Yano, J., E. Lilly, M. Barousse, and P. L. Fidel, Jr. 2010. Epithelial cell-derived s100 calcium-binding proteins as key mediators in the hallmark acute neutrophil response during candida vaginitis. Infection and Immunity 78(12):5126-5137.
Yano, J., J. K. Kolls, K. I. Happel, F. L. Wormley, Jr., K. L. Wozniak, and P. L. Fidel, Jr. 2012. The acute neutrophil response mediated by s100 alarmins during vaginal candida infections is independent of the th17-pathway. PloS One 7(9):e46311. doi: 46310.41371/journal.pone.0046311.
Yeater, K. M., J. Chandra, G. Cheng, P. K. Mukherjee, X. Zhao, S. L. Rodriguez-Zas, K. E. Kwast, M. A. Ghannoum, and L. L. Hoyer. 2007. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology 153(Pt 8):2373-2385.
Zimmer, D. B., E. H. Cornwall, A. Landar, and W. Song. 1995. The s100 protein family: History, function, and expression. Brain Research Bulletin 37:417-429.
MICROBIOTA-TARGETED THERAPIES: AN ECOLOGICAL PERSPECTIVE34
Katherine P. Lemon,35,36,*Gary C. Armitage,37David A. Relman,38,39,40and Michael Fischbach41,42,*
Abstract
The connection between disease and the disruption of homeostatic interactions between the host and its microbiota is now well established. Drug developers and clinicians are starting to rely more heavily on therapies that directly target the microbiota and on the ecology of the microbiota to
________________
34 From K. P. Lemon, G. C. Armitage, D. A. Relman, M. A. Fischbach, Microbiota-Targeted Therapies: An Ecological Perspective. Sci. Transl. Med. 4, 137rv5 (2012). Reprinted with permission from AAAS.
35 Department of Molecular Genetics, The Forsyth Institute, Cambridge, MA 02142, USA.
36 Division of Infectious Diseases, Children’s Hospital Boston, Harvard Medical School, Boston, MA 02115, USA.
37 Division of Periodontology, School of Dentistry, University of California, San Francisco, San Francisco, CA 94143, USA.
38 Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, CA 94305, USA.
39 Department of Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
40 Veterans Affairs Palo Alto Health Care System, Palo Alto, CA 94304, USA.
41 Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, San Francisco, CA 94158, USA.
42 California Institute of Quantitative Biosciences, University of California, San Francisco, San Francisco, CA 94158, USA.
* To whom correspondence should be addressed. Email: klemon@forsyth.org (K.P.L.); fischbach@fischbachgroup.org (M.A.F.)
understand the outcomes of these treatments. The effects of those microbiotatargeted therapies that alter community composition range in scale from eliminating individual strains of a single species (for example, with antibacterial conjugate vaccines) to replacing the entire community with a new intact microbiota (for example, by fecal transplantation). Secondary infections linked to antibiotic use provide a cautionary tale of the unintended consequences of perturbing a microbial species network and highlight the need for new narrow-spectrum antibiotics with rapid companion diagnostics. Insights into microbial ecology will also benefit the development of probiotics, whose therapeutic prospects will depend on rigorous clinical testing. Future probiotics may take the form of a consortium of long-term community residents: “a fecal transplant in a capsule.” The efficacy of microbiota-targeted therapies will need to be assessed using new diagnostic tools that measure community function rather than composition, including the temporal response of a microbial community to a defined perturbation such as an antibiotic or probiotic.
The human microbiota consists of hundreds of microbial species that inhabit distinct body sites, which can be thought of as ecosystems (see review by Costello et al., 2012). The healthy human state is a homeostasis between the microbiota and the host. Maladies such as Crohn’s disease, chronic periodontitis, and bacterial vaginosis are characterized by a disruption of this homeostasis, a state known as dysbiosis (Tamboli et al., 2004). We envision a future in which new therapeutics and diagnostics enable the management of our microbiota to treat and prevent disease (Kolter, 2009).
Two major challenges face the development of microbiota-targeted therapies. First, changes in the composition or function of the microbiota must be proven to cause a disease or contribute to its pathology. Correlative studies are a good start, but without a causative link there is little reason to expect that a microbiota-targeted therapy will cure a disease or alleviate its symptoms. Second, we may need to be satisfied with a less detailed understanding of how microbiota-targeted therapies work than we currently expect for drug development. To date, a threshold requirement for developing a new drug is an understanding of its molecular mechanism at the level of a protein-ligand interaction. Should this requirement be applied to microbiota-targeted therapies? A central lesson of ecology is that perturbations often ripple through an ecosystem, leading to unexpected outcomes (Costello et al., 2012). Likewise, the desired effect might not occur at the level of inhibiting a specific enzyme but by restructuring a microbial community in a specific way. To advance microbiota-directed therapeutics, we will need to define new endpoints (for example, microbial community composition or function), develop therapies to achieve them, and design diagnostics to evaluate our efforts.
What Is the Goal of Therapeutically Perturbing a Microbial Community?
A clear therapeutic goal is a prerequisite for designing a drug; in a practical sense, the goal will determine the diagnostic used to judge the therapy’s success in preclinical studies and clinical trials. The goal of a therapy often takes the form of a clinical endpoint or a surrogate endpoint, and the distinction between them is especially important for microbiota-targeted therapies. A clinical endpoint is the long-term goal of a therapy, such as preventing death from heart disease or eliminating the signs and symptoms of Crohn’s disease (Lesko and Atkinson, 2001). If the goal of a microbiota-targeted therapy is a clinical endpoint (for example, clearing Clostridium difficile–associated diarrhea), then achieving it is equivalent to clinical success.
However, clinical endpoints are often impossible to measure in preclinical studies because of the use of animal models rather than human subjects. In addition, certain clinical endpoints can be impractical to measure in a clinical trial (for example, the prevention of death from heart disease). In these cases, a surrogate endpoint—a marker whose level predicts the therapy’s effect on the clinical endpoint—is used (Lesko and Atkinson, 2001). For example, a reduction in serum cholesterol has been used as a surrogate endpoint for preventing death from heart disease. The goal of many microbiota-targeted therapies will be a surrogate endpoint, such as shifting the gut community from a Crohn’s disease–associated state to a “healthy” state. If these therapies are to succeed, their surrogate endpoints must be predictive of the clinical endpoint; a correlation between the two is not enough.
What Is a Healthy Microbiome?
The goal of many microbiota-targeted therapeutics will be to restore a healthy host–microbiota homeostasis. Metrics for assessing the state of the microbiota can be sorted into two categories: global measurements and specific markers. However, as discussed later, the health of a community may have more to do with its ability to resist pathogen invasion (a functional property) than its static composition (a structural property).
Two commonly used global metrics derive from ecology: alpha diversity (within-community diversity) and beta diversity (between-community diversity). The relationship between alpha diversity and health differs among body sites; for example, increased alpha diversity in the gut is associated with a decreased risk of necrotizing enterocolitis in premature infants (Mshvildadze et al., 2010; Neu and Walker, 2011; Wang et al., 2009), whereas decreased alpha diversity in the vaginal community is associated with a decreased risk of bacterial vaginosis (Ravel et al., 2011). Although the relationship between beta diversity and health is not as well explored, the example of cystic fibrosis is instructive: Increased beta diversity is observed in later-stage disease after the oropharyngeal microbiota is invaded by Pseudomonas aeruginosa and subjected to increased antibiotic
exposure (Klepac-Ceraj et al., 2010). It is not yet known whether the increased beta diversity in cystic fibrosis is a cause or a consequence of antibiotic exposure and pathogen invasion.
In addition to diversity measures associated with health and disease, specific taxa are emerging as indicators of disease states. In many microbiota-related ailments, the disease state is marked by a change in the level of normal community members rather than the presence of a foreign pathogen. For example, a decrease in the common gut resident Faecalibacterium prausnitzii is associated with ileal Crohn’s disease (Sokol et al., 2008; Willing et al., 2009), and the suspected causative agents of gingivitis and periodontitis are normal members of healthy periodontal communities (Hayashi et al., 2006; Li et al., 2004; Tanner et al., 2002; Wilson et al., 1993). Two key challenges for microbiota markers are to define the relationship between the marker species/genes and disease pathophysiology, especially if it is direct, and to determine how broadly these apply within the human population.
What Scale of Perturbation Is Appropriate?
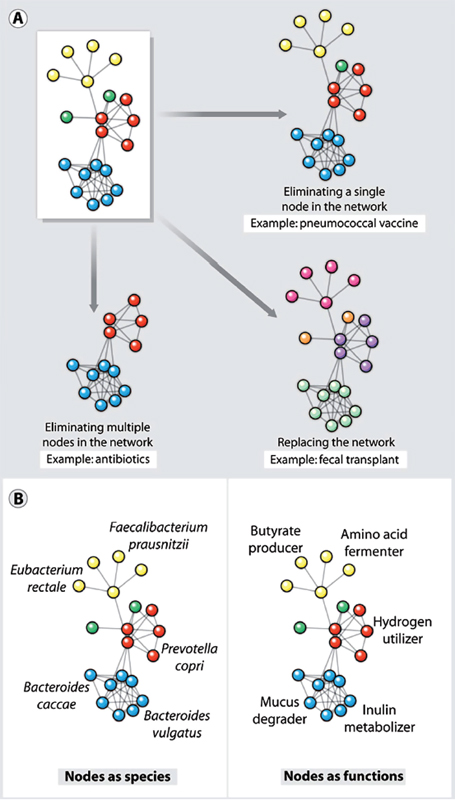
Viewing a human-associated microbial community as a network of interacting species, the scale of perturbation could range from “surgically” excising a single node to eliminating the entire network and replacing it with a new one (Figure A10-1A). There is precedent for interventions at both ends of this spectrum. Antibacterial conjugate vaccines represent the simplest perturbations; these agents stimulate the immune system to remove specific strains of a single species from the community. Fecal transplantation, which lies at the opposite end of the spectrum, consists of a colonic lavage to dislodge the resident community followed by the infusion of a donor community (20 to 30 g of donor feces suspended in 100 ml of water) during endoscopy or colonoscopy. Fecal transplantation appears to be effective in clearing recurrent C. difficile infections that are refractory to antibiotic treatment (Gough et al., 2011), but it is not yet clear whether a perturbation this extreme is required to displace a tenacious strain of C. difficile from the gut community.
As we highlight in the examples below, the tendency of ecological perturbations to ripple through a community can make the effects of even a simple perturbation difficult to predict. A major challenge for the future will be to create predictive models of microbial community ecology that can anticipate—and ultimately circumvent—these ripple effects (Costello et al., 2012; Robinson et al., 2010).
Targeted Removal of Specific Strains from the Upper Respiratory Tract Microbiota
Several widely used vaccines target bacterial mutualists or commensals that are occasionally pathogenic (termed “pathobionts”). For pathobionts, colonization is the first step in infection; these strains colonize many people and cause deadly infections in a subset. Two examples of such vaccines are conjugate vaccines against Haemophilus influenzae type b (Hib) and Streptococcus pneumoniae that protect against infection and clear or prevent colonization (Figure A10-1A). Once the leading cause of bacterial meningitis in children < 5 years old, Hib infections have been nearly eliminated by the use of a conjugate vaccine (Adams et al., 1993; CDC, 1996). Before the vaccine’s introduction in the early 1990s, Hib colonized 3 to 5% of healthy children; in immunized populations, the vaccine has virtually eradicated Hib from the pharyngeal microbiota. It has apparently been replaced by less virulent H. influenzae strains (Mohle-Boetani et al., 1993; Takala et al., 1991).
Similarly, the introduction in 2000 of the seven-valent pneumococcal conjugate vaccine against S. pneumoniae resulted in a 77% decline in severe invasive disease (CDC, 2008; Whitney et al., 2003). After vaccination, nonvaccine serotypes of S. pneumoniae (most of which were less virulent than the vaccine serotypes) became more prevalent in the pharyngeal community, and overall S. pneumoniae colonization rates did not decline significantly. Thus, immunizations against Hib and S. pneumoniae have led to their replacement by less virulent members of the same species and appear to have caused few other effects in the ecosystem. This phenomenon may prove to be a common outcome of the targeted removal of a pathobiont, because many pathobionts have closely related, nonpathogenic relatives that can occupy the same niche. (Dethlefsen et al., 2007). An important caveat is that initial concerns about a possible increase in Staphylococcus aureus carriage after the use of the S. pneumoniae vaccine may yet prove to be valid (Regev-Yochay et al., 2008).
The success of vaccines against pathobionts underscores the need for a better understanding of the immunological aspects of host–microbiota interactions and the potential for success in developing new microbiota-targeted vaccines (see reviews by Blumberg and Powrie, 2012, and Hooper et al., 2012). It also illustrates that strain-level changes in community composition can have a clinically important impact. Because these changes are not usually detectable by 16S rRNA (ribosomal RNA) sequence analysis, there is a growing need for culture-independent diagnostics that enable species- and strain-level identification of the microbiota. Finally, our ability to surgically remove a single taxon from a community has come far in advance of our ability to understand the ways in which communities respond to such perturbations, including the role of microbe-microbe interactions in determining which species fill a niche opened by a perturbation (Figure A10-2A).
Antibiotics Cause Collateral Damage to Microbiota
Antibiotics save countless lives and are essential to clinical practice, but they profoundly perturb human-associated microbial communities (Blaser, 2011; Costello et al., 2012; Dethlefsen and Relman, 2011). Antibiotic treatment often achieves the goal of eradicating infection with help from the immune system. However, it can have the ecologically undesirable side effect of killing mutualistic (that is, helpful) bacterial symbionts, creating a rare opportunity for a hardy pathogen—whose growth usually is suppressed by these mutualists—to become a dominant species in the community (for example, alternative state 2 in Figure A10-3) (Costello et al., 2012). Two examples of such “secondary infections” are C. difficile–associated diarrhea and yeast infections (candidiasis), both of which are associated with antibiotic use (Kelly and LaMont, 2008; Spinillo et al., 1999). In such situations, the goal of a microbiota-targeted therapy would be to perturb the new “unhealthy” alternative stable state of the community sufficiently to shift it to a “healthier” alternative stable state (Figure A10-3).
The clinicians who prescribe antibiotics and the researchers who develop them face four challenges, which we focus on here and in the following section: (i) Can clinically useful adjunctive therapies be developed to prevent secondary infections? (ii) Can physicians be encouraged to use antibiotics with narrower activity spectra to minimize the collateral damage to bacterial mutualists? (iii) Should diagnostics be designed to identify the etiologic agent of infection and then actively monitor the microbiota for signs of a secondary infection? Clinicians routinely monitor patients for adverse effects of antibiotics on the kidneys and liver, but apart from counseling patients to be alert for symptoms of C. difficile–associated diarrhea and candidiasis, they have no good way to monitor the state of the microbiota. (iv) Can targeted antibiotics that cause minimal perturbation to the healthy microbiota be developed?
FIGURE A10-1 Microbial communities as networks. (A) Shown are three types of perturbation to a network of microbial species such as that found in the microbiota at various body sites. The microbiota can be perturbed by excision of a single species (node) by a vaccine or a species-specific antibiotic, by elimination of multiple nodes or a subnetwork by an antibiotic, or by replacement of a whole network using microbiota transplantation. (B) Two ways of modeling a microbial community as a network. (Left) Nodes as species, and edges as interactions among species. Species networks can be constructed directly from metagenomic sequence data, but they lack functional information. (Right) Nodes as functions, and edges as interactions among functions. Function networks can generate hypotheses about the mechanism of microbiota–host interactions, but they require mapping genes to functions or a panel of direct functional measurements.
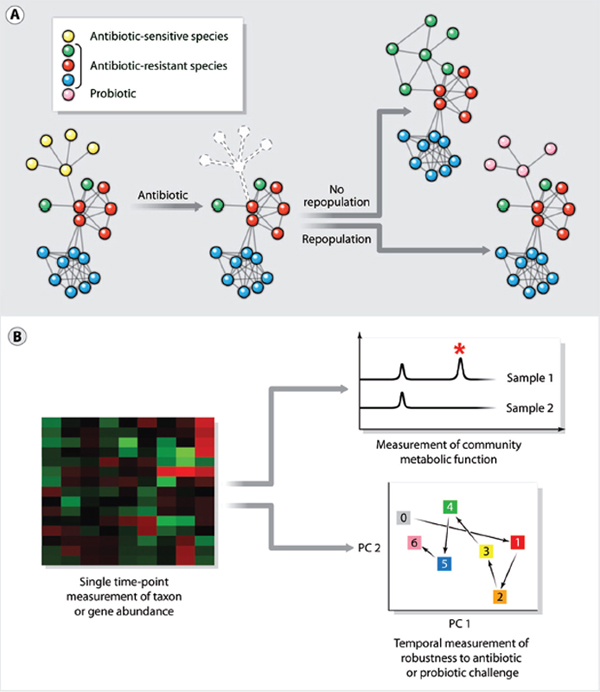
FIGURE A10-2 New opportunities in treatment and diagnostics. (A) Antibiotics save countless lives, but when they kill mutualistic (that is, helpful) microbiota that normally check the growth of pathogens, a secondary infection can ensue. Repopulating antibiotic-treated patients with probiotics is a promising strategy to prevent secondary infections. (B) Given that the human microbiota has many normal taxonomic compositions, it might be easier to develop markers of a normal or healthy community in terms of functional attributes like resistance and resilience. A single time-point measurement of taxon and gene abundance (left) is limited in its ability to provide functional information. Diagnostics based on direct measurements of metabolites (right, top) and temporal measurements of robustness to antibiotic or probiotic challenge (right, bottom) would enable community function to be assessed directly.
Passive Recovery Versus Active Repopulation
In a real sense, the current approach to antibiotic treatment is the equivalent of treating a weed-infested garden with a herbicide, and then leaving it fallow in the hope that the desired crops take root. This is an example of the ecological phenomenon of secondary succession, which describes changes in a community’s species after a disturbance. One way to influence the course of secondary succession is to seed the newly cleared ecosystem with one or more pioneer species (Figure A10-2A) (Costello et al., 2012).
An intriguing example of success with ecosystem repopulation in the context of the microbiome is the use of fecal transplantation to treat refractory recurrent C. difficile–associated diarrhea. The incidence of C. difficile infections is rising rapidly; between 1996 and 2005, the number of annual reported cases in the United States tripled and the number of deaths in England rose nearly sevenfold to 3,393 (Kelly and LaMont, 2008). Nearly all C. difficile infections are healthcare associated, occurring in patients who are either hospitalized or were recently discharged (CDC, 2012). A recent study reported that, in patients with multiple
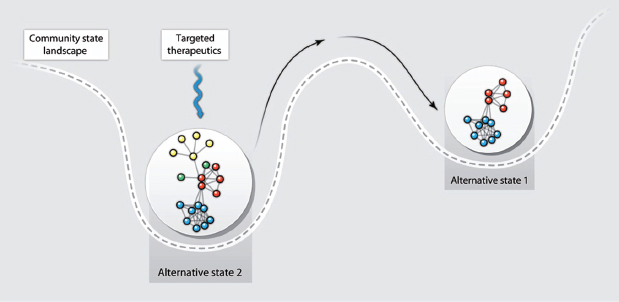
FIGURE A10-3 Microbiota-targeted therapy can shift a community to a healthier stable state. A microbial community’s response to a microbiota-targeted therapy can be illustrated with a stability landscape diagram (1). The ball containing the network represents the microbial community, and the shift in its horizontal position within the landscape represents movement between alternative stable states. The depth of a basin indicates the probability that the community will stay in that specific state in response to perturbation and, therefore, reflects the degree of perturbation needed to shift the community to an alternative stable state, for example, from state 1 to state 2. In this illustration, a therapeutic perturbation that removes some nodes (yellow and green) from the community network is sufficient to shift the community to an alternative, and in this case healthier, stable state (state 1).
recurrences, fecal transplantation resolved the symptoms of 34 of 34 patients who were infected with conventional strains of C. difficile and 32 of 36 patients who were infected with a C. difficile strain from a particularly virulent group known as ribotype 027 (Mattila et al., 2012). At present, fecal transplantation is only used to treat patients for whom secondary antibiotic therapy has repeatedly failed; however, its potential use as an adjunctive or follow-up therapy to the initial course of anti–C. difficile antibiotics could markedly reduce the rate of recurrence, which is currently 20 to 30%. Further in the future, one might imagine autologous fecal transplantation with the patient’s stored preantibiotic fecal microbiota as a way to prevent or treat C. difficile–associated diarrhea (Figure A10-2A).
Apart from this example, the literature suggests that a shift in clinical practice from passive recovery to active repopulation of microbiota during and after antibiotic treatment will require proof of efficacy and pharmaceutical-grade products (Thomas and Greer, 2010). A recent meta-analysis of randomized controlled trials of adjunctive probiotic therapy for the prevention of antibiotic-associated diarrhea in children suggests a possible benefit of high-dose probiotics (Johnston et al., 2011). In contrast, there are not yet sufficient data to support concurrent probiotic therapy during antibiotic treatment in adults to prevent C. difficile– associated diarrhea (Cohen et al., 2010). In both cases, there is a strong need for large randomized controlled trials with a consensus definition of the disease state and clinical endpoints, and standardized probiotic constituents and dosing.
Similarly, will future antibiotic treatment of pathobiont infections be followed by therapeutic repopulation with a mutualistic microbiota? For example, after treatment of S. aureus infections, the microbiota recovers passively but S. aureus is not necessarily eradicated, potentially leaving the patient at higher risk for a new S. aureus infection than someone who is not colonized. In certain high-risk populations, efforts have been made to eradicate S. aureus carriage, especially for methicillin-resistant S. aureus, through the use of a topical antibiotic to the anterior nares (mupirocin) and chlorohexidine body washes (Chow and Yu, 1989; Fritz et al., 2012). However, the current approach to decolonization makes no attempt to influence secondary succession. A future challenge will be to determine what role pathobionts such as S. aureus play in a healthy microbiota and whether a pathobiont-free consortium can be used to replace the original community.
Narrow-Spectrum Antibiotics: Policy, Diagnostics, and New Agents
Numerous evidence-based clinical practice guidelines and antibiotic stewardship programs promote the use of narrow-spectrum antibiotics. For example, a children’s hospital in Kansas introduced a clinical practice guideline for the empiric treatment (that is, before the etiologic agent of infection is identified) of uncomplicated community-acquired pneumonia that favored the use of ampicillin, a relatively narrow-spectrum antibiotic, over ceftriaxone, which has a much
broader activity spectrum. The result was a shift from empiric use of ceftriaxone in 72% of cases to empiric use of ampicillin in 63% of patients, with no significant change in the failure rate of the initial antibiotic (Newman et al., 2012). Similarly, a 2011 national clinical practice guideline recommends ampicillin for hospitalized children with uncomplicated community-acquired pneumonia (Bradley et al., 2012).
For patients with severe acute infections, the empiric use of broad-spectrum antibiotics remains a life-saving measure. In these situations, the switch to equally effective narrow-spectrum agents is limited by the fact that current diagnostics require 24 to 72 hours to cultivate, identify, and determine the antibiotic susceptibilities of the bacterium causing the infection. The increased use of narrow-spectrum antibiotics will require companion diagnostics that are not only more rapid and sensitive but also just as specific as current cultivation-based methods. Nucleic acid–based tests for pathogen-specific genes seem to meet these criteria, but their exquisite sensitivity raises a question that is both ecological and practical in nature: If such a test comes back positive, how can a clinician be certain that the pathogen is still alive, and if so, that it is the etiologic agent of infection and not a contaminant (for samples from sterile sites/compartments) or a bystander (for samples from nonsterile sites)?
In addition to providing equally effective treatment of susceptible bacterial pathogens, narrow-spectrum antibiotics minimize the collateral damage to microbiota community structure, decrease the selective pressure for the spread of antibiotic resistance genes within the microbiota, and lower the probability of acquiring resistant strains. Much of this is predicted by ecological studies showing that perturbations render a community more susceptible to invasion (Costello et al., 2012). Further research on the impact of antibiotics on the microbiota is needed to determine how the loss of antibiotic-susceptible species leads to unintended changes in the species network (Little et al., 2008); even a narrow-spectrum antibiotic may remove a well-connected node from a network, which could have an unexpectedly large impact on community structure.
How Should New Antibiotics Be Designed
Two key insights from ecology are likely to change the way future antibiotics are designed and developed. First, a long-standing paradigm has been that for activity spectrum, “broader is better.” The unintended consequences of broad-spectrum antibiotics provide an impetus for supplementing their position in the physician’s toolbox with targeted agents. Two recent efforts have proven that antibiotics can be designed with the narrowest of spectra: Both inhibit S. aureus but not closely related bacteria. One molecule, a dehydrosqualene synthase inhibitor, blocks the production of the glycolipid virulence factor staphyloxanthin, the yellow pigment from which the species name of S. aureus is derived. This antibiotic—which was originally developed as a candidate cholesterol
biosynthesis inhibitor for use in humans—does not kill S. aureus but makes the bacterium more sensitive to innate immune clearance in an animal model of infection (Liu et al., 2008). The other molecule, an FtsZ inhibitor, has been proposed to bind FtsZ in a region that is polymorphic among Firmicutes; the result is an antibiotic with activity against various species of Staphylococcus and Bacillus subtilis, but not against S. pneumoniae and Enterococcus faecalis (Haydon et al., 2008). To be used as single-agent therapies very early in the course of treating an infection, these drugs would require rapid companion diagnostics as described above.
Second, although antibiotics are often designed and tested against bacteria in liquid culture, pathogens often live in structured, surface-associated communities known as biofilms (López et al., 2010). Numerous infections are caused by bacteria living in biofilms: These include dental caries, periodontal infections, otitis media, endocarditis, osteomyelitis, artificial joint infections, pulmonary infections in cystic fibrosis, and device-associated infections (Darouiche, 2004; Furukawa et al., 2006). Given that existing antibiotics are rarely able to clear the biofilm from the affected site, patients often require weeks to months of therapy and surgical/mechanical intervention. To more effectively treat biofilm-associated infections, agents that promote the dispersion of biofilms or block their formation altogether are needed. A recent promising discovery comes from the observation that the commonly studied soil bacterium B. subtilis produces a diffusible factor that promotes the disassembly of its own biofilms, which is a mixture of D-amino acids (Kolodkin-Gal et al., 2010). The generality of this result was demonstrated by showing that D-amino acids can prevent biofilm formation by the gram-positive pathogen S. aureus and the gram-negative pathogen P. aeruginosa. Studies into the mechanism by which D-amino acids act could open the door to a new class of agents that reverse or prevent biofilm formation. This would make their constituent pathogens easier to treat with conventional antibiotics, many of which are more active against free-living bacteria than bacteria in a biofilm (Stewart and Costerton, 2001).
What Is the Path Forward for Probiotic Therapies
Probiotics are another class of microbiota-targeted therapies whose properties are intimately connected to ecological principles. One key question for the design of future probiotics is whether they should be short-term or long-term residents of a community. Most current probiotics (for example, species of Lactobacillus and Bifidobacterium) are transient members of the gut community, present for only a few days (Denou et al., 2008). Among the advantages of short-term colonization are a lower risk of unintended consequences and the ability to determine empirically which probiotic is most effective through rounds of trial and observation. However, long-term residents (for example, species of Bacteroides and Clostridium) would be capable of modulating community structure and
function in ways that short-term residents could not, including signaling to the host via receptors expressed in gut epithelial cells that may have evolved to monitor chemical cues from common long-term residents (see review by Holmes et al., 2012, and review by Nicholson et al., 2012). If future probiotic therapies supplying long-term residents become widely used, an important challenge—made famous by ecological studies of agriculture—will be to ensure that no single strain is used in too broad a swath of the human population; a limited choice of probiotic strains could inadvertently create a population-level vulnerability to disease (Hughes et al., 2008).
In addition to residence time, another key property of a probiotic therapy is the number of bacterial species it comprises. Most existing probiotics consist of one or a few strains, and the constituents of those that are simple consortia appear to have been chosen on the basis of the dictum of “more is better” rather than for scientific reasons. At the other end of the complexity spectrum, fecal transplants consist of an intact, highly complex community of hundreds of species. Will new probiotics occupy the middle of this spectrum? Two possibilities are synthetic communities containing ~20 species, chosen to represent the major taxa of common gut bacteria (Faith et al., 2011), and, more ambitiously, a defined mixture of 50+ species chosen to mimic the healthy state of one of the common “enterotypes” (Arumugam et al., 2011)—a simplified fecal transplant in a capsule.
Currently, most probiotics target the gut, but the principles of designing and testing gut probiotics may also apply to the skin, oral, vaginal, and upper respiratory tract communities. For example, for atopic dermatitis, a topical ointment with a probiotic Corynebacterium species that blocks colonization by S. aureus may be just as effective as an antibiotic or an immunosuppressant (Kong et al., 2012). Likewise, probiotic toothpastes and mouthwashes could be highly effective in reversing or preventing caries and periodontal disease (Zahradnik et al., 2009).
The widespread clinical use of probiotics will require proof of efficacy and safety from large randomized controlled trials with a consensus definition of the disease state, pharmaceutical-grade products, and standardized dosing (Cohen et al., 2010; Johnston et al., 2011; Thomas and Greer, 2010). A few notable studies have led the way in showing a positive effect of probiotics on disease outcome (Shanahan, 2010). Clear successes have been seen with the use of probiotics for preventing necrotizing enterocolitis in preterm neonates (Dshpande et al., 2010) and acute diarrhea in children (Sur et al., 2011), and promising data exist for preventing antibiotic-associated diarrhea and C. difficile–associated diarrhea in adults (Gao et al., 2010; Hempel et al., 2012). Probiotics are beneficial for ulcerative colitis and pouchitis, and although the data look less promising for Crohn’s disease, differences in study design (the use of different bacterial strains and underpowered studies) make the results difficult to interpret and compare (Isaacs and Herfarth, 2008). A hint of benefit has been seen for irritable bowel syndrome, but a recent meta-analysis highlighted that most studies to date have been poorly
designed (Brenner et al., 2009), emphasizing the need for better study design in the future. Notably, low success rates in individual trials need not be a reason to abandon the effort; as long as a subset of patients are clear responders, much could be learned from pursuing the biological basis of their response, just as a subset of responders have revealed the molecular basis for a response to targeted kinase inhibitors in oncology trials (Jänne et al., 2009).
For probiotics with proven safety and efficacy, elucidating the molecular mechanisms by which they exert their effects will also be important for convincing the academic and clinical communities of their importance. Notably, a probiotic does not need to cause a change in the composition of the community to exert an effect, because community function (or that of the host) may be altered without changing community membership. Indeed, a recent report shows that a mixture of probiotics does not alter the composition of the gut microbiota, but it changes their metabolic function (McNulty et al., 2011).
Will Microbiota Diagnostics Lead or Follow Therapeutics?
The microbiota has two characteristics that make it an excellent source of diagnostics (Figure A10-2B). First, information richness: Bacterial cells are exquisitely sensitive to their environment, often changing in number or activating transcriptional circuits in response to extracellular cues. The community roster (metagenome) and state (metatranscriptome, metaproteome, and metametabolome) will therefore be sensitive readouts of, inter alia, inflammation, and metabolism (Blumberg and Powrie, 2012; Holmes et al., 2012; Hooper and Littman, 2012; Nicholson et al., 2012). Second, ease of collection and analysis: Fecal samples, saliva, and nasal swabs are easier to provide than blood. For a patient at risk for developing Crohn’s disease, mailing a monthly fecal sample from home would be simpler and more cost effective than an annual colonoscopy. Tools being developed for cancer genotyping could be adapted for microbiota analysis, so the big challenge will be less technical in nature and more scientific: What question should be asked of the sample?
A simple combination of perturbation and measurement, such as used in the glucose tolerance test, should be the goal of some diagnostics for microbiota (Figure A10-2B). For example, administering a probiotic or antibiotic and then measuring community composition at a few longitudinal time points would determine the community’s resistance to perturbation and its resilience (ability to return to its original state). Although single time-point measurements of community composition are informative, perturbation-based diagnostics will be especially important for the microbiota, which has many “normal” taxonomic compositions. It might be easier to define normal or healthy in terms of functional attributes like resistance and resilience rather than by a single measurement of composition.
An emerging theme that links ecology to the latest studies of the microbiota is the need to map a community’s composition to its function (Figure A10-1B).
The mapping is rarely obvious, because closely related taxa can have distinct functions and unrelated species can have surprisingly similar functions. Nevertheless, a mechanistic understanding of how composition determines function is critical to developing useful diagnostics and therapeutics. The host may be indifferent to which species are present, but the community’s ability to liberate nutrients from the diet and resist pathogen invasion are vital.
Two questions thus arise: First, at what level of granularity should a function be interrogated? Is it enough to know that a certain gut species produces short-chain fatty acids, or is the distinction between propionate and butyrate important? Second, what will function-based diagnostics look like? One possibility, which takes advantage of the wealth of expertise in analyzing metagenomic samples from the microbiome, would be to develop ways of predicting functions from the taxa and genes present in a community. An alternative would be to develop simple tests, akin to a serum creatinine test for renal function, which could quickly assess an important metabolic index of community function from an easily obtained sample (Holmes et al., 2012; Nicholson et al., 2012). Whether diagnostics lead or follow therapeutics, an emphasis on measuring functions rather than taxa will help build what is becoming an increasingly critical bridge between ecology and therapeutics.
Efforts to make microbial ecology easier to model and more predictable will pay dividends, boosting our ability to understand and manipulate the human microbiota. Not surprisingly, industrial and academic drug discovery groups rarely have an ecologist on staff. That may need to change; rigorous ecological analysis will be an essential component of designing microbiota-targeted therapies, measuring their efficacy, and assessing their unintended effects on the community.
Acknowledgments
We thank L. Costello, B. Bohannon, J. Gordon, V. Klepac-Ceraj, the members of the Forsyth Host-Microbiota Focus Group, and the three reviewers for helpful comments. Funding: Research in the authors’ laboratories is supported by a Medical Research award from the W. M. Keck Foundation (M.A.F.), a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.), NIH DP2 OD007290 (M.A.F.), NIH DE020751 (K.P.L.), a Children’s Hospital Boston, Office of Faculty Development Career Development Fellowship (K.P.L.), a Distinguished Clinical Scientist Award from the Doris Duke Charitable Trust (D.A.R.), an NIH Pioneer Award DP1OD000964 (D.A.R.), and the Thomas C. and Joan M. Merigan Endowment at Stanford University (D.A.R.).
References
E. Costello, K. Stagaman, L. Dethlefsen, B. J. M. Bohannon, D. A. Relman, The application of ecological theory towards an understanding of the human microbiome. Science, 336, 1255–1262 (2012).
C. P. Tamboli, C. Neut, P. Desreumaux, J. F. Colombel, Dysbiosis in inflammatory bowel disease. Gut 53, 1–4 (2004).
R. Kolter, A visit to the paediatrician in the not-so-distant future. Environ. Microbiol. Rep. 1, 12–16 (2009) (10.1111/j.1758-2229.2008.00010.x).
L. J. Lesko, A. J. Atkinson Jr., Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: Criteria, validation, strategies. Annu. Rev. Pharmacol. Toxicol. 41, 347–366 (2001).
M. Mshvildadze, J. Neu, J. Shuster, D. Theriaque, N. Li, V. Mai, Intestinal microbial ecology in premature infants assessed with non–culture-based techniques. J. Pediatr. 156, 20–25 (2010).
J. Neu, W. A. Walker, Necrotizing enterocolitis. N. Engl. J. Med. 364, 255–264 (2011).
Y. Wang, J. D. Hoenig, K. J. Malin, S. Qamar, E. O. Petrof, J. Sun, D. A. Antonopoulos, E. B. Chang, E. C. Claud, 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 3, 944–954 (2009).
J. Ravel, P. Gajer, Z. Abdo, G. M. Schneider, S. S. Koenig, S. L. McCulle, S. Karlebach, R. Gorle, J. Russell, C. O. Tacket, R. M. Brotman, C. C. Davis, K. Ault, L. Peralta, L. J. Forney, Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl. 1), 4680–4687 (2011).
V. Klepac-Ceraj, K. P. Lemon, T. R. Martin, M. Allgaier, S. W. Kembel, A. A. Knapp, S. Lory, E. L. Brodie, S. V. Lynch, B. J. Bohannan, J. L. Green, B. A. Maurer, R. Kolter, Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ. Microbiol. 12, 1293–1303 (2010).
H. Sokol, B. Pigneur, L. Watterlot, O. Lakhdari, L. G. Bermúdez-Humarán, J. J. Gratadoux, S. Blugeon, C. Bridonneau, J. P. Furet, G. Corthier, C. Grangette, N. Vasquez, P. Pochart, G. Trugnan, G. Thomas, H. M. Blottière, J. Doré, P. Marteau, P. Seksik, P. Langella, Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736 (2008).
B. Willing, J. Halfvarson, J. Dicksved, M. Rosenquist, G. Järnerot, L. Engstrand, C. Tysk, J. K. Jansson, Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm. Bowel Dis. 15, 653–660 (2009).
F. Hayashi, M. Okada, Y. Soda, K. Miura, K. Kozai, Subgingival distribution of Campylobacter rectus and Tannerella forsythensis in healthy children with primary dentition. Arch. Oral Biol. 51, 10–14 (2006).
J. Li, E. J. Helmerhorst, C. W. Leone, R. F. Troxler, T. Yaskell, A. D. Haffajee, S. S. Socransky, F. G. Oppenheim, Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97, 1311–1318 (2004).
A. C. Tanner, P. M. Milgrom, R. Kent Jr., S. A. Mokeem, R. C. Page, C. A. Riedy, P. Weinstein, J. Bruss, The microbiota of young children from tooth and tongue samples. J. Dent. Res. 81, 53–57 (2002).
M. Wilson, D. Lopatin, G. Osborne, J. B. Kieser, Prevalence of Treponema denticola and Porphyromonas gingivalis in plaque from periodontally-healthy and periodontally-diseased sites. J. Med. Microbiol. 38, 406–410 (1993).
E. Gough, H. Shaikh, A. R. Manges, Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53, 994–1002 (2011).
C. J. Robinson, B. J. Bohannan, V. B. Young, From structure to function: The ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74, 453–476 (2010).
Centers for Disease Control and Prevention (CDC), Progress toward elimination of Haemophilus influenzae type b disease among infants and children—United States, 1987–1995. MMWR Morb. Mortal. Wkly. Rep. 45, 901–906 (1996).
W. G. Adams, K. A. Deaver, S. L. Cochi, B. D. Plikaytis, E. R. Zell, C. V. Broome, J. D. Wenger, Decline of childhood Haemophilus influenzae type b (Hib) disease in the Hib vaccine era. JAMA 269, 221–226 (1993).
J. C. Mohle-Boetani, G. Ajello, E. Breneman, K. A. Deaver, C. Harvey, B. D. Plikaytis, M. M. Farley, D. S. Stephens, J. D. Wenger, Carriage of Haemophilus influenzae type b in children after widespread vaccination with conjugate Haemophilus influenzae type b vaccines. Pediatr. Infect. Dis. J. 12, 589–593 (1993).
A. K. Takala, J. Eskola, M. Leinonen, H. Käyhty, A. Nissinen, E. Pekkanen, P. H. Mäkelä, Reduction of oropharyngeal carriage of Haemophilus influenzae type b (Hib) in children immunized with an Hib conjugate vaccine. J. Infect. Dis. 164, 982–986 (1991).
Centers for Disease Control and Prevention (CDC), Invasive pneumococcal disease in children 5 years after conjugate vaccine introduction—Eight states, 1998–2005. MMWR Morb. Mortal. Wkly. Rep. 57, 144–148 (2008).
C. G. Whitney, M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, A. Schuchat; Active Bacterial Core Surveillance of the Emerging Infections Program Network, Decline in invasive pneumococcal disease after the introduction of protein–polysaccharide conjugate vaccine. N. Engl. J. Med. 348, 1737–1746 (2003).
L. Dethlefsen, M. McFall-Ngai, D. A. Relman, An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449, 811–818 (2007).
G. Regev-Yochay, D. Bogaert, R. Malley, P. W. M. Hermans, R. H. Veenhoven, E. A. M. Sanders, M. Lipsitch, E. Rubinstein, Does pneumococcal conjugate vaccine influence Staphylococcus aureus carriage in children? Clin. Infect. Dis. 47, 289–291 (2008).
R. Blumberg, F. Powrie, Microbiota, disease, and back again to health: A metastable journey. Sci. Transl. Med. 4, 137rv7 (2012).
L. V. Hooper, D. R. Littman, A. J. Macpherson, Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012).
M. Blaser, Antibiotic overuse: Stop the killing of beneficial bacteria. Nature 476, 393–394 (2011).
L. Dethlefsen, D. A. Relman, Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U.S.A. 108, (Suppl. 1), 4554–4561 (2011).
A. Spinillo, E. Capuzzo, S. Acciano, A. De Santolo, F. Zara, Effect of antibiotic use on the prevalence of symptomatic vulvovaginal candidiasis. Am. J. Obstet. Gynecol. 180, 14–17 (1999).
C. P. Kelly, J. T. LaMont, Clostridium difficile—More difficult than ever. N. Engl. J. Med. 359, 1932–1940 (2008).
Centers for Disease Prevention (CDC), Vital signs: Preventing Clostridium difficile infections. MMWR. Morb. Mortal. Wkly. Rep. 61, 157–162 (2012).
E. Mattila, R. Uusitalo-Seppälä, M. Wuorela, L. Lehtola, H. Nurmi, M. Ristikankare, V. Moilanen, K. Salminen, M. Seppälä, P. S. Mattila, V. J. Anttila, P. Arkkila, Fecal transplantation, through colonoscopy, is effective therapy for recurrent Clostridium difficile infection. Gastroenterology 142, 490–496 (2012).
D. W. Thomas, F. R. Greer; American Academy of Pediatrics Committee on Nutrition; American Academy of Pediatrics Section on Gastroenterology, Hepatology, and Nutrition, Probiotics and prebiotics in pediatrics. Pediatrics 126, 1217–1231 (2010).
B. C. Johnston, J. Z. Goldenberg, P. O. Vandvik, X. Sun, G. H. Guyatt, Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst. Rev. 11, CD004827 (2011).
S. H. Cohen, D. N. Gerding, S. Johnson, C. P. Kelly, V. G. Loo, L. C. McDonald, J. Pepin, M. H. Wilcox; Society for Healthcare Epidemiology of America; Infectious Diseases Society of America, Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31, 431–455 (2010).
J. W. Chow, V. L. Yu, Staphylococcus aureus nasal carriage in hemodialysis patients. Its role in infection and approaches to prophylaxis. Arch. Intern. Med. 149, 1258–1262 (1989).
S. A. Fritz, P. G. Hogan, G. Hayek, K. A. Eisenstein, M. Rodriguez, E. K. Epplin, J. Garbutt, V. J. Fraser, Household versus individual approaches to eradication of community-associated Staphylococcus aureus in children: A randomized trial. Clin. Infect. Dis. 54, 743–751 (2012).
R. E. Newman, E. B. Hedican, J. C. Herigon, D. D. Williams, A. R. Williams, J. G. Newland, Impact of a guideline on management of children hospitalized with community-acquired pneumonia. Pediatrics 129, e597–e604 (2012).
J. S. Bradley, C. L. Byington, S. S. Shah, B. Alverson, E. R. Carter, C. Harrison, S. L. Kaplan, S. E. Mace, G. H. McCracken Jr., M. R. Moore, S. D. St Peter, J. A. Stockwell, J. T. Swanson; Pediatric Infectious Diseases Society and the Infectious Diseases Society of America, Executive summary: The management of community-acquired pneumonia in infants and children older than 3 months of age: Clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin. Infect. Dis. 53, 617–630 (2011).
A. E. Little, C. J. Robinson, S. B. Peterson, K. F. Raffa, J. Handelsman, Rules of engagement: Interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 62, 375–401 (2008).
C. I. Liu, G. Y. Liu, Y. Song, F. Yin, M. E. Hensler, W. Y. Jeng, V. Nizet, A. H. Wang, E. Oldfield, A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319, 1391–1394 (2008).
D. J. Haydon, N. R. Stokes, R. Ure, G. Galbraith, J. M. Bennett, D. R. Brown, P. J. Baker, V. V. Barynin, D. W. Rice, S. E. Sedelnikova, J. R. Heal, J. M. Sheridan, S. T. Aiwale, P. K. Chauhan, A. Srivastava, A. Taneja, I. Collins, J. Errington, L. G. Czaplewski, An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321, 1673–1675 (2008).
D. López, H. Vlamakis, R. Kolter, Biofilms. Cold Spring Harb. Perspect. Biol. 2, a000398 (2010).
R. O. Darouiche, Treatment of infections associated with surgical implants. N. Engl. J. Med. 350, 1422–1429 (2004).
S. Furukawa, S. L. Kuchma, G. A. O’Toole, Keeping their options open: Acute versus persistent infections. J. Bacteriol. 188, 1211–1217 (2006).
I. Kolodkin-Gal, D. Romero, S. Cao, J. Clardy, R. Kolter, R. Losick, D -Amino acids trigger biofilm disassembly. Science 328, 627–629 (2010).
P. S. Stewart, J. W. Costerton, Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138 (2001).
E. Denou, R. D. Pridmore, B. Berger, J. M. Panoff, F. Arigoni, H. Brüssow, Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 190, 3161–3168 (2008).
E. Holmes, J. Kinross, G. R. Gibson, R. Burcelin, W. Jia, S. Pettersson, J. K. Nicholson, Therapeutic modulation of microbiota-host metabolic interactions. Sci. Transl. Med. 4, 137rv6 (2012).
J. Nicholson, E. Holmes, J. Kinross, R. Burcelin, G. Gibson, W. Jia, S. Pettersson, Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012).
A. R. Hughes, B. D. Inouye, M. T. Johnson, N. Underwood, M. Vellend, Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623 (2008).
J. J. Faith, N. P. McNulty, F. E. Rey, J. I. Gordon, Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 333, 101–104 (2011).
M. Arumugam, J. Raes, E. Pelletier, D. Le Paslier, T. Yamada, D. R. Mende, G. R. Fernandes, J. Tap, T. Bruls, J. M. Batto, M. Bertalan, N. Borruel, F. Casellas, L. Fernandez, L. Gautier, T. Hansen, M. Hattori, T. Hayashi, M. Kleerebezem, K. Kurokawa, M. Leclerc, F. Levenez, C. Manichanh, H. B. Nielsen, T. Nielsen, N. Pons, J. Poulain, J. Qin, T. Sicheritz-Ponten, S. Tims, D. Torrents, E. Ugarte, E. G. Zoetendal, J. Wang, F. Guarner, O. Pedersen, W. M. de Vos, S. Brunak, J. Doré; MetaHIT Consortium, M. Antolín, F. Artiguenave, H. M. Blottiere, M. Almeida, C. Brechot, C. Cara, C. Chervaux, A. Cultrone, C. Delorme, G. Denariaz, R. Dervyn, K. U. Foerstner, C. Friss, M. van de Guchte, E. Guedon, F. Haimet, W. Huber, J. van Hylckama-Vlieg, A. Jamet, C. Juste, G. Kaci, J. Knol, O. Lakhdari, S. Layec, K. Le Roux, E. Maguin, A. Mérieux, R. Melo Minardi, C. M’Rini, J. Muller, R. Oozeer, J. Parkhill, P. Renault, M. Rescigno, N. Sanchez, S. Sunagawa, A. Torrejon, K. Turner, G. Vandemeulebrouck, E. Varela, Y. Winogradsky, G. Zeller, J. Weissenbach, S. D. Ehrlich, P. Bork, Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
H. H. Kong, J. Oh, C. Deming, S. Conlan, E. A. Grice, M. A. Beatson, E. Nomicos, E. C. Polley, H. D. Komarow; NISC Comparative Sequence Program, P. R. Murray, M. L. Turner, J. A. Segre, Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012).
R. T. Zahradnik, I. Magnusson, C. Walker, E. McDonell, C. H. Hillman, J. D. Hillman, Preliminary assessment of safety and effectiveness in humans of ProBiora3, a probiotic mouthwash. J. Appl. Microbiol. 107, 682–690 (2009).
F. Shanahan, Probiotics in perspective. Gastroenterology 139, 1808–1812 (2010).
G. Deshpande, S. Rao, S. Patole, M. Bulsara, Updated meta-analysis of probiotics for preventing necrotizing enterocolitis in preterm neonates. Pediatrics 125, 921–930 (2010).
D. Sur, B. Manna, S. K. Niyogi, T. Ramamurthy, A. Palit, K. Nomoto, T. Takahashi, T. Shima, H. Tsuji, T. Kurakawa, Y. Takeda, G. B. Nair, S. K. Bhattacharya, Role of probiotic in preventing acute diarrhoea in children: A community-based, randomized, double-blind placebo-controlled field trial in an urban slum. Epidemiol. Infect. 139, 919–926 (2011).
X. W. Gao, M. Mubasher, C. Y. Fang, C. Reifer, L. E. Miller, Dose–response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and Clostridium difficile-associated diarrhea prophylaxis in adult patients. Am. J. Gastroenterol. 105, 1636–1641 (2010).
S. Hempel, S. J. Newberry, A. R. Maher, Z. Wang, J. N. V. Miles, R. Shanman, B. Johnsen, P. G. Shekelle, Probiotics for the prevention and treatment of antibiotic-associated diarrhea: A systematic review and meta-analysis. JAMA 307, 1959–1969 (2012).
K. Isaacs, H. Herfarth, Role of probiotic therapy in IBD. Inflamm. Bowel. Dis. 14, 1597–1605 (2008).
D. M. Brenner, M. J. Moeller, W. D. Chey, P. S. Schoenfeld, The utility of probiotics in the treatment of irritable bowel syndrome: A systematic review. Am. J. Gastroenterol. 104, 1033–1049 (2009).
P. A. Jänne, N. Gray, J. Settleman, Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat. Rev. Drug Discov. 8, 709–723 (2009).
N. P. McNulty, T. Yatsunenko, A. Hsiao, J. J. Faith, B. D. Muegge, A. L. Goodman, B. Henrissat, R. Oozeer, S. Cools-Portier, G. Gobert, C. Chervaux, D. Knights, C. A. Lozupone, R. Knight, A. E. Duncan, J. R. Bain, M. J. Muehlbauer, C. B. Newgard, A. C. Heath, J. I. Gordon, The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 3, 106ra106 (2011).