to Seti@Home or Galaxy Zoo have created new information in aggregate. The number of real discoveries and innovations coming from these crowd applications is increasing, and crowd discovery or citizen science is emerging as an innovative new model for research. This also means that the infrastructure for data-enabled research must support these new crowd applications.
During the discussion today, we heard about enabling digital research infrastructure, and the importance of interoperability between its systems and components. We heard about semantic webs and the need for diverse relationships among the data. We focused on the need for human computer interfaces, portals, ways of accessing the data, and ways of using the data. We discussed the different characteristics of the data, about privacy and the sharing of data. We also heard about gaps—gaps between academia and industry, and gaps between earlier generations and the millennial generation who are using data in a very different way.
All of these issues are critical to consider in developing a viable infrastructure to support data-enabled research, and they will inform what we will discuss in this session. Our panelists will examine what works, how we might assess the effectiveness and success of our
infrastructure, and what we are doing. We will also discuss what needs to be done now, and what needs to be studied so that we can continue to accelerate research-driven discovery and innovation in the future.
Victoria Stodden
-Columbia University-
My work focuses on the intersection of computational science and legal and policy issues.
Aspects related to scientific communication are critical to this discussion. As scientists, one of our first duties is to be deeply skeptical. Our first assumption when we see other people’s scientific research or the presentation of a scientific fact is that it is not correct. Then we think more and say, “Well, maybe this is right,” or we have managed to address the errors in this area or in that area. We move the science forward in that way, and it is a core defining principle of how we should address these issues as technology changes our scientific communication, our modes of research, and the different modalities that we use. At the core, it is about getting at error control.
While it is true that the data deluge is hitting various places and giving us new questions and new solutions, this is not the only way science is being affected. It is also true that fields themselves are becoming transformed.
In my field of statistics, the Journal of the American Statistical Association is one of the flagship publications. I took a look at the proportion of computational articles in the journal beginning in 1996. That year a little less than half of the articles, 9 of 20, were computational, and none of them talked about where to get the codes so that I could actually verify those results.
Ten years later, in 2006, 33 of 35 articles published in that journal were computational. Nine percent of those gave me a link to get the code or pointed me to a package. By 2009, all of the articles were computational. Of those, only 16 percent tell me how I can actually get into the code. Therefore, if someone is publishing in this journal as a statistician, he or she is almost certainly publishing computational work. When computational research is done, however, what is communicated in the paper is typically not enough to allow us to understand how the results were generated. In almost all cases, we need to get into the code and the data.
Without access to the code or the data, we are engendering a credibility crisis in computational science. We have enormous amounts of data being collected. What is typical is that the intellectual and scientific contributions are encoded in the software, but the software is not shared. Such things as data filtering, how the data were prepared for analysis, and how the analysis was done, which can all be very deep intellectual contributions, are in the software and are not easily captured in the paper.
Both the data and the code are important. Much of the discussion here is about the data, although code did sometimes come up. It is equally important that they both be shared and be open, which they are not. This is at the root of our credibility crisis in computational science.
How should we proceed? As a scientist, the first thought is to fall back on the scientific method, and how the problems of openness and communication of discoveries have both been
addressed in the other two branches of the scientific method, the deductive and the empirical branches.
In the deductive branch in mathematics and logic, people have worked out what it means to have a proof, to really communicate the thinking behind the conclusions that are being published. Similarly, in the empirical sciences, there are standards that have been developed, such as controlled experiments and the machinery of hypothesis testing, and how they are communicated. In a methods section, there is a very clear way that these results are to be written for publication, designed so that other people can reproduce the thinking and the results themselves.
In computational science, we are now struggling with this issue: how to communicate the innovations that are happening in computational science in such a way that they will meet the standards that have been established in the deductive and empirical branches of science.
My approach is to understand these issues in terms of the reproducibility of computational science results, and this gives me the imperative to share the code and the data. We have seen many interesting examples of how reuse can be facilitated and what happens when someone actually shares open data. This gives rise to a host of issues about ontologies, standards, and metadata. This is framed within the context of reproducibility. The reason that we are putting the data and the code online is to make sure that the results can be verified and reproduced.
Here is an example. In 2007, a series of clinical trials were started at Duke University that have since been terminated, but it took a few years to terminate them. They were based on computational science results in personalized medicine that had been published in prestigious journals, such as Nature Medicine. Researchers at the M.D. Anderson Cancer Center tried to replicate the computational work that had gone into the underlying science, and uncovered serious flaws undetected by peer review. The study was plagued with a myriad of issues, such as flipped drug labels in the dataset and errors of alignment between observations in treatment and control groups—errors that are simple to make. The clinical trials were canceled in late 2010, after patients had been assigned into treatment and control groups and had been given drugs. One of the principal investigators resigned from Duke at the end 2010. The point is that we have to assume that errors are everywhere in science, and our focus should be on how we address and root out those errors.
There was a discussion earlier of how the data deluge is a larger manifestation of issues that have been seen before. Also, Dr. Hey gave the example of Brahe and Kepler and how what must have been a data deluge in their context ended up engendering significant scientific discoveries. In that sense, there is nothing fundamentally new here. We are doing the same thing in a methodological sense as we have always done, but we are doing it on a much larger scale. The scope of the questions that we are addressing has changed. In that sense, the nature of the research has changed.
Dr. Hey told us that we need more skills to address this concern. Dr. Friend then said we need verifiability, a point that I have also attempted to make in this talk. This means that the infrastructure and incentives need to adapt to the research reality even though the process of science is not changing in any fundamental way. That in turn means that it will be important to develop tools for reproducibility and collaboration. For example, some presenters also talked about provenance- and workflow-tracking tools and openness in the publication of discoveries.
In short, there are many different efforts that are needed. The solutions in this area will not be something that comes down as an executive order and then all scientists are suddenly open with their data and code. The problems are much too granular, and so the solutions must emerge from within the communities and within the different research and subject areas.
Vis Trails is a scientific workflow management system. It was developed by a team from the University of Utah that is now moving to New York University. It tracks the order in which scientists call their functions and the parameter settings that they have when generating the results. These workflows can then be shared as part of the publication, and they can be regenerated as necessary. Vis Trails also promotes collaboration.
Some recent work by David Donoho and Matan Gavish was presented for the first time in a symposium on reproducibility and computational science, held at the American Association for the Advancement of Science. They have developed a system for automatically storing results in the cloud in a verifiable way as they are generated, and creating an identifier that is associated with each of the published results. For example, if a paper contains a figure, then we would be able to click on it and see how it was done and reproduce the results, as the means to do this are automatically in the cloud.
Another useful tool is colwiz, a name derived from “collective wisdom.” Its purpose is to manage the research process for scientists.
One of the major problems with reproducibility is that, unless a scientist is using these specialized tools, there is no automatic mechanism for researchers to save their steps as they advance. After they have finished an experiment and written the paper, they may find that going back and reproducing the experiment is even more painful than going through it the first time. Thus, tools like colwiz could help both with communicating scientific discovery and with reproducibility.
These issues are all related to the production of scientific data and results. There are also some aspects related to publication and peer review. It is a lot of work to request reviewers, who are already overworked, to review code or data and incorporate them into the review process. Maybe we will get there one day for computational work, but certainly not now.
The journals Molecular Systems Biology and the European Molecular Biology Organization are publishing the correspondence between the reviewer and the authors, anonymously but openly, along with the actual published results. This is one approach that is being tried to be more open and transparent.
One of the reasons for this practice is that there is a great inequality in the power of the reviewers and the authors. In particular, reviewers can ask for additional experiments and more exploration of data from the person who is trying to get the paper published. Particularly for prestigious journals, reviewers have a lot of power. These journals are trying to balance this power by allowing readers to see the dialogue that took place between editors and authors before a publication.
Furthermore, many journals now have supplemental materials sections in which datasets and code can be housed and made available for experimentation. The sections are not reviewed and so far have had varying amounts of success.
In the February 11, 2011, issue of Science, there was an editorial emphasizing that, in addition to data, Science is now requiring that code be available for published computational results. That is extremely forward thinking on the part of Science. Science is folding this into its existing policy, which is that if someone contacts an author after publication and asks for the data—and now the code—the author must make it available.
An approach that other journals have taken is to employ an associate editor for reproducibility. The Journal of Biostatistics and the Biometrical Journal do that. The associate editor for reproducibility will regenerate and verify the results, and if the editor can produce the same results that are in the manuscript, the journal will Kitemark the published article with a “D” for data or “C” for code. In this case the journal can advertise that readers can have confidence in the results, because they have been independently verified.
There are also new journals that are trying to address the lack of credit authors get for releasing and putting effort into code or data or for attaching metadata. They are trying to address the issue of incentives, and their focus is on open code and open data. Open Research Computation offers data notes, for example, and BioMed Central has research notes.
PubMed Central and open access are older concepts embedded within the infrastructure of the National Institutes of Health (NIH). But why does it stop with NIH? Could we have a Pub Central for all areas and allow people to deposit their publications when they publish, similar to the NIH policy?
There has been much discussion about the peer-review data management plan at the National Science Foundation (NSF). This is a very important step even though it has been called an unfunded mandate. It is an important experiment in that it creates the possibility of gathering information about how much it will cost and how data should be managed. It is, in a way, a survey of researchers on how they are conceiving of these issues. Maybe the costs are less than NSF worries about, or maybe they are more, but at least we will be able to get a sense of this.
One report that I was involved with along with John King was for the NSF Office of Cyberinfrastructure on virtual communities. We advocated reproducibility as part of the way forward for the collaborative, very high-powered computing problems that we are addressing.
As part of the fallout from the problem I mentioned with the Duke University clinical trials, the Institute of Medicine convened a committee to review omics-based tests for predicting patient outcomes in clinical trials. “Omics” refers to genomics, proteomics, and so on. The committee is chaired by Gil Omenn, and part of its mandate concerns issues of replicability and repeatability and how the articles published in Nature Medicine that led to the clinical trials could have gotten through with what were, in hindsight, such glaring errors.
There seems to be a hesitation on the part of some funding agencies to fund the software development or infrastructure necessary to address reproducibility and many of the other issues that we have discussed so far. Let me give an example of an e-mail that was sent to a group e-mail. The author of the e-mail was talking about how his group develops open-source software for research. He is a prominent researcher, who is very well known and very influential. His group develops open-source tools, and it is very difficult for him to get funding even when applying to NIH programs that are targeted at promoting software development and maintenance. In particular, he said, “My group developed an open-source tool written in Java.
We started with microarrays and extended the tool to other data. There were 25,000 downloads of this tool last year. So we submitted a grant proposal. Two reviewers loved it. The third one did not because he or she felt it was not innovative enough. We proposed three releases per year, mapped out the methods we would add, included user surveys, user support, and instructional workshops. We had 100 letters of support.”
This quote is from the negative review: “This is not science. This is software development. This should be done by a company.” We can see that there seems to be a bifurcation in understanding the role that software plays in the development of science.
One idea for the funders of research might be: Why not fund a few projects to be fully reproducible to see what barriers they run into? Is the problem that they do not have repositories where they can deposit their data? Is the problem that they encounter issues of maintaining the software? Where are the problems? Let us do a few experiments to see the stumbling blocks that they encounter.
On the subject of citation and contributions, as we incorporate databases and code, we need to think about how to reward these contributions. Many contributions to databases now are very small, and there are databases where 25,000 people have contributed annotations. Hence, there are questions about how to cite and how to reward this work. What is the relative worth between, for example, a typical article with a scientific idea versus software development versus maintaining the databases? Typically the last two have not been well rewarded, and our discussion here calls that practice into question.
I will end with a figure from a survey I did of the machine-learning community (Figure 3-11). These are the barriers that people said that they faced most dramatically when they were sharing code and sharing data.
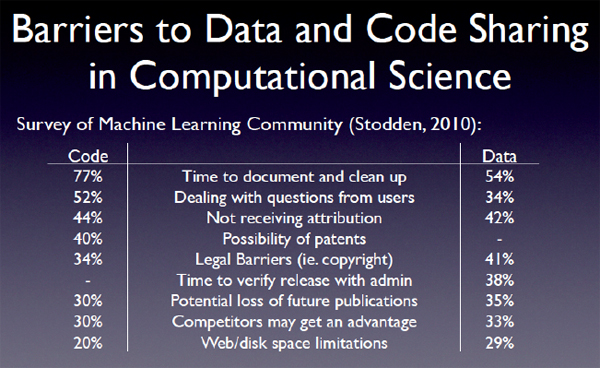
FIGURE 3-11 Barriers to data and code sharing in computational science.
SOURCE: Victoria Stodden
This page intentionally left blank.
Walter L. Warnick
-Department of Energy-
I am the director of the Office of Scientific and Technical Information, which manages many of the scientific and technical information operations of the Department of Energy (DOE). Our goal is to maximize the value of the research and development (R&D) results coming from the department.
To put this into perspective, each government agency has an organization that manages information. Those organizations have gotten together and formed an interagency group called CENDI. Bonnie Carroll is the executive secretary of CENDI. The National Science Foundation is represented in CENDI by Phil Bogden, and the National Institutes of Health (NIH) is represented by Jerry Sheehan. I represent the DOE, and all the other agencies have representatives too, including the Library of Congress, Government Printing Office, Department of Agriculture, Department of the Interior, and practically every other organization that has a big R&D program. Ninety-eight percent of the government’s R&D is represented, and several organizations that do not have R&D programs are also in CENDI.
The results of the U.S. government R&D investment, which amounts to about $140 billion a year, are shared via different formats. There is the text format, which includes journals, e-prints, technical reports, conference proceedings, and more. There is nontext data, which includes numeric datasets, visualizations such as geographic information systems, nonnumeric data such as genome sequences, and much more. And there are other formats, including video and audio.
Each format is in a state of change and presents its own set of challenges. With journal articles, for example, the big issue within the government is public open access versus proprietary access. I think we all agree that the gold standard for text-based scientific technical information is the peer-reviewed journal article. Many highly respected journals are available only by proprietary subscription access. NIH has pioneered a transition to make journal literature publicly accessible. The effort has attracted a lot of attention, and it is still getting a lot of attention within the government. Principal investigators are asked to submit journal-accepted manuscripts for publication in the NIH public-access tool, PubMed Central.
What is significant now is that the America COMPETES Reauthorization Act, which became law in December 2010, calls upon the Office of Science and Technology Policy (OSTP) to initiate a process to determine if public access to journal literature sponsored by government should be expanded. For now, NIH is the only agency that makes a requirement of public access to journal articles. The DOE and other agencies are already empowered by law to adopt that requirement, but we do not have to adopt it, and as a matter of practice we do not. I think that OSTP will soon formulate a committee, which the COMPETES Act calls for, to get input from stakeholders, consider the issues, and develop some recommendations.
Beyond the journal literature, there are gray literature issues, and integrating them with journal literature is important. Gray literature includes technical reports, conference proceedings, and e-prints. It is typically open access, but not all of it is. All of the DOE’s
classified research is reported in gray literature, and of course that is closely held, but I am talking here about the open access part of DOE’s offerings. Gray literature is often organized into single-purpose databases. For example, in the DOE we have something called the Information Bridge, which has 300,000 technical reports produced by DOE researchers from 1991 to the present. It is all full-text searchable. The average report is 60 pages long, so they are fairly detailed. Many other agencies have similar resources. Other databases handle e-prints, conference proceedings, patents, and more.
DOE pioneered novel and inexpensive ways to make multiple databases act as if they are an integrated whole, one example of which is Science.gov. Science.gov posts the publications of all the agencies that are in CENDI, so it is a very large virtual collection of databases. It is all searchable, and a single query brings back results ranked by relevance. It has won awards for being easy to use and is an example of transparency in government.
The DOE’s largest virtual collection that integrates gray literature, and some journal literature, is WorldwideScience.org, which is a computer-mediated scientific knowledge discovery tool. WorldwideScience.org makes the knowledge published by, or on behalf of, the governments of 74 countries, including the United States, all searchable by a single query. The amount of content is huge, about 400 million pages. A user can enter a query in any one of nine languages, and the system will search all the databases in the language of the database and then bring back the list of hits in the language requested. It is new, and it is growing very rapidly under the supervision of the international WorldWideScience Alliance.
We also manage nontext sources—the numeric data, genome sequences, and so forth. The main questions are to what extent should such sources be made accessible and for how long. Some agencies are grappling with the issue by formulating data policies. Some agencies require principal investigators to propose data-management plans. The America COMPETES Reauthorization Act calls upon OSTP to initiate a process to encourage agencies to consider these issues, and it is the same part of the act that I mentioned previously that addresses journal literature. Hence, committees stemming from the act are handling both text items and nontext items.
Everything we do entails cost. Whether it is just sharing information or doing analysis of the information, there is always a cost. Here is a way that I talk to my funding sources about cost. Imagine a graph whose vertical axis is the pace of scientific discovery and whose horizontal axis is the percentage of funding for sharing of scientific knowledge (see Figure 3-12). I think everybody agrees that science advances only if knowledge is shared. Therefore, let us postulate an imaginary situation in which no one shared any knowledge. That would take us to the origin of this graph, because there would be no funds expended for the sharing, but there would be no real advance in science either. At the other end of the x-axis, at the 100 percent mark, if we spent all our money sharing and none of it doing bench research, soon your pace of scientific discovery would draw down close to zero too.
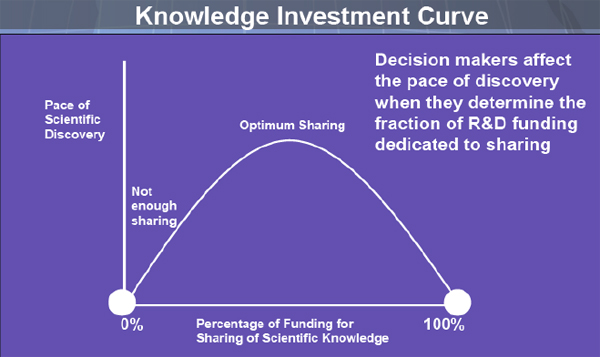
FIGURE 3-12 Knowledge investment curve.
SOURCE: Walter Warnick
We have two data points on this graph, both lying on the x-axis. In between the two points, there is a curve, which we call the Knowledge Investment Curve. We do not know the shape of the curve, but it is likely to have a maximum. The point is that decision makers affect the pace of discovery when they determine the fraction of R&D funding dedicated to sharing. That is the argument I make to my funders.
The point of the Knowledge Investment Curve is to make funders realize that while they can dedicate funds to buy computers, hire more researchers, or build a new facility, they should also weigh that investment against the benefits of getting information out better, sharing it with more people, making searches more precise, and doing the kinds of analyses and data mining we have talked about here today.
It would require a significant research program to calculate what the shape of this curve is and where that optimum is, but we know that such an optimum exists somewhere. Furthermore, the optimum is not the minimum, which is another message I give to the funding sources. If we think that the purpose of an information organization is to be a repository where information goes in, seldom comes back out, and seldom sees the light of day, that is not the optimal expenditure for sharing.
DISCUSSANT: My question to Dr. Warnick and Dr. Stodden is related to the knowledge investment curve presented by Dr. Warnick. In a sense, the peak of that graph is the amount of funding spent on infrastructure that enables research versus the amount of funding spent on research. Where do you think that should fall for any example you choose?
DR. WARNICK: We would probably reach a consensus that there is not enough money being spent on, for example, sharing of knowledge and analysis, and the development of some of the tools that were discussed earlier. As to how much below the optimum it is, let me give an example.
Consider the National Library of Medicine as an excellent example of an information management organization. The funding for that organization exceeds the funding for all the other information organizations combined. I am not suggesting that the National Library of Medicine is overfunded, but I will say that the others are underfunded.
DR. STODDEN: I absolutely agree. I think it became much harder than it has been traditionally to share our work. As our science becomes more data intensive and involves code, those two areas add extra expenses involved with sharing that are not wholly taken care of in funder budgets. The science itself, through technology, has changed, and it is making ripple effects through our funding agencies, which have not quite caught up yet, I think.
DISUSSANT: Dr. Hey talked about the new data-intensive work as a new paradigm, yet so much of the discussion has been about things like reproducibility in the traditional sense, but with code and data added, or sharing in the traditional sense, but with code and data added. So where does thinking about new paradigms or new ways of doing things come in? Where do you see that falling in the spectrum of who is responsible and how that affects this whole question?
DR. WARNICK: Even the sharing part is being subjected to new paradigms. Just to make a point, the infrastructure behind Science.gov and WorldwideScience.org is something that we see very rarely in everyday experience on the Web, and it was developed and matured as a result of some government investment.
To take your point directly about the other kinds of analysis that were discussed earlier, however, since the government is providing $140 billion of funding for research, then the analysis that gets more mileage out of that research ought to be funded by the government too. Of course, the government always welcomes the idea that the private sector can take and utilize these results, but it must be the funding agencies that do the initial work. I think that the reason why people have not heard the Department of Energy mentioned in this discussion before now is because we were doing very little in this regard compared to the National Institutes of Health (NIH) and the National Science Foundation, and that ought to change.
DR. STODDEN: My perspective within academia is that processes are changing for hiring, promotion, and work committees. The scientists who are clued into these issues of reproducibility, open code, and open data seem to be a little more interested than people who are carrying on in a different paradigm. Academia is conservative in the sense that things change slowly. Therefore, it takes time for these practices to percolate through.
DR. BERMAN: The issue of the gap really intrigues and concerns me, because in our real life, if we want to find a restaurant, we can go to Yelp. We can find which restaurants are near us, what is available, who likes them, and so on. There is an application for that. We can get this information on an iPhone.
Consider taking all of our scientific products and putting them in that world. Is there a place where we can find scientific databases and see who liked them? Can we see who added metadata to them in a very user-friendly way? Can we access them easily?
We are starting to see crossover between the academic world and the world of commercial applications. Phil Bourne has a project called SciVee, where we can show how to do different kinds of experiments or give a talk on a data YouTube-style. We can imagine using many of these commercial types of applications and technologies in academia. Some interesting questions arise: What does it mean if we have a data collection and many people like it? Does that mean it is a good data collection? Does that mean it is an economically sustainable data collection? We should not utilize one set of tools for our academic work and another set of tools for applications in the real world without bridging the gap.
MR. UHLIR: There is a proposed act in Congress, the Federal Research Public Access Act, that broadens the NIH PubMed Central grantee deposit policy to include other agencies with annual research and development budgets of $100 million or more, although I do not know if it is going to actually become law.
Also, in the list of peer reviews presented earlier, there is one other model that was missing: postpublication review. It is not a traditional peer review. It is an open peer review, it is moderated, and it is ongoing. There are two kinds of this model. One is just commentary, and one is papers generated in response to a big paper. The model I am thinking of is the European Geosciences Union’s Journal of Atmospheric Chemistry and Physics in Munich. I do not know how many other journals do that, but it is yet another model for a review. Even if the code and the data are not available, people can ask very pointed questions that can be answered by the researchers. That is a potentially valuable way of reviewing results.
In response to Dr. Hendler’s comment, there have been several people who have made some intermediate suggestions, such as Dr. Friend’s journal of models being published. But the fundamental problem is that we have moved all the print journals wholesale onto the Web without hardly changing the model at all. To obtain good models, one would deconstruct the scholarly communication process used in the print paradigm and reconstruct it in a way that makes sense on the Web. Thus, we are repeating everything we have done before and not really thinking about what the Web allows us to change in order to achieve greater efficiency. I think the print journal system itself is an outmoded way of communicating. I am sure there are all kinds of new paradigms, but I will leave it at that.
This page intentionally left blank.
































































