Perhaps one of the most important changes we can make is to supersede the 20th-century metaphor of war for describing the relationship between people and infectious agents. A more ecologically informed metaphor, which includes the germs’-eye view of infection, might be more fruitful. Consider that microbes occupy all of our body surfaces. Besides the disease-engendering colonizers of our skin, gut, and mucous membranes, we are host to a poorly cataloged ensemble of symbionts to which we pay scant attention. Yet they are equally part of the superorganism genome with which we engage the rest of the biosphere.
—Joshua Lederberg, “Infectious History” (2000)
MICROBIAL ECOLOGY IN STATES OF HEALTH AND DISEASE
Introduction
Individually and collectively, resident microbes play important roles in host health and survival. Shaping and shaped by their host environments, these microorganisms form intricate communities that are in a state of dynamic equilibrium.
________________
1 The planning committee’s role was limited to planning the workshop, and the workshop summary has been prepared by the workshop rapporteurs (with the assistance of Charlee Alexander, Rebekah Hutton, and Katherine McClure) as a factual summary of what occurred at the workshop. Statements, recommendations, and opinions expressed are those of individual presenters and participants and are not necessarily endorsed or verified by the Forum, the Institute of Medicine, or the National Research Council, and they should not be construed as reflecting any group consensus.
This ecologic and dynamic view of host–microbe interactions is rapidly redefining our view of health and disease. It is now accepted that the vast majority of microbes are, for the most part, not intrinsically harmful, but rather become established as persistent, co-adapted colonists in equilibrium with their environment, providing useful goods and services to their hosts while deriving benefits from these host associations. Disruption of such alliances may have consequences for host health, and investigations in a wide variety of organisms have begun to illuminate the complex and dynamic network of interactions—across the spectrum of hosts, microbes, and environmental niches—that influence the formation, function, and stability of host-associated microbial communities (Dethlefsen et al., 2007; Turnbaugh et al., 2007; Robinson et al., 2010; IOM, 2012).
From the microbiota2 on the surface of our skin to those that inhabit the mucus-covered lining of our gut, we are deeply embedded in a microbial world— an observation that extends to most, if not all, plant and animal life on Earth. By the time we reach adulthood, more than 100 trillion microorganisms—including Archaea, Bacteria, Fungi, Protozoa, and Viruses—inhabit specialized environmental niches in and on our body surfaces, forming complex communities that contribute to the nutrition, defense, and development of the intricate, microbedominated ecosystems that we humans call “ourselves.” Indeed, we are more accurately viewed as superorganisms—compilations composed of human and microbial cells that are “yoked into a chimera of sorts’” (Lederberg, 2000; Hooper and Gordon, 2001; Xu and Gordon, 2003).
Recent studies of the human gut microbiota have suggested intriguing associations between “dysbiosis” (a general term3 denoting alterations to the composition and dynamics of our microbiota) and a variety of chronic conditions not thought to have a microbial etiology—including severe acute malnutrition, obesity, cardiovascular disease, asthma, adult-onset diabetes, and the inflammatory bowel diseases (Ley et al., 2005; Petersen et al., 2008; Han et al. 2012; Karlsson et al., 2012, 2013; Qin et al., 2012; Ridaura et al., 2013; Smith et al., 2013; Tang et al., 2013). Scientists have yet to determine whether these associations reflect a causal relationship, and many have urged caution about “overselling” the importance of these initial observations. Still, the dramatic rise in the global pervasiveness of many of these apparently noncommunicable diseases over the past half-century has fueled intense interest in the possibility that local and global alterations in our microbial ecology may be contributing to the
________________
2 For the purposes of this workshop overview, microbiota is a collection of microorganisms—including Archaea, Bacteria, Fungi, Protozoa, and Viruses—that exist in the same place at the same time (see Robinson et al., 2010). The terms resident, endogenous, or indigenous microorganisms describe host-associated microbiota.
3 Coined by Mechnikoff in the early 1900s, dysbiosis describes a state of microbial imbalance in the gut and now refers to a change in the structural and/or functional configuration of the microbiota that produces a disruption in the homeostasis between a host and its indigenous microbes (Gordon, 2012).
development or progression of a wide variety of complex diseases (Blaser and Falkow, 2009).
In addition to the varied landscapes provided by animals and plants, microbial communities inhabit Earth’s soil, water, and air, where they drive important geochemical and biological processes. Indeed, microbial communities form the “heart of all ecosystems” (Shade et al., 2012), and their exploration promises to transform our understanding of the natural world (IOM, 2006, 2009, 2012; Shade et al., 2012). Taking a more holistic view of “who [and what] we are” that includes consideration of the role that our resident microbiota plays in influencing states of health and disease may also revolutionize clinical approaches to the diagnosis, treatment, and, ultimately, prevention of disease (Shade et al., 2012).
Statement of Task
On March 18 and 19, 2013, the Institute of Medicine’s (IOM’s) Forum on Microbial Threats hosted a public workshop, in Washington, DC, to explore the scientific and therapeutic implications of microbial ecology in states of health and disease. Participants explored host–microbe interactions in humans, animals, and plants; emerging insights into how microbes may influence the development and maintenance of states of health and disease; the effects of environmental change(s) on the formation, function, and stability of microbial communities; and research challenges and opportunities for this emerging field of inquiry. This meeting built and expanded upon many of the topics explored at a 2002 Forum workshop, The Infectious Etiology of Chronic Diseases (IOM, 2004).
Organization of the Workshop Summary
This workshop summary was prepared by the rapporteurs for the Forum’s members and includes a collection of individually authored papers and commentary. The contents of the unattributed sections of this summary report provide a technical context for the reader to appreciate the presentations and discussions that occurred over the 2 days of this workshop and do not represent the views of the members of the Forum on Microbial Threats, its sponsors, or the IOM.
The summary is organized into sections as a topic-by-topic distillation of the presentations and discussions that took place at the workshop. Its purpose is to present information from relevant experience, to delineate a range of pivotal issues and their respective challenges, and to offer differing perspectives on the topic as discussed and described by the workshop participants. Manuscripts and reprinted articles submitted by workshop participants may be found, in alphabetical order by author, in Appendix A.
Although this workshop summary provides a description of the individual presentations, it also reflects an important aspect of the Forum’s philosophy. The workshop functions as a dialogue among representatives from different sectors
and perspectives that allows them to present their views about which areas, in their opinion, merit further study. This report only summarizes the statements of participants over the course of the workshop. This summary is not intended to be an exhaustive exploration of the subject matter, nor does it represent the findings, conclusions, or recommendations of a consensus committee process.
HUMANS, ANIMALS, AND PLANTS IN A MICROBIAL WORLD
Co-evolution, co-adaptation, and codependency are all features of our relationships with our indigenous microbiota.
—Blaser and Falkow (2009)
Like all forms of life on Earth, microorganisms exist within often complex communities. In every habitat studied—from acidic hot springs to the external and internal surfaces of plants, animals, and humans—microorganisms interact with and influence one another and their environment (IOM, 2012). As noted by David Relman, chair of the Forum on Microbial Threats, the important biology accomplished by the net actions of interacting microbial communities is really the norm on our planet, yet we are only just beginning to explore and appreciate the principles that might define these communities.
Most studies of the microbial world, until fairly recently, isolated microorganisms from their natural ecological settings and studied in sterile monoculture.4 Guided by a reductionist strategy of reductionism that relied upon the isolation and growth of single microbial species in pure culture, this approach limited observations to a narrow range of species that could be isolated and grown under controlled conditions5 (IOM, 2012). Molecular, sequence-based approaches6—pioneered by environmental microbiologists in the late 20th century—ultimately alleviated these constraints and allowed scientists to characterize communities of bacteria and Archaea in a wide range of environments. These studies dramatically expanded our understanding of the natural world by revealing the vast, and previously unseen, diversity of the microbial world (Pace, 1997; Whitman et al., 1998; Handelsman, 2004). Today, genomic methods are being
________________
4 Most studies of microbes were performed on organisms isolated in the laboratory and apart from their natural environmental contexts.
5 Culturing single cells of a particular microbial type is a useful approach to learn about the biology of a particular organism. It is an unnatural environment for most microorganisms because cells are grown in isolation and under controlled conditions. However, by some estimates greater than 99 percent of the microbial world is or may be unculturable (Robinson et al., 2010).
6 Because all cell-based organisms possess rRNA genes, these gene sequences were used as a culture-independent means for organism detection. Researchers used sequences shared by all rRNA it is an unnatural environment for most microorganisms because to amplify each rRNA gene sequence present in a sample and then analyzed subtle differences between rRNA gene sequences to infer the types of organisms present. Today, sequence-based techniques can be applied directly to DNA isolated from an environmental sample to characterize entire genomes of organisms present (Eisen, 2007).
developed to extend these ecological surveys to fungi and viruses (see Virgin et al., 2009; Findley et al., 2013).
Over the past decade, investigations of the ecology of host-associated microbial communities in a variety of contexts have flourished because of the lower cost, increased speed, and greater capacities of nucleic acid sequencing and other analytic techniques, coupled with advances in bioinformatics and computational biology. In addition to metagenomic surveys of the taxa and genomic content present in a microbial community (discovering “who is there?”), it is increasingly possible to examine their function (“what are they doing and why are they doing it?”) by probing gene expression (metatranscriptomics), protein synthesis (proteomics), and production of small molecular weight compounds (metabolomics). Coupling these techniques with concepts developed in the field of macroecology, researchers are able to create a rich, multidimensional picture of the ecology of microbial communities (Robinson et al., 2010; Boyle and Gill, 2012). Terminology commonly used in these studies are defined in Table WO-1.
Microbes in a Microbial World: Exploring Host–Microbe Ecosystems
The idea that disruption of host–microbe interactions or alterations to community structure may lead to disease raises fundamental questions about the ways in which host–microbe, and microbe–microbe, associations are formed and maintained. Such questions have been the basis of a rich body of research on symbiotic interactions, in which disparate organisms form beneficial (mutualistic), neutral (commensal), or harmful (parasitic) associations that often persist over the lifetime of the host. While such symbiotic associations were once considered to be exceptions, symbioses are now known to be the rule in biotic and abiotic systems. Microbial symbionts are not only a normal part of the life cycle of plants and animals, they are often integral to host development and evolution (Fraune and Bosch, 2010; Gilbert et al., 2012).
Studies of symbioses in a wide variety of organisms have provided important insights into the factors that drive the formation, function, and stability of host–microbe associations and a means to pursue the deeper question of whether universal rules and mechanisms govern these processes (IOM, 2009, 2012; Fraune and Bosch, 2010). As noted by Forum member Margaret McFall-Ngai of University of Wisconsin, Madison, “Nature and evolution have done phenomenal experiments from which we can learn, and although humans often forget that they are part of the environment, we are . . . highly linked, and, more importantly, we are products of our evolutionary history.” McFall-Ngai went on to observe that comparative investigations of host–microbe associations in a variety of systems will help to define the “very basic rules by which animals and plants interact with microorganisms.”
TABLE WO-1 Microbial Ecology Definitions
| Term | Definition |
| Biogeography | The study of biodiversity in space and time |
| Diversity | A measure of how much variety is present in a community, irrespective of the identities of the organisms present; consists of richness and evenness |
| Evenness | The distribution of individuals across types |
| Function | An activity or “behavior” associated with a community (e.g., nitrogen fixation or resistance to invasion) |
| Invasion | An ecological event characterized by the establishment of a foreign organism in a new community and the persistence and spread of this organism |
| Metagenomics | A culture-independent method used for functional and sequence-based analysis of total environmental (community) DNA (note that this is not the same as amplifying, cloning, and sequencing the 16S rRNA-encoding gene, although metagenomic sequences, such as those generated via modern sequencing methods, can be probed for 16S rRNA-encoding genes or other phylogenetic markers) |
| Microbiome | The gene complement of a community |
| Microbiota/community | A collection of microorganisms existing in the same place at the same time |
| Resilience | The rate at which a community recovers to its native structure following a perturbation |
| Resistance | The ability of a community to resist change to its structure after an ecological challenge |
| Richness | Number of types (e.g., species) in a community |
| Similarity | A measure that determines the similarity of two or more communities, typically based on shared members, total richness, and sometimes abundance of members |
| Structure | The composition of the community and the abundance of individual members |
| Temporal stability | The ability of a community to maintain its native structure |
SOURCE: Robinson et al. (2010).
Examples of Host–Microbe Communication, Colonization, and Development
Symbiotic associations can be quite specific, as illustrated in the following examples, suggesting co-evolution of host and microbe over long periods of time. This shared evolutionary past is also reflected in the chemical dialog that mediates these associations (McFall-Ngai et al., 2013). In addition to long-term selective forces that hone microbe–host interdependencies, indigenous microbes are subject to shifting environmental conditions over the lifetime of the host. Early patterns of niche colonization and community assembly can strongly— and in some cases, irreversibly—influence the developing host environment and microbiota.
The bacterium and the squid Studied for more than 25 years, the persistent association between the Hawaiian bobtail squid Euprymna scolopes and the gram-negative, luminescent bacterium Vibrio fischeri continues to reveal important insights into host–microbe associations. An early and exclusive association between the squid and a single species of bacteria (V. fischeri) triggers tissue maturation within the squid’s body cavity to form a specialized light-emitting organ. Luminescence emitted by the bacteria resembles moonlight and starlight filtering through ocean waters, camouflaging the squid from predators swimming below (Nyholm and McFall-Ngai, 2004).
Bacterial colonization begins within hours of the squid’s hatching (Figure WO-1). The squid acquires V. fischeri from its environment, and upon the initiation of colonization the juvenile squid selectively recruits V. fischeri in a glycan-rich mucus, separating it from the rich mixture of seawater microbes. It has now been reported that first contact within the squid-vibrio symbiosis triggers profound molecular and chemical changes that are crucial for bacterial colonization7 (Kremer et al., 2013). Once colonized, the squid undergoes dramatic morphological changes—including programmed cell death that eliminates “symbiont-recruiting” structures and the remodeling of tissue to favor the maintenance of V. fischeri within the mature light organ. Host tissue recognition of two non-specific bacterial products—peptidoglycan and lipopolysaccharide—triggers theses developmental events in the squid (Nyholm and McFall-Ngai, 2004). These molecules are members of a broad class of microbe-associated molecular patterns8 (MAMPs) that have now been shown to trigger developmental processes in a wide variety of animals and plants (see Koropatnick et al., 2004, and McFall-Ngai et al., 2013).
Plant–microbe interactions in the rhizosphere Like the development of the bobtail squid’s light organ, one of the best-characterized plant–microbe interactions features a symbiotic association that triggers tissue development in leguminous plants. Nitrogen-fixing bacteria called rhizobia, attracted by plant-secreted flavonoid compounds, colonize legume roots and release chemicals called nodulation (Nod) factors. These factors trigger gene expression within plant roots that results in the uptake of bacteria by plant tissues to form root nodules. The captured bacteria provide a critical nutrient for the plant and in
________________
7 This exquisitely sensitive response to the host’s specific symbiotic partner includes the upregulation of a host endochitinase, whose activity hydrolyzes polymeric chitin in the mucus into chitobiose, thereby priming the symbiont and also producing a hemoattractant gradient that promotes V. fischeri migration into host tissues. Thus, the host responds transcriptionally upon initial symbiont contact, which facilitates subsequent colonization (Kremer et al., 2013).
8 Microbe-associated molecular patterns (MAMPs) are essential structures on features of microbes that are recognized by the innate immune system (Koropatnick et al., 2004). They are recognized by toll-like receptors (TLRs) and other pattern-recognition receptors (PRRs) in plants and animals. MAMPs are often referred to as pathogen-associated molecular patterns (PAMPs). However these motifs are shared among microbes.
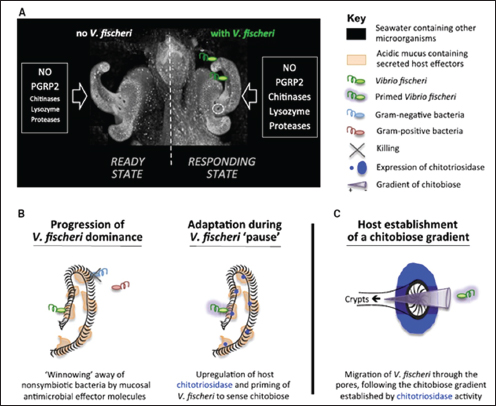
FIGURE WO-1 Model for early colonization. (A) The initial contact of V. fischeri with host tissues induces the expression of several genes (e.g., proteases, chitinases such as EsChitotriosidase, and lysozyme) whose products, when supplemented with components already present in the mucus (NO and EsPGRP2), affect the chemistry of the mucus matrix, shaping the specificity and preparing for future colonization events. (B) Course of events that allow selective colonization by V. fischeri. Left: Antimicrobial compounds (e.g., lysozyme and PGRP2) are activated by acidic proteases in the low-pH environment and participate in the selective exclusion of nonsymbiotic bacteria. Right: While V. fischeri cells are “pausing” in the aggregate, the upregulation of EsChitotriosidase in the ciliated field of the light organ hydrolyzes chitin into chitobiose, which prepares V. fischeri to sense and be attracted toward chitobiose. (C) EsChitotriosidase, which is highly expressed close to the pores and optimally active at low pH, degrades chitin produced by the host into chitobiose, thereby establishing a chitobiose gradient extending out of the pores. Primed V. fischeri cells are attracted by the chitobiose gradient and migrate through the pores (Mandel et al., 2012).
SOURCE: Mandel et al. (2012) in Kremer et al. (2013).
return are fed by the plant’s roots. The roots of most higher-plant species form similar, flavonoid-mediated symbiotic associations with mycorrhizal fungi (IOM, 2006, 2009; Desbrosses and Stougaard, 2011).
Distinct microbiota colonize the rhizosphere (root–soil interface) and endosphere (endophytic compartment within plant root tissues), both of which differ in composition from those found in the surrounding soil. Plant roots provide a structured and nonhomogeneous habitat for tens of thousands of species of microbes. Within this complex environment, microhabitats of nutrient, water, pH, and oxygen gradients shape—and are shaped by—root-associated microbial communities (Ramirez-Puebla et al., 2013). Explorations of this complex environment have illuminated a variety of small molecules (including MAMPs) and chemical signaling networks that mediate plant–microbe and microbe–microbe interactions. The intricate chemical and genetic “cross-talk” between plants and their associated microbiota supports a variety of functions—including plant growth, nutrition, productivity, carbon sequestration, phytoremediation,9 and protection (Figure WO-2) (Badri et al., 2009; Berendsen et al., 2012; Bulgarelli et al., 2012).
Community Assembly and Dynamics in a Model Vertebrate
Microbial colonization is a crucial event in the development of the vertebrate gut, and early colonization events appears to be important to the normal maturation and functioning of the immune and digestive systems. In a recent study in the Proceedings of the National Academy of Sciences of the United States of America, Everard et al. (2013) appear to have demonstrated a causal relationship between Akkarmansia carriage and obesity. Speakers Karen Guillemin and Brendan Bohannan, both of the University of Oregon, described wide-ranging work with the zebrafish Danio rerio, which Guillemin described as “a model vertebrate that is allowing us to explore the complex systems biology of host–microbe interacting systems” (Dr. Guillemin’s contribution may be found on pages 323-346 in Appendix A; Dr. Bohannan’s contribution may be found on pages 164-184 in Appendix A). Zebrafish have several advantages as a model system, Guillemin explained: their guts and immune systems resemble those of other mammals, including humans; they develop rapidly; they are transparent, so their digestive tract can be easily visualized; they are readily amenable to genetic manipulation; and, perhaps most importantly, germ-free zebrafish have been developed, along with methods to associate them with different bacterial communities.
Patterns of colonization Guillemin described her use of germ-free zebrafish and methods to associate them with different bacterial communities. “What our models allow us to do is to build up these systems and look at increasing complexity, starting with the germ-free animal,” she said. “We can then associate them with very
________________
9 The use of green plants to decontaminate polluted soil or water.
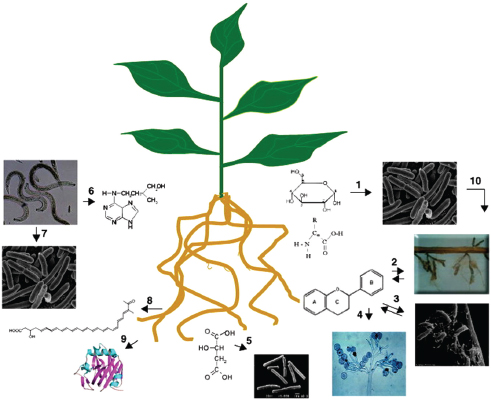
FIGURE WO-2 Illustration of the chemical communication that exists between plant roots and other organisms in the complex rhizosphere. Plant roots secrete a wide range of compounds; among those are sugars and amino acids that are engaged in attracting (chemotaxis) microbes (1), flavonoids act as signaling molecules to initiate interactions with mycorrhiza (AM fungi) (2), rhizobium and (3) pathogenic fungi (oomycetes) (4), aliphatic acids (e.g., malic acid) are involved in recruiting specific plant growth promoting rhizobacteria (Bacillus subtilis) (5), nematodes secrete growth regulators (cytokinins) that are involved in establishing feeding sites in plant roots (6), and nematodes secrete other compounds (organic acids, amino acids, and sugars) involved in attracting bacteria and in bacterial quorum sensing (7). Knowledge of the roles of other types of compounds, such as fatty acids (8) and proteins (9), secreted by roots in the rhizosphere and other multipartite interactions (10), remains unknown.
SOURCE: Badri et al. (2009).
simple communities, or we can allow them to be colonized with complex natural communities . . . [to] look at this whole spectrum of complexity” (Figure WO-3). The recently developed light sheet microscope provides Guillemin and coworkers with high-resolution, real-time imaging of the colonization and growth of bacteria in the developing zebrafish gut (Taormina et al., 2012).
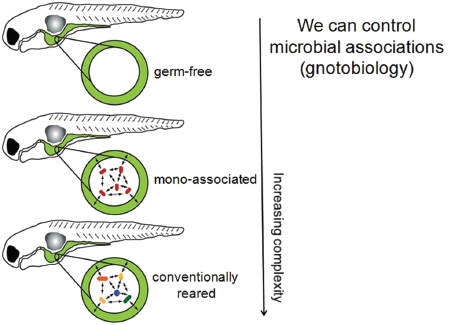
FIGURE WO-3 Host–microbe systems biology. In germ-free model systems, microbial associations may be manipulated to explore host–microbe interactions of increasing complexity. Gnotobiology comprises the study of germ-free plants and animals, as well as living things in which specific microorganisms, added by experimental methods, are known to be present.
SOURCE: Guillemin (2013).
Using this method in germ-free animals, the researchers observed that “early colonizers”—those bacteria that first reach the digestive tract—tend to dominate the developing community (Guillemin and Parthasarathy, unpublished). “We’re exploring the possibility that those first colonizers might have access to certain privileged niches within the gut, and . . . whether there are changes in bacterial physiology upon colonization, perhaps in conjunction with changes in the host environment upon colonization,” she said.
Dynamic interactions Bacterial colonization of the zebrafish gut triggers an innate immune response by the host, in the form of neutrophils—which are not present in the intestinal tracts of germ-free fish, Guillemin noted (Bates et al., 2007). Through experiments in which different species of bacteria that naturally populate the zebrafish gut were individually introduced to germ-free animals, she and coworkers found that the host response (as measured by neutrophil influx) was both varied and species specific; some isolates were very pro-inflammatory, inducing a large number of neutrophils, while others had very little effect on the
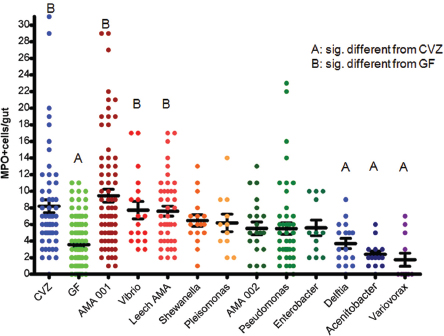
FIGURE WO-4 Monoassociation with zebrafish gut bacterial isolates elicits a wide range of neutrophil influx (MPO+ cells/gut).
NOTE: CVZ = conventionally raised; GF = germ free.
SOURCE: Guillemin (2013).
population of neutrophils (Figure WO-4). Positive, neutral, and negative correlations were observed for the number of neutrophils and specific strains of bacteria.
These findings led the researchers to ask whether host response to a community of bacteria would be a predictable sum of responses to its constituent species or an unpredictable “emergent property” of a complex system. To investigate these possibilities, researchers colonized germ-free fish with individual and paired combinations of Aeromonas, Vibrio, and Shewanella strains. When paired with Aeromonas, Vibrio dominated the community and shaped the neutrophil response. When paired with Vibrio, Shewanella growth was suppressed as compared with monoculture, while Vibrio growth was unchanged; however, neutrophil response appeared to be dictated by Shewanella, despite its minority.
“We’re starting to learn that there are some really interesting kinds of interactions that we can detect between members of the microbiota, and that those relationships impact the host,” Guillemin concluded. “The sum of those interactions could not simply be predicted by each individual member’s effect alone. There are emergent properties that come out of even these very simple systems.”
Drivers of community variation Identifying the mechanisms that produce variation among communities is a fundamental goal of ecology and an important step toward revealing how host-associated microbiota influence host health (Costello et al., 2012). In addition to the several advantages of the zebrafish model described by Guillemin, Bohannan noted that these aquarium fish can readily be raised in the large numbers and controlled environments required to study community variation.
Inquiry into the sources of variation in the zebrafish gut microbiome are informed by ideas developed over decades by plant and animal ecologists. According to one influential theoretical framework, three basic processes drive community variation over short time frames and coarse taxonomic resolutions:
• Dispersal (movement in space),
• Ecological drift (the stochastic loss of organisms from a community and their replacement from within or without), and
• Ecological selection (the differential fitness among organisms due to the community or environment) (Vellend, 2010).
Bohannan’s work explores how these forces interact to drive community variation, in an effort to predict their dynamics, and, ultimately, ways to manage those dynamics that benefit host health, he explained. The zebrafish system enabled him and coworkers to conduct a longitudinal study10 to track gut colonization in germ-free, nearly isogenic,11 fish hatched into a common source of nonsterile water, thereby eliminating potential contributions to microbial community variation from dispersal or selection based on host genotype. Twelve fish were sampled at each of six stages of development from hatching to sexual maturity; sequencing of gut bacterial 16S rDNA determined the species present in each individual microbial community (Figure WO-5).12
Analyses of the abundance of colonizing microbial species in the gut relative to those available in the surrounding water demonstrated both positive and negative selection for certain taxa, Bohannan noted (Figure WO-6).
Bohannan reported that mathematical projections of variation caused entirely by drift accounted for more than 60 to 80 percent (approximately) of the variation observed among individual fish at various time points in the study. The remainder he attributed to nongenetically encoded host factors, such as adaptive immunity, or to interactions among the colonizing microbes—a possibility they are now testing in mutant zebrafish that lack functioning adaptive or innate immune function. One way to interpret changes in the fit of the “drift” model relative to observed
________________
10 Study in which subjects are followed over time with continuous or repeated monitoring of study variables or outcomes.
11 Organisms having the same or closely similar genotypes.
12 For the experiments discussed by Bohannan, operational taxonomic unit (OTU) variation was defined at 97 percent.
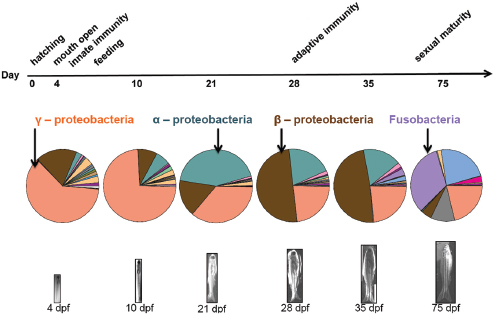
FIGURE WO-5 The gut microbial community composition changes over developmental time.
SOURCE: Bohannan (2013).
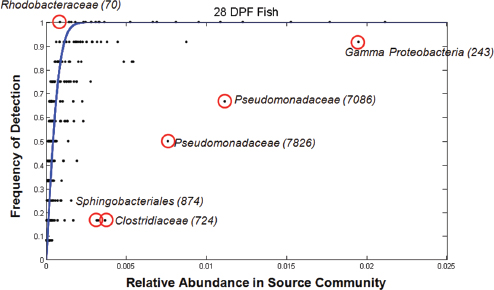
FIGURE WO-6 Positive and negative selection of colonizing microbial species in the gut. The outliers that lie above the blue line are likely taxa that are positively selected regardless of their relative abundance in the source community. Those below the line are taxa that are likely negatively selected despite their commonness in the source community.
SOURCE: Bohannan (2013).
variation over the course of the experiment is that the strength of the selective pressures on the microbiota varies with particular developmental milestones, including the initiation of innate or adaptive immunity and the onset of sexual maturity; he and coworkers are currently testing this hypothesis.
Microbiota Composition and Function in a Plant Root System
Speaker Gerald Tuskan and his team from Oak Ridge National Laboratory (ORNL) use a member of the Salicaceae family as a model system to explore plant–microbe interactions in the rhizosphere and endosphere (Dr. Tuskan’s contribution may be found on pages 412-435 in Appendix A).13 Tuskan’s research originally focused on the Populus14 genome. The catalyst for his research began with an interest in using trees as a source of cellulose for biofuel and of lignin for carbon fibers, and as living “sinks” for carbon dioxide to offset the effects of climate change (Tuskan et al., 2006). When modified plant genotypes transplanted into an “open” environment did not behave as they did in the greenhouse or in the laboratory, Tuskan and colleagues discovered that “it wasn’t solely about the host plant.” The difference was the microbiome, Tuskan said.
In his presentation, Tuskan described his group’s efforts to characterize the sources of genetic diversity among these microbial communities and to relate community composition to function (Figure WO-7). “We want to understand the communication, the chemical signaling that occurs between the bacteria, the fungi, and the host, to see if we can then ultimately reconstruct the community in a predictive manner,” Tuskan explained. By studying the genetic and functional diversity of the Populus microbiome, the investigators ultimately hope to understand its influence on cell wall synthesis and to use this knowledge to increase cellulose and lignin production.
Over the course of several seasons, Tuskan and coworkers sampled Populus microbiomes of multiple host genotypes across a range of microenvironments along two southeastern rivers and among 1,000 individual genotypes collected throughout the Pacific Northwest and transplanted to one of four common gardens.15 Sequencing the microbial communities in their extensive sample set revealed that about 70 percent of the identified OTUs were shared by both rhizosphere and endosphere—although the abundance of a given OTU often varied between the two compartments (Gottel et al., 2011). After separating out differences according to compartment, the second largest driver of diversity was host genotype. The least important variables were year and season: “When we
________________
13 Microbial communities that live within the tissues (endophytic compartment) of plant roots.
14 Populus is a genus of 25–35 species of deciduous flowering plants in the family Salicaceae, native to most of the Northern Hemisphere. English names variously applied to different species include poplar, aspen, and cottonwood.
15 Controlled setting for experiments in which one or more organisms are moved from one environment to another.
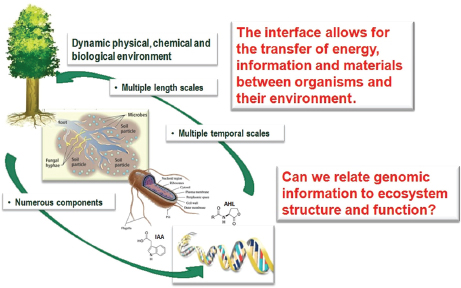
FIGURE WO-7 The dynamic interfaces that exist between plants, microbes, and the environment.
SOURCE: Tuskan (2013).
went back to the same trees and the same root systems over multiple seasons over multiple years we isolated basically the same broad microbiome both from the rhizosphere and the endosphere,” Tuskan reported.
In terms of community diversity, “the endosphere turns out to be a subcomponent of the rhizosphere,” Tuskan concluded (Gottel et al., 2011). This observation supports a conception of microbiota acquisition that he called the “lottery hypothesis” where plants recruit microbial inhabitants of the root endosphere from the wider range of microbes present in the immediate environment of the rhizosphere—which, in turn, represents a subset of the far larger pool of potential endosymbionts available in the various environments in which the plant grows. Tuskan likened this scenario to that of shoppers in open markets that are populated with many kiosks selling similar goods: rather than sampling all of the kiosks, shoppers typically make their purchases from the first or second merchant they encounter. A similar pattern emerged from their analysis of fungal diversity in the Populus endosphere and rhizosphere, reinforcing the notion that the Populus selects members of its endophytic community from the rhizosphere.
Tuskan described several experiments investigating how individual and communities of microorganisms may contribute to important functions. Based on the culture, isolation, and sequencing of about 2,800 representative bacteria from three compartments of the Populus environment—the nearby soil, the soil in contact with the root system, and the endosphere—the researchers determined
that the endosphere microbiota was rich in Pseudomonas species, according to Tuskan. From these Pseudomonas isolates,16 they identified the individual genes that could confer functional advantages to a colonizing bacterium through their effects on such processes as quorum sensing17 and biofilm formation.
In addition, certain endophytic bacteria are known to partner with the mycorrhizal fungi that form symbiotic associations with plant roots. Through pairwise inoculation experiments with Populus fungal isolates, the ORNL team identified “mycorrhizal helper bacteria” among their collections of Populus bacterial isolates. “When we take those helper bacteria in combination with the mycorrhizal fungi and we inoculate the host we see positive differential effects upon the host,” he explained. “There appears to be a combinatorial interaction between the bacteria, the fungi, and then ultimately the host genotype.” Based on this observation, the researchers have identified regions of the Populus genome that control colonization by Laccaria bicolor, a mycorrhizal fungus, and ultimately, a gene locus favoring colonization by the bacterium.
Bringing together the knowledge they have gained from these analyses, Tuskan’s group can now assess how promising combinations of host genotypes possessing favorable signaling and colonization profiles interact with the numerous bacteria–fungi combinations populating the endosphere (Weston et al., 2012). According to Tuskan, 21 days after inoculating Populus cuttings with five phylogenetically diverse bacterial isolates representing distinct functional types, some plants outgrew controls by as much 30 percent. In these cases, he said, “We see a favorable enhancement in growth, which we had predicted based on functional assays, quorum sensing, biofilm formation, chemotaxis, etc., as well as the host genotype” (Figure WO-8). The diverse nature of these reconstructed communities suggests that the functional abilities of the constituents of the endosphere microbiota in Populus—and not the specific bacterial and fungal taxa—are crucial to host growth and biomass productivity, he concluded. “There’s really not much to be learned [through pyrosequencing] other than phylogenetic taxonomy. We need to study individual organisms and their interaction with their host in order to gain predictive power in terms of function at both the bacterial and fungal levels.”
Coevolved Metabolic Networks
Speaker Angela Douglas of Cornell University discussed the implications of animal signaling networks that evolved within the context of preexisting interactions with the microbial world (Dr. Douglas’ contribution may be found on
________________
16 As an illustration of the importance of going beyond taxonomic definitions to understanding the genomic content of host-associated microbes, Tuskan noted that for the 21 isolates of Pseudomonas, there were on the order of 3,000 shared among all isolates (core genes) but more than 11,000 genes unique across isolates (the pan genome).
17 Cell–cell communication system that allows bacteria to monitor population density and control of specific genes in a density-dependent manner (IOM, 2012).
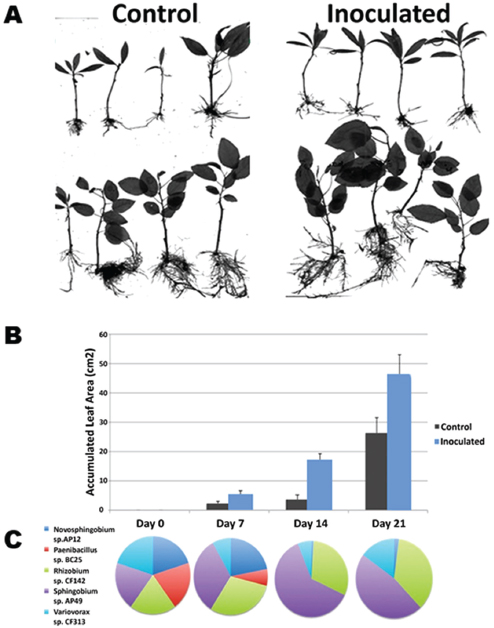
FIGURE WO-8 Constructed communities based on functional diversity. Five phylogenetically diverse bacterial isolates that each brings an alternate set of function types. The Populus host—both trichocarpa (panel A, top) and deltoides (panel A, bottom)—were inoculated and monitored over a 21-day period. Both species exhibited increased growth versus the control over the 21 days (panels A and B). Community composition changed over time (panel C).
SOURCE: Tuskan (2013).
pages 207-224 in Appendix A). Animals “were multiorganismal before they were multicellular” she said, and their evolution was accompanied by a “dramatic flowering of variation of new genes.” Accounting for approximately 16 percent of genes in the human genome, these genes are largely associated with cell signaling and communication and are hotspots for disease-related genes (Figure WO-9) (Domazet-Loso and Tautz, 2008). Consequently all animals—including humans— may share certain fundamental principles of interaction with their resident microbiota, Douglas said.
To explore these principles, Douglas and her group make use of insect symbioses, which, she has noted “offer various clear-cut exemplars of processes
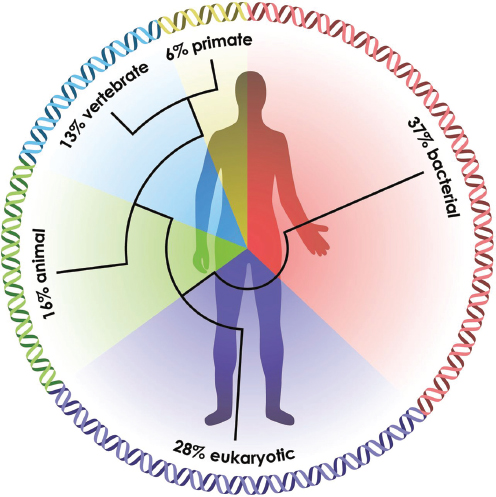
FIGURE WO-9 The ancestry of humans reflected in the genomic signature. A phylogenetic analysis of the human genes reveals the relative percentage of the genome that arose at a series of stages in biological evolution (Domazet-Loso and Tautz, 2008).
SOURCE: McFall-Ngai et al. (2013).
underlying interactions between animals and their resident microbiota,” including “spectacular examples of coevolution with major consequences for the health and well-being of the animal host” (Douglas, 2011).
Her workshop presentation featured studies of two such model systems: in the fruit fly Drosophila melanogaster and in the pea aphid Acyrthosiphon pisum. Douglas observed that compared to a typical mammal, the fruit fly—like most other insects investigated—has a much less diverse microbiome (Douglas, 2011; Wong et al., 2011, 2013). Consisting of 20 to 50 taxa, it is dominated by two species of Acetobacter (particularly A. pomorum) and three species of Lactobacillus (particularly L. fructivorans). Germ-free flies achieve similar body weights compared to conventional flies, but store energy as triglycerides and glucose at much higher levels (Ridley et al., 2012, 2013). “Our interpretation of these data is that carbohydrate and energy sensing and signaling networks in the animal are clearly modulated by the microbiota,” she said.
Douglas has used the pea aphid in order to explore how different bacteria may interact with the nutritional biology of its host. The pea aphid is dependant upon its bacterial endosymbiont, Buchnera aphidicola, for the supply of “essential amino acids”—those amino acids that contribute to protein synthesis but that the animal cannot synthesize on its own. The essential amino acids are in short supply in the aphid diet of plant sap. Housed within specialized cells known as bacteriocytes, the bacterium engages in a dynamic flow of nutrients with the surrounding cytoplasm, Douglas observed. From a survey of metabolism-related genes in B. aphidicola, the researchers inferred the number of reactions and metabolites involved in its exchange with aphids and used that information to characterize the “metabolic network” that connects the two organisms (Figure WO-10) (Thomas et al., 2009; MacDonald et al., 2011). Further studies (Wilson et al., 2010; Poliakov et al., 2011) have revealed the loss of Buchnera genes encoding reactions that are also present in the insect and the dedicated expression of the compensatory host genes in the host cells, she added. “We now have evidence of shared metabolic pathways [between aphid and bacterium] in the synthesis of 5 out of the 10 essential amino acids,” Douglas reported. This metabolite exchange between animal and resident microbiota is underpinned by coevolved animal and microbial metabolic networks, a phenomenon that “may be general to many more complex types of associations,” concluded Douglas.
Hibernation-Related Dynamics of a Vertebrate Gut Microbiome
Mammals found in all classes hibernate during periods of unfavorable environmental conditions, during which they experience profound changes in physiology, morphology, and behavior (Carey et al., 2003). According to speaker Hannah Carey of the University of Wisconsin, Madison, the examination of the transitions between extremes of feeding and fasting in an animal model of hibernation may
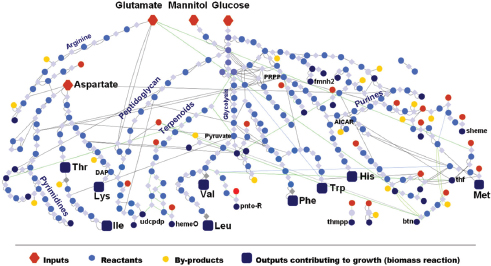
FIGURE WO-10 The metabolic network of Buchnera aphidicola APS1, illustrating the flow of carbon from the main precursors (red diamonds: glucose, mannitol, etc.) to essential amino acids (blue squares: Thr, Lys, etc.).
SOURCE: Thomas et al. (2009).
reveal insights into host–gut microbe dynamics (Dr. Carey’s contribution may be found on pages 184-206 in Appendix A). Dietary and environmental changes experienced by animals during this annual cycle are extreme but can also be “viewed as a normal, recurring perturbation that is well tolerated by the host and its microbial symbionts” (Carey et al., 2013).
Carey’s group studies the 13-lined ground squirrel Ictidomys tridecemlineatus, which undergoes an annual hibernation cycle (see Figure WO-11). The squirrels are lean when they emerge from hibernation in the spring and feed voraciously throughout the summer until they reach near-obesity in the fall, she said. These squirrels voluntarily stop eating as hibernation begins. Much remains to be learned about the appetite signals that control the squirrels’ feeding behavior, which coincides with a shift from homeothermy (maintaining a constant, high body temperature) to heterothermy (alternating periods of high body temperature with periods of lower temperatures closer to the surrounding environment). During hibernation, the animals undergo bouts of torpor, during which metabolism is extremely slow and body temperatures dip toward the freezing point, punctuated with periods of arousal to normal (~37°C) body temperature.
Once normal digestive processes cease, the squirrel’s intestinal mucosa atrophy. “The animals aren’t eating, so they don’t need a robust mucosa to finish digestion and undergo absorption of the food,” Carey explained. However, she added, the gut must still act as a barrier to restrict bacteria and their metabolites
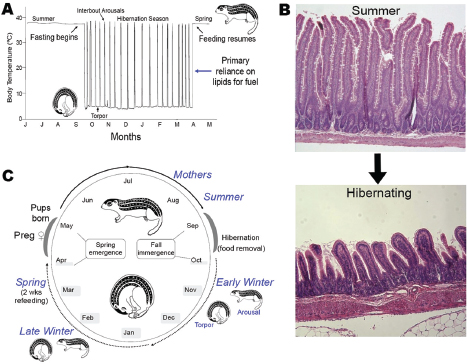
FIGURE WO-11 Annual cycles of body temperature, gut morphology, and feeding behavior. Panel A, body temperature during annual hibernation cycle. Panel B, winter fasting induces mucosal atrophy. Food processing and absorptive functions are largely absent. However, there is a continued need for barrier function and a mutualistic relationship with bacteria. Panel C, annual hibernation cycle.
SOURCE: Carey (2013).
to the lumen and thereby prevent disease. To explore how the gut microbiota responds to the extreme transitions during the annual hibernation cycle, she and her group compared the sequences of 16S rRNA genes from the microbiota occupying the squirrels’ intestinal lumen during summer, early winter, late winter, and spring (Carey et al., 2013) (Figure WO-11). Their results demonstrate that intestinal microbes also follow a diet-dependent annual cycle, with different taxa dominating when the animal is feeding than when it relies on stored energy reserves during its hibernation fast.
These investigators are currently studying whether a similar pattern occurs among the microbiota of the intestinal mucosa, which is more intimately associated with the host, Carey said. They are also interested in the possible role of the microbiota in maintaining host energy and protein stores. “It has long been suggested in the literature of hibernation that protein conservation, which is absolutely important in hibernation, is helped by nitrogen recycling due to gut
microbes,” Carey observed, “but there is very little data that demonstrate that directly.”
Several physiological features of hibernation have implications for gut–microbiome interactions, Carey noted. In the squirrel, as well as in other animals, the gut becomes more permeable during hibernation, which can increase translocation of bacterial products across the epithelial surface, where they may cause widespread deleterious immune effects (Carey, 1990; Carey et al., 1992). “There is a pretty significant remodeling of the intestinal immune system during hibernation, perhaps in response to this leakier gut . . . [or to] moderate levels of bacterial product movement across the epithelium,” she reported. These changes—including an increase in the numbers of intra-epithelial lymphocytes considered to be the “first line of defense” against bacterial pathogens; an elevation in anti-inflammatory cytokines; and an apparent increase in the expression of proteins that maintain tight junctions between the cells of the intestinal epithelium—may represent a compensatory response to limit the enhanced permeability that accompanies winter fasting, she observed.
Carey concluded her presentation with the observation that many intriguing questions remain, and “ultimately we would all like to . . . get beyond the species and look at the actual functional output of these hibernator microbiomes. Metagenomic analyses, coupled with mRNA, protein, and metabolite profiling, will be required to illuminate how metabolic pathways of the community as a whole, and within specific bacterial species, are modified by winter fasting and the return of dietary substrates in the spring” (Carey et al., 2013).
An Ecological View of Health and Disease
A central lesson of ecology is that perturbations often ripple through an ecosystem, leading to unexpected outcomes.
—Lemon et al. (2012)
The human body is host to many, many, highly complex microbial ecosystems. Distinct microbial populations colonize different body sites, such as the mouth, skin, vagina, and gut (Human Microbiome Project Consortium, 2012) (Figure WO-12). At the level of bacterial species and strains, the exact mix present on any given individual is as “unique as a fingerprint”—reflecting the influence of factors ranging from genetics and life history to diet (Spor et al., 2011; Human Microbiome Project Consortium, 2012; Yatsunenko et al., 2012).
The structure of our microbiota reflects our long history of evolution within a microbial world. Of the estimated 100 bacterial phyla18 on Earth, only 4 dominate the human microbiota (Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria). When compared to those found in other vertebrates, the structure of the
________________
18 Groups of organisms ranking above a class and below a kingdom.
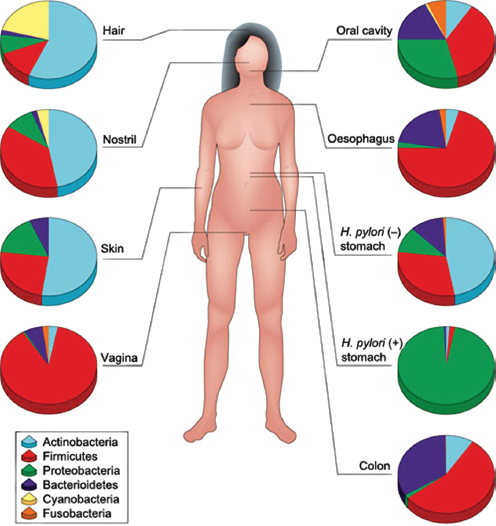
FIGURE WO-12 Compositional differences in the microbiota by anatomical site. Highthroughput sequencing has revealed substantial intraindividual microbiota variation at different anatomical sites, and interindividual variation at the same anatomical sites. However, higher-level (for example at the level of phyla) taxonomic features display temporal (longitudinal) stability in individuals at specific anatomical sites. Such site-specific differences and the observed conservation between human hosts provide an important framework to determine the biological and pathological significance of a particular microbiota composition. The figure indicates the relative proportion of sequences determined at the taxonomic phylum level at eight anatomical sites. Certain features, such as the presence (+) or absence (–) of Helicobacter pylori, can lead to permanent and marked perturbations in community composition.
SOURCE: Cho and Blaser (2012).
human microbiota is similar but distinct, suggesting that host species coevolve with, and adapt to, their microbial inhabitants (Ley et al., 2006; Dethlefsen et al., 2007).
Despite the variation of community structure between individuals, each human microbiome—the collection of genes encoded by members of a microbiota—exhibits shared functional attributes that influence host physiology, immunity, and metabolism (Human Microbiome Project Consortium, 2012). A recent study of the human gut revealed more than 3.3 million microbial genes—about 150 times as many genes than are carried in the human genome (Qin et al., 2010).19 Many of these genes encode biosynthetic and metabolic enzymes that the host is lacking, “thereby greatly expanding the host’s own biochemical and metabolic ability” (Sommer and Bäckhed, 2013). In addition to nutrition and digestion, services provided by our microbiota include detoxification of xenobiotics;20 promotion of growth and differentiation of mucosal tissues; stimulation and shaping of innate and adaptive immune systems; and defense against pathogen invasion through inhibition and competition (Round and Mazmanian, 2009; Hooper et al., 2012; Nicholson et al., 2012).
The human microbiome may have systemic as well as local effects on host physiology. According to McFall-Ngai et al. (2013), our microbiota contribute to as much as one-third of the metabolites21 found in our bloodstream (McFall-Ngai, 2013). Distributed throughout the body by the circulatory system, microbial metabolites have the potential to influence distant microbiota and organs (Nicholson et al., 2012). Gut bacteria, for example, play a key role in the conversion of choline—a nutrient found in high-fat foods—to TMAO (trimethylamine N-oxide), and elevated blood levels of this metabolite have been linked to increased plaque development in arteries and an elevated risk of heart disease (Tang et al., 2013).
“Transplantation” of entire microbial communities from one host to another has also revealed tantalizing links between the microbial ecology of the gut and states of health and disease. Germ-free mice22 that receive, via fecal transplantation,23 the gut microbiota of obese mice gain twice as much body fat
________________
19 These microbial genes were detected in fecal samples obtained from 124 individuals and suggest the presence of 1,000–1,150 prevalent bacterial species. Each individual gut harbored at least 160 species (Qin et al., 2012).
20 A chemical compound (such as a drug, pesticide, or carcinogen) that is foreign to a living organism.
21 Metabolites are small molecules produced during the metabolism of food or other compounds. Microbial metabolites such as short chain fatty acids (SCFAs), bile acids, choline metabolites, vitamins, and lipids (including liposaccharide [LPS], peptidoglycan [PG], and triglycerides) play critical roles in shuttling information between host and microbial cells (Nicholson et al., 2012).
22 Germ-free animals are animals that have no microorganisms living in or on them. Such animals are raised within germ-free isolators in order to control their exposure to viral, bacterial, and parasitic agents.
23 The engraftment of gut microbiota (derived from feces) of a healthy human donor into a recipient (Borody and Khoruts, 2012).
as those that receive communities from lean mice—a phenotype thought to result from an enhanced capacity of the “obese” microbiota to extract energy from the host’s diet (Turnbaugh et al., 2006, 2007). In humans, fecal transplantations have resolved otherwise recurrent and potentially fatal cases of severe diarrhea caused by Clostridium difficile infections. The success of this approach has been attributed to the “restoration” of a healthy gut microbial ecosystem that can resist displacement by pathogens such as C. difficile (Borody and Khoruts, 2012). The possibility that health may be the product of community-level microbial processes and traits—including stability, resistance, and resilience24—only underscores the importance of adopting an ecological perspective to illuminate the complex interactions and interrelationships between a host and its microbiome (Relman, 2012).
Current Studies of the Human Microbiome
Researchers have long suspected that microbes “indigenous” to the human body may be helpful or harmful to the host depending upon the ecological context (see Dubos et al., 1965; Savage, 1977; Mackowiak, 1982). Over the past 7 years, two high-profile projects—the National Institutes of Health’s (NIH’s) Human Microbiome Project and the European Union’s Metagenomics of the Human Intestinal Tract (metaHIT) (see Box WO-1)—have largely focused on identifying the types, genetic potential, and functions of bacteria associated with specific sites on the human body (Bäckhed et al., 2012). These efforts have produced a variety of important data sets, computational tools, and study methods and significantly advanced our understanding of the microbiome of “healthy”25 adult humans from developed countries (Duskko Ehrlich et al., 2010; Procter, 2011). However, “the composition and functional characteristics of a healthy microbiome remain to be precisely defined” (Bäckhed et al., 2012).
As noted by many of this workshop’s participants, investigators are just beginning to explore the ecological contexts in states of health and disease. As summarized in this section, researchers at the workshop described efforts to build upon initial characterizations of the structure and function of the microbiome by mapping the spatial and temporal dynamics of host-associated microbiota; elucidating the means by which host genetics, life history, and environmental exposures may influence community structure and function; and expanding our view of our microbiota to include fungi and viruses. Workshop presentations on the inflammatory bowel diseases (IBDs) underscore the complexity of interactions between the host and microbiome, and environmental factors that may underlie complex and chronic disease states.
________________
24 The capacity of a system to recover from disruption.
25 The Human Microbiome Project studied adult subjects lacking evidence of disease (including the absence of inflammatory diseases). For an extensive list of exclusion criteria see the Human Microbiome Project Consortium (2012).
BOX WO-1
Human Microbiome Research Projects
NIH: The Human Microbiome Project (HMP) is a 5-year, $157 million undertaking launched by the NIH in 2007 (Buchen, 2010) to sequence the microbial communities of several hundred people in order to define commonalities and patterns, and to determine a core microbiome if one exists. The first stage of the project was focused on the metagenomes of the human skin, nose, mouth, gut, and vagina of 300 healthy volunteers and has since expanded, sampling additional body sites. Beyond describing the human microbiota, the HMP seeks to understand aspects of communities such as function, including whether alterations to the microbiome can be correlated to changes in human health. Another project goal is to sequence 3,000 genomes from both cultured and uncultured bacteria, plus viral and small eukaryotic microbes isolated from human body sites.
Metagenomics of the Human Intestinal Tract (MetaHIT) is a project financed by the European Commission that seeks to establish associations between the genes of the human intestinal microbiota and health and disease. Launched in 2008, this 5-year, 21.2M![]() project gathers 13 partners from academia and industry, from a total of eight countries (China, Denmark, France, Germany, Italy, Netherlands, Spain, and the United Kingdom). Focused on two disorders of increasing importance in Europe, inflammatory bowel disease (IBD) and obesity, MetaHIT has (1) established an extensive reference catalog of microbial genes present in the human intestine and bioinformatics tools to store, organize, and interpret this information; (2) developed tools to determine which genes of the reference catalog are present in different individuals and at what frequency; (3) gathered cohorts of individuals, some sick and some healthy; (4) determined for most which genes they carry; and (5) developed methods to study the function of bacterial genes associated with disease aiming to understand the underlying mechanisms and host–microbe interactions.
project gathers 13 partners from academia and industry, from a total of eight countries (China, Denmark, France, Germany, Italy, Netherlands, Spain, and the United Kingdom). Focused on two disorders of increasing importance in Europe, inflammatory bowel disease (IBD) and obesity, MetaHIT has (1) established an extensive reference catalog of microbial genes present in the human intestine and bioinformatics tools to store, organize, and interpret this information; (2) developed tools to determine which genes of the reference catalog are present in different individuals and at what frequency; (3) gathered cohorts of individuals, some sick and some healthy; (4) determined for most which genes they carry; and (5) developed methods to study the function of bacterial genes associated with disease aiming to understand the underlying mechanisms and host–microbe interactions.
SOURCES: http://commonfund.nih.gov/index.aspx; http://www.hmpdacc.org/reference_genomes/reference_genomes.php; The Human Microbiome Jumpstart Reference Strains Consortium, 2010; Robinson et al., 2010; and http://www.metahit.eu. (all URLs accessed January 9, 2014).
Formation and Maintenance of the Human Gut Microbiome
In a dynamic process that appears to be strongly influenced by their environment and genetic makeup, humans—like other organisms studied and reported on in this workshop—develop a resident gut microbiome shortly after birth (Figure WO-13) (Domínguez-Bello et al., 2011; Clemente et al., 2012). As previously noted, the primary microbial community inoculum at birth is the mother’s microbiome.26 Following vaginal delivery, an infant’s microbiome
________________
26 Recent research suggests that the microbial communities of an infant’s mecomium may also play a role in microbial colonization during the first month of life (Moles et al., 2013).
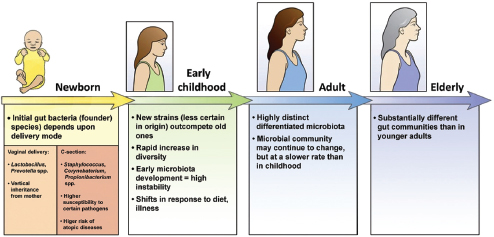
FIGURE WO-13 The development of the microbiota from the first inoculum as an infant through continued change, modified by diet, genetics, and environment, throughout life.
SOURCE: Domínguez-Bello et al. (2011).
initially resembles its mother’s vaginal microbiome, with Lactobacilli dominating in the infant gut. Babies born by Caesarean section are initially colonized by species associated with skin and dominated by taxa including Staphylococcus and Propionibacterium spp.—a composition that persists for several months following birth (Domínguez-Bello et al., 2010). Initial assembly of the infant gut microbiome may have implications for the early and long-term health of the host. The infant microbiome provides resistance to pathogen invasion, furnishes developmental cues, and influences immunological functions. Recent research has demonstrated that colonization during the neonatal period strongly influences mucosal immune development (reviewed in Costello et al., 2012; Lozupone et al., 2012).
Just how differences in microbial community composition in the early stages of life may influence disease susceptibility at later stages of life is not well understood. Investigators have recently described intriguing associations between the composition of the microbiota of children born by Caesarean section and their subsequent development of asthma (Couzin-Frankel, 2010). These results suggest that early colonization events might be deterministic—imparting substantial and lasting effects on the immune system—irrespective of the composition of the mature microbiome. The ways in which factors such as the environment (including diet) and life events (illness, puberty, and pregnancy) influence the community composition of the human gut from birth to old age is an area of active investigation (Domínguez-Bello et al., 2011; Lozupone et al., 2012; Maynard et al., 2012).
Following initial colonization events, the gut microbiota of human adults undergoes consecutive changes in composition and function, increasing in diversity and stability until a relatively constant adult gut community is established (Lozupone et al., 2012). The microbiota of “healthy” adults exists in dynamic
equilibrium; that is, these communities of microorganisms are generally stable, but not static, over space and time. Time series data suggest indigenous microbial communities, like other ecosystems, progress toward ecological climax,27 after which their composition rarely varies in the absence of disturbances like changes in diet, antibiotic therapy, or pathogen invasion (Clemente et al., 2012). The host maintains this equilibrium—or homeostasis—through its mechanical, chemical, and immunological control of the microbiota (Eberl, 2010).
Certain life events, such as pregnancy, are known to dramatically shift the composition and function of an individual’s microbiota. By the third trimester, a healthy pregnant woman’s gut microbiota has changed markedly, perhaps in response to immunological changes known to inhibit rejection of the fetus. In its structure and function, the late-pregnancy microbiome resembles that of an individual who exhibits weight gain and inflammation associated with type 2 diabetes. In the context of late pregnancy, what might otherwise be termed dysbiosis is, in reality, “healthy,” because it promotes storage of extra calories in adipose tissues, and thereby, contributes to the late growth spurt of the fetus in the 6–8 weeks before birth (Koren et al., 2012).
The workshop presentations summarized below provide insights on the interactive course of host–microbiota establishment in mammals and the ensuing dynamic equilibrium in which each partner influences the other—a process that may profoundly influence human health.
Mother’s Milk and the Infant Microbiome
“Human milk is the only food that evolved to make humans healthy,” according to speaker David Mills of the University of California, Davis, as he introduced the workshop to this “very complex fluid that is remarkably understudied in our opinion” (Dr. Mills’ contribution may be found on pages 356-382 in Appendix A). Carbohydrates, protein, lipids, and macro- and micronutrients contained in human milk nourish the infant, he noted, while other elements, including immunoglobulins,28 lysozyme, lactoferrin, and free fatty acids, are known to shape the microbiota of the infant gut, mostly by killing or removing organisms (Sela and Mills, 2010).
After lactose and lipids, the most prominent component of human milk is a collection of large oligosaccharides—free soluble carbohydrates consisting mostly of 3 to 15 monosaccharide units (although larger structures exist) linked through a variety of glycosidic bonds29 (Garrido et al., 2012). Human milk also
________________
27 Community in an equilibrium composition (Gonzales et al., 2011).
28 Also known as antibodies, immunoglobulins are any of numerous proteins produced by B lymphocytes in response to the presence of foreign molecules.
29 Although these linkages could result in a vast array of oligosaccharide species, only about 200 different species are present in human breast milk. According to Mills, this suggests that “nature selected the smaller amount of oligosaccharides that are present.”
contains glycoconjugates of oligosaccharides, attached to lipid and protein molecules, Mills reported. Because infants do not possess the metabolic or enzymatic capacity to degrade oligosaccharides, he said, investigators have long sought to understand its contribution to human fitness. Various hypotheses, including contributions to immune system and neurological development, and “pathogen deflection” by providing pathogen binding sites that resemble the intestinal epithelium, have been advanced to explain the presence of these oligosaccharides in human milk. Mills and coworkers have explored the possibility that the oligosaccharides in human breast milk enrich specific populations of microorganisms in the infant gut leading to early colonization events in the newborn infant that are beneficial to the infant.
Mills’ group focuses on bifidobacteria, a universal member of the infant microbiome and a common probiotic in dairy foods. “Complex milk glycans enhance bifidobacteria colonization,” he stated. “All of the data that we have generated so far seems to support a rationale for why that would be.” For example, the researchers determined that one of the bifidobacteria species and subspecies routinely found in infants, Bifidobacterium longum subspecies infantis (henceforth B. infantis), preferentially consumes the specific small-mass oligosaccharides that are most abundant in human milk, while its close relative in the adult gut, B. longum subspecies longum, does not—a preference that can be traced to unique genomic features of B. infantis (Sela and Mills, 2010; Zivkovic et al., 2011; Garrido et al., 2012). This preferential dietary requirement apparently provides a competitive advantage to B. infantis in successfully colonizing the infant gut in the days to weeks after birth.
Mills and coworkers hope to apply their understanding of this host–microbe interaction to improve nutrition for preterm infants, who frequently suffer dysbiosis. According to Mills, providing preterm infants with human milk (instead of formula lacking oligosaccharides) and B. infantis as a probiotic30 enabled the bacteria to become established in the gut. The group is now trying to “mine” similar oligosaccharides from bovine milk—in the form of whey, a waste product of cheese production—that might be used to supplement formula in combination with B. infantis.
Role of the Mammalian Immune System in Host–Microbe Equilibria
An ecological understanding of host–microbe relationships invites a reconsideration of the role of the host immune system in states of health and disease. Speaker Gérard Eberl, of the Institut Pasteur, has challenged the long-accepted view of the immune system as a discriminating killer of pathogens and proposed instead that it “does not react to combat evil, but merely shapes the microbial
________________
30 Live microorganisms that confer a health benefit on the host.
environment to allow the organism to live with the microbes” (Eberl, 2010) (Dr. Eberl’s contribution may be found on pages 224-248 in Appendix A).
In his presentation to the workshop, Eberl characterized the mammalian immune system as an important tool, shaped over the course of coevolution with microbial communities, over many thousands of years, to maintain equilibrium in their shared ecosystem, the “superorganism.” Microbial species interact with the host along a continuum that ranges from mutualism to pathogenicity, he argued, and their position on this spectrum can shift, depending on the context in which the interaction takes place (see Figure WO-14). For example, he said, “a mutualist in the gut can be good one day, but if you go into surgery . . . [and] it goes systemic, it can kill you.”
A healthy immune response is both plastic and diverse, Eberl observed, reflecting the complexity of the microbial world and the necessity to establish a dynamic equilibrium through adaptation; if it is too weak, the microbiota overgrows and invades host tissues; if it is too strong, the host suffers inflammatory “collateral damage.” An example of the latter has been observed in a Drosophila mutant lacking regulation of a key inflammatory pathway such that it kills most species of endogenous microbes—except for a single toxic strain that eventually kills the fly (Ryu et al., 2008). This same pathway is also associated with inflammatory bowel disease in man.
A growing body of evidence suggests that the gut microbiota plays a key role in establishing this adaptive ability in the mammalian immune system, according to Eberl (Ohnmacht et al., 2011). However, his group has found evidence to suggest that in mammals, the immune system also acts early in development to influence the composition of the microbiota. They discovered that a key initiator of inflammation known as RORγt31 is first expressed in the fetal gut, prior to any encounter with microbes (Eberl, 2012). Eberl observed that while many animals develop immune cells in response to microbial exposure, mammals are unique in preparing for these colonization events during fetal development. “Only mammals have lymph nodes,” he said, an adaptation that has enabled them to anticipate their inevitable colonization by microorganisms.
The developmental program that leads to the formation of lymph nodes—as well as specialized tissues called Peyer’s patches in the fetal gut—replicates the inflammatory process of adults. The cells that induce the development of lymph nodes, so called Lymphoid Tissue inducer (LTi) cells, produce high levels of the inflammatory cytokines interleukin (IL)-17 and IL-22 and are also found scattered in the intestine before and after birth, Eberl reported. Although the intestine has yet to be exposed to microbes, he said, “we think that it starts at very high levels of IL-17 and IL-22 because again it is going to be colonized, and we think
________________
31 The nuclear hormone receptor retinoid-related orphan receptor gamma-t (RORãt) induces a proinflammatory program in lymphoid cells, culminating in the expression of interleukin-6 (IL-6), IL-17, IL-22, granulocyte-macrophage colony-stimulating factor, and tumor necrosis factor (Ohnmacht et al., 2011).
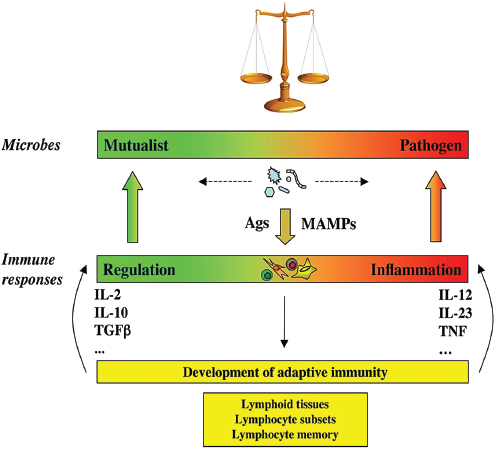
FIGURE WO-14 The continuum of microbial states and immune responses: a dynamic equilibrium. Microbes, including members of the symbiotic microbiota, are not inherently mutualistic or pathogen, but navigate between shades of mutualism and parasitism. Facing the microbes, the immune system is not designed to discriminate between mutualists or pathogens, but merely to react to signals, including MAMPs and antigens (Ags). The nature of the immune response is not purely regulatory or inflammatory, but more generally adjusts to the nature of the trigger it faces, like a spring that is pulled by the intensity of the microbial challenge. Furthermore, the immune system has the capacity to evolve when challenged, through generation of different types of lymphocyte subsets such as Th1, Th2, Th17, Treg, Th22 cells, and follicular T helper cells, and generation of memory lymphocytes and lymphoid tissues. This adds another level of adaptability to the immune system and provides it with the necessary flexibility to maintain homeostasis of the superorganism.
NOTE: MAMPs, microbe-associated molecular patterns.
SOURCE: Eberl (2010).
this is a very strong selective pressure on those microbes, which will be able to productively colonize the intestine.” Shortly before weaning, IL-17 and IL-22 levels drop dramatically in response to negative feedback from the newly established microbiota, only to rise in the event of infection or trauma. He noted that the period of weaning appears “extremely important, as it is a period in which the immune system is still developing, being shaped by the microbiota, with consequences that can be measured later during adult life.”
The intestinal mucosal immune system Mechanisms of host–microbe homeostasis have been extensively studied at the mucosal surfaces of the gut, which are constantly exposed to a diverse and dynamic community of microorganisms and form an interactive barrier that defends the body against invasion by commensal and pathogenic microorganisms through a variety of physical, chemical, and immune-mediated mechanisms (Sansonetti, 2004). At equilibrium, commensal bacteria contribute to barrier function directly, by competitively excluding pathogens from the host-secreted and nutrient-rich mucus. Commensals may also operate indirectly, by expressing antimicrobial peptides (AMPs) and proinflammatory molecules that modulate the hosts’ immune response, as well as by inducing regulatory immune responses (Sansonetti, 2004; Kamada et al., 2013). These highly specialized barrier defenses restrict these microbial communities to the intestinal lumen, where nutrient uptake occurs. Immune system activity also minimizes the incidence of systemic inflammation that would normally occur in the presence of so many bacterial products. Should these tiered host defenses fail, microorganisms might be able to exploit the host for additional resources or trigger damaging systemic inflammatory responses (see Dethlefsen et al., 2007; Costello et al., 2012; Hooper et al., 2012; Maynard et al., 2012). Environmental or genetic factors may also disrupt homeostasis, leading to dysbiosis and a resulting loss of the protective functions of commensal microbes (Figure WO-15).
Indigenous Microbes and the Ecology of Chronic Diseases
In his keynote remarks, Martin Blaser, of New York University, acknowledged the “world of diversity” that has been revealed by early studies of our microbiomes and emphasized the ancient origins of our resident microbial communities (Dr. Blaser’s contribution may be found on pages 110-153 in Appendix A). As a result of our long, shared, evolutionary history, he observed, these microbial associations are largely benign or beneficial (Blaser, 2006). Many of our indigenous microbes are commonly observed in the majority of human populations that have been studied to date and persist throughout our lifetimes. These enduring relationships are mediated by a variety of signaling mechanisms—through which, according to Blaser, our microbiota “know how to talk to the host and to receive signals back, to engage in a very lively conversation” that fosters homeostasis and community stability. Although robust and resilient, the host–microbiota
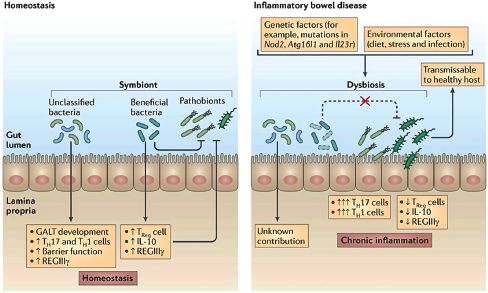
FIGURE WO-15 The intestinal mucosa in states of health and disease. During homeostasis (left), the gut microbiome has important roles in the development of intestinal immunity. Beneficial subsets of commensal bacteria tend to have anti-inflammatory activities. Pathogens can be directly suppressed by beneficial commensal bacteria, partly through the induction of regulatory immune responses, involving regulatory T (Treg) cells, interleukin-10 (IL-10) and regenerating islet-derived protein 3-gamma (REGIIIγ). In inflammatory bowel disease (IBD) (right), a combination of genetic factors and environmental factors are thought to result in disruption of the microbial community structure (dysbiosis), leading to chronic inflammation.
SOURCE: Figure and adapted text from Kamada et al. (2013).
equilibrium can be perturbed, he said. Such perturbations, and their apparent relationship with a range of chronic diseases, were the focus of his presentation.
Helicobacter pylori
Blaser began his presentation with the evolving story of Helicobacter pylori—a bacterium indigenous to the human stomach. In 1984, Dr. Barry Marshall, an Australian physician, demonstrated that the presence of H. pylori in the human stomach could be associated with chronic and acute stomach ailments (in this case, acute but self-limited gastritis). In a demonstration of the causal link between bacterial infection of the stomach and the development of gastritis, Marshall ingested a solution teeming with H. pylori organisms and later developed acute symptomatic gastritis. This act attracted substantial attention and was one factor that overturned decades of medical dogma, asserting that
microorganisms could not survive in the stomach and attributing the chronic, “noncommunicable” condition of peptic ulcer disease to stress and lifestyle factors. Marshall and his collaborator Robin Warren were jointly awarded the 2005 Nobel Prize in Physiology or Medicine for the discovery of H. pylori, its linkage to chronic gastritis, and effective treatments for peptic ulcers through the treatment of H. pylori infection (Pincock, 2005). Marshall and Warren demonstrated that patients with ulcers could be successfully treated with antibiotics to eliminate H. pylori and markedly reduce the recurrence rates of ulcers in these patients.
Blaser noted that for greater than 90 percent of individuals, H. pylori causes a persistent, asymptomatic infection that is acquired early in life and remains “a ‘silent’ infection” throughout the individual’s lifetime. Underlying this largely peaceful coexistence is an ancient and coevolved relationship—extending back at least 100,000 years—during which time H. pylori has become intertwined with the physiology of a healthy host and able to survive in the highly acidic and turbulent gastric environment (Atherton and Blaser, 2009).
H. pylori has been a ubiquitous colonist present in the stomachs of people in every part of the world. Over the course of the 20th century, and coincident with the rise of practices that may reduce H. pylori acquisition and persistence, including smaller family size, better nutrition, cleaner water, and widespread use of antibiotics (Blaser et al., 2008), the bacterium all but disappeared from the stomachs of people in developed countries, stated Blaser. Within the same time period and populations, gastric cancer rates have declined dramatically (Howson et al., 1986). The strength of this association is such that H. pylori infection in adulthood is considered to be the leading risk factor for gastric cancer worldwide (Forman et al., 1994; Peek and Blaser, 2002). Concurrently, rates for other types of cancer—including cancers of the gastroesophageal junction (GEJ)32—have steadily increased over the past half-century. The incidence of esophageal adenocarcinoma, the fastest-rising cancer in the United States and other developed countries, has increased six-fold over the course of the past quarter century. The inverse association between the presence of H. pylori and the development of this cancer and its premalignant lesions—as now shown in multiple studies—suggests that stomach colonization with H. pylori early in life may confer a protective effect (Chow et al., 1998; Corley et al., 2008; Islami and Kamangar, 2008).
Comparisons of the H. pylori–colonized “ancient” stomach to that of the “postmodern” H. pylori–free stomach have identified several key physiological differences with potential health consequences (Figure WO-16) (Blaser and Atherton, 2004; Blaser et al., 2008; Blaser and Falkow, 2009). Blaser observed that individuals lacking H. pylori were at increased risk of developing asthma in childhood (Chen and Blaser, 2007, 2008; Reibman et al., 2008; Arnold et al., 2011). This is but one of many examples of the influences of the gut microbiome. He also noted inverse correlations between the presence of H. pylori and risks for
________________
32 The lower part of the esophagus that connects to the stomach.
FIGURE WO-16 Physiological differences observed between the ancient (H. pylori–colonized) stomach and the postmodern (H. pylori–free) stomach. Leptin and ghrelin are gastric hormones that have a role in energy homeostasis; in the stomach, the hormone somatostatin inhibits the secretion of gastrin.
NOTE: GEJ, gastroesophageal junction; GERD, gastric esophageal reflux disease; PUD, peptic ulcer disease.
SOURCE: Adapted from Blaser et al. (2008).
obesity, celiac disease, stroke, and even tuberculosis reactivation (Blaser et al., 2008; Perry et al., 2010; Cox and Blaser, 2013).
The apparent influence of H. pylori on human physiology may be associated with a protein called CagA. The bacterium injects CagA into the epithelial cells of the human stomach, where it is phosphorylated and subsequently interacts with several major signal-transduction pathways that affect host cell morphology, growth, and programmed cell death (apoptosis) (Blaser and Atherton, 2004). Blaser observed that, given these wide-ranging effects, the relationship between H. pylori and disease is not likely to be simple. Just as most people infected with H. pylori never develop gastric cancer, “there are obviously other risk factors [for gastric cancer] … involving host responses, cofactors, and aging as well,” he said. “It’s a complex biology.” Nevertheless, Blaser stated, “I believe that the evidence shows that Helicobacter is mostly beneficial early in life and is mostly harmful late in life…. The consequence of that is that some years from now doctors … [could] be giving Helicobacter pylori back to children to replace their ancient microbe that
has been lost, and then at some age—30, 40, 50—eradicate it, so as to have the early-life benefit and not the late-in-life cost.”
Disappearing Microbiota
According to the “disappearing microbiota” hypothesis, H. pylori may be an indicator organism for disruptions to other ancient host–microbe associations (Blaser, 2006; Blaser and Falkow, 2009). “Changing human ecology, beginning in the late 19th century, has dramatically altered the transmission and maintenance of our indigenous microbiota,” Blaser observed. The resulting shifts in microbial community composition may influence human physiology, which in turn affects risk for various diseases. “The loss of ancestral bacteria that usually are acquired early in life is especially important, because it affects a developmentally critical stage,” he added.
Accumulating evidence suggests that the human gut microbiota becomes established during the first 3 years of life and is shaped by a variety of factors, including host genetics, diet, and other environmental exposures (Lozupone et al., 2012; Yatsunenko et al., 2012). Blaser highlighted recent research that suggests that this pattern of community development is consistent across distinct cultural traditions—Amerindians in Venezuela, Malawians in Africa, and Americans in the United States (Figure WO-17). Compared to adults in Venezuela and Africa, the gut microbiota of American adults, have less phylogenetic diversity. Blaser noted that this difference may reflect the different diets of these populations and has led some to speculate that factors related to “Westernization” may be altering the bacterial diversity landscape of humans in developed nations (Yatsunenko et al., 2012).
“Maternal status is very important for the composition of the resident microbiota of the next generation,” Blaser observed. Mothers transmit their microbiota to their children in a variety of ways, beginning with organisms acquired during passage through the birth canal and continuing through close contact and breastfeeding (Cho and Blaser, 2012). Several factors associated with modern life are likely to have altered this process—including maternal exposure to antiseptics and antibiotics, changes in the maternal diet, cesarean section, bathing, and bottle-feeding—but none, perhaps, as profoundly as antibiotic treatment during the child’s early life, observed Blaser (Figure WO-18). In the United States, five out of the eight most frequently prescribed medications for children are antibiotics (Chai et al., 2012). Furthermore, according to Blaser’s estimates, the average American child under 2 years of age currently receives more than one course of antibiotics per year.
Antibiotic therapy and obesity Suspecting that the use of “drugs as powerful as antibiotics must have a cost,” Blaser is exploring the connections between early-life exposure to antibiotics and the rising epidemic of human obesity. He
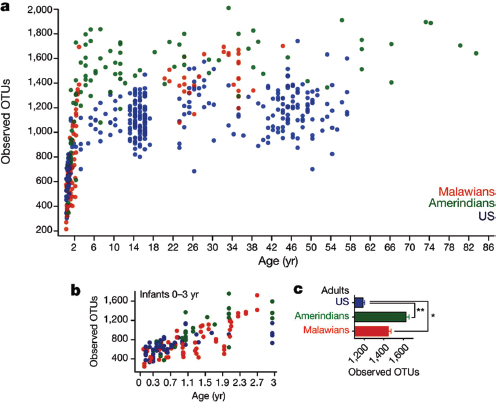
FIGURE WO-17 Bacterial diversity increases with age in populations from distinct cultural traditions. Panel A shows the number of observed operational taxonomic units (OTUs) sharing ≥97 percent nucleotide sequence identity plotted against age for all subjects; Panel B shows the number of OTUs during the first 3 years of life; and Panel C shows the number of OTUs for adults. Mean ± s.e.m. are shown in Panel C. *P<0.05, **P<0.005 (ANOVA with Bonferroni post-hoc test).
NOTE: Taxonomic level of sampling that is similar to the species level, which is more difficult to calibrate. OTU is used to describe variations in 16S ribosomal RNA sequences. Dissimilarity of <1 percent has commonly been used to define an OTU, but <3 percent (similarity ≥97 percent) and <5 percent have also been used (Cho and Blaser, 2012).
SOURCE: Yatsunenko et al. (2012).
noted that over the past two decades obesity rates have risen worldwide, including a doubling of the percentage of obese adults in the United States. Based on the observation that sub-therapeutic antibiotic treatment (STAT) of livestock stimulates weight gain and increases feeding efficiency (particularly among animals treated at young ages), Blaser and colleagues modeled these phenomena in experiments on mice (Cox and Blaser, 2013).
When continuously exposed to STAT since birth, mice gained weight at the same rate as controls, but developed a higher percentage of body fat (Cho et al., 2012). Such phenotypic effects are preceded, by several weeks, by significant
changes in the composition of the fecal microbiota of the STAT mice. Coincident with these changes, key genes involved in fat metabolism were up-regulated in the livers of STAT mice, Blaser reported. This research suggests that in STAT mice, the liver receives increased quantities of short-chain fatty acids produced by microbes in the intestine and converts them into fat; these fat calories are then transported to adipose tissue for storage. Current studies suggest that early-life antibiotic exposures in the mouse model are sufficient for lifelong physiological changes (Cho et al., 2012).
These results led Blaser’s group to investigate whether a series of short, therapeutic-dose pulses of antibiotics administered in early life—an experience shared by many U.S. children—could produce sufficient change in the mouse gut microbiome to alter body composition; as demonstrated in the mouse model, preliminary results suggests lifelong effects. Blaser observed that studies in other fields may help to illuminate these results. Similar patterns have been observed in forest ecosystems, he noted, which tend to recover from infrequent disturbances unless a second disturbance follows before stability is restored (Paine et al., 1998).
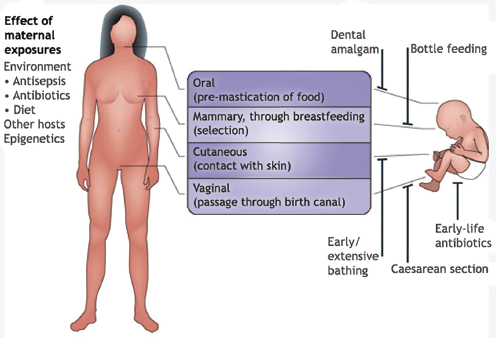
FIGURE WO-18 Factors modifying mother-to-child microbial transmission. Through live birth, mammals have important opportunities for mother-to-child microbial transmission through direct surface contact. However, many modern practices can reduce organism and gene flow; several examples are illustrated.
SOURCE: Cho and Blaser (2012).
Blaser expressed concern that current levels of antibiotic exposure in children may cause “collateral damage” to their metabolic and immune systems that may have lifelong repercussions that include obesity, other chronic diseases, and increased susceptibility to resistant organisms and potentially to epidemic infections.33 Forum member Jesse Goodman, of the Food and Drug Administration, observed that antimicrobial therapy in humans may be associated with socioeconomic, health-related, and other unmeasured factors that could potentially confound determining the etiology of obesity and other chronic conditions. Goodman wondered whether such confounders were under examination through epidemiological studies or clinical trials. Blaser noted that few such studies have been performed, but a birth cohort study of 14,000 English children born in 1991–1992 revealed a significant association between antibiotic use in the first 6 months of life and measures of adiposity at 3.8 and 7 years of age (Trasande et al., 2013).34 He observed that the same cohort also provided evidence of a link between birth by cesarean section and increased risk of obesity (Blustein et al., 2013). Ultimately, large-scale epidemiological studies are needed to determine the meaning of such observations and of findings from model systems, Blaser observed. In the meantime, he added, “we’re working on mice, because we can control many of the important risk factors [for obesity].”
Estrogen-related cancers Blaser concluded his presentation by discussing research exploring whether the human microbiome may contribute to the fact that “only a proportion of individuals exposed to environmental carcinogens or carrying a genetic predisposition to cancer develop [the] disease” (Plottel and Blaser, 2011). He and his colleague Claudia Plottel suggest that estrogen-metabolizing bacteria may contribute to the development of estrogen-related cancers in host organisms—an influence not restricted to the tissues colonized by these bacteria. Epidemiological evidence consistent with this scenario includes the finding that among women positive for breast cancer susceptibility genes BRCA-1 and -2, those born before 1940 (coincident with the dawn of the antibiotic era) tended to develop breast cancer less frequently and later in life than those born after 1940 (King et al., 2003) (Figure WO-19).
Blaser observed that like breast cancer, endometrial cancer is a “disease of development,” occurring at significantly higher rates in wealthy countries where antibiotic use has been prevalent for the longest period of time (Munstedt et al., 2004). Plottel and Blaser hypothesize that the “estrobolome”—the estrogen-metabolizing genes expressed by indigenous bacteria—modulates circulation of estrogens within the liver, which in turn affects estrogen levels throughout the
________________
33 For example, Chang et al. (2008) suggested that decreased microbial diversity due to antibiotic treatment may contribute to the development of “antibiotic-associated” Clostridium difficile infection and diarrhea. Disruption of the gut microbial community by antibiotics may favor colonization by the pathogen.
34 See http://www.bristol.ac.uk/alspac (accessed January 9, 2014).
FIGURE WO-19 Influence of birth cohort on risk of breast cancer among BRCA-1 and BRCA-2 mutation carriers. Age by age, breast cancer risks for mutation carriers born after 1940 were significantly higher than risks for mutation carriers in the same families born before 1940 (log rank P<.0001).
SOURCE: King et al. (2003).
body35 (Plottel and Blaser, 2011). Investigation of the estrobolome hypothesis will measure bacterial deconjugation of estrogen in the human gut and its effect on estrogen levels in the blood and other tissues and determine the risks associated with estrogen exposure at measured levels.
Expanding Perspectives on Host–Microbe Interactions
While considerable research on the role of the microbiome in human health and disease has focused on the resident bacteria of the human gut, studies of other mucosal surfaces and the skin, and of other resident microbes, have broadened our understanding of the complex interplay between the host, environment, and microbial communities. The presentations summarized below reflect expanding
________________
35 Estrogens leave the liver as bile conjugates and circulate to the gastrointestinal tract, from which they are excreted—unless they are deconjugated through the action of the resident bacteria, causing estrogen to be reabsorbed, Blaser explained.
knowledge of the structure and function of indigenous microbial communities, as well as nascent efforts to uncover the physiological and genetic mechanisms with which host and microbe engage each other, and the consequences of those interactions for states of human health and disease.
Community Ecology and the Human Vaginal Microbiome
Studies of the vaginal microbiome are redefining concepts such as “normal” and “healthy,” noted speaker Larry Forney of the University of Idaho (Dr. Forney’s contribution may be found on pages 292-323 in Appendix A). They have also provided insights into community stability, because most women are free of vaginal disease most of the time—despite frequent disturbances such as menses, sexual intercourse, and other sexual and hygienic practices. This microbial community resilience suggests coevolution of the vaginal microbiome with its host.
Forney observed that more than a century ago, Lactobacillus spp. was identified as a key member of the vaginal microbiome, that and its presence has been associated with vaginal health ever since. Bacterial production of lactic acid from glycogen supplied by the host is thought to prevent invasion—and therefore infection—of the vagina by nonindigenous microbes, he explained. As a result, a vaginal pH of 4.5 has long been equated with health. But as Forney and coworkers have discovered, many non-Caucasian women self-described as “healthy” have a vaginal pH significantly higher than 4.5 (Ravel et al., 2011). “This is really a call for personalized medicine,” he argued. “We cannot take a simple rule like a healthy vagina is one with a pH less than 4.5, which may be true for many Caucasian women, and apply it to Hispanic and to black women equally well.”
Indeed, Forney observed, much of the common wisdom regarding the vaginal microbiome is unfounded. The vast majority of studies conducted to date have focused on women of reproductive age, and have tended to advance the view that all women have “more or less the same bacteria in their vaginal microbiome,” and that with the exception of menses, these bacterial communities are stable. By contrast, he and coworkers have conducted research that disputes these conclusions and raises a number of fascinating questions regarding the composition and function of the vaginal microbiome, the response of the microbiome to disturbance, and the contribution of these combined factors to states of health and disease (Hickey et al., 2012; Ma et al., 2012).
Forney’s group conducted a phylogenetic examination of the vaginal microbiome in a cross-sectional survey of nearly 400 healthy, asymptomatic women equally representing Asian, Black, Caucasian, and Hispanic ethnic groups (Ravel et al., 2011). Their results reveal five distinct types of microbial communities, all of which contained significant numbers of lactic acid–producing bacteria—but not necessarily lactobacilli (Figure WO-20). According to Forney, this finding suggests that the function of the vaginal microbiome is conserved, while species
FIGURE WO-20 Phylogenetic surveys of the vaginal microbiome of healthy women identified five distinct clusters. Across community states, function may be conserved as all include significant numbers of lactic acid–producing bacteria. Panel A is a heat map of percentage abundance of microbial taxa found in the vaginal microbial communities of 394 reproductive-age women. Panel B is a representation of vaginal bacterial community groups within each ethnic group of women. The number of women in each ethnic group is in parentheses.
SOURCE: Ravel et al. (2011).
composition varies. Noting that a recent study implicated lactic acid as a signaling molecule for the immune system (Witkin et al., 2011), Forney observed that the specific role of lactic acid in maintaining vaginal health by lowering pH should be revisited.
Another study by Forney and coworkers that examined variation of vaginal bacterial communities in 32 women over the course of 16 weeks, found that each woman’s vaginal microbiome was unique to her, and that no single paradigm of community ecology could explain the wide range of variation they observed both within and between subjects (Gajer et al., 2012). Ultimately, he continued, “[i]t appears as though there are different rules for different women that determine the composition of the vaginal microbiota over time, and in response to disturbance.” Therefore, “we need to think critically and reevaluate what is meant by ‘healthy and normal.’”
Ecosystems research suggests that stable communities resist invasion, and these findings may be applicable to vaginal health and disease, Forney observed. A disturbed community itself could elicit signs and symptoms of disease, or it could be vulnerable to invasion by infectious organisms, he explained. The former scenario seems to describe bacterial vaginosis, a condition estimated to affect more than one-quarter of American women and is associated with an increased risk of complications such as sexually transmitted diseases and preterm birth. Calling efforts to understand bacterial vaginosis “the curse of Koch’s postulates,”36 he remarked that the long history of failed efforts to find a single causative agent for this condition supports the theory that an ecological problem underlies this disease. It may also illustrate Forney’s observation that differences in vaginal microbiota composition between women in different ethnic groups over short time periods may have important consequences that should be accounted for in disease diagnosis and risk assessment.
Forney noted that microbial communities that differ in species composition may also differ in community stability. If the different states identified by Forney and colleagues reflect different risks to invasion or disease, he observed, then for a particular woman disease risk will also vary with time “depending upon what day it is and what the events are that are driving the changes we are seeing,” Forney observed. To better understand the relationships between community composition, stability, and disease, we need to appreciate “what functions are being performed by the microbial communities that are important to maintaining health and the ecological interactions necessary to maintain them,” he concluded.
________________
36 Koch’s postulates must be satisfied in order to state that a particular microbe causes a specific infectious disease. They include the following: (1) The parasite occurs in every case of the disease in question and under circumstances which can account for the pathological changes and clinical course of the disease; (2) The parasite occurs in no other disease as a fortuitous and nonpathogenic parasite; (3) After being fully isolated from the body and repeatedly grown in pure culture, the parasite can induce the disease anew (Koch, 1891; Rivers, 1937; Fredericks and Relman, 1996).
Host Defense and Immunomodulation of Mucosal Candidiasis
Continuing the discussion, begun by Eberl, of the contextual nature of host–microbe interactions related to immunity, speaker Paul Fidel, of Louisiana State University, discussed the range of human immune responses to the fungus Candida albicans, a commensal inhabitant of mucosal tissues (Dr. Fidel’s contribution may be found on pages 249-273 in Appendix A). This member of the normal flora, in certain circumstances, can cause a range of pathogenic conditions generally known as “yeast infections.” These include oropharyngeal candidiasis (OPC, commonly known as oral thrush), denture stomatitis, and vulvovaginal candidiasis (VVC), which may be acute or recurrent. As discussed by Fidel, immune responses to a particular microorganism vary by body site but also in magnitude between individuals.
In its guise as an opportunistic pathogen,37C. albicans expands beyond its commensal niche on the mucosal surface by extending hyphae38 into the upper layers of surrounding epithelial cells. Fidel observed that it can also form biofilms that, in certain contexts, may lead to pathogenicity. Whether C. albicans is a commensal inhabitant or pathogen largely depends upon host environmental factors. Although OPC and denture stomatitis are primarily associated with immunocompromised individuals—people with HIV infection, those undergoing chemotherapy, transplant patients, and the elderly—Fidel noted that VVC frequently occurred in otherwise healthy women.
As Fidel and coworkers have discovered, the means by which host tissues in these different sites defend against Candida infection are strikingly different, with distinct consequences for the manifestation and treatment of disease (Yano et al., 2012). In the case of OPC, mucosal tissues defend against fungal invasion through an adaptive immune response involving CD4 T cells.39 Despite considerable efforts to find a similar protective role for adaptive immunity against VVC, it was suspected—and then demonstrated—that this disease did not arise from insufficient defense by the host against invasion, but rather by an immune “overreaction” to the presence of a commensal—“collateral damage,” as Eberl described it (Fidel, 2007; Yano et al., 2010, 2012).
Interactions between the vulvovaginal mucosal epithelium and C. albicans have evolved in the direction of maintaining a beneficial symbiosis, Fidel explained. In this context, high levels of inflammation associated with an adaptive immune response would cause far more harm than good, he said. Communication between host and microbe preempts the adaptive immune response and preserves
________________
37 An organism that exists harmlessly as part of the normal human body environment and does not become a health threat until it enters the body via wounds; when the body’s immune system is weakened; or when there is a disturbance of a normally benign host–microbe relationship.
38 Slender tubes that develop from germinated spores and form the structural parts of the body of a fungus.
39 Fidel noted that HIV patients lack these cells, and in these patients CD8 cells mount the immune response.
the peace. “It turns out that epithelial cells themselves have the ability to inhibit the growth of Candida,” he continued. “This is a very noninflammatory process. It actually enables asymptomatic colonization to remain and keep the organism in check.” Details of the interaction between Candida and vaginal epithelial cells in conditions of “health” and “disease” are shown in Figure WO-21.
“Symptomatic infection of vaginal candidiasis is associated with an acute inflammatory response, not of the adaptive immune system, but of the innate immune system,” Fidel stated. “The whole idea of protection and susceptibility to infection has gone through a paradigm shift over the past several years now, going away from some deficiency in adaptive immunity to issues of innate immunity being responsible for both protection and susceptibility.” But, as his research demonstrates, both models—involving the same host and microbe—can exist simultaneously in different contexts.
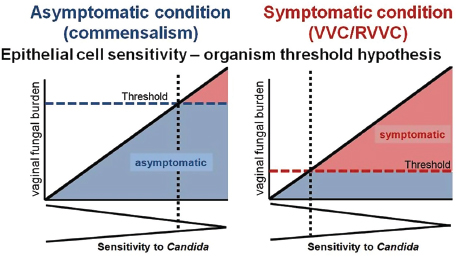
FIGURE WO-21 Epithelial cell sensitivity—organism threshold hypothesis. In women with no history of VVC (left panel), their vaginal epithelial cells are insensitive to Candida and fail to induce the inflammatory response. These women remain asymptomatic even in the presence of high numbers of Candida (threshold number to initiate pathological response rarely breached) that are held in check by noninflammatory processes. In women with RVVC (right panel), vaginal epithelial cells are extremely sensitive to Candida and induce the inflammatory response. These women are susceptible to symptomatic infection following exposure to even small numbers of Candida (low organism threshold) and with less ability to hold them in check. The thresholds represent an arbitrary organism number of the upper limit for vaginal fungal burden that would initiate symptomatic infection.
SOURCE: Yano et al. (2012).
Host–commensal interactions in the presence of pathogens According to speaker Yasmine Belkaid of the National Institute of Allergy and Infectious Diseases, complex interactions between resident and infectious microbes and the host immune system are likely to occur in barrier sites such as the gut, skin, and lungs (Belkaid et al., 2013) (Dr. Belkaid’s contribution may be found on pages 93-109 in Appendix A). In the gut, where most such interactions have been studied, acute infections have been shown to induce dysbiosis and lasting shifts in microbial community composition, while commensals have been shown to promote immune responses against pathogens, she noted. Little is known about similar interactions involving other barrier sites, such as the skin—the largest organ of the body and the most exposed surface—with distinct microbiota and pathogen exposures.
In order to explore how resident microbes influence immunity and inflammation in the skin, Belkaid and coworkers injected germ-free and “normal” mice with the skin parasite Leishmania major (Naik et al., 2012). The germ-free mice mounted a significantly weaker immune response to the pathogen and were less able to control the infection. This response, they discovered, was due to reduced production of pathogen-killing inflammatory cytokines by T cells in the skin of the germ-free mice. If the same mice were colonized with a single bacterial species from the “normal” skin microbiome before pathogen exposure; however, the germ-free mice mounted a competent immune response, as did normal mice treated with antibiotics to abolish their gut microbiome. Based on these observations, the researchers concluded that skin-colonizing microbes influenced the local response to infection. This response was driven by mechanisms that are distinct and independent from the immunity-promoting mechanisms of the gut flora. This finding should provoke further exploration of the role of niche-specific microbial communities in local immune responses and the potential for cross-talk between host sites, Belkaid added (see Belkaid and Naik, 2013).
The skin microbiome acts like a topical adjuvant, according to Belkaid, tuning the level of activation and function of T cells in their immediate environment. Further experiments have revealed that skin commensals exert this effect through an innate immune signaling pathway that has been linked to several chronic inflammatory and skin disorders, including arthritis, asthma, and psoriasis (Hand et al., 2012).
Having examined the ability of the microbiota to promote host immunity to pathogens, Belkaid then considered how an acute infection with a pathogen may alter an otherwise healthy relationship between a host and its microbiota. The gastrointenstinal (GI) tract is one of the most frequent sites of infection—there is an “average of 2 GI acute diarrheal illnesses per child [per year] in America and 12 or more in children in developing countries.” When the physical barrier of the intestinal epithelium is breached by pathogens, the immune system responds not only to their presence, but also to that of commensal bacteria that gain access to otherwise inaccessible host tissues, she explained.
She and coworkers examined how acute gastrointestinal infection of mice by the protozoan parasite Toxoplasma gondii influenced their subsequent immune response to commensal microbes (Hand et al., 2012). Using a transgenic mouse as a source of T cells specific for a single commensal antigen (rather than the diversity of commensal antigens present in wild-type mice), they found that these T cells became activated to participate in an inflammatory immune response when transferred into T. gondii-infected mice, but not when transferred into uninfected mice. “In the context of [pathogen] infection, a T cell that is specific for a commensal is going to become an effector cell at the same level that the effector cells that are generated against a pathogen,” she observed.
Belkaid reported that in addition to this immediate immune response, commensal microbiota-specific T cells also generate immune memory cells, much as do pathogen-specific T cells (Hand et al., 2012). This work has led to the development of a model of the potential consequences of commensal-specific memory T cells (Figure WO-22). Having T cells that recognize commensals could help control subsequent infections. However, Belkaid noted, if the host has a defect in immune regulation, or in barrier integrity, the uncontrolled immune responses to commensals could lead to inflammatory disorders such as Crohn’s disease.40 These responses could also “increase the whole inflammatory setup of the host” and potentially contribute to the etiology of other inflammatory disorders such as allergy and IBD, according to Belkaid. “This is likely to happen not only at the level of the GI tract, but [also] potentially of the lung or at the level of the skin.” Every breach at these barrier sites is likely to increase the pool of commensal-specific memory cells that can be reactivated upon secondary challenge, she said. Understanding the consequences of this buildup of immunity against commensals could lead to insights into the etiology of microbiota-associated inflammatory disorders, concluded Belkaid.
Exploring the Virome
While the bacterial microbiota have been increasingly cataloged, revealing vast numbers of unculturable bacterial species, investigators are also exploring other components of our microbiota—which include all the organisms that infect and live in and on us. The virome encompasses a wide diversity of organisms— from the retroviral elements that “infest our chromosomes” to the myriad chronic infections that are a part of the normal flora of humans. As noted by speaker Herbert “Skip” Virgin, of Washington University, evidence suggests that our virome contributes to “who we are and how we respond to our environment” (Dr. Virgin’s contribution may be found on pages 445-471 in Appendix A). He described recent
________________
40 Crohn’s disease is one of the inflammatory bowel diseases (IBDs) and is characterized by chronic inflammatory disease of the intestines.
research exploring the human enteric virome that has yielded fascinating insights into its community composition, dynamics, and role in health and disease.
By way of introduction to the persistent viruses, Virgin stated that “[a]ll of us are chronically infected with multiple viruses.” Each person is infected with approximately 10 different types of persistent virus, which our immune systems contain but cannot eliminate through constant surveillance (Figure WO-23)
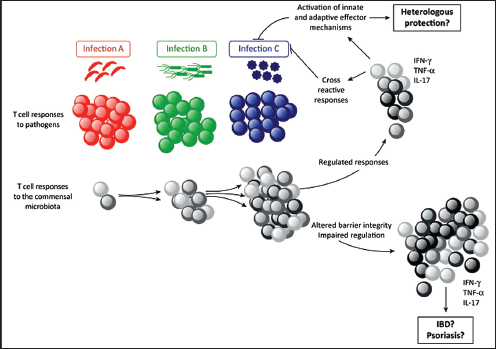
FIGURE WO-22 Potential consequences of commensal-specific memory T cells. During infection at barrier sites (gut, skin, airway, and genital mucosa), immune responses against the invading agent can be associated with specific T cell responses against a large number of coincident commensal antigens. These commensal-specific effector T cell responses can persist as memory cells that upon subsequent infection will be recalled as secondary commensal-specific effectors, alongside the priming of a novel immune response to the invasive pathogen. Therefore, each infection at barrier surfaces represents an additional opportunity for the reactivation of commensal-specific T cells. Given the extraordinary number of commensal antigens (an estimated 20 million antigens/microbiota), these responses may represent a significant proportion of memory T cells. If properly controlled, then commensal-specific T effector/memory cells could contribute to protection against infections by promoting innate and adaptive effector mechanisms assist in the clearance of the pathogen. Further, commensal memory responses could be protective because of cross reactivity with pathogen-derived antigens. By contrast, in situations where commensal-specific T cells become dysregulated because of impaired regulatory pathways and/or barrier function, these T cells could drive chronic pathology such as inflammatory bowel disease or psoriasis.
SOURCE: Belkaid et al. (2013).
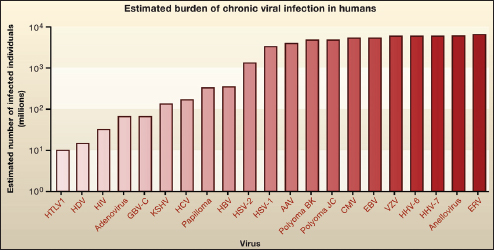
FIGURE WO-23 Chronic viral infections in humans. The number of humans infected with different chronic viruses is estimated from the percentage of humans carrying a given virus, assuming that the world population is 6.75 billion. The estimates in the graph are approximate as they apply data on prevalence in the limited populations studied to date and to the global human population. (See also Table 1 in Virgin et al. [2009]).
SOURCE: Virgin et al. (2009).
(Virgin et al., 2009). Many persistent viruses inhabit mucosal tissues, where they interact with indigenous and invasive bacteria, as well as with the host, he noted. These include the herpesviruses, which appear to have coevolved with vertebrates. “Every single species, including us, has had its own herpesviruses,” he noted. “So these things have been studying us, and we have been studying them evolutionarily for a very, very long period of time—probably in the range of 100 million years or so.”
Through studies of latent herpesvirus infections in mice,41 Virgin and coworkers determined that both viruses were capable of activating their host’s innate immune system, enabling the host to survive challenges with Listeria monocytogenes or Yersinia pestis that killed uninfected animals (Barton et al., 2007). Viral latency42 was required to confer this protection, because the same bacterial pathogens killed mice infected with a mutant form of the virus that causes acute infection. In another study, infection of mice with a different latent herpesvirus “armed” the natural killer (NK) cells of the innate immune system,
________________
41 Murine gammaherpesvirus 68 and murine cytomegalovirus—herpesviruses that are genetically highly similar to the human pathogens Epstein–Barr virus and human cytomegalovirus (Barton et al., 2007).
42 A type of persistent infection in which viral production ceases after initial infection but the viral genome is not fully eradicated from the host. The virus can “reactivate,” emerging from latency to produce viral progeny.
to defend the host against an otherwise lethal lymphoma challenge (White et al., 2010). These research results suggest that “the fundamental nature of the immune response is determined by this latent virus, or maybe other parts of the host virome,” according to Virgin.
In these examples, the combined action of virus and host genomes proved advantageous to the host, other such interactions could have negative consequences, Virgin pointed out. As he and coworkers have discovered, latent viral infection profoundly affects host gene expression (Canny et al., 2013). According to Virgin, “[T]he basic nature of the transcription in the organs of an animal is defined by whether the animal is latently infected with a herpesvirus.” In particular, he and coworkers noted changes in loci that were associated with a predisposition for several chronic diseases, including celiac disease, Crohn’s disease, and multiple sclerosis. “A number of these genes are changed in the latently infected animal, suggesting—but not proving—that your susceptibility to disease might also be altered by your endogenous virome,” he observed (Canny et al., 2013).
Virgin speculated that “perhaps there is an immunophenotype, at least in the innate immune system, which is the individual’s normal immune responsiveness, and that represents the summation of genetic, developmental, and environmental factors … [and] viruses are one of those environmental factors.” Virgin observed that by influencing immunophenotype—whether through contributions to immunity, inflammation, or disease processes—the virome (as well as our bacterial, fungal, parasite, and phage constituents of the microbiome)—may be helping to define the field upon which pathogens or invasive species play their roles in causing disease.
To illustrate the concept of the immunophenotype, Virgin described studies his research team performed on the interaction between an enteric virus that infects mice, murine norovirus (MNV), and a hypomorphic mutation in a gene, known as Atg16L1, associated with increased susceptibility to Crohn’s disease in humans (Cadwell et al., 2008, 2010). Mutation of Atg16L1 produces a pathological effect on the microbe-regulating Paneth cells of the intestinal epithelium in mice similar to what is observed in humans—but not under all circumstances. In the murine system, there was clearly an environmental trigger for the gene-associated pathology. Through both histological and gene-expression comparisons of Atg16L1-bearing tissues before and after infection with MNV, the investigators determined that the Crohn-like pathology resulted from a combination of the virus plus the susceptibility gene, in the presence of commensal bacteria. “When you put the virus and the gene together, you get a new phenotype,” he said, adding that inflammatory diseases are unlikely to be exclusively attributable to the host’s genetic susceptibility or to a microbial pathogen, but instead to the synergistic effects of genes being expressed (or silenced) in both the host and the bacterial microbiome and virome.
Given the great diversity of both viruses that infect humans and of alleles in the human population, one could expect a number of different outcomes related
to disease. This makes sense for many complex diseases—such as the inflammatory bowel diseases—in which genome-wide association studies suggest that the total risk conferred by the observable genotypic difference is less than 20 percent. In such cases, genome does not equal phenome,43 unless all the components of our metagenome (including the virome) are accounted for, Virgin argued. The “lack of quantitative definition of the virome … could be contributing to human phenotypic variation [observed for many chronic diseases] not explained to date by variations in our own genome,” he said.
Recent work from Virgin’s research group illustrated the uncharacterized nature of the enteric virome. Simian immunodeficiency virus (SIV) is a close relative of HIV that, in rhesus monkeys (and under certain as yet undefined conditions), progresses to an AIDS-like disease (Handley et al., 2012). Disease progression is associated with the breakdown of the intestinal epithelial lining, which allows bacteria to invade host tissues and prompts immune activation. While “translocation” of bacteria is arguably a better predictor of progression to AIDS than other more commonly used measures (such as CD4 count and viral RNA levels), previous studies limited to bacterial 16S rRNA sequences had failed to link the intestinal bacterial microbiome to AIDS-related epithelial damage. When Virgin’s group conducted similar studies of the enteric virome, using molecular methods that search for both viruses and bacteria, they discovered a significantly larger range of virus species and genera—including more than 23 previously unknown viruses from pathogenic genera—in the intestines of primates with pathogenic lentivirus infection.
“In pathogenic SIV infection there is a huge expansion of the enteric virome,” Virgin reported. Some of these viruses may cause disease; others may translocate into the blood; some may damage the intestinal epithelium, enabling molecules associated with pathogens and recognized by cells of the innate immune system to move into host tissues, causing systemic inflammation that is the hallmark of AIDS. Based upon these observations, Virgin and coworkers are now studying such possible effects of AIDS-associated virome expansion in humans and primates.
Phage-Mediated Immunity at Mucosal Surfaces
Speaker Forest Rohwer of San Diego State University discussed another member of the virome: viral parasites of bacteria known as bacteriophage (Dr. Rohwer’s contribution may be found on pages 383-400 in Appendix A). Some investigators have projected that there are approximately 1031 phages worldwide, making these viruses among the world’s most numerous biological entities (Youle et al., 2012). Recent research by Rohwer and colleagues suggests that bacteriophages populating mucosal surfaces provide a form of immunity to
________________
43 Sum total of its phenotypic traits.
its animal host. In his presentation, Rohwer described a series of observations he and coworkers have made that led them to characterize bacteriophages as the “hired killers of the immune system.”
Mucosal surfaces form the initial barrier between animals and the environment, Rohwer noted. Mucus covers the entire living surface of some organisms, such as coral, and in humans, mucus lines the lungs, sinus and oral cavities, and the gastrointestinal tract. A variety of microorganisms can be found in mucosal surfaces, which provides the microbes with structure and nutrients. After discovering that mucosal surfaces harbor unusually high concentrations of phages relative to their bacterial prey, Rohwer and colleagues conducted experiments that revealed this association to be quite specific (Barr et al., 2013). The phages stick to mucins, the large, entangled glycoproteins that form the mucus matrix, he said; this serves to concentrate the phages on mucosal surfaces where they are able to infect and kill incoming bacteria. The specificity of this interaction appears to extend to the phage itself, Rohwer continued.
About a quarter of all phages display immunoglobulin-like domains, known as “decoration proteins,” on their capsid surfaces. Like immunoglobulins,44 decoration proteins are highly variable (Minot et al., 2012). “We know that these decoration proteins, for the most part, don’t matter in the lab,” he added. “We can take them off, and the phages do just fine.” Rohwer and his coworkers hypothesized that selection for the presence of decoration proteins might involve their adherence to mucins. As Rohwer reported, binding experiments using phage knockout mutants demonstrated that decoration proteins were required for phagemucin adhesion, and mathematical modeling indicated that mucus-bound phages are 15 times more likely than their free-living counterparts to encounter a bacterial cell (Barr et al., 2013).
As reported by Rohwer, phages are further concentrated in the mucus layer through lysogeny of infected bacteria.45 Through a phenomenon known as “superinfection exclusion”—the inability of a cell infected by one virus to be infected by another virus of the same type—only incoming bacteria not bearing the mucus-adherent phages will be killed, observed Rohwer. Figure WO-24 depicts these interactions at the mucosal surface, which comprise a system Rohwer and coworkers have termed the Bacteriophage Adherence to Mucus Immunity model (Barr et al., 2013). By his description, it functions as an “acquired, adaptive, and specific immune system.”
________________
44 Also known as antibodies, immunoglobulins are any of numerous proteins produced by B lymphocytes in response to the presence of foreign molecules.
45 The biological process in which a bacterium is infected by a bacteriophage that integrates its DNA into that of the host such that the host is not destroyed.
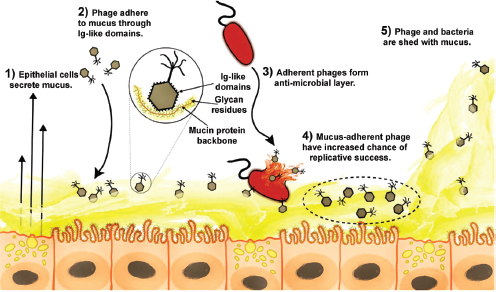
FIGURE WO-24 The Bacteriophage Adherence to Mucus Immunity model. (1) Mucus is produced and secreted by the underlying epithelium. (2) Phages bind variable glycan residues displayed on mucin glycoproteins via variable capsid proteins (e.g., Ig-like domains). (3) Phage adherence creates an antimicrobial layer that reduces bacterial attachment to and colonization of the mucus, which in turn lessens epithelial cell death. (4) Mucus-adherent phages are more likely to encounter bacterial hosts, thus are under positive selection for capsid proteins that enable them to remain in the mucus layer. (5) Continual sloughing of the outer mucus provides a dynamic mucosal environment.
SOURCE: Barr et al. (2013).
Host–Mycobiome Interactions in Gut Homeostasis and Pathogenesis
The fungal contribution to the host–microbe metagenome, like that of viruses, remains largely unknown, according to speaker David Underhill of Cedars-Sinai Medical Center, Los Angeles (Dr. Underhill’s contribution may be found on pages 435-445 in Appendix A). Given their ubiquity46 and importance as human pathogens, Underhill noted that it would seem likely that fungi are represented in the human microbiome and, in that capacity, interact with the immune system in ways that may influence human health. Indeed, he pointed out, antibodies recognizing a common fungal cell wall component, mannan, are currently used to identify cases of Crohn’s disease. “This suggests that in inflammatory bowel disease, there is novel exposure to fungi,” observed Underhill (Underhill and Braun, 2008).
________________
46 Fungi are estimated to comprise nearly one-quarter of Earth’s biomass.
In order to assess the presence of fungi in the gut microbiome, Underhill’s lab developed an approach to detect and characterize fungal ribosomal DNA from several sites in the mouse gastrointestinal tract; using this approach they were able to identify more than 200 species of fungi, representing more than 50 genera (Iliev et al., 2012). Employing a cell wall-binding fluorescent probe, they were able to locate fungal cells in intimate associations with bacteria in the mouse colon, and in the feces from a wide range of mammals, including humans. Treatment with fluconazole, a fungal-specific drug, was shown to abolish these populations. The researchers also observed the appearance of anti-mannan antibodies in a mouse model of chemically-induced colitis. This observation suggests that the immune system interacts with endogenous fungi when the gut is disturbed.
Dectin-1, the cell wall–binding probe that Underhill’s group used to visualize endogenous fungi, is an innate immune receptor expressed by macrophages, dendritic cells, and neutrophils, he explained. Suspecting that Dectin-1 serves to recognize fungi in the gut, the researchers further explored its influence on inflammation in the presence of fungi. Dectin-1 knockout mice experienced increased levels of inflammation by every known measure, Underhill reported; in a chemically-induced mouse model of colitis, the knockout mice suffered more severe disease that featured fungal invasion of host tissue, which did not occur in wild-type mice (Iliev et al., 2012). “We think this is ultimately what contributes to the increased inflammation disease of these mice,” he added. “In the absence of Dectin-1, the innate immune cells … in the tissue [surrounding the colon] are unable to clear these fungi as quickly.” This conclusion was supported by the finding that, following induction of colitis, treating the knockout mice with fluconazole resolved their symptoms, but not those of wild-type mice, he added. Extending this observation to humans, Underhill and coworkers surveyed Dectin-1 gene sequences among patients with ulcerative colitis, and discovered a specific polymorphism associated with a two-fold increased risk for very severe disease. “The people who have the risk allele progressed to surgery much faster [than those who did not],” he reported. “There may be some predictive utility here.”
Host–Microbe Interactions in the Inflammatory Bowel Diseases
The term inflammatory bowel disease is used to describe a constellation of conditions associated with chronic or recurrent inflammation of the gastrointestinal tract. Two clinically defined forms of IBD are ulcerative colitis and Crohn’s disease, which “are chronic remittent or progressive inflammatory conditions that may affect the entire gastrointestinal tract and the colonic mucosa, respectively, and are associated with an increased risk for colon cancer” (Kaser et al., 2010). Extra-intestinal manifestations, including liver problems, arthritis, skin manifestations, and eye problems, also occur in Crohn’s disease patients. As one of the five most prevalent gastrointestinal diseases in the United States, IBD accounts
for more than $1.7 billion in health care costs, annually.47 According to the U.S. Centers for Disease Control and Prevention (CDC) patients often require medical care throughout their lifetime because there are no medical cures available for IBD. Once limited to the industrialized world, Crohn’s disease (Figure WO-25) and ulcerative colitis are emerging as global diseases (Molodecky et al., 2012).
When compared to that of “normal” individuals IBD patients exhibit abnormalities in their gut microbiome (Figure WO-26), including
- reduced or altered levels of dominant bacteroidetes and firmicutes phyla populations;
- an overall decrease in microbes known to be protective of gut tissues; and
- increased representation by proteobacteria, some members of which are recognized by the epithelium as pathogens, leading to inflammation.
These alterations, discussed at the workshop, represent a disruption in the normal homeostatic relationship that exists between the intestinal epithelium, the immune system, and the gut microbiome—each component of which is influenced by a variety of genetic and environmental factors. The presentations summarized below explore the complex interplay of these factors in the pathogenesis of IBD.
The Influence of the Mucosal Immune System and Gut Microbiota on IBD
Recent discoveries in genetics, microbiology, and immunobiology have helped to illuminate the complex interactions between host, microbiota, and environmental factors that contribute to the development and persistence of IBD (Figure WO-27). According to speaker Richard Blumberg of Harvard Medical School, research suggests that the altered microbiota in IBD patients is largely a secondary effect of conditions leading to IBD, rather than its primary cause (Lupp et al., 2007) (Dr. Blumberg’s contribution may be found on pages 153-164 in Appendix A). Indeed, various experimental regimes (including chemical treatment) can simulate the effects of IBD on the intestinal microbiota, he observed. That is not to discount the importance of dysbiosis, Blumberg continued; it may be inflammatory in and of itself, and thereby perpetuate IBD.
To identify the primary cause of IBD, Blumberg and coworkers sought to examine the coordinated development of the intestinal tract and its microbiome and ask the question, “When does IBD start?” In the course of these studies, Blumberg’s research team noted that germ-free mice suffered higher rates of mortality from chemically-induced colitis, when compared to conventionally raised mice (Olszak et al., 2012). This lethal inflammatory response was associated with
________________
47 See http://www.cdc.gov/ibd (accessed December 5, 2013).
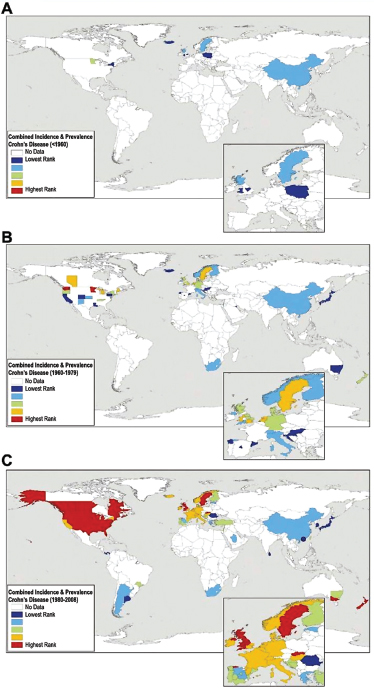
FIGURE WO-25 Worldwide Crohn’s disease incidence rates and/or prevalence for countries reporting data (A) before 1960, (B) from 1960 to 1979, and (C) after 1980. Incidence and prevalence values were ranked into quintiles representing low (dark and light blue) to intermediate (green) to high (yellow and red) occurrence of disease. Maps are based on a systematic literature review of 262 studies (1950–2010).
SOURCE: Molodecky et al. (2012).
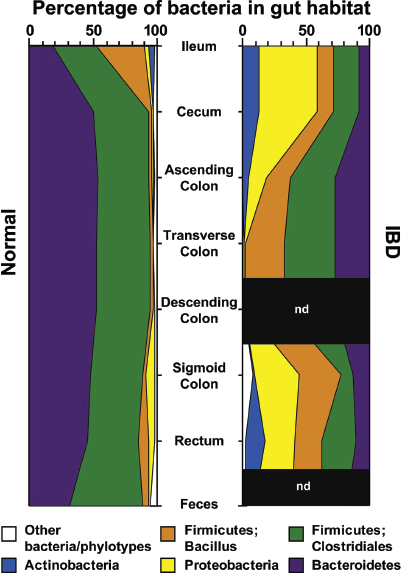
FIGURE WO-26 Bacterial phyla identified in the human gut microbiota. Distribution of predominant bacterial phylotypes in the human intestinal tract. The graphs depict relative abundance as a function of location along the cephalocaudal axis of the distal gut. DNA sequences were compiled from the studies of Eckburg et al. (2005) and Frank et al. (2007). Phylogenetic classifications were made by parsimony insertion of aligned sequences into a 16S rRNA tree provided by the Greengenes database (DeSantis et al., 2006), using ARB (Ludwig et al., 2004).
NOTES: IBD, samples from patients with either Crohn’s disease or ulcerative colitis (Frank et al., 2007); Normal, data from healthy young adults from Eckburg et al. (2005) plus the non-IBD controls reported in Frank et al. (2007); n.d., not done. A total of 18,405 16S rRNA sequences were analyzed (5,405 IBD, 13,000 normal).
SOURCE: Peterson et al. (2008).
natural killer T cells48 (NKT cells), which they found in increased concentrations in the intestinal epithelium and underlying lamina propria of germ-free mice. Other researchers (e.g., Jostins, 2012) have linked NKT cells to human IBD, and, Blumberg added, “there is a lot of reason to believe that NKT cells are a critical part of the biology of these disorders.”
Blumberg observed that the natural microbiota appear to protect conventionally raised mice from the damaging inflammation typical of IBD. Seeking clues to the process by which this protection arises, the researchers discovered that germ-free mice colonized with conventional microbes within the first 2 weeks of life did not suffer from NKT cell-mediated colitis. On the other hand, germ-free mice whose guts were colonized as adults, germ-free mice, and newborn conventional mice treated with antibiotics were susceptible to colitis (Olszak et al., 2012). “What this shows is that an environmental event that degrades or diminishes or abrogates the important microbial cue in the first 2 weeks of life allows the animal to become susceptible to an environmental trigger later in life that leads to the activity of this disease,” he said.
Protection against NKT-mediated colitis appears to involve a molecule called CXCL16, reported Blumberg. This molecule is produced by the cells of the intestinal epithelium and orchestrates infiltration of intestinal tissues by NKT cells. Blumberg observed that this process also takes place in the mucosal tissues of the lung. In the lung, infiltration by NKT cells has been linked to asthma in germ-free mouse models—a development that can be prevented by exposing neonates to a conventional microbiota during their first two weeks of life (Olszak et al., 2012) (Figure WO-28).
Blumberg also noted that such findings recall the hygiene hypothesis, which suggests that early-life exposures to a wide variety of microorganisms determine later-life susceptibility to immune-mediated diseases. First suggested by David Strachan to explain the relationship between household size, birth order, and hay fever in a birth cohort during 1 week in 1958 and followed for 23 years (Strachan, 1989), this hypothesis was later extended to include autoimmune diseases (Bach, 2002). Blumberg noted an increasing amount of epidemiological data that suggests that microbial exposures early in life may influence the later development of diseases such as asthma and IBD (Shaw et al., 2010; Ege et al., 2011). Blumberg concluded by noting that worldwide rates of IBD incidence and prevalence—like those of asthma—are on the rise (Benchimol et al., 2009;
________________
48 NKT cells are a unique group of T cells that, in contrast to classical T cells that recognize peptides, sense lipids. Lipids derived from host cells and microbes are presented to NKT cells by the CD1d molecule. “The NKT-CD1d pathway is present and operative in the intestines and very important in the management of pathogens and commensals,” said Blumberg. By responding to host and pathogen-derived lipids, NKT cells are able to initiate and control a variety of different inflammatory events. NKT cells are critical because they function as the earliest phases of the immune response—at the cusp between innate and adaptive immunity.
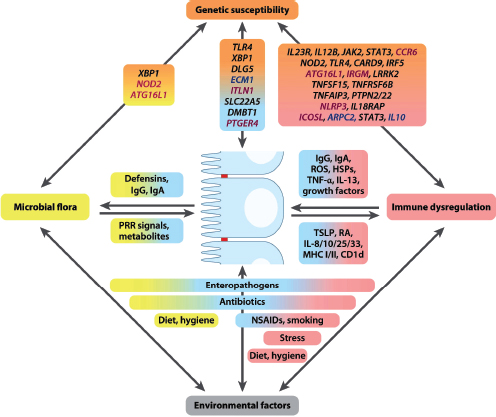
FIGURE WO-27 IBD as a multifactorial disorder. The development and course of IBD are affected by several factors including genetic susceptibility of the host, the intestinal microbiota, other environmental factors, and the host immune system. In addition, these factors cross-regulate each other in multiple ways, as shown. IBD-associated genes are summarized by molecular pathways with genes belonging to the same pathway arranged next to each other in one line. Polymorphisms in genes specific for associations shared between both diseases are shown in black text.
NOTES: HSPs, heat shock proteins; MHC, major histocompatibility complex; NSAIDs, nonsteroidal anti-inflammatory drugs; PRR, pattern-recognition receptor; RA, retinoic acid; ROS, reactive oxygen species; TSLP, thymic stromal lymphopoietin.
SOURCE: Kaser et al. (2010).
Molodecky et al., 2012). Children in the first 5 to 10 years of life appear to be the most vulnerable to environmental events associated with these diseases (Benchimol et al., 2009).
Genetics of Susceptibility in Microbiota-Associated Complex Diseases
Based on family studies, metagenomic analyses of Crohn’s disease and ulcerative colitis have confirmed earlier assumptions that genetic predisposition
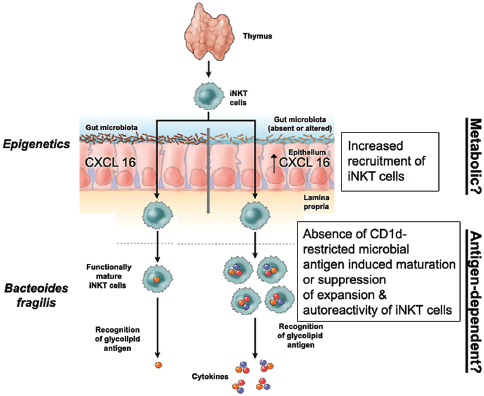
FIGURE WO-28 Potential mechanisms of NKT cell control in mucosal tissues by commensal microbiota during early life. Exposure to antigen independent (metabolic?) and antigen-dependent (bacterially-derived sphingolipids?) factors derived from the symbiotic microbiota determine the homeostatic set-point of NKT cell pathways associated with mucosal tissues in organs such as the colon and lung. These include those that affect the infiltration of these tissues with NKT cells and the degree to which they are activated by self and nonself lipid antigens. Such signals are critically received during early life and associated with durable (epigenetic) changes that provide protection from later life environmental triggers that are capable of activating NKT cells and causing inflammation and disease. In the absence of these essential microbial exposures during early life (as associated with early-life use of antimicrobials), these protective signals are not provided.
SOURCE: Adapted from Coppieters and Elewaut (2012).
plays an important role in IBD, and suggest that a vast array of possible factors— directly or indirectly—contribute to risk for these complex diseases (Khor et al., 2011; Jostins et al., 2012; Graham and Xavier, 2013). Speaker Ramnik Xavier of the Harvard Medical School described his perspective on the intricate relationship between host genetics and the microbiota that he and fellow investigators are gradually unbundling (Dr. Xavier’s contribution may be found on pages 471-488 in Appendix A). Following perturbation by exposure to antibiotics, early-life
events, or changes in microflora, the extent to which an individual can reverse the ensuing disease state is strongly influenced by genetic makeup, Xavier noted. By examining the functional pathways and immune responses that underlie IBD, he and his colleagues hope to reveal host genetic and microbial factors that increase or decrease risk for disease.
Reduced taxonomic diversity in the gut microbiome is now recognized as one of the hallmarks of IBD, along with characteristic increases and decreases in specific bacterial clades that may define specific subtypes of these disorders, Xavier said. Examining the functional implications of these shifts, he and colleagues discovered that they were accompanied by striking declines in carbohydrate and amino acid metabolism, short-chain fatty acid synthesis, and DNA maintenance, accompanied by increased secretory and invasive activity (Morgan et al., 2012). Xavier observed that IBD-associated microbes collectively can take up increasing amounts of host products, abrogate immune system control of inflammation (leading to further damage), and adhere more readily to the damaged epithelia. Most importantly, he continued, the functional picture of the IBD-associated microbiota is that of a community more tolerant to oxidative stress—a feature that links them to many risk loci identified in humans known to control oxidative metabolism, suggesting communication between endogenous microbes and the host genome (Figure WO-29).
To illustrate his research group’s approach to understanding the many and diverse mechanisms that underlie these interactions, Xavier described his team’s interest in autophagy—a constitutive and conserved process present from yeast to humans and active in every cell type—that describes the engulfment and degradation of cytoplasmic contents by lysosomes, a process associated with Crohn’s disease and other inflammatory disorders (Kuballa et al., 2012). “There are an increasing number of disease associations with several of the autophagy genes,” Xavier noted. “In addition to Crohn’s disease, there is now emerging evidence that some of the neurodegenerative diseases are associated with polymorphisms in autophagy genes, and we and others have more recently identified that some genes might also be associated with increased risk of chronic infections such as tuberculosis.” Autophagy serves a range of adaptive purposes including generating energy, healing inflammation, and controlling antigen presentation, he explained, but clearly it has the potential, if defective, to cause disease. One of the autophagy genes associated with susceptibility to Crohn’s disease is Atg16L1, also discussed by Virgin as a component of an “immunophenotype” model of disease. As described in Virgin’s presentation, mice expressing this gene in their epithelial tissues have abnormal Paneth cells and are defective in immune defenses in the presence of an enteric virus.
Recognizing that host gene polymorphisms can influence the host–microbe relationship opens opportunities for treating IBD and other complex microbiotaassociated diseases such as psoriasis and obesity, Xavier concluded. According
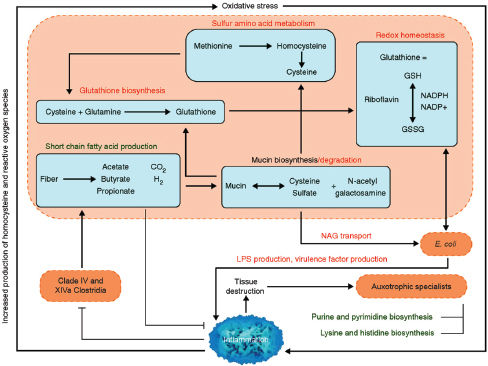
FIGURE WO-29 Proposed metabolic roles of the gut microbiome in IBD. Host-mediated processes (blue text) create an environment of oxidative stress in the intestine, which is more favorable to Enterobacteriaceae (increased abundance) than to clades IV and XIVa Clostridia (decreased abundance). This study’s inferred IBD metagenomes include broadly increased oxidative metabolism, decreased SCFA production, and increased mucin degradation relative to healthy subjects. These processes all occur within microbes and rely on transport of small molecules to and from the lumen. The resulting tissue-destructive environment provides nutrients such as nucleotides and amino acids, which allow for increased growth of auxotrophic “specialists.” Bacterial clades of interest are indicated in orange, bacterially mediated processes increased in IBD in red, and processes that decrease in green. Metabolic pathways differential in our IBD communities are contained in blue boxes.
NOTES: GSH and GSSG indicate reduced and oxidized forms of glutathione; LPS, lipopolysaccharide; NAG, N-acetyl galactosamine.
SOURCE: Morgan et al. (2012).
to Xavier, IBD research has progressed from identifying host gene associations, to examining the consequences of metabolites and the effects of diet, to monitoring the functional effects of gene expression, and recognizing disease-associated shifts in microbiome composition. Today, new technologies are needed to elucidate the mechanisms that connect human genetics and microbiome composition, resulting in complex diseases (Graham and Xavier, 2013).
CHALLENGES AND OPPORTUNITIES
We owe much of our biology and our individuality to the microbes that live on and in our bodies—a realization that promises to radically alter the principles and practice of medicine, public health, and basic science.
—Relman (2012)
The human metagenome is orders of magnitude more manipulatable than the human genome. This difference provides the opportunities to intercede to prevent and treat illness, if only we knew what was important!
—Blaser et al. (2013)
We have only just begun to appreciate the exquisitely complex relationships between microbial ecology and community dynamics in states of health and disease. The wealth of data and insights emerging from early studies of our resident microbes have complemented our growing knowledge of the human genome and furthered efforts to develop a holistic and dynamic view of health and disease states (Grice and Segre, 2012). The emerging picture is dauntingly complex, in which every individual is a unique host–microbial ecosystem—a dynamic assemblage of human and microbial cells that can be modulated throughout our lifetimes in ways that influence health and disease risk factors (Gonzalez et al., 2011; Relman, 2012). Many believe that this more nuanced conception of health and disease will move us closer to a vision of personalized medicine (Virgin and Todd, 2011; Holmes et al., 2012).
Exploration of the human microbiome is just one extension of the work initiated by environmental microbiologists over a half a century ago to reveal the vast diversity and complexity of the microbial world around us (Robinson et al., 2010). The same tools and techniques used to explore the microbial communities that live in and on us are also being used in a variety of fields to explore the role of microbial communities in driving biosphere processes—from the conversion of energy from the sun to the bioremediation of polluted landscapes. Research to identify factors influencing the formation, function, and stability of microbial communities in a wide range of biotic and abiotic systems will have unanticipated benefits for human, animal, plant, and ecosystem health and well-being (IOM, 2012).
Toward Ecological Therapeutics
Given the ecological parallels between assembly of the human microbiome and assembly of other ecological communities, we suggest that human medicine has more in common with park management than it does with battlefield strategy. To effectively manage a plant or animal community requires a multipronged approach of habitat restoration, promotion of native species, and targeted removal of invasives.”
—Costello et al. (2012)
Unlike the host genome, our microbiome is dynamic, raising the possibility that it can be modified—with regard to its diversity, abundance, or resilience or in order to promote specific interactions between and among its microbial communities and the host—for therapeutic purposes (Lemon et al., 2012). Disease prevention and treatment would nurture the microbiome as a provider of critical services to the host. Changes in the microbiome associated with different stages of life or life events may present critical windows of opportunity for therapeutic interventions (Figure WO-30). Careful monitoring of community composition or function (via small molecules or other biomarkers) may better inform clinical decision making and help to devise strategies that favor host health (see Costello et al., 2012; Holmes et al., 2012).
Therapies designed to shift the microbiome from states associated with host disease to those that promote host health could take many forms, including
- prebiotics, which are substrates that promote targeted growth of a limited
- number of (beneficial) members of the microbiota;
- probiotics, which are live microorganisms that confer a health benefit on the host;
- small molecules and biological drugs targeted to the microbiota or to microbiota-responsive host factors;
- narrow-spectrum antibiotics to minimize collateral damage to the mutualist members of the microbiota, while decreasing selective pressure for the spread of antibiotic-resistant strains; and
- antibacterial conjugate vaccines that stimulate the immune system to remove only specific strains of a single species from the microbiota (see Sonnenburg and Fischbach, 2011; Lemon et al., 2012).
While noting that we have much to learn about whether and how to manipulate established microbial communities to achieve such goals, several speakers described efforts to lay a foundation for future microbiota-directed therapies— and in one case, to successfully treat potentially lethal dysbiosis through “ecological” intervention.
Therapeutic Molecules from the Human Microbiome
The chemical “conversations” that take place between host and microbe, and among members of the human microbiome, offer a promising source of molecules with therapeutic properties, according to speaker Michael Fischbach of the University of California, San Francisco (Dr. Fischbach’s contribution may be found on pages 273-291 in Appendix A).
FIGURE WO-30 Therapeutic modulation of the gut microbiota: from the cradle to the grave. The changing relationship between the gut microbiota and the human host throughout life offers a series of time windows for therapeutic intervention. During infancy and early childhood, a healthy gut microbiota helps to avoid dysbiosis that can lead to disease later in life. In adulthood, corrective strategies to deal with emergent dysbiosis and associated diseases such as inflammatory bowel disease, obesity, and type 2 diabetes include modulating the microbiota using probiotics and prebiotics, antibiotics, bariatric surgery, or “drugging the microbiota.” Drugs targeted to microbial enzymes could also be used to boost the efficacy and reduce the toxicity of therapeutics. For extreme cases of gut-related disease, fecal transplantation or helminth therapy may prove beneficial. In the future, it may be possible to prevent or treat abnormalities of the gut microbiota using modified organisms engineered through genetic or synthetic biology approaches.
SOURCE: Holmes et al. (2012).
FIGURE WO-31 Natural products with medicinal relevance.
SOURCE: Newman et al. (2003).
Indeed, he noted, many current drugs derived from natural products49— including most antibiotics, the antihypercholesterolemic lovastatin, and the cancer drug doxorubicin, among others—are likely to have evolved to modulate interactions among microorganisms and between microbes and their hosts (Figure WO-31). Long a mainstay of drug discovery, natural products can now be identified and characterized with new technologies such as genomics and bioinformatics. These inquiries not only reveal novel and potentially useful compounds, he observed, but also provide “a new window into the biology of the organisms that produce these molecules, and maybe new ways of understanding why they make them.”
The process of discovering natural products traditionally began with an environmental sample, such as soil, from which a wealth of small molecules—produced by microbes—could be isolated and characterized. Today, however, insight into the genetic architecture underlying the synthesis of these molecules presents the opportunity to rapidly identify sets of natural product genes in microbial genomes and connect them to the molecules they ultimately manufacture (Walsh and Fischbach, 2010). “If we … just went through all the bacterial genomes in the database and asked ourselves can we find all the sets
________________
49 Chemical compound or substance produced by a living organism.
of genes that probably are involved in making a small molecule … [then] we could make a prediction about what those molecules might be,” Fischbach asserted. Such a search is feasible because of shared features of natural product gene sets that make them readily identifiable, he explained: clusters of genes— often located on mobile elements—that permit “assembly line” synthesis of small molecules.
Genomic databases provide abundant raw material for such searches, Fischbach said. “There are lots of genomes and metagenomes that have been sequenced, and the tools that we have developed are quite good at going through [these data] and finding sets of genes that encode a small molecule,” he elaborated. He added that many well-characterized organisms that produce natural products such as antibiotics possess additional natural product gene sets that remain to be characterized (Figure WO-32). “This means that we probably do not need to go to the corners of the earth to find new molecules,” he continued. “Maybe we should just go to our backyard or our freezer collection and find a way of tickling these organisms so they turn on the production of molecules they probably don’t normally make under the artificial condition of cultivation in the lab.”
In Fischbach’s lab, the search for novel small molecules begins with computational analyses of genome sequences. “We have a training set of about 750 known biosynthetic gene clusters with known small molecule products, and we’ve devised a very simple machine-learning algorithm that has recovered an enormous set of biosynthetic gene clusters,” he said. Fischbach’s lab has begun to focus on the human microbiome because “many of the most interesting biosynthetic gene clusters that we found were not in exotic soil and marine microorganisms; they were in [human] gut and skin and oil bacteria.” Experiments on candidate gene clusters have borne out the hypothesis that these molecules mediate host–microbe interactions and have led to the discovery of several intriguing small molecules that the Fischbach lab is currently investigating.
These initial findings represent just a tiny fraction of the untapped potential of small molecules produced by the human microbiome, noted Fischbach. “If I were to ask you, ‘What are the top 10 bacterial strains by abundance in your gut?,’ you would probably be able to figure the answer out by looking through some recent publications,” Fischbach remarked. “But if I asked you what are the top 10 molecules in your gut and how do they compare one person to the next, you probably would not know, and I certainly don’t know—but I think it’s going to be just as important a question if these molecules indeed mediate signaling among microorganisms and between them and the host,” he concluded. “There will be some interesting things to learn over the next few years.”
FIGURE WO-32 Genomics can reveal cryptic natural products. Saccharopolyspora erythraea is a well-characterized bacterial species that is cultivated on an industrial scale to produce the potent antibiotic, erythromycin. Analysis of this bacterium’s genome revealed a number of additional gene clusters that potentially encode as yet unknown natural products.
SOURCE: Adapted from Oliynk et al. (2007).
Antimicrobial Peptides: Innate Defense Against Infection
The arsenal of proteins produced by the innate immune system represents another untapped resource for understanding and potentially manipulating host–microbe interactions (Figure WO-33). AMPs, an important component of innate immunity, have played a fundamental role in the evolution of multicellular life in a microbial world and represent an important paradigm for therapeutics, according to speaker Michael Zasloff of Georgetown University (Zasloff, 2002a,b) (Dr. Zasloff’s contribution may be found on pages 489-496 in Appendix A).
AMPs protect the epithelial surfaces of all multicellular organisms (Zasloff, 2002a). Although diverse in composition, they share certain basic structural properties, most notably an affinity for the negatively charged outer surfaces of microbial cytoplasmic membranes. Among the more than 500 different antimicrobial peptides that have been discovered in organisms from insects to humans, most have a broad spectrum of activity and are capable of killing bacteria, fungi, and viruses. Most organisms display several different types of peptides on various epithelial surfaces, which together comprise a broad-spectrum antimicrobial
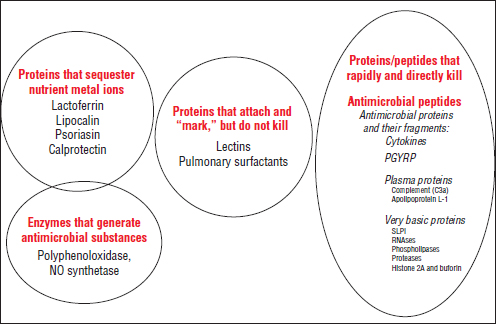
FIGURE WO-33 The secreted anti-infective protein/peptide arsenal of innate immunity.
SOURCE: Zasloff (2013).
cocktail. “Anywhere on the body where a bacterial or fungal organism could creep in from the outside, we find an array of antimicrobial peptides,” Zasloff observed. Zasloff also noted that AMPs appear to be highly adapted to the host organism’s macroenvironment, as well as to the microenvironment of the specific tissues where they are expressed: “They are different, how they’re regulated is different, what stimulates their expression is different, [and] their constitutive levels are different.”
In the years since their initial discovery in the African clawed frog, Xenopus laevis (Zasloff, 1987), several AMPs have been developed as topical antibiotics (Zasloff, 2002b). Although AMPs are less prone to microbial resistance than conventional antibiotics and are highly active in vitro, in animal models of infection these molecules often require effective doses that are so high as to be unsafe. Nevertheless, the ubiquity, diversity, and effectiveness of AMPs continue to intrigue researchers.
AMPs have a variety of attributes through which they help mold and influence the microbiome. They are often produced on an ongoing basis, although some are induced by tissue injury and contribute to tissue defense and repair. Zasloff described these broad-ranging effects in sites such as the mammalian tongue and hair follicles. Both sites feature a complex array of both constitutive and inducible AMPs, which may contribute to the fact that although these sites seem vulnerable to pathogens, they in fact rarely become infected. He also
discussed the activation of the human AMP known as LL37 by vitamin D, as the means by which sunlight exposure stimulates antimicrobial defense of the skin and blood cells, noting that this phenomenon underlies phototherapy as a cure for tuberculosis infection of the skin (Liu et al., 2006).50 AMPs are also of interest because of their involvement in tissue repair, Zasloff noted, for example, that LL37 interacts with receptors that in turn stimulate angiogenesis51 (Koczulla et al., 2003).
Several diseases in humans are associated with impaired AMP function (Zasloff, 2002a). They include cystic fibrosis, in which a disturbed mucus barrier impedes the effectiveness of AMPs; shigellosis, in which the pathogen appears to suppress AMP expression, thereby increasing the likelihood of dysentery; and eczema, in which a dysfunctional inflammatory process suppresses AMP expression (Ong et al., 2002). Understanding the mechanisms by which AMPs shape the microbial ecology of their host and the processes underlying AMP-associated diseases offers another route to therapeutic discovery, Zasloff said.
To illustrate how much is left to be learned about host–microbe interactions, Zasloff concluded his presentation with his observations about the remarkable wound-healing capacity of dolphins that had suffered a severe shark bite (Figure WO-34) (Zasloff, 2011): “If one of us were bitten by a shark … we most certainly would die of sepsis. The shark has all sorts of Vibrio in its mouth and [shark bites] are among the most difficult infections to treat in man.” However, continued Zasloff, dolphins in the ocean will “heal without infection or inflammation and with virtually no blood loss.” Another way of isolating antimicrobial agents or identifying mechanisms for tissue repair is to study animals such as this, which appear to be “a warm blooded mammal [that has achieved] the most perfect solution to its coexistence with microbes.”
Diagnostic Information from the Skin Microbiome
Human skin surfaces are complex ecosystems that provide diverse environments for our resident microorganisms, observed speaker Julie Segre, of the National Human Genome Research Institute (Dr. Segre’s contribution may be found on pages 401-412 in Appendix A). In her discussion of recent phylogenetic surveys of bacterial (Grice et al., 2009) and fungal (Findley et al., 2013) diversity within and across multiple skin sites in healthy humans, Segre highlighted how these data are identifying opportunities to better diagnose and treat microbiotarelated skin disorders. These findings are also providing insights into the role of endogenous microbes in disease states and the microbial interdependencies required to maintain healthy skin.
________________
50 Zasloff noted that Neils Finsen received the Nobel prize in 1903 for the development of phototherapy as a cure for tuberculosis infection of the skin.
51 The formation of new blood vessels, especially blood vessels that supply oxygen and nutrients to cancerous tissues.
FIGURE WO-34 Wound healing in a severely wounded bottlenose dolphin. Injured animal was first observed on February 13, 2009. Photos were provided by Trevor Hassard who headed the rescue team at Tangalooma. Scale bar in Panel A corresponds to about 30 cm on the animal’s surface.
SOURCE: Zasloff (2011).
Genomic surveys of the skin microbiome detected far greater microbial diversity—on the genera and species levels—than had previously been appreciated by studies limited to culturable organisms, Segre observed. Both bacterial and fungal diversity were found to be markedly site dependent—a finding that may help to explain the predilection of skin disorders for stereotypical sites. “Dermatologists have known for a long time that part of the diagnosis of a rash is about where it is on your body,” she observed; for example, “eczema is always on the inside of the elbow and psoriasis is on the outside.” Different forces appear to shape that diversity, she reported. While different bacterial communities were associated with oily, dry, or moist skin, fungal communities assorted themselves by body part. She also noted that they observed a dramatic shift in the skin microbiota associated with individuals before and after puberty (Oh et al., 2012), suggesting that there are transitions that we go through as humans that may be times at which we could “reset” our microbiota.
To illustrate the ways in which genomics is providing information that clinicians can use fairly rapidly, Segre discussed her lab’s work on atopic dermatitis (AD), more commonly known as eczema, a condition associated with Staphylococcus aureus infection that affects approximately 15 percent of children in the United States (Kong et al., 2012). Over the past 30 years, the prevalence of atopic dermatitis has doubled, Segre stated, and it is widely suspected that this trend reflects changes in the skin microbiome resulting from decreased exposure to common infections in early life, in accordance with the hygiene hypothesis (Strachan, 1989). These children experience periodic “flares” of inflammation and intense itching on the insides of their elbows and behind their knees. In addition, Segre observed, approximately half of the children afflicted with severe eczema go on to develop asthma.
Through a clinical trial enrolling children with moderate to severe eczema, Segre and coworkers evaluated bacterial community diversity in the affected skin areas at baseline, during a flare, and after its resolution (Kong et al., 2012). During a flare, bacterial diversity plummets, and the amount of S. aureus dramatically increases, Segre reported. These observations may contribute to the generation of research hypotheses regarding potential preventable causes of flares; they may also be used as a signal to provide an early warning of incipient flares, and thus more effective management of this skin condition, she explained (Figure WO-35). Eczema, she noted, is currently treated with a variety of therapies, only some of which are effective for any given patient. According to Segre, it may eventually be possible to determine which therapies work best based on the species or genomic composition, and to treat disease earlier—possibly reducing their risk of progression to asthma. “We don’t have to wait until we are all the way through these stories to come up with the mechanism if there are biomarkers we can use, based on the microbial diversity, to tailor therapies for patients or to predict who is about to have a skin flare,” she concluded.
Fecal Microbiota Transplantation for Recurrent Clostridium difficile Infection
Over the past two decades, Clostridium difficile infections in humans have become among the most common hospital-acquired infections in the United States. Linked to more than 14,000 deaths (U.S.) each year, C. difficile infections have also become more difficult to treat (Kelly, 2013). According to speaker Josbert Keller of the University of Amsterdam, most C. difficile cases respond to antimicrobial therapy, but approximately one-quarter of patients experience a recurrent infection, which in turn raises their risk of subsequent relapses (Dr. Keller’s contribution may be found on pages 347-355 in Appendix A). “Clostridium difficile [infection] can only occur in a bowel with a disturbed intestinal flora,” he observed. “[Cases] typically occur after antibiotic treatment, and the longer such patients are hospitalized, the greater their risk of developing a C. difficile infection, with symptoms ranging from diarrhea to potentially deadly
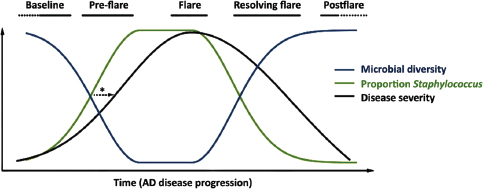
FIGURE WO-35 Atopic dermatitis (AD) progression hypothesis. Episodes of increased disease severity are accompanied by changes in microbial diversity. According to Segre, biomarkers for changes that occur during the preflare stage (*) may help patients to predict incipient flares and clinicians to better tailor therapies to individual patient needs
SOURCE: Kong et al. (2012).
fulminant colitis,” he said. For patients with a second recurrence of C. difficile infection after antibiotic treatment, subsequent courses of antibiotics—typically vancomycin—have a success rate of less than 50 percent. In those patients, the prolonged disturbed bowel flora seems to have lost its restorative capacity (Chang et al., 2008), and the antibiotics fail to kill the pathogen’s spores. In addition, an ineffective immune response may also contribute to the pathogenesis of recurrent C. difficile infection (Kyne et al., 2001).
Over the course of the past 50 years, more than 300 patients with recalcitrant C. difficile infections have been effectively treated by fecal microbiome transplantation (Figure WO-36) (Borody and Khoruts, 2012; Kelly, 2013).52 While procedures have varied significantly, “What is consistent in all those reported patients is that there are very high success rates of about 90 percent,” Keller stated. He and colleagues, having already successfully treated several patients with this technique, launched a clinical trial to compare fecal microbiome transplantation with a standard vancomycin regime (van Nood et al., 2009, 2013). The trial was halted early because of poor outcomes among patients who received vancomycin—more than two-thirds failed to resolve their infections, as compared with more than 80 percent of patients who received a single infusion of donor feces. Most of the patients in the antibiotic-treated group were subsequently treated with donor feces infusions, which cured infections in 15 out of 18 cases (Figure WO-37) (van Nood et al., 2013).
Patients who received donor feces infusions reported few side effects, Keller said, and most were cured within a day of treatment and remained free of
________________
52 The use of diluted mixtures of fecal matter from healthy animals has long been used by veterinarians as a treatment for stomach ailments, particularly in racehorses (McKenna, 2011).
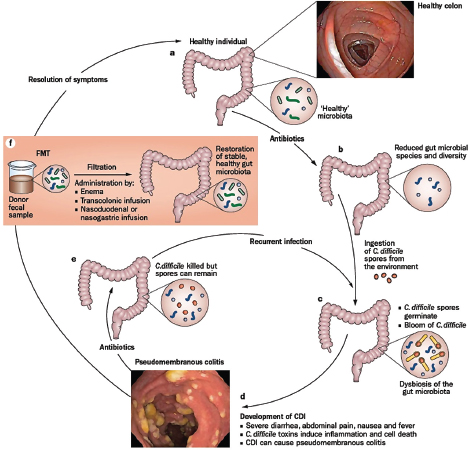
FIGURE WO-36 Fecal microbiota transplantation for patients with recalcitrant Clostridium difficile infection (CDI). CDI causes severe diarrhea, intestinal inflammation, and cell death as a result of toxin-mediated infection with the pathogenic bacteria. Patients with CDI are typically treated with antibiotics, which not only kill the pathogenic C. difficile but also exhibit activity against the dominant colonic microbiota phyla. Incomplete antibiotic eradication of C. difficile and persistent disturbance of healthy gut microbiota can result in recurrent CDIs. Transplantion of fecal microbiota from a healthy donor into an individual with CDI can restore the healthy gut microbiota in the patient’s diseased colon, thereby probably preventing further outgrowth of C. difficile and leading to resolution of symptoms.
SOURCE: Borody and Khoruts (2012).
C. difficile during the 10-week follow-up period. Before-and-after comparisons of patient bowel flora revealed that the procedure increased microbiota diversity to resemble that of the donor, he reported. While Keller expressed some concern about careful screening of the donor feces to prevent the transmission of an infectious disease from a donor to recipient, he concluded that fecal microbiota transplantation was a safe (if unappealing) treatment for C. difficile infection.
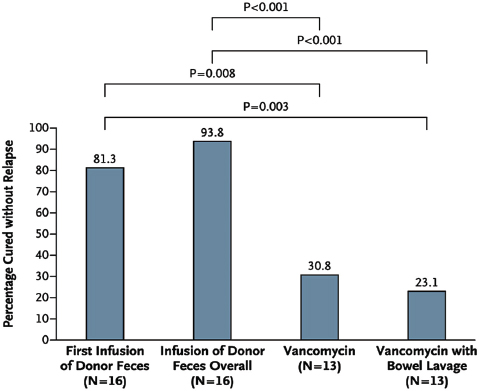
FIGURE WO-37 Rates of cure without relapse during 10 weeks of follow-up for recurrent Clostridium difficile infection. Shown are the proportion of patients who were cured by the infusion of donor feces (first infusion and overall results), by standard vancomycin therapy, and by vancomycin therapy plus bowel lavage.
SOURCE: van Nood (2013).
Keller also noted that researchers are investigating the potential for fecal microbiota transplants to treat other diseases, including IBD, obesity, and metabolic syndrome. However, he cautioned, for diseases other than recurrent C. difficile infection, fecal transplantation is a very experimental approach, and much work remains to be done before it is ready for clinical application.
Keller expressed his hope that a more appealing probiotic alternative to fecal transplantation would soon become available. A mixture of 10 different facultatively aerobic and anaerobic bacteria diluted in sterile saline has been used as a treatment for chronic relapsing C. difficile infection (Tvede and Rask-Madsen, 1989). He also discussed a recent proof of principle study that used a synthetic stool mixture or human probiotic called RePOOPulate. Administration of this multispecies mixture (33 purified isolates of bacteria from a healthy donor) resolved C. difficile infections in two patients, each of whom had failed to recover after at least three courses of antibiotics. Sequencing performed before
and 6 months post treatment suggests that the strains present in this multispecies community of bacteria had stably colonized the colon (Petrof et al., 2013).
Regulatory Considerations
The various potential and actual interventions described during the workshop, Relman noted, raise complex questions about oversight and prudent regulation. For example, efforts to alter microbiota function by introducing microbial communities, rather than a specific species, will be difficult to monitor, he said. “You are no longer saying we either did or did not have this organism, or did or did not see a fall in this particular well-understood end point,” Relman explained. In these cases, biomarkers and surrogate endpoints will need to be developed to gauge the effectiveness of treatment, he concluded.
Calling fecal transplantation a “great test case,” Goodman nonetheless cautioned about potential harms inherent in unrefined microbiota-based approaches to medicine, including the introduction of antibiotic-resistant bacteria through probiotic therapy. “I think that this is one of these emerging areas where the trick is going to be to get the right balance between protecting patients and letting the field … move forward,” he observed. In the case of fecal transplantation, Goodman agreed with Keller’s prior assessment that it provides a starting point for developing more precise treatments, based on an understanding of the therapeutic effects of specific microbial strains.53
Even with this knowledge, there are risks, as Relman pointed out, through the example of the use of “bacterial interference” therapy in the 1960s, a practice premised on the idea of using an “interfering” nonpathogenic strain of Staphylococcus aureus to prevent colonization of the nasal mucosa by pathogenic strains. Doctors successfully used interference therapy to curtail epidemics of disease caused by pathogenic strains of S. aureus in nurseries, the recurrence of boils in older persons, and persistent nasal carriage of pathogenic strains of S. aureus in adults (Boris et al., 1964; Light et al., 1965; Mackiowak, 1982). In 1972, Houck et al. (1972) reported that a 5.9 percent (38) of newborns treated as part of a bacterial interference program later developed disease thought to be related to the “interfering” strain of S. aureus, including pustules, conjunctivitis, and abscess. One newborn died from septicemia and meningitis (Houck et al., 1972).
________________
53 On May 2, 2013, the Food and Drug Administration, Center for Biologics Evaluation and Research, and the National Institutes of Health’s National Institute of Allergy and Infectious Diseases, jointly hosted a public workshop titled “Fecal Microbiota for Transplantation” to exchange information with the medical and scientific community about the regulatory and scientific issues associated with fecal microbiota for transplantation. For more information, see http://www.fda.gov/BiologicsBloodVaccines/NewsEvents/WorkshopsMeetingsConferences/ucm341643.htm (accessed January 9, 2014).
Rethinking the Antimicrobial Arsenal
Dramatic changes in human ecology over the past 40 years may have longstanding consequences for the transmission and selection of our microbiota (Table WO-2). As noted in many of the workshop presentations, correlations between changes to our microbial ecology and the recent, rapid rise of many chronic diseases may reveal the limitations of clinical medicine’s “war” on microbes that has been pursued since the articulation and promotion of the germ theory of disease in the 19th century.54 Rather than wage a war on germs, we should be conserving—and possibly replacing—our ancestral microbiota, noted Blaser. This notion was raised in presentations and discussions throughout the workshop, culminating in a lively conversation on the need to consider the collateral damage to the beneficial microbes associated with antibiotic use; whether and how we should restrict the use of antimicrobial interventions—from antibiotics and antiseptics to the widespread use of hand sanitizers—and how to convey such messages to a germ-phobic public.
Amplifying Blaser’s suggestion that current levels of antibiotic exposure in children may lead to lifelong repercussions such as asthma, obesity, and increased susceptibility to infectious diseases, Belkaid speculated that “the sum of the infection and the antibiotic treatment is very likely to be [much] more severe than just antibiotic treatment.” She advised that trials be devised to assess these risks over the long term. Blaser agreed, stating that “[t]he experiments have to be done, the epidemiologic studies have to be done, the clinical studies have to be done, the animal models have to be done, and they are being done.” In the meantime, Blaser added, it should not be assumed that antibiotic use comes without a biological cost—that they are “innocent until proven guilty. The public is still [waging a] war against germs, and we have to educate the public better, that there are good germs as well as bad ones,” he concluded.
Using antimicrobials on the skin “is an absurdity,” said Belkaid. “I think there is absolutely no reason to remove what we have evolved with to be a perfectly viable way of coexisting.” The skin is an important entry point for allergens and has broad inflammatory capabilities, so its natural barriers should not be disturbed, she said. “I think being exposed to commensals or microbes is a perfectly healthy way of living.” The use of all antimicrobials, including preservatives, should be questioned, Zasloff added, but public fear mongering should be avoided.
Illustrating the different ways that microbes are currently perceived by the public, Segre commented on the “great divide between Activia® yogurt and Purell® hand sanitizer” noting that “in this country we want to colonize our guts
________________
54 The germ theory of disease, consolidated in the 19th century by Louis Pasteur and Robert Koch, understood each acute infectious disease to be caused by a single pathogen. Isolation of the pathogen via “Koch’s postulates” provided standard confirmation. Once isolated, the pathogen could be targeted with antimicrobial therapies, with antibiotics becoming the weapon of choice in the 20th century. The suitable metaphor for expressing the relationship between microbes and humans as understood at the time was thus “warfare” (Lederberg, 2000; IOM, 2006).
TABLE WO-2 Changes in Human Ecology That Might Affect Microbiota Composition
| Change | Consequence |
| Clean water | Reduced fecal transmission |
| Increase in Caesarean sections | Reduced vaginal transmission |
| Increased use of preterm antibiotics | Reduced vaginal transmission |
| Reduced breastfeeding | Reduced cutaneous transmission and a changed immunological environment |
| Smaller family size | Reduced early-life transmission |
| Widespread antibiotic use | Selection for a changing composition |
| Increased bathing, showering, and use of antibacterial soaps | Selection for a changing composition |
| Increased use of mercury-amalgam dental fillings | Selection for a changing composition |
SOURCE: Blaser and Falkow (2009).
and sterilize our skin.” The public needs to understand the difference between protecting vulnerable patients from life-threatening infection and everyday hygiene, Segre contended. Though Americans have largely manufactured their “need” for hand sanitizers, she strongly advocated hand washing. “We are concerned about oral-fecal transmission,” she explained. “That is really what we need to be worried about in terms of the normal human health … [that] you are washing your hands after going to the bathroom, before you prepare food, or eat food.”
Manipulation of Our Microbiomes
Given mounting evidence of the biological costs of antibiotic therapy—long considered a medical triumph—workshop participants were at best cautiously optimistic regarding prospects for the strategic manipulation of microbiota to improve host health. While the use of probiotics to treat IBD (as described by Keller), and perhaps the early-life replacement of H. pylori (as described by Blaser), or prebiotic creams to maintain the skin microbiota (as mentioned by Segre) have been proposed, some participants expressed overall skepticism with the notion of altering microbiota. “The idea that maybe we can intervene somehow and manage this system in a very effective way to sort of re-create what should have happened naturally—I hope we can,” Forney stated. “But we are talking about trying to tweak something that has developed over hundreds of thousands of years, with all the unsuccessful experiments becoming extinct.”
Virgin disagreed with the premise that we are “trapped by our own evolution.” On the contrary, he said, “We can make cells do things that they can’t do ‘naturally’ by manipulating signaling pathways, once we understand that, I think the idea that you can reprogram things is perfectly legitimate.” McFall-Ngai
agreed, on the basis that human life histories are varied and plastic, and our relationship with our microbiota is a developmental one.
Douglas presented another view, stating that we should be careful not to be too “Panglossian”55 about our relationship with our microbiota. “It could well be that in some instances, what we’re seeing is that the animal host or human host is actually addicted to the microbiota,” she said. “Perhaps we can’t do without them—not because they are beneficial, but because animal signaling and metabolic systems have evolved in the context of the universal presence of these microorganisms. We should discriminate quite carefully between instances of addiction versus instances of benefit.”
“We certainly are not smart enough to think that we could deliberately reprogram the entire [microbiota],” Relman said—moreover, he added, such a feat could only be achieved through a series of manipulations, akin to successional stages. Rohwer agreed, observing that science is far better at tearing things apart than putting them together. “If you really want to reestablish a complex ecosystem, it may be very complicated,” he concluded.
Such hypothetical debates typify this emerging field of inquiry, during what Relman called “a lush time in the early phase of a science before the reality sets in and we come back down to some reasonable level of balance.” In the meantime, he observed, “It is a pretty exciting time in this area of work, and it’s made exciting by the fact that we have the means of discovering complexity that offers untold possible explanations and mechanisms. But we don’t know enough about any of it to realize that much of it may be wrong.”
Advancing Research
Several workshop participants emphasized the importance of assembling multidisciplinary teams and to train the next generation of scientists. Goodman advocated the training of generalist scientists, as well as specialists, who understand clinical and population science in a broad context, as well as basic research, and can coordinate members in an interdisciplinary team and evaluate the overall value of their work. Otherwise, he worried, “big science” may be done that is either irrelevant or that drains funding and talent from better opportunities.
There is great potential for collaboration—indeed, it will be essential. “Understanding this level of biological complexity will require the involvement of statisticians, computational biologists, geneticists, pathogenesis experts, virologists, bacteriologists, and parasitologists in an integrated fashion to identify mechanistically important interactions,” Virgin and Todd observe (2011). As many also noted, existing organizational and funding structures fail to reward participants in multidisciplinary efforts, either in terms of promotion or grants.
________________
55 Blindly or naively optimistic, after Pangloss, an optimistic character in Voltaire’s Candide (1759).
Xavier, who said he had been relatively successful in funding multidisciplinary studies, nonetheless observed, “When you submit large program projects, they get reviewed by individual people who read individual sections. They do not see how the entire grant comes together. You are reviewed by an individual investigator looking at his or her area of expertise without paying any attention to the interaction of the team. This often brings the grant down because the view is that you are now doing enough in that area that the reviewer is an expert in. It is very clear by reading the comments that this person paid no attention to how synergy was brought together in this application.”
Exploration of our microbial ecology will also continue to benefit from the techniques, concepts, and principles developed through investigations in a wide variety of biotic and abiotic systems, and several participants noted the need to break down the arbitrary divide between clinical microbiology and the work of environmental microbiologists, ecologists, veterinary scientists, and myriad other disciplines. Relman advocated for a greater emphasis on transdisciplinarity, which occurs when “experts from one discipline adopt the perspectives of other disciplines in formulating questions and in designing experimental plans for addressing them” (Relman, 2012). Bohannan noted that transdisciplinarity may require a reconfiguration of the review process. “I think there are assumptions made about what sort of science is likely to be fruitful in terms of human health. There is a hard line drawn that separates NIH from other funding agencies. For people like me who bridge those questions, it means it is difficult to get really innovative grants reviewed.”
Looking Ahead
The impact of our growing appreciation of the microbial world around us extends well beyond human health and disease. As observed by several workshop participants, new tools and approaches have inspired fresh approaches to the study of host–microbe–environment interactions in a variety of organisms and settings. These novel approaches are being exploited to investigate an increasingly broad range of topics—from the influence of the microbiota on the origins of animals to defining what constitutes an “individual” organism and exploring global principles of ecology (Gilbert et al., 2012; Shade et al., 2012; McFall-Ngai et al., 2013). The complex relationships between microbial community diversity, function, stability, and resilience are relevant to problems confronting scientists in a variety of contexts. As noted by Shade et al. (2012), “[m]icrobial communities are at the heart of all ecosystems”—thus, biomedical, environmental, agricultural, and bioenergy research all share “a common challenge: to predict how functions and composition of microbial communities respond to disturbances” (Shade et al., 2012).
Arnold, I. C., N. Dehzad, S. Reuter, H. Martin, B. Becher, C. Taube, and A. Müller. 2011. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. Journal of Clinical Investigation 121(8):3088-3093.
Atherton, J. C., and M. J. Blaser. 2009. Coadaptation of Helicobacter pylori and humans: Ancient history, modern implications. Journal of Clinical Investigation 119(9):2475-2487.
Bach, J. F. 2002. The effect of infections on susceptibility to autoimmune and allergic diseases. New England Journal of Medicine 347:911-920.
Bäckhed, C., M. Fraser,, Y. Ringel, M. E. Sanders, R. B. Sartor, P. M. Sherman, J. Versalovic, V. Young, and B. B. Finlay. 2012. Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host and Microbe 12(5):611-622.
Badri, D. V., T. L. Weir, D. van der Lelie, and J. M. Vivanco. 2009. Rhizosphere chemical dialogues: Plant–microbe interactions. Current Opinion in Biotechnology 20(6):642-650.
Barr, J. J., R. Auro, M. Furlan, K. L. Whiteson, M. L. Erb, J. Pogliano, A. Stotland, R. Wolkowicz, A. S. Cutting, K. S. Doran, P. Salamon, M. Youle, and F. Rohwer. 2013. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proceedings of the National Academy of Sciences of the United States of America. DOI:10.1073/pnas.1305923110.
Barton, E. S., D. W. White, J. S. Cathelyn, K. A. Brett-McClellan, M. Engle, M. S. Diamond, V. L. Miller, and H. W. Virgin. 2007. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447(7142):326-329.
Bates, J. M., J. Akerlund, E. Mittge, and K. Guillemin. 2007. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host & Microbe 2(6):371-382.
Belkaid, Y., and S. Naik. 2013. Compartmentalized and systemic control of tissue immunity by commensals. Nature Immunology 14:646-653.
Belkaid, Y., N. Bouladoux, T. W. Hand. 2013. Effector and memory T cell responses to commensal bacteria. Trends in Immunology 34(6):299-306.
Benchimol, E. I., A. Guttmann, A. M. Griffiths, L. Rabeneck, D. R. Mack, H. Brill, J. Howard, J. Guan, and T. To. 2009. Increasing incidence of paediatric inflammatory bowel disease in Ontario, Canada: Evidence from health administrative data. Gut 58(11):1490-1497.
Berendsen, R. L., C. M. J. Pieterse, and P. A. H. M. Bakker. 2012. The rhizosphere microbiome and plant health. Trends in Plant Science 17(8):478-486.
Blaser, M. J. 2006. Who are we? EMBO Reports 7(10):956-960.
Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: Biology and disease. Journal of Clinical Investigation 113(3):321-333.
Blaser, M. J., and S. Falkow. 2009. What are the consequences of the disappearing human microbiota? Nature Reviews Microbiology 7(12):887-894.
Blaser, M. J., Y. Chen, and J. Reibman. 2008. Does Helicobacter pylori protect against asthma and allergy? Gut 57(5):561-567.
Blaser, M., P. Bork, C. Fraser, R. Knight, and J. Wang. 2013. The microbiome explored: Recent insights and future challenges. Nature Reviews Microbiology 11(3):213-217.
Blustein, J., T. Attina, M. Liu, A. M. Ryan, L. M. Cox, M. J. Blaser, and L. Trasande. 2013. Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. International Journal of Obesity. doi: 10.1038/ijo.2013.49. [Epub ahead of print] Bohannan, B. 2013. Session III: The application of ecological concepts to host-associated microbial communities. Presentation at the Forum on Microbial Threats Workshop, Microbial Ecology in States of Health and Disease, Washington, DC, Institute of Medicine, Forum on Microbial Threats. March 19.
Boris, M., T. F. Sellers, H. F. Eichenwald, J. C. Ribble, and H. R. Schinefeld. 1964. Bacterial interference: Protection of adults against nasal Staphylococcus aureus infection after colonization with a heterologous S. aureus strain. American Journal of Diseases of Children 108(3):252-261.
Borody, T. J., and A. Khoruts. 2012. Fecal microbiota transplantation and emerging applications. Nature Reviews Gastroenterology and Hepatology 9:88-96.
Boyle, N. R., and R. T. Gill. 2012. Tools for genome-wide strain design and construction. Current Opinion in Biotechnology 23(5):666-671.
Buchen, L. 2010. Microbiology: The new germ theory. Nature 468(7323):492-495.
Bulgarelli, D., M. Rott, K. Schlaeppi, E. Ver Loren van Themaat, N. Ahmadinejad, F. Assenza, P. Rauf, B. Huettel, R. Reinhardt, E. Schmelzer, J. Peplies, F. O. Gloeckner, R. Amann, T. Eickhorst, and P. Schulze-Lefert. 2012. Revealing structure and assembly cues for arabidopsis root-inhabiting bacterial microbiota. Nature 488(7409):91-95.
Cadwell, K., J. Y. Liu, S. L. Brown, H. Miyoshi, J. Loh, J. K. Lennerz, C. Kishi, W. Kc, J. A. Carrero, S. Hunt, C. D. Stone, E. M. Brunt, R. J. Xavier, B. P. Sleckman, E. Li, N. Mizushima, T. S. Stappenbeck, and H. W. Virgin IV. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal paneth cells. Nature 456(7219):259-263.
Cadwell, K., K. K. Patel, N. S. Maloney, T. C. Liu, A. C. Ng, C. E. Storer, R. D. Head, R. Xavier, T. S. Stappenbeck, and H. W. Virgin. 2010. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16l1 phenotypes in intestine. Cell 141(7):1135-1145.
Canny, S. P., G. Goel, T. A. Reese, X. Zhang, R. Xavier, and H. W. Virgin. 2013. Latent gammaherpesvirus 68 infection induces distinct transcriptional changes in different organs. Journal of Virology. Epub ahead of print.
Carey, H. V. 1990. Seasonal changes in mucosal structure and function in ground squirrel intestine. American Journal of Physiology—Regulatory, Integrative and Comparative Physiology 259(2):R385-R392.
_____. 2013. Session I: Annual cycles of extreme dietary change shape gut microbiota and their hibernator hosts. Presentation at the Forum on Microbial Threats Workshop, Microbial Ecology in States of Health and Disease, Washington, DC, Institute of Medicine, Forum on Microbial Threats. March 18.
Carey, H. V., W. A. Walters, and R. Knight. 1992. Effects of fasting and hibernation on ion secretion in ground squirrel intestine. American Journal of Physiology—Regulatory, Integrative and Comparative Physiology 263(6):R1203-R1208.
_____. 2013. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. American Journal of Physiology Regulatory, Integrative, and Comparative Physiology 304(1):R33-R42.
Carey, H. V., M. T. Andrews, and S. L. Martin. 2003. Mammalian hibernation: Cellular and molecular responses to depressed metabolism and low temperature. Physiological Reviews 83:1153-1181.
Chai, G., L. Governale, A. W. McMahon, J. P. Trinidad, J. Staffa, and D. Murphy. 2012. Trends of outpatient prescription drug utilization in US children, 2002-2010. Pediatrics 130(1):23-31.
Chang, J. Y., D. A. Antonopoulos, A. Kalra, A. Tonelli, W. T. Khalife, T. M. Schmidt, and V. B. Young. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. Journal of Infectious Diseases 197(3):435-438.
Chen, Y., and M. J. Blaser. 2007. Inverse associations of Helicobacter pylori with asthma and allergy. Archives of Internal Medicine 167(8):821-827.
_____. 2008. Helicobacter pylori colonization is inversely associated with childhood asthma. Journal of Infectious Diseases 198(4):553-560.
Cho, I., and M. J. Blaser. 2012. The human microbiome: At the interface of health and disease. Nature Reviews Genetics 13(4):260-270.
Cho, I., S. Yamanishi, L. Cox, B. A. Methe, J. Zavadil, K. Li, Z. Gao, D. Mahana, K. Raju, I. Teitler, H. Li, A. V. Alekseyenko, and M. J. Blaser. 2012. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488(7413):621-626.
Chow, W. H., M. J. Blaser, W. J. Blot, M. D. Gammon, T. L. Vaughan, H. A. Risch, G. I. Perez-Perez, J. B. Schoenberg, J. L. Stanford, H. Rotterdam, A. B. West, and J. F. Fraumeni, Jr. 1998. An inverse relation between caga+ strains of Helicobacter pylori infection and risk of esophageal and gastric cardia adenocarcinoma. Cancer Research 58(4):588-590.
Clemente, J. C., L. K. Ursell, L. W. Parfrey, and R. Knight. 2012. The impact of the gut microbiota on human health: An integrative view. Cell 148(6):1258-1270.
Coppieters, K. T., and D. Elewaut. 2012. Natural killer T cells: Born in the thymus, raised in the gut. Gastroenterology 143(2):293-296.
Corley, D. A., A. Kubo, T. R. Levin, G. Block, L. Habel, W. Zhao, P. Leighton, G. Rumore, C. Quesenberry, P. Buffler, and J. Parsonnet. 2008. Helicobacter pylori infection and the risk of Barrett’s oesophagus: A community-based study. Gut 57(6):727-733.
Costello, E. K., K. Stagaman, L. Dethlefsen, B. J. Bohannan, and D. A. Relman. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336(6086):1255-1262.
Couzin-Frankel, J. 2010. Bacteria and asthma: Untangling the links. Science 330(6008):1168-1169.
Cox, L., and M. A. Blaser. 2013. Pathways in microbe-induced obesity. Cell Metabolism 17:883-894.
DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16s rRNA gene database and workbench compatible with arb. Applied and Environmental Microbiology 72(7):5069-5072.
Desbrosses, G. J., and J. Stougaard. 2011. Root nodulation: A paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10(4):348-358.
Dethlefsen, L., M. McFall-Ngai, and D. A. Relman. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449(7164):811-818.
Domazet-Loso, T., and D. Tautz. 2008. An ancient evolutionary origin of genes associated with human genetic diseases. Molecular Biology and Evolution 25(12):2699-2707.
Dominguez-Bello, M. G., E. K. Costello, M. Contreras, M. Magris, G. Hidalgo, N. Fierer, and R. Knight. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America 107(26):11971-11975.
Dominguez-Bello, M. G., M. J. Blaser, R. E. Ley, and R. Knight. 2011. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology 140(6):1713-1719.
Douglas, A. E. 2011. Lessons from studying insect symbioses. Cell Host Microbe 10(4):359-367.
Dubos, R., R. W. Schaedler, R. Costello, and P. Hoet. 1965. Indigenous, normal, and autochthonous flora of the gastrointestinal tract. Journal of Experimental Medicine 122:67-76.
Dusko Ehrlich, S. 2010. [metagenomics of the intestinal microbiota: Potential applications]. Gastroenterologie Clinique et Biologique 34(Suppl 1:)S23-S28.
Eberl, G. 2010. A new vision of immunity: Homeostasis of the superorganism. Mucosal Immunology 3(5):450-460.
_____. 2012. Development and evolution of RORγt+ cells in a microbe’s world. Immunological Reviews 245(1):177-188.
Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308(5728):1635-1638.
Ege, M. J., M. Mayer, A. C. Normand, J. Genuneit, W. O. Cookson, C. Braun-Fahrlander, D. Heederik, R. Piarroux, and E. von Mutius. 2011. Exposure to environmental microorganisms and childhood asthma. New England Journal of Medicine 364(8):701-709.
Eisen, J. A. 2007. Environmental shotgun sequencing: Its potential and challenges for studying the hidden world of microbes. PLoS Biology 5(3).
Everard, A., C. Belzer, L. Geurts, J. P. Ouwerkerk, C. Druart, L. B. Bindels, Y. Guiot, M. Derrien, G. G. Muccioli, N. M. Delzenne, W. M. de Vos, and P. D. Cani. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America 110(22):9066-9071.
Fidel, P. L., Jr. 2007. History and update on host defense against vaginal candidiasis. American Journal of Reproductive Immunology 57(1):2-12.
Findley, K., J. Oh, J. Yang, S. Conlan, C. Deming, J. A. Meyer, D. Schoenfeld, E. Nomicos, M. Park, H. H. Kong, and J. A. Segre. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498(7454):367-370.
Forman, D., P. Webb, and J. Parsonnet. 1994. H. pylori and gastric cancer. Lancet 343(8891):243-244.
Frank, D. N., A. L. St. Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America 104(34):13780-13785.
Fraune, S., and T. C. Bosch. 2010. Why bacteria matter in animal development and evolution. Bioessays 32(7):571-580.
Fredricks, D. N., and D. A. Relman. 1996. Sequence-based identification of microbial pathogens: A reconsideration of Koch’s postulates. Clinical Microbiology Reviews 9:18-33.
Gajer, P., R. M. Brotman, G. Bai, J. Sakamoto, U. M. Schutte, X. Zhong, S. S. Koenig, L. Fu, Z. S. Ma, X. Zhou, Z. Abdo, L. J. Forney, and J. Ravel. 2012. Temporal dynamics of the human vaginal microbiota. Science Translational Medicine 4(132):132ra152.
Garrido, D., D. Barile, and D. A. Mills. 2012. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Advances in Nutrition 3(3):415S-421S.
Gilbert, S. F., J. Sapp, and A. I Tauber. 2012. A symbiotic view of life: We have never been individuals. Quarterly Review of Biology 87(4):325-341.
Gonzalez, A., J. C. Clemente, A. Shade, J. L. Metcalf, S. Song, B. Prithiviraj, B. E. Palmer, and R. Knight. 2011. Our microbial selves: What ecology can teach us. EMBO Reports 12(8):775-784.
Gordon, J. I. 2012. Honor thy gut symbionts redux. Science 336(6086):1251-1253.
Gottel, N. R., H. F. Castro, M. Kerley, Z. Yang, D. A. Pelletier, M. Podar, T. Karpinets, E. Uberbacher, G. A. Tuskan, R. Vilgalys, M. J. Doktycz, and C. W. Schadt. 2011. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Applied Environmental Microbiology 77(17):5934-5944.
Graham, D. B., and R. J. Xavier. 2013. From genetics of inflammatory bowel disease towards mechanistic insights. Trends in Immunology 34(8):371-378.
Grice, E. A., and J. A. Segre. 2012. The human microbiome: Our second genome. Annual Review of Genomics and Human Genetics 13:151-170.
Grice, E. A., H. H. Kong, S. Conlan, C. B. Deming, J. Davis, A. C. Young, G. G. Bouffard, R. W. Blakesley, P. R. Murray, E. D. Green, M. L. Turner, and J. A. Segre. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324(5931):1190-1192.
Guillemin, K. 2013. Session II: Interactions and functions of the gut microbiota in a model vertebrate host. Presentation at that Forum on Microbial Threats Workshop, Microbial Ecology in States of Health and Disease, Washington DC, Institute of Medicine, Forum on Microbial Threats. March 18.
Han, M. K., Y. J. Huang, J. J. Lipuma, H. A. Boushey, R. C. Boucher, W. O. Cookson, J. L. Curtis, J. Erb-Downward, S. V. Lynch, S. Sethi, G. B. Toews, V. B. Young, M. C. Wolfgang, G. B. Huffnagle, and F. J. Martinez. 2012. Significance of the microbiome in obstructive lung disease. Thorax 67(5):456-463.
Hand, T. W., L. M. Dos Santos, N. Bouladoux, M. J. Molloy, A. J. Pagan, M. Pepper, C. L. Maynard, C. O. Elson, 3rd, and Y. Belkaid. 2012. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337(6101):1553-1556.
Handelsman, J. 2004. Metagenomics: Application of genomics to uncultured microorganisms. Microbiology and Molecular Biology Reviews 68(4):669-685.
Handley, S. A., L. B. Thackray, G. Zhao, R. Presti, A. D. Miller, L. Droit, P. Abbink, L. F. Maxfield, A. Kambal, E. Duan, K. Stanley, J. Kramer, S. C. Macri, S. R. Permar, J. E. Schmitz, K. Mansfield, J. M. Brenchley, R. S. Veazey, T. S. Stappenbeck, D. Wang, D. H. Barouch, and H. W. Virgin. 2012. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151(2):253-266.
Hickey, R. J., X. Zhou, J. D. Pierson, J. Ravel, and L. J. Forney. 2012. Understanding vaginal microbiome complexity from an ecological perspective. Translational Research 160(4):267-282.
Holmes, E., J. Kinross, G. R. Gibson, R. Burcelin, W. Jia, S. Pettersson, and J. K. Nicholson. 2012. Therapeutic modulation of microbiota-host metabolic interactions. Science Translational Medicine 4(137):137rv136.
Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292(5519):1115-1118.
Hooper, L. V., D. R. Littman, and A. J. Macpherson. 2012. Interactions between the microbiota and the immune system. Science 336(6086):1268-1273.
Houck, P. W., J. D. Nelson, and J. L. Kay. 1972. Fatal septicemia due to Staphylococcus aureus 502a. Report of a case and review of the infectious complications of bacterial interference programs. American Journal of Diseases of Children 123(1):45-48.
Howson, C. P., T. Hiyama, and E. L. Wynder. 1986. The decline in gastric cancer: Epidemiology of an unplanned triumph. Epidemiologic Reviews 8:1-27.
Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486(7402):207-214.
Iliev, I. D., V. A. Funari, K. D. Taylor, Q. Nguyen, C. N. Reyes, S. P. Strom, J. Brown, C. A. Becker, P. R. Fleshner, M. Dubinsky, J. I. Rotter, H. L. Wang, D. P. McGovern, G. D. Brown, and D. M. Underhill. 2012. Interactions between commensal fungi and the c-type lectin receptor dectin-1 influence colitis. Science 336(6086):1314-1317.
IOM (Institute of Medicine). 2004. The infectious etiology of chronic diseases: Defining the relationship, enhancing the research, and mitigating the effects: Workshop summary. Washington, DC: The National Academies Press.
_____. 2006. Ending the war metaphor: The changing agenda for unraveling the host-microbe relationship: Workshop summary. Washington, DC: The National Academies Press.
_____. 2009. Microbial evolution and co-adaptation: A tribute to the life and scientific legacies of Joshua Lederberg: Workshop summary. Washington, DC: The National Academies Press.
_____. 2012. The social biology of microbial communities: Workshop summary. Washington, DC: The National Academies Press.
Islami, F., and F. Kamangar. 2008. Helicobacter pylori and esophageal cancer risk: A meta-analysis. Cancer Prevention Research 1(5):329-338.
Jostins, L., S. Ripke, R. K. Weersma, R. H. Duerr, D. P. McGovern, K. Y. Hui, J. C. Lee, L. P. Schumm, Y. Sharma, C. A. Anderson, J. Essers, M. Mitrovic, K. Ning, I. Cleynen, E. Theatre, S. L. Spain, S. Raychaudhuri, P. Goyette, Z. Wei, C. Abraham, J. P. Achkar, T. Ahmad, L. Amininejad, A. N. Ananthakrishnan, V. Andersen, J. M. Andrews, L. Baidoo, T. Balschun, P. A. Bampton, A. Bitton, G. Boucher, S. Brand, C. Buning, A. Cohain, S. Cichon, M. D’Amato, D. De Jong, K. L. Devaney, M. Dubinsky, C. Edwards, D. Ellinghaus, L. R. Ferguson, D. Franchimont, K. Fransen, R. Gearry, M. Georges, C. Gieger, J. Glas, T. Haritunians, A. Hart, C. Hawkey, M. Hedl, X. Hu, T. H. Karlsen, L. Kupcinskas, S. Kugathasan, A. Latiano, D. Laukens, I. C. Lawrance, C. W. Lees, E. Louis, G. Mahy, J. Mansfield, A. R. Morgan, C. Mowat, W. Newman, O. Palmieri, C. Y. Ponsioen, U. Potocnik, N. J. Prescott, M. Regueiro, J. I. Rotter, R. K. Russell, J. D. Sanderson, M. Sans, J. Satsangi, S. Schreiber, L. A. Simms, J. Sventoraityte, S. R. Targan, K. D. Taylor, M. Tremelling, H. W. Verspaget, M. De Vos, C. Wijmenga, D. C. Wilson, J. Winkelmann, R. J. Xavier, S. Zeissig, B. Zhang, C. K. Zhang, H. Zhao, M. S. Silverberg, V. Annese, H. Hakonarson, S. R. Brant, G. Radford-Smith, C. G. Mathew, J. D. Rioux, E. E. Schadt, M. J. Daly, A. Franke, M. Parkes, S. Vermeire, J. C. Barrett, and J. H. Cho. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491(7422):119-124.
Kamada, N., S. U. Seo, G. Y. Chen, and G. Nunez. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nature Reviews: Immunology 13(5):321-335.
Karlsson, F. H., F. Fak, I. Nookaew, V. Tremaroli, B. Fagerberg, D. Petranovic, F. Bäckhed, and J. Nielsen. 2012. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nature Communications 3:1245.
Karlsson, F. H., V. Tremaroli, I. Nookaew, G. Bergström, C. J. Behre, B. Fagerberg, J. Nielsen, and F. Bäckhed. 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498:99-103.
Kaser, A., S. Zeissig, and R. S. Blumberg. 2010. Inflammatory bowel disease. Annual Review of Immunology 28:573-621.
Kelly, C. P. 2013. Fecal microbiota transplantation—An old therapy comes of age. New England Journal of Medicine 368(5):474-475.
Khor, B., A. Gardet, and R. J. Xavier. 2011. Genetics and pathogenesis of inflammatory bowel disease. Nature 474(7351):307-317.
King, M. C., J. H. Marks, and J. B. Mandell. 2003. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302(5645):643-646.
Koch, R. 1891. A further communication on a remedy for tuberculosis. British Medical Journal 1(1568):125-127.
Koczulla, R., G. von Degenfeld, C. Kupatt, F. Krotz, S. Zahler, T. Gloe, K. Issbrucker, P. Unterberger, M. Zaiou, C. Lebherz, A. Karl, P. Raake, A. Pfosser, P. Boekstegers, U. Welsch, P. S. Hiemstra, C. Vogelmeier, R. L. Gallo, M. Clauss, and R. Bals. 2003. An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. Journal of Clinical Investigation 111(11):1665-1672.
Kong, H. H., J. Oh, C. Deming, S. Conlan, E. A. Grice, M. A. Beatson, E. Nomicos, E. C. Polley, H. D. Komarow, P. R. Murray, M. L. Turner, and J. A. Segre. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Research 22(5):850-859.
Koren, O., J. K. Goodrich, T. C. Cullender, A. Spor, K. Laitinen, H. K. Bäckhed, A. Gonzalez, J. J. Werner, L. T. Angenent, R. Knight, F. Bäckhed, E. Isolauri, S. Salminen, and R. E. Ley. 2012. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150(3):470-480.
Koropatnick, T. A., J. T. Engle, M. A. Apicella, E. V. Stabb, W. E. Goldman, and M. J. McFall-Ngai. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306(5699):1186-1188.
Kremer, N., E. E. R. Philipp, M-C. Carpentier, C. A. Brennan, L. Kraemer, M. A. Altura, R. Augustin, R. Häsler. E. A. C. Heath-Heckman, S. M. Peyer, J. Schwartzman, B. A. Rader, E. G. Ruby, P. Rosenstiel, and M. J. McFall-Ngai. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host & Microbe 14(2):183-194.
Kuballa, P., W. M. Nolte, A. B. Castoreno, and R. J. Xavier. 2012. Autophagy and the immune system. Annual Review of Immunology 30:611-646.
Kyne, L., M. Warny, A. Qamar, and C. P. Kelly. 2001. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet 357(9251):189-193.
Lederberg, J. 2000. Infectious history. Science 288(5464):287-293.
Lemon, K. P., G. C. Armitage, D. A. Relman, and M. A. Fischbach. 2012. Microbiota-targeted therapies: An ecological perspective. Science Translational Medicine 4(137):137rv135.
Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America 102(31):11070-11075.
Light, I. J., J. M. Sutherland, and J. E. Schott. 1965. Control of a staphylococcal outbreak in a nursery, use of bacterial interference. JAMA 193:699-704.
Liu, P. T., S. Stenger, H. Li, L. Wenzel, B. H. Tan, S. R. Krutzik, M. T. Ochoa, J. Schauber, K. Wu, C. Meinken, D. L. Kamen, M. Wagner, R. Bals, A. Steinmeyer, U. Zugel, R. L. Gallo, D. Eisenberg, M. Hewison, B. W. Hollis, J. S. Adams, B. R. Bloom, and R. L. Modlin. 2006. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311(5768):1770-1773.
Lozupone, C. A., J. I. Stombaugh, J. I. Gordon, J. K. Jansson, and R. Knight. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489(7415):220-230.
Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ArbRB: A software environment for sequence data. Nucleic Acids Research 32(4):1363-1371.
Lupp, C., M. L. Robertson, M. E. Wickham, I. Sekirov, O. L. Champion, E. C. Gaynor, and B. B. Finlay. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of enterobacteriaceae. Cell Host Microbe 2(3):204.
Ma, B., L. J. Forney, and J. Ravel. 2012. Vaginal microbiome: Rethinking health and disease. Annual Review of Microbiology 66:371-389.
MacDonald, S. J., G. H. Thomas, and A. E. Douglas. 2011. Genetic and metabolic determinants of nutritional phenotype in an insect-bacterial symbiosis. Molecular Ecology 20(10):2073-2084.
Mackowiak, P. A. 1982. The normal microbial flora. New England Journal of Medicine 307:83-93.
Mandel, M. J., A. L. Schaefer, C. A. Brennan, E. A. C. Heath-Heckman, C. R. Deloney-Marino, M. J. McFall-Ngai, and E. G. Ruby. 2012. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Applied and Environmental Microbiology 78:4620-4626.
Maynard, C. L., C. O. Elson, R. D. Hatton, and C. T. Weaver. 2012. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489(7415):231-241.
McFall-Ngai, M., M. G. Hadfield, T. C. Bosch, H. V. Carey, T. Domazet-Loso, A. E. Douglas, N. Dubilier, G. Eberl, T. Fukami, S. F. Gilbert, U. Hentschel, N. King, S. Kjelleberg, A. H. Knoll, N. Kremer, S. K. Mazmanian, J. L. Metcalf, K. Nealson, N. E. Pierce, J. F. Rawls, A. Reid, E. G. Ruby, M. Rumpho, J. G. Sanders, D. Tautz, and J. J. Wernegreen. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America 110(9):3229-3236.
McKenna, M. 2011. Swapping germs. Scientific American 305(6):34-36.
Minot, S., S. Grunberg, G. D. Wu, J. D. Lewis, and F. D. Bushman. 2012. Hypervariable loci in the human gut virome. Proceedings of the National Academy of Sciences of the United States of America 109(10):3962-3966.
Moles, L., M. Gómez, H. Heilig, G. Bustos, S. Fuentes, W. de Vos, L. Fernández, J. Rodríguez, and E. Jiménez. (2013) Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE 8(6):e66986. doi:10.1371/journal.pone.0066986
Molodecky, N. A., I. S. Soon, D. M. Rabi, W. A. Ghali, M. Ferris, G. Chernoff, E. I. Benchimol, R. Panaccione, S. Ghosh, H. W. Barkema, and G. G. Kaplan. 2012. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142(1):46-54 e42.
Morgan, X. C., T. L. Tickle, H. Sokol, D. Gevers, K. L. Devaney, D. V. Ward, J. A. Reyes, S. A. Shah, N. LeLeiko, S. B. Snapper, A. Bousvaros, J. Korzenik, B. E. Sands, R. J. Xavier, and C. Huttenhower. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biology 13(9):R79.
Munstedt, K., P. Grant, J. Woenckhaus, G. Roth, and H. R. Tinneberg. 2004. Cancer of the endometrium: Current aspects of diagnostics and treatment. World Journal of Surgical Oncology 2:24.
Naik, S., N. Bouladoux, C. Wilhelm, M. J. Molloy, R. Salcedo, W. Kastenmuller, C. Deming, M. Quinones, L. Koo, S. Conlan, S. Spencer, J. A. Hall, A. Dzutsev, H. Kong, D. J. Campbell, G. Trinchieri, J. A. Segre, and Y. Belkaid. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337(6098):1115-1119.
Newman, D. J., G. M. Cragg, and K. M. Snader. 2003. Natural products as sources of new drugs over the period 1981-2002. Journal of Natural Products 66(7):1022-1037.
Nicholson, J. K., E. Holmes, J. Kinross, R. Burcelin, G. Gibson, W. Jia, and S. Pettersson. 2012. Host-gut microbiota metabolic interactions. Science 336(6086):1262-1267.
Nyholm, S. V., and M. J. McFall-Ngai. 2004. The winnowing: Establishing the squid-Vibrio symbiosis. Nature Reviews Microbiology 2(8):632-642.
Oh, J., S. Conlan, E. C. Polley, J. A. Segre, and H. H. Kong. 2012. Shifts in human skin and nares microbiota of healthy children and adults. Genomic Medicine 4(10):77.
Ohnmacht, C., R. Marques, L. Presley, S. Sawa, M. Lochner, and G. Eberl. 2011. Intestinal microbiota, evolution of the immune system and the bad reputation of pro-inflammatory immunity. Cell Microbiology 13(5):653-659.
Oliynyk, M., M. Samborskyy, J. B. Lester, T. Mironenko, N. Scott, S. Dickens, S. F. Haydock, and P. F. Leadlay. 2007. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea nrrl23338. Nature Biotechnology 25(4):447-453.
Olszak, T., D. An, S. Zeissig, M. P. Vera, J. Richter, A. Franke, J. N. Glickman, R. Siebert, R. M. Baron, D. L. Kasper, and R. S. Blumberg. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336(6080):489-493.
Ong, P. Y., T. Ohtake, C. Brandt, I. Strickland, M. Boguniewicz, T. Ganz, R. L. Gallo, and D. Y. Leung. 2002. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. New England Journal of Medicine 347(15):1151-1160.
Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276(5313):734-740.
Paine, R. T., M. J. Tegner, and E. A. Johnson. 1998. Compounded perturbations yield ecological surprises. Ecosystems 1(6):535-545.
Peek, R. M., Jr., and M. J. Blaser. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature Reviews Cancer 2(1):28-37.
Perry, S., B. C. de Jong, J. V. Solnick, M. de la Luz Sanchez, S. Yang, P. L. Lin, L. M. Hansen, N. Talat, P. C. Hill, R. Hussain, R. A. Adegbola, J. Flynn, D. Canfield, and J. Parsonnet. 2010. Infection with Helicobacter pylori is associated with protection against tuberculosis. PloS ONE 5(1):e8804.
Peterson, D. A., D. N. Frank, N. R. Pace, and J. I. Gordon. 2008. Metagenomic approaches for defining the pathogenesis of inflammatory bowel disease. Cell Host Microbe 3(6):417-427.
Petrof, E. O., G. B. Gloor, S. J. Vanner, J. S. Weese, D. Carter, M. C. Daigneault, E. M. Brown, K. Schroeder, and E. Allen-Vercoe. 2013. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “Repoopulating” the gut. Microbiome 1(1):1-12.
Pincock, S. 2005. Nobel prize winners Robin Warren and Barry Marshall. Lancet 366(9495):1429.
Plottel, C. S., and M. J. Blaser. 2011. Microbiome and malignancy. Cell Host Microbe 10(4):324-335.
Poliakov, A., C. W. Russell, L. Ponnala, H. J. Hoops, Q. Sun, A. E. Douglas, and K. J. van Wijk. 2011. Large-scale label-free quantitative proteomics of the pea aphid-buchnera symbiosis. Molecular & Cellular Proteomics 10(6):M110 007039.
Qin, J., R. Li, J. Raes, M. Arumugam, K. S. Burgdorf, C. Manichanh, T. Nielsen, N. Pons, F. Levenez, T. Yamada, D. R. Mende, J. Li, J. Xu, S. Li, D. Li, J. Cao, B. Wang, H. Liang, H. Zheng, Y. Xie, J. Tap, P. Lepage, M. Bertalan, J. M. Batto, T. Hansen, D. Le Paslier, A. Linneberg, H. B. Nielsen, E. Pelletier, P. Renault, T. Sicheritz-Ponten, K. Turner, H. Zhu, C. Yu, M. Jian, Y. Zhou, Y. Li, X. Zhang, N. Qin, H. Yang, J. Wang, S. Brunak, J. Dore, F. Guarner, K. Kristiansen, O. Pedersen, J. Parkhill, J. Weissenbach, P. Bork, and S. D. Ehrlich. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464(7285):59-65.
Qin, J., Y. Li, Z. Cai, S. Li, J. Zhu, F. Zhang, S. Liang, W. Zhang, Y. Guan, D. Shen, Y. Peng, D. Zhang, Z. Jie, W. Wu, Y. Qin, W. Xue, J. Li, L. Han, D. Lu, P. Wu, Y. Dai, X. Sun, Z. Li, A. Tang, S. Zhong, X. Li, W. Chen, R. Xu, M. Wang, Q. Feng, M. Gong, J. Yu, Y. Zhang, M. Zhang, T. Hansen, G. Sanchez, J. Raes, G. Falony, S. Okuda, M. Almeida, E. LeChatelier, P. Renault, N. Pons, J. M. Batto, Z. Zhang, H. Chen, R. Yang, W. Zheng, S. Li, H. Yang, J. Wang, S. D. Ehrlich, R. Nielsen, O. Pedersen, K. Kristiansen, and J. Wang. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490(7418):55-60.
Ramirez-Puebla, S. T., L. E. Servin-Garciduenas, B. Jimenez-Marin, L. M. Bolanos, M. Rosenblueth, J. Martinez, M. A. Rogel, E. Ormeno-Orrillo, and E. Martinez-Romero. 2013. Gut and root microbiota commonalities. Applied and Environmental Microbiology 79(1):2-9.
Ravel, J., P. Gajer, Z. Abdo, G. M. Schneider, S. S. Koenig, S. L. McCulle, S. Karlebach, R. Gorle, J. Russell, C. O. Tacket, R. M. Brotman, C. C. Davis, K. Ault, L. Peralta, and L. J. Forney. 2011. Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America 108(Suppl 1):4680-4687.
Reibman, J., M. Marmor, J. Filner, M. E. Fernandez-Beros, L. Rogers, G. I. Perez-Perez, and M. J. Blaser. 2008. Asthma is inversely associated with Helicobacter pylori status in an urban population. PLoS ONE 3(12):e4060.
Relman, D. A. 2012. The human microbiome: Ecosystem resilience and health. Nutrition Reviews 70(Suppl 1):S2-S9.
Ridaura, V. K., J. J. Faith, F. E. Rey, J. Cheng, A. E. Duncan, A. L. Kau, N. W. Griffin, V. Lombard, B. Henrissat, J. R. Bain, M. J. Muehlbauer, O. Ilkayeva, C. F. Semenkovich, K. Funai, D. K. Hayashi, B. J. Lyle, M. C. Martini, L. K. Ursell, J. C. Clemente, W. Van Treuren, W. A. Walters, R. Knight, C. B. Newgard, A. C. Heath, and J. I. Gordon. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341(6150):1241214.
Ridley, E. V., A. C. Wong, S. Westmiller, and A. E. Douglas. 2012. Impact of the resident microbiota on the nutritional phenotype of drosophila melanogaster. PLoS ONE 7(5):e36765.
Ridley, E. V., A. C. Wong, and A. E. Douglas. 2013. Microbe-dependent and nonspecific effects of procedures to eliminate the resident microbiota from Drosophila melanogaster. Applied and Environmental Microbiology 79(10):3209-3214.
Rivers, T. M. 1937. Viruses and Koch’s postulates. Journal of Bacteriology 33:1-12.
Robinson, C. J., B. J. Bohannan, and V. B. Young. 2010. From structure to function: The ecology of host-associated microbial communities. Microbiology and Molecular Biology Reviews 74(3):453-476.
Round, J. L., and S. K. Mazmanian. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology 9(5):313-323.
Ryu, J. H., S. H. Kim, H. Y. Lee, J. Y. Bai, Y. D. Nam, J. W. Bae, D. G. Lee, S. C. Shin, E. M. Ha, and W. J. Lee. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in drosophila. Science 319(5864):777-782.
Sansonetti, P. J. 2004. War and peace at mucosal surfaces. Nature Reviews Immunology 4(12):953-964.
Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annual Review of Microbiology 31:107-133.
Sela, D. A., and D. A. Mills. 2010. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends in Microbiology 18(7):298-307.
Shade, A., H. Peter, S. D. Allison, D. L. Baho, M. Berga, H. Burgmann, D. H. Huber, S. Langenheder, J. T. Lennon, J. B. Martiny, K. L. Matulich, T. M. Schmidt, and J. Handelsman. 2012. Fundamentals of microbial community resistance and resilience. Frontiers in Microbiology 3:417.
Shaw, S. Y., J. F. Blanchard, and C. N. Bernstein. 2010. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. American Journal of Gastroenterology 105(12):2687-2692.
Smith, M. I., T. Yatsunenko, M. J. Manary, I. Trehan, R. Mkakosya, J. Cheng, A. L. Kau, S. S. Rich, P. Concannon, J. C. Mychaleckyj, J. Liu, E. Houpt, J. V. Li, E. Holmes, J. Nicholson, D. Knights, L. K. Ursell, R. Knight, and J. I. Gordon. 2013. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339(6119):548-554.
Sommer, F., and F. Bäckhed. 2013. The gut microbiota—Masters of host development and physiology. Nature Reviews Microbiology11(4):227-382.
Sonnenburg, J. L., and M. A. Fischbach. 2011. Community health care: Therapeutic opportunities in the human microbiome. Science Translational Medicine 3(78):78ps12.
Spor, A., O. Koren, and R. Ley. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature Reviews Microbiology 9:279-290.
Strachan, D. P. 1989. Hay fever, hygiene, and household size. British Medical Journal 299(6710): 1259-1260.
Tang, W. H., Z. Wang, B. S. Levison, R. A. Koeth, E. B. Britt, X. Fu, Y. Wu, and S. L. Hazen. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. New England Journal of Medicine 368(17):1575-1584.
Taormina, M. J., M. Jemielita, W. Z. Stephens, A. R. Burns, J. V. Troll, R. Parthasarathy, and K. Guillemin. 2012. Investigating bacterial-animal symbioses with light sheet microscopy. Biological Bulletin 223(1):7-20.
Thomas, G. H., J. Zucker, S. J. Macdonald, A. Sorokin, I. Goryanin, and A. E. Douglas. 2009. A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola. BMC Systems Biology 3:24.
Trasande, L., J. Blustein, M. Liu, E. Corwin, L. M. Cox, and M. J. Blaser. 2013. Infant antibiotic exposures and early-life body mass. International Journal of Obesity 37(1):16-23.
Turnbaugh, P. J., R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis, and J. I. Gordon. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027-1031.
Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon. 2007. The human microbiome project. Nature 449(7164):804-810.
Tuskan, G. 2013. Session I: Plant-microbe interactions in root endophyte and rhizosphere communities of Populus. Presentation at the Forum on Microbial Threats Workshop, Microbial Ecology in States of Health and Disease, Washington, DC, Institute of Medicine, Forum on Microbial Threats. March 18.
Tuskan, G. A., S. Difazio, S. Jansson, J. Bohlmann, I. Grigoriev, U. Hellsten, N. Putnam, S. Ralph, S. Rombauts, A. Salamov, J. Schein, L. Sterck, A. Aerts, R. R. Bhalerao, R. P. Bhalerao, D. Blaudez, W. Boerjan, A. Brun, A. Brunner, V. Busov, M. Campbell, J. Carlson, M. Chalot, J. Chapman, G. L. Chen, D. Cooper, P. M. Coutinho, J. Couturier, S. Covert, Q. Cronk, R. Cunningham, J. Davis, S. Degroeve, A. Dejardin, C. Depamphilis, J. Detter, B. Dirks, I. Dubchak, S. Duplessis, J. Ehlting, B. Ellis, K. Gendler, D. Goodstein, M. Gribskov, J. Grimwood, A. Groover, L. Gunter, B. Hamberger, B. Heinze, Y. Helariutta, B. Henrissat, D. Holligan, R. Holt, W. Huang, N. Islam-Faridi, S. Jones, M. Jones-Rhoades, R. Jorgensen, C. Joshi, J. Kangasjarvi, J. Karlsson, C. Kelleher, R. Kirkpatrick, M. Kirst, A. Kohler, U. Kalluri, F. Larimer, J. Leebens-Mack, J. C. Leple, P. Locascio, Y. Lou, S. Lucas, F. Martin, B. Montanini, C. Napoli, D. R. Nelson, C. Nelson, K. Nieminen, O. Nilsson, V. Pereda, G. Peter, R. Philippe, G. Pilate, A. Poliakov, J. Razumovskaya, P. Richardson, C. Rinaldi, K. Ritland, P. Rouze, D. Ryaboy, J. Schmutz, J. Schrader, B. Segerman, H. Shin, A. Siddiqui, F. Sterky, A. Terry, C. J. Tsai, E. Uberbacher, P. Unneberg, J. Vahala, K. Wall, S. Wessler, G. Yang, T. Yin, C. Douglas, M. Marra, G. Sandberg, Y. Van de Peer, and D. Rokhsar. 2006. The genome of black cottonwood, Populus trichocarpa (torr. & gray). Science 313(5793):1596-1604.
Tvede, M., and J. Rask-Madsen. 1989. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 1(8648):1156-1160.
Underhill, D., and J. Braun. 2008. Current understanding of fungal microflora in inflammatory bowel disease pathogenesis. Inflammatory Bowel Diseases 14(8):1147-1153.
van Nood, E., P. Speelman, E. J. Kuijper, and J. J. Keller. 2009. Struggling with recurrent Clostridium difficile infections: Is donor faeces the solution? Eurosurveillance 14(34).
van Nood, E., A. Vrieze, M. Nieuwdorp, S. Fuentes, E. G. Zoetendal, W. M. de Vos, C. E. Visser, E. J. Kuijper, J. F. Bartelsman, J. G. Tijssen, P. Speelman, M. G. Dijkgraaf, and J. J. Keller. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. New England Journal of Medicine 368(5):407-415.
Vellend, M. 2010. Conceptual synthesis in community ecology. Quarterly Review of Biology 85(2):183-206.
Virgin, H. W., and J. A. Todd. 2011. Metagenomics and personalized medicine. Cell 147(1):44-56.
Virgin, H. W., E. J. Wherry, and R. Ahmed. 2009. Redefining chronic viral infection. Cell 138(1):30-50.
Walsh, C. T., and M. A. Fischbach. 2010. Natural products version 2.0: Connecting genes to molecules. Journal of the American Chemical Society 132(8):2469-2493.
Weston, D. J., D. A. Pelletier, J. L. Morrell-Falvey, T. J. Tschaplinski, S. S. Jawdy, T. Y. Lu, S. M. Allen, S. J. Melton, M. Z. Martin, C. W. Schadt, A. A. Karve, J. G. Chen, X. Yang, M. J. Doktycz, and G. A. Tuskan. 2012. Pseudomonas fluorescens induces strain-dependent and strain-independent host plant responses in defense networks, primary metabolism, photosynthesis, and fitness. Molecular Plant-Microbe Interactions 25(6):765-778.
White, D. W., C. R. Keppel, S. E. Schneider, T. A. Reese, J. Coder, J. E. Payton, T. J. Ley, H. W. Virgin, and T. A. Fehniger. 2010. Latent herpesvirus infection arms NK cells. Blood 115(22):4377-4383.
Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: The unseen majority. Proceedings of the National Academy of Sciences of the United States of America 95(12):6578-6583.
Wilson, A. C., P. D. Ashton, F. Calevro, H. Charles, S. Colella, G. Febvay, G. Jander, P. F. Kushlan, S. J. Macdonald, J. F. Schwartz, G. H. Thomas, and A. E. Douglas. 2010. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Molecular Biology 19(Suppl 2:)249-258.
Witkin, S. S., S. Alvi, A. M. Bongiovanni, I. M. Linhares, and W. J. Ledger. 2011. Lactic acid stimulates interleukin-23 production by peripheral blood mononuclear cells exposed to bacterial lipopolysaccharide. FEMS Immunology and Medical Microbiology 61(2):153-158.
Wong, A. C., J. M. Chaston, and A. E. Douglas. 2013. The inconstant gut microbiota of Drosophila species revealed by 16s rRNA gene analysis. ISME Journal. doi: 10.1038/ismej.2013.86.
Wong, C. N., P. Ng, and A. E. Douglas. 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environmental Microbiology 13(7):1889-1900.
Xu, J., and J. I. Gordon. 2003. Honor thy symbionts. Proceedings of the National Academy of Sciences of the United States of America 100(18):10452-10459.
Yano, J., E. Lilly, M. Barousse, and P. L. Fidel, Jr. 2010. Epithelial cell-derived s100 calcium-binding proteins as key mediators in the hallmark acute neutrophil response during Candida vaginitis. Infection and Immunity 78(12):5126-5137.
Yano, J., M. C. Noverr, and P. L. Fidel, Jr. 2012. Cytokines in the host response to candida vaginitis: Identifying a role for non-classical immune mediators, s100 alarmins. Cytokine 58(1):118-128.
Yatsunenko, T., F. E. Rey, M. J. Manary, I. Trehan, M. G. Dominguez-Bello, M. Contreras, M. Magris, G. Hidalgo, R. N. Baldassano, A. P. Anokhin, A. C. Heath, B. Warner, J. Reeder, J. Kuczynski, J. G. Caporaso, C. A. Lozupone, C. Lauber, J. C. Clemente, D. Knights, R. Knight, and J. I. Gordon. 2012. Human gut microbiome viewed across age and geography. Nature 486(7402):222-227.
Youle, M., M. Haynes, and F. Rohwer. 2012. Scratching the surface of biology’s dark matter. In Viruses: Essential agents of life, edited by G. Witzany. Springer.
Zasloff, M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cdnaDNA sequence of a precursor. Proceedings of the National Academy of Sciences of the United States of America 84(15):5449-5453.
_____. 2002a. Antimicrobial peptides in health and disease. New England Journal of Medicine 347(15):1199-1200.
_____. 2002b. Antimicrobial peptides of multicellular organisms. Nature 415(6870):389-395.
_____. 2011. Observations on the remarkable (and mysterious) wound-healing process of the bottlenose dolphin. Journal of Investigative Dermatology 131(12):2503-2505.
_____. 2013. Session IV: Innate antimicrobial mechanisms in disease prevention and treatment. Presentation at the Forum on Microbial Threats Workshop, Microbial Ecology in States of Health and Disease, Washington, DC, Institute of Medicine, Forum on Microbial Threats. March 19.
Zivkovic, A. M., J. B. German, C. B. Lebrilla, and D. A. Mills. 2011. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America 108(Suppl 1):4653-4658.




























































































