






































Below is the uncorrected machine-read text of this chapter, intended to provide our own search engines and external engines with highly rich, chapter-representative searchable text of each book. Because it is UNCORRECTED material, please consider the following text as a useful but insufficient proxy for the authoritative book pages.
4 Potential Technologies for the BioWatch Program Having been provided with an overview of the BioWatch program and input from public health officials that run local BioWatch pro- grams and use its information for critical, potentially high-regret1 deci- sion making, the workshop participants discussed the current state of the art, exploring the potential uses of four families of technology in the BioWatch program, and considering how these technologies might be strategically combined or deployed to optimize their contributions to an effective environmental detection capability. For each of these four sessions, the writer of a commissioned paper presented an over- view of the technology being discussed (see Appendixes GâJ). Each presentation was followed by three or four talks on potential next- generation technologies and then an open discussion among the work- shop participants. Each of the panels in the four sessions focused on autonomous, ful- ly automated, end-to-end systems that would be technology readiness level (TRL) 6-plus ready over three time frames: by 2016, by 2020, and beyond 2020. Panel members were also asked to identify the major in- tegration issues in making these systems field-deployable and capable of detecting organisms of interest with the required sensitivity and specificity to meet the needs expressed earlier by public health offi- cials. This chapter summarizes those presentations and the ensuing open discussions among the workshop participants. Key points during the four sessions include 1 In this context, âhigh-regretâ refers to a situation in which a decision maker would en- counter large negative consequences for being wrong. 41
42 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH ï· Potential challenges to consider: o Integration of multiple components or technologies (âdetec- tion is doneâ). o Effectively maximizing sample concentration and minimizing sample contamination (sample to environmental background). o High-quality databases, bioinformatics tools, and assay vali- dation are needed for all technologies. ï· A multidimensional approach is encouraged: o Technologies using immunoassays and mass spectrometry are fast; nucleic-acid approaches and genome sequencing can provide specificity and identify novel threats. o Orthogonal testing increases the reliability of a BioWatch Actionable Result (BAR) and confidence in the system. ï· Extensive and repeated field testing will be necessary for a de- tection system. ï· It is important to involve the end user (public health) in system development and testing. ï· The Department of Homeland Security (DHS) was encouraged to look beyond the familiar technologies and development pathways. AUTONOMOUS DETECTION SYSTEMS USING NUCLEIC-ACID SIGNATURES In the first of the four sessions, Raymond Mariella, Jr., a senior sci- entist at Lawrence Livermore National Laboratory (LLNL), provided an overview of his commissioned paper reviewing the state-of-the-art tech- nologies available for detecting organisms using nucleic acid signatures (see Appendix G). The following topics were then discussed by the pan- elists: lessons that could be learned from other industries in autonomous detection and how those lessons might apply to BioWatch; the evolution of assays to include and exclude various threats; the current state-of-the- art system used by the U.S. Postal Service (USPS) and next-generation autonomous detection system based on multiplex polymerase chain reac- tion (PCR) assays; and systems concepts and integration issues. Nucleic-Acid Signatures at Three Levels of Readiness In his interpretation of the three tiers of readiness, Mariella said that any system that would be deployable at TRL 6-plus by 2016 would have
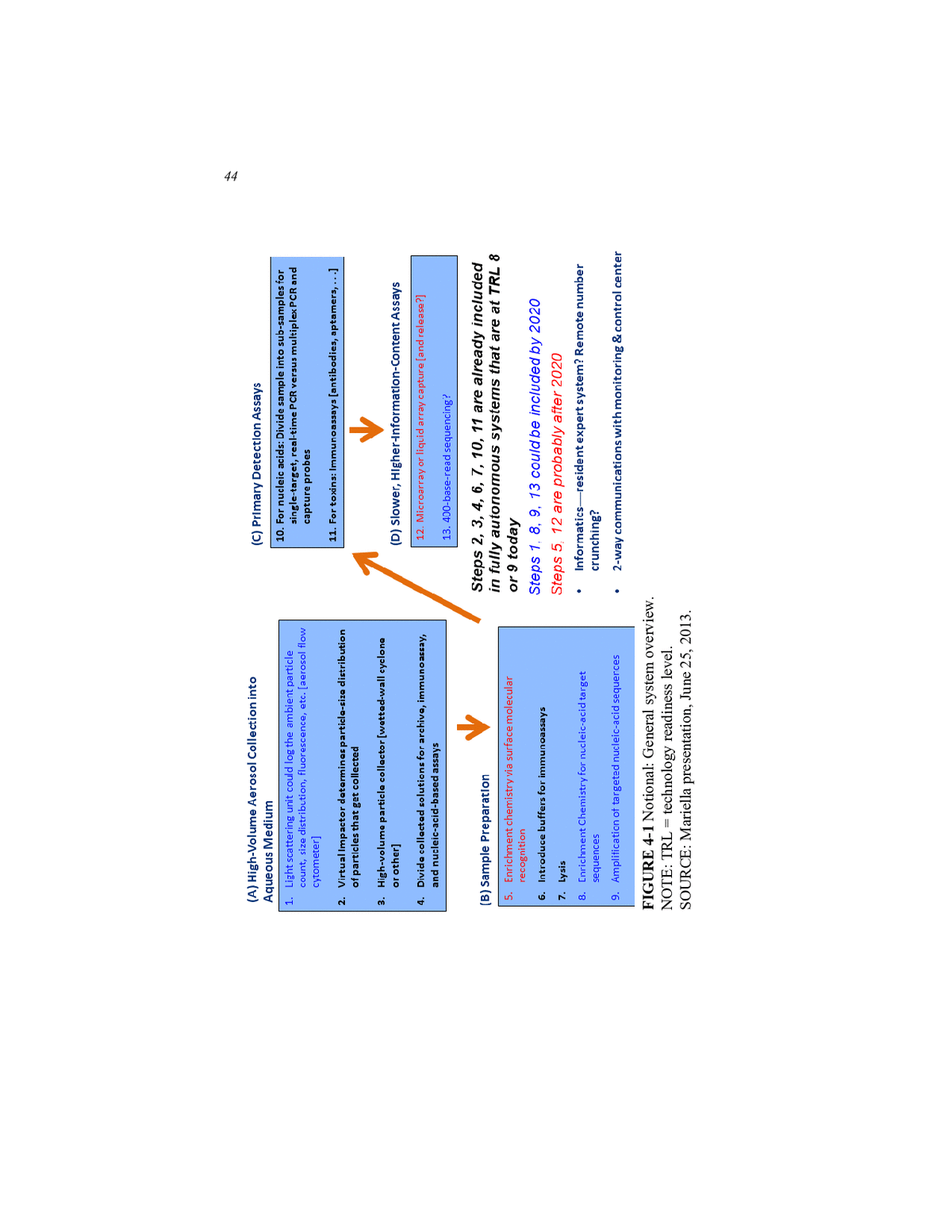
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 43 to consist of off-the-shelf components and be running today. For Tier 2â those ready by 2020âit should be possible to improve sample prepara- tion and add another level of identification to the automated assays, per- haps through some sort of targeted amplification of virulence regions. For Tier 3 systems, those available after 2020, technology developers still have years of vigorous research ahead to create and validate assays, develop the necessary informatics software and data interfaces, and per- form extensive integration work. Mariella then provided his notional overview of the system compo- nents and their states of readiness (see Figure 4-1). For Tier 1 readiness, the technologies that are available today and are at TRL 8 or TRL 9 (see Appendix E for TRL descriptions) include those for high-volume aerosol collection into an aqueous medium, sample collection, and primary de- tection. For collection, virtual impact collectors can reduce sample con- tamination and determine the particle size distribution of the particles that it collects. It should also be a simple matter to add a video camera to the system to monitor external events that might explain a positive sig- nal. Wetted-wall and other high-volume particle collectors can increase sample yield and deposit particles directly into aqueous media that can be divided into aliquots for multiplexed analyses. Collecting samples directly into a liquid would also increase the odds that any microorganisms would remain alive for subsequent viability testing. Automated systems that in- clude PCR for nucleic acid analysis, which require the lysis of spores to release nucleic acids, are currently the only technologies ready for 2016 deployment. For primary detection, Tier 1âready technologies can divide samples into aliquots for single-target, real-time PCR or can conduct mul- tiplexed assays using bead-based capture probes, Mariella said. Tier 2 systems could incorporate a light-scattering unit that would log the ambient particle count, size distribution, background fluores- cence, and other information that provides context for any positive sig- nal. Enrichment chemistries and amplification methods for nucleic acid target sequences could improve sample preparation in ways that allow for virulence assays that would increase the signal-to-noise ratio of the system. It may also be possible by 2020 to add 400-base sequencing of targeted sequences to an autonomous system and to incorporate resident expert systems for preliminary data analysis and two-way communica- tions with monitoring. One potential technology that Mariella mentioned that could be available after 2020 would use a surface molecular recogni- tion reagent or a pool of reagents to capture organisms of interest to en- rich samples for subsequent analysis (Hou et al., 2013).
44 FIGURE 4-1 Notional: General system overview. NOTE: TRL = technology readiness level. SOURCE: Mariella presentation, June 25, 2013.
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 45 Today, Mariella said, two systems are ready for deployment: the Au- tonomous Pathogen Detection System (APDS) developed largely by LLNL (Regan et al., 2008) and the Microfluidic Bioagent Autonomous Networked Detector (M-BAND) developed by Microfluidic Systems Inc., a subsidiary of PositiveID (Sanchez et al., 2011). Mariella said that both systems have been tested successfully in the field. âIf you want to put something in the field this afternoon, you have those two to choose from,â he said. He characterized a suite of aerosol-characterizing equip- ment as being mature and at TLR-8 or higher, including an inexpensive simple aerodynamic particle sizer and more expensive systems that use fluorescence measurements to characterize particles. The most advanced system that is ready for deployment uses an aerosol flow cytometer to capture particles and feed them into a mass spectrometer for further characterization. Mariella quickly described the use of selective reagents for capturing pathogenic bacteria and viruses using antibodies, aptamers, synthetic peptides, or nanolipoproteins. In his opinion, he said, it should be feasi- ble to use a panel of 20 or 30 reagents on magnetic beads to capture all of the threat organisms, separating them from the background atmospheric microbial content and thereby improving the performance of the down- stream analytical technologies. He also mentioned the AmpliSeq tech- nology (Murphy et al., 2005; Towler et al., 2008), which can be used to search for targeted amplification of virulence regions or other identifiers, including plasmids in bacteria, which again would provide better signal- to-noise performance. Emulsion or digital PCR could also improve both single and multiplexed amplification and provide intrinsic quantification of starting copy number, offer real-time detection in fewer cycles with less amplification bias, and be available by 2020 (Kiss et al., 2008; Shiroguchi et al., 2012; Vogelstein and Kinzler, 1999; Whale et al., 2013; Woyke et al., 2010). Cindy Bruckner-Lea from the Pacific Northwest National Laboratory commented that one driver of technology that had not been mentioned was the need to analyze indoor samples more efficiently. She suggested that particle detection could be used to trigger the more costly sample analysis. Mariella questioned this suggestion, saying that, in his view, it would not provide the best return on investment.
46 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH Issues and Opportunities for BioWatch Autonomous Detection According to Allen Northrup, sample processing and fluidics are the two areas related to next-generation BioWatch systems that need the most attention. âIn my opinion, detection is done,â he said. âThe point is to get the sample into the detector, and there are plenty of state-of-the- art, FDA [Food and Drug Administration]-approved detection technolo- gies for doing that.â Fluidic control has its challengesâmainly related to situations in which the output of one module may not match the input needs of another moduleâbut industries other than biodefense, particu- larly the food and beverage and pharmaceutical industries, have solved this problem and have created proven commercial systems using fluidics. The key challenges for any system, he said, are to define specificity within a defined limit of detection and to then define sensitivity within the context of that specificity (see Figure 4-2). False-positives and false- negatives, he said, also need to be defined within a predetermined limit of detection, with the caveat that, at the limits, false-positives and false- negatives are inevitable with all analytical systems. âThe question be- comes what can we live with? What is realistic?â PCR is a well-proven, reliable technology that can provide quantita- tive results in a multiplexed system. The challenge for the next- generation BioWatch system, Northrup said, is to automate it and im- prove handling of the complex samples that are collected in the field. âHowever, the thing I learned today is that BioWatch is more than in- struments and threshold cycles,â he said. âIt is a basis of human interac- tion and communication, informed decisions, and ancillary information. Whatever we build and deploy is going to have to have that human interac- tion.â In his opinion, a lab-in-a-box system that inputs a sample and outputs a result will not be well accepted by public health, which will want to see the direct result of any analysis and understand thoroughly the analytical param- eters. Northup recommended that system developers provide public health agencies and laboratories with the subcomponents of their systems for eval- uation and approval early in the development process. To conclude his remarks, Northrup said that the BioWatch communi- ty needs to test new systems repeatedly and be tolerant of early failures. He proposed that BioWatch should be developed as a modular system with significant, ongoing input from public health laboratories concern- ing which processes they would like to see automated. The resulting au- tomated modules should then be provided to the laboratorians for
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 47 FIGURE 4-2 Fundamentals of all instrument analyses. SOURCE: Northrup presentation, June 25, 2013; Northrup Consulting Group. validation alongside currently accepted methods in an iterative process that yields a robust, proven module for incorporation into an automated system for which public health will have already developed familiarity and confidence. Assay Development and Evolution In the next presentation, Ivor Knight, senior vice president and chief technology officer of Canon U.S. Life Sciences, Inc., focused his re- marks on the question: How will the technology evolve signatures as in- creasing knowledge of targets and neighbor strains reveals both false- positive and false-negative situations? As background, he described how such signatures are developed today, starting with identifying genes that are distinctive to a target organism and developing probes or primers to detect those genes. This deductive approach, which has been used success-
48 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH fully for more than 25 years, relies on deep knowledge of the pathogen in question and works well for targeting key pathogenic determinants, such as toxin-coding genes. However, this approach ignores most of the genome as a potential signature candidate. The newer genomic approach, which has been developed in the past decade or so in parallel with the revolutionary advances in genome se- quencing technology, starts with a target organismâs full-genome se- quence data and compares that sequence with the full-genome sequence data from all other organisms. The result is a set of sequences that are unique to the target. This approach requires no knowledge of the patho- gen, and the analysis is done completely in silico, yielding multiple tar- gets. Knight noted that this approach relies on coherence between the in silico world of genome databases and the real world, where false-positives and false-negatives arise. As the number of microbial sequences continues to increase, he said, signature designs will continue to improve, lowering the probability of false-positive and false-negative results. During the past decade, this type of approach has also yielded a new understanding of bacterial and viral genetics. One thing that has become apparent, Knight said, is that there is not a clear-cut situation in which a sequence is found in the target and not in the background genomes. âWhat we have learned is that microbial genomes form a continuous ge- netic background,â he said. Instead of each species having a completely unique genome, species share what Knight called pan-genomes, core ge- nomes that are common to all members of a particular genus of bacteria, with various additions and subtractions of genes and gene families across the species and strains of species in that genus (Hu et al., 2011) (see Fig- ure 4-3). As a result of this new understanding of microbial genomes, it is now necessary to consider pan-genomes and to use multiplexed capabilities to interrogate more potential genomic signatures. âWe need multiple targets to be able to assess differences on this continuous genome space,â Knight said. Moreover, target sets will have to be adaptive, and analyti- cal systems will have to accommodate changing signatures on what Knight suggested could be a quarterly basis. âWeâre going to want to change the signatures so that we can have greater accuracy in our detec- tions,â he said. For the 2020 scenario, Knight said, it will be necessary to sequence not just signature regions but the entire genetic complement of a sample in order to provide a probability-based analysis of the genomic content of the sample.
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 49 FIGURE 4-3 Pan-genomes of the genus Burkholderia. SOURCE: Knight presentation, June 25, 2013 (adapted from Hu et al., 2011). In his opinion, Knight said, PCR-based multiplex technologies using bead arrays and real-time PCR with or without hybridization probes are mature and will be ready to be deployed by 2016, while digital PCR will be ready for deployment by 2020. The big challenges ahead, he said, will involve system integration and workflow. For example, how will reflex- ive testing, in which an initial screen for a single target provides a poten- tial positive result that triggers additional, deeper testing, be integrated into an autonomous system? Knight also noted that consumables han- dling has not yet reached the required level of maturity for it to be ready for widespread deployment in an autonomous system in a way that would enable laboratorians responding to a positive result to instantly trace the history of the consumables in an instrument all the way back to
50 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH their manufacturers. Similarly, systems monitoring and preventive maintenance procedures will need to be developed. Finally, reiterating Northrupâs earlier recommendation, Knight said that all of these systems and procedures must from the beginning be worked out with the laboratorians involved. âIf public health laboratorians are going to be involved in decision making, they are going to want to know and under- stand the quality system that was put in place for that automated system. They are going to want to see that the quality system is the same or simi- lar or analogous to the quality system that they are used to in their la- boratories that has been developed and is tried and true over the years.â State-of-the-Art and Next-Generation Autonomous Detection Systems Next, in a talk on Northrop Grummanâs work in the biodefense field, David Tilles, vice president for CBRNE [chemical, biological, radiologi- cal, nuclear, and explosive] Defense at Northrop Grumman, focused on two systems: the Biohazard Detection System (BDS) that the USPS has been using for more than a decade and the Next Generation Automated Detection System (NG-ADS) that the company has developed and field tested under a Generation 3 Phase 1 contract with the Department of Homeland Security (DHS). NG-ADS, Tilles explained, evolved from LLNLâs APDS device, which Mariella had discussed earlier. BDS, which was developed in the wake of the anthrax letter incident in 2001, screens mail as it enters the USPS network and provides a warn- ing before contaminated mail is distributed through the mail network. In a sense, Tilles said, BDS can be considered âLab-in-a-Box 1.0,â an au- tonomous system that collects samples adjacent to a USPS mail pro- cessing machine. Samples are taken over the course of an hour, during which time between 30,000 and 40,000 letters could have been processed. The USPS can track those letters to isolate them in the event of an alarm. At the end of an hour, the sample moves into a reservoir for PCR testing, while the instrument starts collecting the next sample. Sample prepara- tion and PCR are performed using a disposable cartridge operating in a Cepheid GeneXpert machine, and the results are then reported. So far, the USPS BDS system has conducted more than 11 million tests with no false-positives. Tilles said that the system has been validat- ed by a number of federal agencies outside the USPS, including the De- partment of Defense and the Centers for Disease Control and Prevention (CDC). The USPS also collaborated with the U.S. Armyâs Edgewood
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 51 Chemical Biological Center to create a test bed to assess system perfor- mance under simulated operational conditions. National deployment of the system was completed in 2005, and more than 1,000 detectors have been installed at hundreds of sites. These detectors are networked in a fully integrated support infrastructure that not only transmits data but also monitors the health and status of the instruments; manages consum- ables, spares, and repairs; and conducts trend analysis for continuous improvement. The National Field Monitoring Center, run by Northrup Grumman, oversees operations of the system. NG-ADS, Tilles explained, is similar to BDS in that it is an automat- ed system that collects, processes, and analyzes samples. âBut because of the requirements of the BioWatch mission, there are some pretty signifi- cant differences as well,â he said, particularly in terms of assay flexibil- ity. NG-ADS uses a separate fluidic module to perform sample prep- aration and the Luminex bead array technology to perform multiplexed PCR assays that can detect up to 50 discrete targets from a single sample. Northrop Grumman has built more than 50 units and subjected them to multiple tests at third-party and government laboratories. Tilles charac- terized NG-ADS as being close to, if not at, TRL 8 (see Table 4-1). As it currently stands, NG-ADS is running 22 signatures, including control signatures, to ensure that there are no non-specific reactions occurring. He added that, guided by experience with the BDS and the Postal Ser- vice, NG-ADS has added diagnostic features. In closing, Tilles empha- sized the importance of iterative testing, noting that NG-ADS has undergone extensive testing that revealed some issues that the company has since addressed. Issues in Systems Concepts and Integration Stevan Jovanovich, cofounder and chief technology officer for IntegenX, Inc., discussed a series of potential problems associated with automating workflow in a way that is reliable and meets the sensitivity needs of the BioWatch program. The hardest part of integrating workflow, he said, is matching the input of one module to the output of another in a way that retains the highest possible sensitivity; to do this, an integrated system must process as much of the initial sample as possible and then use mul- tiplexed sample analysis to avoid the need to split a sample into aliquots.
52 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH TABLE 4-1 NG-ADS Key Performance Parameters Maturity TRL 7 Operational Indoor/Outdoor (rain, snow, wind, dust, etc.) environment Temp: â28°C to 50°C Humidity: 5â100% Agents Six agents in current assay, expandable to 20 agents or more Capable of detecting DNA, RNA, and protein threats Sensitivity 100â600 org/m3 air PD / FAR Probability of detection >95%, FAR better than 1 in 109 Size 28" W à 35" D à 70" H Weight ~600 lbs with on-board consumables Power Standard 110/120 VAC, 20A Communications Cellular, 802.11x, Ethernet capable; VPN and other fea- tures for data security Operational >99.5% Time to result 2 hrs from end of sample collection Collection Configurable collection periods up to 24 hrs (or longer); interval compatible with triggered collections NOTE: FAR = false-alarm rate; PD = probability of detection; VAC = volts al- ternating currents; VPN = virtual private network. SOURCE: Tilles presentation, June 25, 2013. Using technology developed to process human clinical samples combined with other technology that the company developed, IntegenX has built several integrated sample-to-sequence next-generation systems that produce results in a few hours. The RapidHIT 200 system, for ex- ample, uses paramagnetic beads to purify DNA and position it for PCR analysis, processing large volumes of sample into nanoliters of beads. A set of modules makes up the integrated processing system. The current instrument, Jovanovich said, can process from 1 to 8 samples in 70 minutes, and he believes that a well-chosen team of investigators engag- ing in a 2- to 3-year effort costing about $15 million could produce a set of instruments that would be ready for extensive field testing.
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 53 Jovanovich said that in his opinion the most cost-effective approach would be to build a modular system based on current technologies with a consistent set of fluidic interfaces. He added that he believes that BioWatch should have layers of instruments, some of which detect particles and others that perform more extensive and deeper testing, in order to pro- vide flexibility and the ability to respond to new threats. He argued against funding âmoon shotsâ when it comes to biodefense and argued instead for focusing on what is doable based on todayâs state-of-the-art technology. He suggested that BioWatch should fund the development of three systems through TRL 7. Discussion Before opening the discussion to the workshop attendees, session moderator John Vitko reviewed what he saw as the key messages deliv- ered by the five panelists: According to Tilles, there are now several ma- ture technologiesâat TRL 7, 8, or even 9 levelsâthat can meet BioWatch requirements. Northrup made a provocative statement that âdetection is done,â that other fields have solved the fluidics problem, and that the uniqueness of the BioWatch system lies in the fact that there is human involvement and that otherwise it is really just another analytical instru- ment. Vitko noted that several of the talks stressed taking a modular ap- proach and also stressed that it is critical to involve public health laboratorians early in the development and testing process for each mod- ule. Vitko said that Knightâs discussion of pan-genomics and what it means to do genetic testing introduced him to concepts he had not thought about before and raised important questions for the future of de- tection based on genomics. Vitko was also struck by Knightâs clear roadmap for developing multiplexed assays by 2016 and direct sequenc- ing by 2020. John Plante from the Chicago Transit Authority (CTA) noted that one of the challenges with testing in a transit environment, which is not quite indoors or outdoors, is that the environment changes over the course of a yearâs time and that instruments will need to be adjusted to accommodate those changes. In addition, the transit environment con- tains significant levels of âbrake dustâ that adheres strongly to surfaces and any instrument would have to be able to tolerate it. Tilles remarked that his team experienced that seasonality as well as the brake dust phe- nomenon when it was testing its system in an urban subway system and that the information they gained from those tests will be valuable going
54 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH forward. He reiterated the importance of conducting extensive cycles of testing and design and retesting and redesigning to develop a field- worthy instrument. Jeffrey Runge said that he appreciated Jovanovichâs prescription for how the government might fund next-generation technologies and asked the other participants if they had any additional suggestions. Northrup said that the program needs to be less myopic about technology and that it should bring nondefense companies into the effort, an idea that Paul Schaudies, with GenArraytion, also stressed in his comments to the pan- el. Tilles echoed Jovanovichâs desire to see a more consistent effort that maintains momentum through the long development process that will enable putting together the necessary workforce and allow for the exten- sive and repeated testing. Jovanovich agreed with Northrup and added that maintaining flexibility and not selecting a winning technology too early in the process would be important to the eventual success of a BioWatch autonomous detector program. Toby Merlin, director of the Division of Preparedness and Emerging Infection at CDCâs National Center for Emerging and Zoonotic Infec- tious Diseases, asked if any of these technologies could be married to orthogonal technology2 to confirm positive results. While Mariella said that this was a big challenge and would probably occur on the 2020 time frame, Tilles said that multiple PCR signatures are, in effect, a low- technology approach to orthogonal testing and that designing in flexibil- ity in the fluidics systems will provide the means of integrating orthogo- nal technologies in next-generation instruments. He also raised the issue that orthogonal detection can increase the possibility of false-negatives while adding substantial cost to the operation of the system. Northrup, agreeing with Tilles, said that orthogonal testing would only be feasible from a cost perspective as a confirmatory test. Knight agreed with that comment. William Raub asked if next-generation instruments will be able to output self-diagnostic information, and both Mariella and Tilles an- swered that this capability will be included in Generation 3 and any fu- ture instruments. In the same vein, Vitko asked if the instruments would retain an aliquot of sample for retesting, and Tilles answered that this capability is included in next generation instruments under development and testing. Northrup noted that the pharmaceutical production industry 2 In other words, mutually independent technologies.
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 55 has extensive experience developing both of these capabilities and that these should not be viewed as challenges for BioWatch. Both Sandra Smole and Erica Pan voiced the opinion that public health does not really care what technology is used but that it does want to know how it works and to have confidence in the technology. âWe want the technology to work and we want to know that weâre going to make good decisions based on that technology,â said Pan. AUTONOMOUS DETECTION SYSTEMS USING IMMUNOASSAYS AND PROTEIN SIGNATURES To start the second technology session, Paul Schaudies, chief execu- tive officer of GenArraytion, Inc., and author of the commissioned paper on immunoassays and protein signatures (see Appendix H), said that, in- stead of reviewing his paper, he wanted to make one point. BioWatch, he said, has been the subject of recent critical reports claiming that the system has a high rate of false-positives related to the detection of Francisella novicida, a relatively harmless species that occurs naturally in the envi- ronment and is closely related to the pathogen Francisella tularensis. But in his opinion, he said, That is the best real-world data demonstrating that the system works be- cause there was a collection, there was an extraction, there was an ampli- fication, and there was a signal. The piece of DNA that was being looked at was not the right one, but that was not a failure of the system. That is a failure of the test. The fact that the BioWatch program is able to collect organisms that are traveling miles in the air at a very dilute concentration is probably the best data supporting that the system would find something or will find something if it is ever released. Following those remarks, four panelists made presentations on tech- nologies for detecting pathogens based on protein signatures. The discus- sions focused on automated multiplexed immunoassays, a disc-based collection and detection system for protein signature determination, a point detection system based on Raman spectroscopy, and a single-molecule array technology for multiplexed detection of proteins and nucleic acids. When he introduced the panel, Schaudies noted that the systems from Luminex, PathSensors, and Battelle are already being tested in the field. The system from Quanterix, he added, is just now being manufactured for eventual field tests.
56 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH Automated Multiplexed Immunoassays Amy Altman, vice president for biodefense and food safety at Luminex Corporation, began by noting that protein signatures can serve as an im- portant detection tool for bacteria and viruses because of the diverse nature of the protein epitopes that cover the surface of these organisms and which can be readily detected individually using antibodies. When used in a mul- tiplexed assay, a panel of antibodies designed to identify the specific com- bination of protein epitopes that characterize a microbial threat can rapidly and sensitively distinguish between pathogenic and nonpathogenic organ- isms and strains of organisms. Immunoassaysâthe technology that uses antibodies to detect specific protein epitopesâare fast, robust, and efficient, Altman said, and they are able to produce results in 10 minutes to 1 hour, depending on the desired limit of detection. Multiple-epitope analysis provides a high level of speci- ficity that can allow for near-neighbor discrimination, and the ability to detect phenotype versus genotype can provide information on toxicity and viability. Immunoassays can also detect toxins directly, as opposed to de- tecting the genetic signature of a toxin. Altman said that it would be hard to engineer an organism to escape detection via a multiplexed immunoas- say because any alterations that change protein epitopes may render an organism nonvirulent. Immunoassays are inexpensive to carry out, largely because they are based on a well-developed technology platform and come without the pa- tent licensing burden associated with PCR. Sample preparation is signifi- cantly less complicated than for PCR analysis, and immunoassays are less sensitive to dirty samples. In addition, multiplexed immunoassays used for bioweapons detection can be carried out with the same platform that is in widespread use for clinical diagnostics. In fact, Altman said, the system she was highlighting in this presentation for use in Northrop Grummanâs NG-ATS is also approved by the FDA for clinical use. The multiplex technology that Luminex uses, which it calls xMAP, re- lies on a set of polystyrene microspheres that are color-coded with a unique ratio of a red dye and an infrared dye so that a 10-by-10 matrix of these beads would yield 100 spectrally distinct bead classes, each of which can be linked to a specific antibody for detecting a specific protein epitope. Altman said that in addition to antibodies, the beads can be linked to en- zyme substrates, receptor ligands, and nucleic acid probes. Luminex, she added, has recently developed a third dye that will extend the systemâs
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 57 capability to the point that it can multiplex 500 different spectrally distinct beads per assay. The companyâs personal computerâsized MAGPIX instrument is ca- pable of performing simultaneous measurement of 50 targets in a single assay at a cost that is one-fourth that of a comparable enzyme-linked immunosorbent assay (ELISA). This modular, FDA-approved instrument can detect single-digit picogram levels of protein as well as 1 million cop- ies of DNA, and it uses LED-based illumination and charge-coupled de- vice (CCD) image capture to improve reliability over older laser-based systems. After describing the mechanics of how the instrument works, Altman said that one of the strengths of a bead-based platform versus a planar array-type technology is the ease of adding new targets as dictated by threat assessments. She added that modern antibody production tech- nology also offers the possibility of improving assay performance by add- ing or updating assay antibiodies once they are developed in response to a new threat or to better detect current threats. Altman also addressed the benefits of multisignature analysis. As an example, she reviewed an assay developed with a commercial partner, Radix BioSolutions, to detect multiple epitopes of the toxin ricin. This 18- plex assay accounts for natural or intentional genetic drift and extends the dynamic range of the assay. It also decreases the false-alarm rate using decision theory software to analyze assay data. In her final remarks, she said that the MAGPIX module has been inte- grated into Northrop Grummanâs NG-ADS unit and has been tested exten- sively by third parties. What remains is for the device to undergo field testing to determine its real-world performance in an environmental test setting. She said that the company believes that the technology could be TRL 6-plus ready by 2016 if it can demonstrate the same immunoassay performance in an autonomous system that it does on the benchtop and if it is confirmed that stable reagents can be deployed and provide stable per- formance over the entire service period. She also noted that while immu- noassays may not have the same level of sensitivity as a PCR-based assay, it is important to think in terms of clinical relevance, and the sensitivity of protein signature detection may be sufficient for the purposes of biothreat detection. Rapid Detection of Pathogens in Aerosol Samples Originally developed at Massachusetts Institute of Technologyâs Lin- coln Laboratories (Rider et al., 2003) and now being further developed at
58 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH PathSensors, the technology uses pathogen-specific antibodies to detect pathogens and toxins collected using the companyâs proprietary aerosol collection technology. According to Ted Olsen, chief executive officer and president of PathSensors, Inc., the system can provide pathogen and toxin detection in less than 5 minutes once the sample has been collected in its disc-based collector and analyzer. The technology is called CANARY, short for Cellular Analysis and Notification of Antigen Risks and Yields, and it detects bacteria, viruses, and protein toxins simultaneously. It uses a genetically engineered B-cell lymphocyte that, in response to protein bind- ing, releases a calcium spike internal to the cell that in turn triggers light emission that can be detected outside the cell. An instrument based on this technology, Olsen said, is already used for mail screening in smaller mail- rooms that do not opt to use the USPS system designed for higher volumes. When loaded into the BIO-FLASH instrument, the center of the com- pact discâsized collector sits in an airstream and, as it spins, collects air in 1 of 16 radial channels, each fitted with a light sensor. Sample collection can run from 20 seconds to hours. Once the sample is collected by inertial impact on the wall of the disc, the disc continues to spin as reagents are released into the channels. Almost as soon as the reagents come in contact with the sample, light emission occurs if the target organism or protein is present, triggering a visual and audible alarm. Olsen explained that the device can be configured to assay for the same pathogen multiple times in the disc, run positive and negative controls, and retain a sample for later recovery and analysis. The instrument also has the ability to upload data to a remote location. Olsen noted in closing that the instrument has undergone extensive third-party validation. Those tests have shown that the system is capable of identifying with 100 percent accuracy the biological agents in aerosol samples containing between 75 and 450 agent-containing aerosol particles per liter of air, which he said meets or exceeds the targets set by the BioWatch program. Detecting Biothreats with Raman Spectroscopy Andrew Bartko, principal scientist at Battelle Memorial Institute, said that the Resource Effective Bio-Identification System (REBS) that Battelle Memorial Institute has been developing collects and identifies bacteria, viruses, and toxins directly from atmospheric aerosols in a nondestructive manner that is compatible with genetic confirmation. In field tests, the REBS system has demonstrated sensitivity below the lower limit set by the
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 59 BioWatch program and has produced no false-positives in actual environ- mental testing. The system, which can be updated to accommodate new threats in 24 hours, collects continuously and stores an environmental par- ticulate history that can be accessed and reexamined if necessary. Bartko said that REBS is not meant to provide an end result but rather to act as an alert signal that will require further confirmation using other methods. The cost per sample analysis is less than 1 cent, and each portable, battery- powered instrument costs less than $100,000. The core technology in REBS is Raman spectroscopy, a method that measures the unique vibrational frequency of molecular bonds. Because of the large number of bonds in a biological material, the resulting spectra are complex, and Bartko and his colleagues have been studying the Raman signals intensively to understand their nuances in order to use them for species-level identification. REBS is now able to identify mixtures of dif- ferent types of threats and report how many of each type of cell is in a giv- en sample. âIn some instances,â he added, âwe have evidence that the cell viability can be indicated by looking at specific features within the spectral features.â Though not currently included in REBS, the spectra could dis- tinguish organisms intentionally grown in laboratory media from those grown in natural nutrients. The bottom line, Bartko said, is that REBS can identify biothreat species with no primers, probes, liquid reagents, or sub- strates; it is capable of being accurate, agile, and economical to run. The working part of the device consists of an inlet stack, an electro- static collector, a metalized polymer tape in a cassette for collecting samples, and the optics system. Particles collected on the tape are illumi- nated by a laser, producing the Raman spectra. The detector quickly scans the spectra for identifying characteristics, and if those characteris- tics are spotted, the sample is irradiated further to increase the signal-to- noise ratio and to enable a more detailed examination of the fine struc- ture of the spectra. Identification of a single particle does not trigger an alarm, but it does increase the probability of identifying subsequent par- ticles that the instrument searches for specifically and correlates to the first particle and the spectral library. The REBS instrument can also de- tect chemical weapons, Bartko noted, adding that tests conducted using simulated releases in a variety of environments, including the Boston subway system, have produced no false-positives and 100 percent identi- fication of the released simulant (see Table 4-2). Looking to the future, Bartko said that REBS is currently at TRL 6, with additional operational tests and evaluation still needed to demonstrate that the system meets or exceeds 99.9 percent reliability. Field tests have
60 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH demonstrated zero false-positives with a 95 percent confidence, but further operational tests are needed to demonstrate a lower false-positive rate, and further work is needed to integrate REBS with other technologies that can confirm the REBS signal. Single-Molecule Array Technology In contrast to the other technologies described in this session, Quanterixâs single-molecule array (SIMOA) system (Rissen et al., 2010) has only been developed for use in human diagnostics and has not yet been applied to the detection of potential biological threat organisms in the envi- ronment. As David Hanlon, director of business development and strategic collaboration at Quanterix, described the technology, its digital nature ena- bles single-molecule resolution. What makes single-molecule detection possible, he said, is the ability to trap individual molecules in femtoliter- sized wells, which are some 2 billion times smaller than a standard ELISA well. The miniscule volume allows for a rapid buildup of fluorescent sig- nal that is readily visible. Like other protein detection schemes, this one uses antibody capture reagents attached to paramagnetic beads. In this case, the beads are sized so that only one bead will fit in a femtoliter well, and a large excess of beads are used so that the beads will bind to at most one target molecule. After the capture beads are allowed to bind with proteins in blood, serum, or other biological matrices, the mixture is flowed over the wells, detection reagent is added, and the wells are sealed with oil. If the target analyte has been captured, the capturing well will fluoresce. As currently designed, this technology can multiplex up to 10 analytes. Tests on clinical samples for a wide range of substrates demonstrate that this technology routinely achieves sensitivities in the femtogram- per-milliliter range, compared with the pictogram-per-milliliter limit of detection for other technologies. In one test, for example, SIMOA was as sensitive as PCR for detecting the human immunodeficiency virus prior to seroconversion. Hanlon noted that with this increased sensitivity, it should be possible to detect the vast number of proteins with potential relevance to human disease that are now beyond the reach of current detection tech- nologies. The company has also recently demonstrated that it can detect DNA without amplification in complex samples such as blood and river water. Quanterix has partnered with a manufacturing company, Hanlon said, and it plans to introduce an instrument for research use in late 2013.
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 61 TABLE 4-2 Summary of Government Testing of REBS Location (Sponsor) Date Type Result Fort Bliss, Texas 6/2009 to Background sampling Zero false-positives (JPM-G) 7/2009 Dugway, Utah 10/2009 TRE simulant ID sensitivityâ25 Piggyback to challenge in realistic ACPLA (JPM-BD) 11/2009 environment Battelle 11/2009 JBTDS identifier TRE 100% identification Columbus, Ohio to samples, ID (JPM-BD) 01/2010 performance with killed BWA Battelle 6/2009 CWA (particle) ID Dual-threat Columbus, Ohio feasibility feasibility proven (JPM-CA) JHU-APL/ECBC 9/2010 JBTDS TRE shoot-off Selected for JBTDS (JPM-BD) with simulants competitive prototyping DPG 2/2011 to ASEC and ABT testing Demonstrated (JPM-CA for CBDS) 7/2011 with irradiated materi- sensitivity, als, unknown biologi- specificity, and cal, near-neighbors, and false-alarm Interferants performance Boston, 10/2012 Simulant release in Zero false- Massachusetts, to present actual environment positives, 100% Subway Test (DHS identification of S&T) simulant release West Jefferson, Ohio 06/2013 Live agent integrated TBD (CRADA w/JPM- to system testing and ABT NBC) 07/2013 NOTE: ABT = ambient breeze tunnel; ACPLA = agent containing particles per liter of air; ASEC = aerosol simulant exposure chamber; BWA = biological warfare agent; CBDS = Chemical and Biological Defense; CRADA = cooperative research and development agreement; CWA = chemical warfare agent; DHS S&T = Depart- ment of Homeland Security Science and Technology Directorate; DPG = Dugway Proving Ground; ECBC = Edgewood Chemical Biological Center; ID = identifica- tion; JBTDS = Joint Biological Tactical Detection Systems; JHU-APL = The Johns Hopkins University Applied Physics Laboratory; JPM-BD = Joint Program Manger Biological Defense; JPM-CA = Joint Program Manager Contamination Avoidance; JPM-G = Joint Program Manager Guardian; JPM-NBC = Joint Program Manager Nuclear Biological and Chemical; TRE = technology readiness evaluation. SOURCE: Bartko presentation, June 26, 2013.
62 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH Discussion Schaudies remarked that one approach to improving the sensitivity of antibody-based technologies without increasing the cost or the time that it takes to conduct the test would be to sonicate a sample using ex- isting technology. What this would do, he explained, is disrupt the cell membrane of the viral coat and create thousands of liposomes, each with one or a few epitopes on it rather than one entityâthe original cell or virus particleâwith thousands of epitopes. Such an approach, he suggested, could produce sensitivities equivalent to nucleic acid tech- nology with the speed of antibody technology. He then challenged DHS to look beyond the pathway that it has been on. He cited the presenta- tions made in this session as examples of new ways of thinking about the technologies that will enable BioWatch to create the next genera- tion of autonomous instruments. âI can make arguments for each one of these technologies to be in a system,â he said. Bartko was asked by several workshop participants if Raman spec- troscopy would work in environments where the biological background was high. He replied that the system can do that now thanks to years of studying Raman spectra from biological samples and learning which features are the most important to look at for the organisms of interest. This information makes up an important part of the software that the instrument uses to determine which particles to investigate for longer periods of time. Panelists agreed with the comments from several participants that these technologies are not meant to replace nucleic acidâbased tests but rather to augment them as part of an orthogonal test paradigm. There was also agreement that different systems may be more appropriate for different testing environments or for testing over different time frames. Schaudies emphasized these points, noting that hours could be saved if immunoassay or Raman approaches were used to rapidly detect a po- tential biothreat and trigger the more sensitive, specific, and costly nu- cleic acidâbased detection. In a final comment, Jeffery Runge noted that attribution is important and that any autonomous detection system must have archiving capabilities. The panelists agreed and responded that their systems all have the ability to store samples for subsequent analysis.
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 63 AUTONOMOUS DETECTION SYSTEMS USING GENOMIC SEQUENCING In introducing the panel for the third technology session, Rita Colwell, Distinguished University Professor at the University of Maryland and the Johns Hopkins University Bloomberg School of Public Health, comment- ed that the previous sessions had made a strong case for the proposition that it is necessary to have a multidimensional view of identification. âEach of these approachesâand weâll hear about the very powerful one of sequencing todayâprovides a dimensional understanding and contributes to accurate identification of microorganisms, which is really what weâre about,â she said. Colwell also said that she believes that the BioWatch program should shift its perspective, partner with the National Institutes of Health (NIH), and bring public health laboratorians into the discussion. She noted that while BioWatch is focused on detecting the worst pathogens, the tools that the program is developing are essentially the same tools that the biomedi- cal community is developing to rapidly diagnose human diseases. The BioWatch community should be building on all of the information that NIH has developed regarding infectious organisms and genomics, she said, particularly with regard to the issue that Ivor Knight raised in his talk about microbial pan-genomes. For this session, the commissioned paper was written by Chris Detter, director of the Bio-Threat/Bio-Defense Program in the Emerging Threats Program Office of the Global Security and Bioscience Division of Los Alamos National Laboratory (LANL), and by Gary Resnick, an independ- ent consultant and guest scientist at LANL (see Appendix I). Panelists re- viewed the current state of sequencing technologies and provided a glimpse of the future of genomic sequencing; discussed informatics issues involved in developing useful information from advanced sequencing technologies; and described real-world applications of next-generation se- quencing and how those might integrate with the type of sampling meth- ods already used in BioWatch. Current and Future State of Genomic Sequencing Chris Detter said that the focus of the commissioned paper was BioWatchâs three primary needs: that any system provide affordable and continuous coverage of at-risk populations, that it have accuracy and preci- sion to effectively support potentially high-regret responses, and that it be
64 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH responsive to the full scope of biothreats. But before addressing those three needs, he briefly reviewed the history of modern sequencing. It started in 1975 with Sanger sequencing, he said, preceded through the era of âauto- matedâ gel sequencing, and then entered the era of the Human Genome Project and its use of Applied Biosystems capillary sequencing, starting in 1999. During the past decade and a half, a variety of technologies have produced a dramatic drop in the cost of sequencing and a dramatic increase in its speed, and some of the newest technologies can perform single- molecule sequencing, which does not require amplification. Each technol- ogy, Detter said, has its own set of error rates and error issues and is suita- ble for a particular set of applications. The workflow for so-called next-generation sequencing is fairly uni- form for both clinical and environmental samples, consisting of a set of preparatory steps followed by sequencing and then a computational analy- sis of the sequencing reads to assemble the entire genome (see Figure 4-4). Detter discussed three applications of next-generation sequencing. The first was rapid sequencing of the amplicons from PCR reactions to identify and characterize pathogens; this amplicon sequencing can readily test hun- dreds of samples. Shotgun sequencing of multiplexed environmental or clinical samples can identify and characterize both known and emerging pathogens. And full-genome sequencing of a single organism, after the organism is isolated and grown in the laboratory, can identify sequences that are associated with specific outbreaks and can rapidly detect the same pathogen in future outbreaks. FIGURE 4-4 Next-generation sequencing workflow. SOURCE: Detter presentation, June 26, 2013.
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 65 In the commissioned paper prepared for the workshop, Detter and Resnick concluded that high-throughput amplicon sequencing could be at TRL 6 by 2016, with some engineering and integration work to fit it into the BioWatch system. Amplicon sequencing would have limited ability to identify emerging threats, particularly RNA viruses, but it should be able to detect all known pathogens. By 2020, improved next-generation se- quencing technologies, in combination with bioinformatics, should enable metagenomic sequencing of entire environmental samples at TRL 6. These systems would be capable of detecting emerging and engineered patho- gens. The main limitation will be the need to prepare sequencing libraries, which will limit the speed of these processes. Beyond 2020, cutting-edge sequencing technologies that do not require amplification should be at TRL 6. They will provide the same capabilities as those that should be ready by 2020, but on a much faster time scale. In his final remarks, Detter noted that sequencing is rapidly becoming the gold standard for biological identification and is causing biologists to revisit the concept of phylogeny in microorganisms. Today, sequencing can cover the entire threat, known and novel; two examples are the discov- ery of SARS (severe acute respiratory syndrome) and the discovery of the MERS-CoV (Middle East respiratory syndrome coronavirus), both of which were done through sequencing. âWe still donât know the full poten- tial of sequencing for biodetection in the field,â Detter said, but the hard- ware and knowledge bases that are available and that are being improved daily are designed from the start to be used in high-throughput applica- tions. He sees these technologies as being dual-use platforms that can share the cost, risk, and actual processes across fields, particularly between forensics and biosurveillance, which share similar adoption paths and processes. Next-generation sequencing has already transformed microbiology and can be expected to transform it even further with NIHâs human microbiome project, the Beijing Genome Instituteâs 10,000-microbial- genome program, and the Joint Genome Instituteâs microbial earth project, Detter noted. All of these efforts will generate a tremendous amount of sequence data about the microbial world that BioWatch should put to use as it develops and adopts its own sequencing technologies for biodetection, Detter said. Bioinformatics and metadata will play a critical role in making use of these databases, and Detter encouraged the community to share knowledge bases and bioinformatics capabilities. Another key to the suc- cessful adoption of sequencing technologies will be the development of autonomous and modular platforms that can be integrated into the
66 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH BioWatch system. âEase of use is going to be critical for wide ac- ceptance,â he said. For any system to be adopted, âit must be responsive to public health officialsâ needs and provide acceptable and actionable infor- mation for their decision-making process.â Challenges and Opportunities in Genome Sequencing Technologies Jeffrey Schloss, director of the Division of Genome Sciences and pro- gram director of Technology Development and Coordination at the Na- tional Human Genome Research Institute (NHGRI), began by noting that most of the next-generation sequencing technologies were developed as a result of the NHGRIâs realization toward the end of the Human Genome Project that sequencing the human genome was only the first step toward realizing the full potential of genomics, not just for the study of human diseases but also in many fields, such as agriculture and microbial ecology. Schloss stressed that much of the increase in sequencing speed has come from improving the workflow in going from Sanger technologies to cur- rent next-generation sequencing technologies. Gone are the days of having to clone every fragment of DNA in a bacterium, then picking a suitable colony and purifying its DNA for sequencing. Instead, all of that pro- cessing is now done in tubes, and while still technically challenging, this new workflow is easier. The hope, he added, is that this process will be- come even easier with further technology development. The end result of the past decade of development, Schloss said, is that in 10 years sequenc- ing output has gone from 100-base pair reads per sequencing read to more than 100 million reads per run, and sequencing has gone from the province of centralized sequencing centers to individual laboratories. Today, there are sequencing machines the size of a personal computer. NHGRI has not supported much research on sample preparation, Schloss said, but the private sector has, and there are now companies that are commercializing integrated sample preparation and library preparation. These types of integrated systems will be important in any deployable sys- tem, such as those envisioned for BioWatch, as well for moving sequenc- ing from the research laboratory to the clinical setting. Unlike earlier DNA sequencing technologies, next-generation methods sample very large numbers of single molecules or ensembles generated from single molecules, and this approach enables new biological insights. For example, Schloss said, researchers are now able to look for sequence variations, rare variants, and haplotypes, which he said will be important to
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 67 BioWatch because it will allow sequencing to identify which bacterium in an environmental mixture has an antibiotic resistance gene. The important point, he said, is that these techniques are digital, enabling the detection of rare signals in a mixture. The question of exactly how rare remains to be answered. By the end of the Human Genome Project in 2003, sequencing 1 hu- man genome took 3 months with 100 capillary array electrophoresis ma- chines working together. By 2009, one machine using sequencing by synthesis on array technology could sequence a human genome in less than 1 month. By 2011, one machine could sequence three genomes simul- taneously in 1 week. And commercial instruments released in the past year reportedly can sequence a human genome in just 24 hours; Schloss said he has not seen those claims verified in practice yet, but they do give an idea of what the future holds in terms of sequencing speed. The other important trend in sequencing has been the precipitous drop in the cost of sequencing, particularly since 2008, when prices began plummeting. Schloss noted, however, that the cost per genome appears to be leveling off, and he said that he suspects that one reason is that the field seems to be converging on a single technology and an accompanying re- duction in competition. Turning to what lies ahead for sequencing, Schloss discussed several technologies now being developed. Free-running polymerase technology, for example, could provide real-time DNA sequencing from single poly- merase molecules using zero-mode fluorescence detection (Eid et al., 2009). This technology demonstrated read lengths of almost 3,000 bases in 2011, and it is reported to be improving rapidly. Because it sequences di- rectly off genomic DNA and does not require library preparation, it can be used to identify methylated or damaged bases that might be of interest (Flusberg et al., 2010). Other new technologies can sequence RNA directly using reverse transcriptase (Vilfan, 2013), and these methods are already identifying novel modifications in the RNA of microbes and are allowing researchers to map and detect these modifications at the single-base level, enabling new biology. An exciting development in sequencing is the use of nanopores to se- quence DNA in real time without the use of enzymes (Clarke et al., 2009; Kasianowicz et al., 1996; Stoddart et al., 2009). Instead, Schloss explained, nanopores detect changes in ion flow through the pore as a single molecule of DNA moves through the pore. He noted that this type of technology would simplify the MicrobioMe project by enabling the rapid, nondestruc- tive sequencing of DNAâand potentially RNAâon a massive scale.
68 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH Turning nanopores from an interesting research idea into a practical se- quencing method has been incredibly challenging, but progress has accel- erated over the past few years to the point that investigators are starting to generate data that resemble sequencing reads from nanopore devices (Ayub and Bayley, 2012). It may also be possible to use nanopores to de- tect proteins and distinguish among different proteins (Soskine et al., 2012). Next-Generation Sequencing of Microbial Populations Predicting the future impact of next-generation sequencing is a daunt- ing challenge given the pace of technology development, said Thomas Cebula, chief technical officer at CosmosID and visiting professor at Johns Hopkins University, but he felt comfortable with the idea that as more peo- ple use these technologies they will develop an increasing number of appli- cations. As an example, he discussed how researchers in cancer biology are now looking to next-generation sequencing as a technology that will change how patients are treated in the near future (Kamalakaran et al., 2013). âThis is the future,â Cebula said. âThe question is, When is the future?â Though the cost of sequencing has fallen significantly in the past dec- ade, a major part of the expense arises from the need to provide multiple coverageâon the order of 30-foldâin order to be able to assemble a ge- nome from the short reads generated by next-generation sequencing tech- nologies. âBut what if you didnât have to assemble the genome to make a call of whatâs in a sample?â Cebula asked. âThat would cut our costs and bring the time frame forward for doing real-time detection.â It would also provide the ability to interrogate the entire microbial genome rather than use representative gene markers, as is currently done, to characterize mi- crobial genomes. The approach that CosmosID is taking is to develop algorithms that take unassembled reads from a metagenomic microbial sample and return, within minutes, identification at or near the strain level. He noted that there are other software packages available that make identifications at the spe- cies level. In his opinion, strain identification is important for forensic pur- poses because it can provide source identification and attribution. In sup- port of this contention, he showed how the CosmosID software package can identify and distinguish between strains of Escherichia coli O157:H7, only one of which was responsible for outbreaks of human illness.
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 69 Metagenomic Sequencing in Biodetection In his presentation, Brian Young, program manager at Battelle Memo- rial Institute, addressed the question, âWhy consider metagenomic se- quencing in autonomous identification?â The major reason, he said, is that it is a single method that can identify a broad spectrum of biothreat agents and that can meet all three levels of the current PCR strategy: primary screening, verification, and subspecies identification. Metagenomic se- quencing can also identify unexpected species, provide an alert when an unknown species is present, and monitor the environmental background and normal variation in that background while searching for target species (see Table 4-3). This type of background monitoring can provide evidence to help determine whether a potential threat organism, such as Francisella, is present because of an intentional release or because of an environmental disturbance. In the case of the latter, the prevalence of other organisms is likely to undergo coincident increases or decreases in response to the disturbance. Metagenomics, Young added, can also provide deep genetic evidence for use in identifying the strain or even the type of the agent, including the identification of virulence and antibiotic resistance. Nucleotide-level dis- crimination may also be useful for forensic attributions. As an example, Young cited work done at Battelle in which morphologically different col- onies of Brucella suis were sequenced, revealing single-nucleotide varia- tions that could be used to identify the different colonies. He echoed Cebulaâs comments that software and bioinformatics packages will be key to developing these types of applications for rapid identification purposes. On the subject of how metagenomic sequencing would fit into an au- tonomous detection system, Young said that it would be similar to the ap- proach used with PCR. The main issues, he said, would be systems integration and testing the integrated systems. The main difference from current PCR systems would be the need to perform data analysis locally because of the time that would be required to transmit the raw datasets to a central processing location. Performing the analysis locally will require the development of computationally efficient software, which Young said will be a challenge. Another challenge to incorporating next-generation se- quencing into BioWatch is that sample preparation must be better auto- mated and mated with air sampling systems. At present, most workflows are not deterministic or diagnostic, which is what BioWatch requires. Op- erational field sensitivities have not yet been established, nor have opera-
70 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH TABLE 4-3 Monitoring Abundances of Target Environmental Organisms in Oil- Exposed Sediment Using Next-Generation Sequencing Percent of Bacterial Sequences Found With and Without (W/O) Exposure to Oil Relation to Oil Rhode Island Alaska Organism Degradation W/O With W/O With Aerobic Pseudoalteromonas gammaproteobacteria; 0.10% 13% 1% 9% spp. several species consid- ered oil degraders Aerobic Alcanivorax spp. gammaprotebacteria; principal carbon 0.40% 2% 0.07% 0.20% source is linear-chain alkanes Facultative anaerobic Escherichia coli gammaproteobacteria; 4% 0.20% 0.60% 0.80% not associated with oil degradation SOURCE: Young presentation, June 26, 2013. tional field specificities or false-positive rates. Furthermore, the cost of sequencers and sequencing consumables are still too high for routine use. The DNA inputs required for sequences are an order of magnitude too high for practical field use without amplification, and adding amplification would essentially turn these instruments into high-end PCR detectors. Metagenomics could prove to be disadvantageous in high-background scenarios in which heavy pollen or mold spore loads would consume se- quencing bandwidth and reduce sensitivity to pathogens. Developing tech- niques for removing eukaryotic DNA background could solve this problem, Young noted. In terms of technology readiness, Young estimated that the integration of aerosol collection with next-generation sequencing, autonomous opera- tion of sample preparation, and autonomous operation of sequencing are âTRL low.â Data analytics, he estimated, are actually more developed and at TRL 7, and he showed results from tests using the CosmosID system as an example of how far data analytics has come over the past few years. In closing, Young said, âWhat Iâd like to leave the audience with is that no matter what technology gets deployed in [autonomous detection], ge- nomics sequencing has to be in the equation somewhere. Youâre going to
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 71 want it in the lab to do the attribution analysis; you are going to want it in your system for the interrogation that ultimately will happen.â As a result, he said, any technology that gets deployed should at the very least preserve a sample or not destroy the sample so that it can be sent to the laboratory for sequencing. Discussion John Vitko asked if any of these technologies can meet the BioWatch goal of reporting within a few hours of sampling. Detter replied that Tier 1 technologies will have a turnaround time of about 10 hours after sample collection is complete. Tier 2 technologies should reduce that to about 6 hours, and Tier 3 should drop that further to below 4 hours. The big un- known, he said, is sample preparation time. Cindy Bruckner-Lea, from the Pacific Northwest National Laboratory, asked what sequencing provides in the proposed Tier 1 device (sequencing of PCR products) that PCR-based detection does not, given the extra cost and time of sequencing the PCR products. Detter said that instead of mere- ly getting a band (indicative of presence/absence of a PCR product), se- quencing identifies the one or more components of that band. He acknowl- edged that it may not be necessary to do this in the field or as the primary application in Tier 1 devices but rather as a confirmatory test. In response to a question from Colwell about the quality of sequence data in public databases, Detter said that there is an effort under way to create what he calls a trusted database. This database would have quality scores associated with the data, and sequences would be checked against near neighbors to generate a set of curated pathogen sequences. The diag- nostics community is leading this effort. Thomas Companion from the Office of Standards in the Science and Technology Directorate at DHS asked how next-generation sequencing technologies will deal with the various structural variations of DNA that are found in organisms and with genome dynamics, such as in the case of organisms that edit their DNA as they use it. Detter replied that he does not see those as issues that are beyond solving with well-curated databases to a level that will be satisfactory for BioWatch purposes. Cebula agreed with that assessment. Schloss added the issues of variability and change are also being addressed by those who are studying complex human diseases. He said that as the research community builds its databases, it will be possible to identify the normal variation seen in environmental samples.
72 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH Raymond Mariella, Jr., asked about the problem of contamination re- sulting from carryover from one run to the next. Detter replied that todayâs sequencers use disposable slides or chips, which solves what was an issue with older capillary sequencers. Schloss said that this issue is being ad- dressed by the clinical community because it has the same concerns about carryover in sample preparation and sequencing. Mariella then asked why there seems to be so little work on viruses, which can also be important biothreat agents. Cebula said that part of the reason is that the taxonomy of viruses is still poorly understood, making it harder to correlate characterization work with a particular species or strain. Detter replied that another advantage of metagenomics is that it will allow for more efficient investigation of viruses using the DNA and RNA se- quencing technologies that he and Schloss described. He added that the Defense Threat Reduction Agency is now working to expand the reference database of viral sequences. Stephen Morse from Columbia University said that while virologists may argue about the fine points of virus classifi- cation, the known viral threat agents can be identified today. He said that he also believes that some of the new technologies offer great potential for better understanding viruses. Allen Northrup asked if BioWatch would be able to afford next- generation sequencing, given the prospects that full deployment would require several hundred million PCR reactions per year. Schloss replied that part of the motivation for using solid-state nanopores is that they would be manufacturable using industry-standard complementary metalâ oxideâsemiconductor (CMOS) technology that would drive down the cost significantly, although he acknowledged that the effort is still in its infan- cy. Young agreed that costs must drop and was of the opinion that further development of new reagentless technologies would significantly affect cost. He added, though, that he found it hard to imagine a robust BioWatch program that does not use sequencing somewhere in the process, a com- ment that Detter seconded. Cindy Bruckner-Lea commented that using sequencing in the laboratory to confirm results from the field makes a great deal of sense. She then asked the panel to comment on the major challenges in sample preparation. Cebula replied that work still remains to be done to develop a reproducible and automated procedure, but that PCR had the same issues to deal with before it could be approved for routine clinical use. Young said that what is really needed is a program to field test various methods to see what hap- pens with environmental samples. âYou donât know what inhibitors youâre going to see, you donât know when youâre looking at minor components in
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 73 a complex background, you donât know how hard or easy itâs going to be to detect that until you get out in field testing,â he said. Thomas Slezak from LLNL asked what the BAR of the future will be in the context of next-generation sequencing, given how difficult it was to agree on a definition with PCR. Young replied that the BAR problem for sequencing will be two-dimensional. One dimension will be the depth of evidence or the number of reads needed, and that is an issue shared with PCR. The second dimension is how far across the genome the analysis will have to proceed before the evidence is convincing enough to declare a BAR. Detter noted that the BAR will not be declared in a vacuum and that field results can be confirmed in the laboratory. AUTONOMOUS DETECTION SYSTEMS USING MASS SPECTROMETRY For the final technology session, A. Peter Snyder and Rabih Jabbour prepared the commissioned paper (see Appendix J). The panelists dis- cussed, quality issues that are critical for mass spectrometry, the use of single-particle aerosol mass spectrometry (SPAMS) for biothreat detec- tion, and the use of a miniature mass spectrometry system for analyzing microorganisms. Current State of Mass Spectrometry Peter Snyder began by explaining that the fundamental principle of all mass spectrometry methods is that they separate gas-phase ions ac- cording to their mass-to-charge ratio. There are many methods for ioniz- ing samples and a variety of analyzers for moving the ions from the region where they are created to a detector, where they produce an am- plified signal. Mass spectrometry is capable of detecting substances at the low-attomole level, Snyder said, and it generates huge datasets in a short period of time. On a Tier 1 time frame, there are two mass spectrometry systems that are mature enough to identify organisms based on their structural compo- sition and that meet the DHS criteria: the Hamilton Sundstrand chemical biological mass spectrometer (HSMS) and the SPAMS. The HSMS sys- tem, which measures the fatty acid content of a microbial sample, has been refined and tested extensively over the past 20 years, but tests have yet to generate quantitative figures of merit for false-positives and the
74 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH probability of false-positives. Doing so, said Snyder, will merely require a larger body of experiments that will take time to complete. The SPAMS instrument is an improvement on the bioaerosol mass spectrometer developed at LLNL, Snyder said. It uses multivariate data analysis that generates a huge dataset of both positive and negative ions from sampled particles after a preliminary ultraviolet fluorescence analy- sis conducted as particles enter the system. Because of this prescreen, the system can handle 10,000 particles per second. SPAMS was tested at an international airport and produced no false-positives in a 7-week period, during which time it tracked approximately 1 million particles. The down- side to this technology is that the mass spectrometer is large and expen- sive; however, operating costs are low and a comprehensive set of per- formance data has generated figures of merit for prediction of false- positives and false-negatives. Tier 2 systems, Snyder said, cannot begin to address the DHS analyt- ical figures of merit. Though there are many mass spectrometers and sample processing technologies that have the potential to meet those fig- ures of merit, none of the current systems are at the point that they are ready for field testing, and all of them will require appropriate engineer- ing to create a robust, autonomous system. Various microfluidic systems, for example, can concentrate pathogens into a relatively small volume of liquid at a potential operating cost that would be suitable for a large-scale deployment in the field. He noted that biochips have been manufactured to include microcolumn liquid chromatography and electrospray ioniza- tion and that work is being done on a microfluidic proteomic reactor that can digest a protein sample efficiently to produce a sample suitable for analysis by mass spectrometry. In a protein-based system, the peptides resulting from a bacterial pro- tein digest would be analyzed by mass spectrometry and the output would be matched to a complete theoretical list of peptides in a bacterial database generated from genomic sequencing data. The U.S. Army Edgewood Chemical Biological Center has been developing this type of technology for its ABOID (Agents of Biological Origin ID) system and has shown that it can identify specific bacteria in a mixture using mass spectrometry proteomics data generated in the laboratory. Snyder noted that this approach can identify bacteria at the genus level when a specific species is not in the database. The advantages of proteomics-based mass spectrometry as a biothreat detector are that no prior information about the sample is re- quired for analysis, Snyder said. In addition, no specific reagents are
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 75 needed in the analytical process, and it can classify an organism when a primer or probe set is not available. Proteomics mass spectrometry, he added, can provide a presumptive identification of a true unknown or- ganism by mapping its phylogenetic relationship with other known path- ogens (Jabbour et al., 2010). He concluded his summary of the field by stating that there is no sys- tem yet envisioned for Tier 3. He voiced the opinion that PCR or an antigen-antibody technology could be interfaced with mass spectrometry to fill this gap in the market. Critical Quality Issues Rudolph Johnson, acting manager, Emergency Response Branch, at CDCâs National Center for Environmental Health, began by saying that from his perspective as someone who is interested in technology that can be put in the field, the two areas of focus for microbial mass spec- trometry are to develop fingerprint spectra for bacteria and to produce interpretable results on a representative number of samples as quickly as possible. Both of these areas are being tackled by the clinical micro- biology community, he said, and that community has the money to conduct the necessary research. According to Johnson, the BioWatch community needs to be more concerned about quality in the field set- ting than in the laboratory, using environmental samples rather than clinical samples. One of the advantages mass spectrometry has over some of the other technologies that have been discussed is that it is both mature and robust. Mass spectrometry itself is about a century old, and most instrumentation is conserved between applications, Johnson said. Mass spectrometry is a large field, and most of the research effort is aimed at improving the sample preparation, ionization methods, and database analysis, or at de- veloping new applications for the existing detectors, rather than new mass spectrometers. âIf you are looking for a new testing platform, you need to consider mass spectrometry,â Johnson said. He said that mass spectrometry is portable, although he noted that âportableâ does not mean âstand-alone,â as mass spectrometry is usually done with human input. One unusual feature of mass spectrometry is that a smaller sample usually produces better sensitivity and specificity. Sample quality is important, then, to the veracity of the data from a mass spectrometer, particularly in terms of competition for ions during the ion- ization stage. Johnson offered a caveat regarding the selection of instru-
76 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH ments, specifically concerning low- versus high-resolution systems. âThe one thing I want to tell you about mass spectrometry is that better is the enemy of good enough,â he said. âIf you are looking at mass spectrome- try, look for something simple.â Lower-resolution systems, he explained, are more flexible, less expensive to purchase and to operate, and more reliable. Data processing is an area in which advances have brought big changes in the field, and Johnson stressed that the goal for BioWatch should not be data interpretation but rather fingerprint identification, a much simpler task. He said that bacterial databases are now being used in hospitals for routine diagnosis, which suggests that these databases have reached a level of maturity and reliability that bodes well for the biodetection field. He also remarked that these databases are transferable between instruments, which is an important development. In closing, Johnson said that mass spectrometry has reached a stage where it is largely a black box for the average user. However, it is still important to understand the workings of these instruments and their key quality issues. Otherwise, he said, âyou will end up with poor-quality data, and the system that you deploy in the field will not be effective.â The questions that every user needs to answer include ï· How do you verify instrument performance daily? ï· What are your positive and negative controls? ï· How do you characterize fingerprint spectra, and who controls database entries? ï· What is a minimum threshold criteria for a positive result? ï· What is your matrix? Instruments placed in different locations will have different matrices. ï· How do you compensate for charge competition? Single-Particle Aerosol Mass Spectrometry Eric Gard, scientist in the Defense Biology Division at LLNL, began by saying that one of the advantages of SPAMS is that it eliminates the need to process bulk samples of aerosols, a process that adds sample preparation delays into the analysis pipeline. Instead, aerosol particles are pulled directly from the atmosphere and analyzed independently in a time that is of the order of milliseconds. This short time frame allows for multiple measurements that can improve detection sensitivity and detec- tion statistics (Steele et al., 2008).
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 77 The collector for this device is an aerodynamic lens that focuses par- ticles and directs them past tracking lasers that provide information on size, speed, and location of particles as they move through the system. A fluorescent stage excites the particles and measures fluorescence emis- sions as an indicator of whether a particle is likely to be biological. Based on measurements of a particleâs speed, the instrument knows when a prescreened particle reaches the center of the ion source, and at that point it delivers a high-powered laser pulse that produces both posi- tive and negative ions that are then analyzed by the mass spectrometer. Gard explained that he and his colleagues had what he characterized as a fantastic opportunity over 3 years as different sponsors allowed them to put their instrument through a variety of field tests. As Snyder men- tioned, the system was first deployed at an international airport for ap- proximately 7 weeks to test background monitoring in a realistic environ- ment. The second field deployment, which occurred at Fort Irwin, evalu- ated how it would perform in a dusty environment combined with heavy equipment emissions. Finally, the Applied Physics Laboratory at Johns Hopkins University conducted independent, laboratory-based simulation tests in the presence of high concentrations of highly interfering back- grounds. Over the course of running 24 hours per day in full autonomous mode for 1 week on two separate occasions, the instrument was able to detect relevant target concentrations of up to 100,000 particles per liter of air within the size range of interest. Detection time was on the order of 1 minute with a greater than 90 percent probability of detection. There was one false-positive during the 14 days of testing, and the instrument failed one time when the inlet clogged as a result of the extremely high particle concentration used in the test. From these data, Gard concluded that this technology is at TRL 5, perhaps approaching TRL 6. Describing the weaknesses of SPAMS, Gard said that the harshness of the ionization method reduces the information content of the finger- print, restricting it largely to metabolic signals. The system does not have predictive capability for signatures in this mass range; therefore, the sys- tem needs to be trained for each signature, given the lack of understand- ing of how genomic and proteomic information translates into metabolic products. Finally, the cost and size of this system are too great for wide- spread deployment. The strengths of the system are that it has a broad range of potential signatures, given the generality of the ionization technique, and that analysis times are in the microsecond range with detection times of less than a minute. Contextual information provided by the system in terms of
78 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH particle size, concentration, temporal profiles, spatial distributions, and the potential to see other additives can indicate whether a release was intentional. In addition, SPAMS can detect other threats, such as chemi- cal and radiological releases. With its low false-alarm rate and the ability to generate data rapidly, SPAMS could be used to initiate modeling and low-level data-gathering activities in preparation for a potential BAR event, Gard said. In the near term, he argued in closing, the atmospheric context that this technology produces and the speed with which it produces that information can sup- port decision making and response activities that support data from high- confidence detectors. By preserving the aerosol characteristics of the sample, this technology allows one to âtranslateâ between the biology and its implications in terms of transport and potential consequences. This information would fill what Gard said is an important gap in the BioWatch system. âI think this intermediate detection specificity has not really been looked at in terms of how it could be useful within the net- work and supporting the decision makers,â he said. Miniature Mass Spectrometry System for Microorganisms Mass spectrometers do not have to be large, said Zheng Ouyang, as- sociate professor at Purdue University, but mass spectrometers cannot access samples directly and therefore need to be interfaced with sample preparation purification and injection modules that add to the size and complexity of the total system. For example, an instrument that he and his colleagues developed is about the size of a large box of tissues and weighs about 4 kilograms (Gao et al., 2008). However, this device func- tions at atmospheric pressure, and so any biomarker larger than a fatty acid molecule would not be volatile enough to enter the mass spectrome- ter for analysis. He said that he does not believe it is possible to miniatur- ize a universal mass spectrometry platform, but it is possible to miniatur- ize a system for a specific application. As an example, he described a system that his group developed for monitoring therapeutic drug levels from a drop of blood in about 60 seconds. How might this approach be applied to the analysis of microorgan- isms in aerosol droplets? Zheng proposed starting with a disposable fur- nace filter rated for microorganisms fitted inside a small cartridge. Air would be drawn through the filter, which would collect particles. After a suitable collection time, the cartridge would be inserted into the machine for paper spray mass spectrometry analysis (Yang et al., 2012).
POTENTIAL TECHNOLOGIES FOR THE BIOWATCH PROGRAM 79 Before discussing what might be observed using such an approach, Zheng said that there are more than 40 research systems available today that can do ambient ionizationâthat is, ionization without any sample preparationâup from 2 that were available in 2005. More than 10 compa- nies have produced miniature mass spectrometers, but most of these can only handle gas samples. The key questions, he said, are whether any of these systems can identify biomarkers that can be efficiently sampled and ionized by ambient ionization methods and that can be used for high- specificity characterization and identification of bacteria. He believes that lipid profiles may be the answer, and he presented data experiments demonstrating real-time identification of bacteria grow- ing in a biofilm using both positive and negative ion modes to create li- pid fingerprints (Song et al., 2009). These experiments demonstrated that it is feasible to differentiate subspecies, too. More recent work has shown that the same lipid fingerprint is obtained from organisms collected on filter paper and analyzed using paper spray mass spectrometry. According to Zheng, none of these methods is ready for Tier 1, but he believes that with further work, lipid-based identification using ambi- ent ionization on small mass spectrometers could be Tier 2 or Tier 3. What is needed to start this development effort, he said, is the provision of standard samples of microorganisms to generate a large body of spec- tra in order to optimize statistical data analysis methods. From there, it would be necessary to optimize sampling ionization conditions, charac- terize matrix effects and detection limits, and determine if the method can identify multiple microorganisms in the same sample. Discussion Raymond Mariella, Jr., asked if either fatty acid esters or the low mass-to-charge ratios measurable with relatively simple instruments have the ability to generate fingerprints that can meet the BioWatch criteria of detecting a thousand organisms of interest against a background of 1 to 10 million other organisms with a false-positive rate of 10â7 or better. Gard replied that for the system he has been working with, the answer is no, and that is one reason that he sees that particular method as an or- thogonal technology that would provide atmospheric context and other information that would benefit BioWatch. Gard also said that he has not seen any mass spectrometry system that can meet those specifications. Mariella agreed that these systems would have value for orthogonal de-
80 TECHNOLOGIES TO ENABLE AUTONOMOUS DETECTION FOR BIOWATCH tection, but he was curious if they could serve as primary detection sys- tems as well. Rabih Jabbour said that proteomic mass spectrometry is more likely to reach the needed level of specificity, and Johnson added that what he likes about whole-organism protein mass spectrometry is that this field is moving forward without requiring government investment. He predicted that these instruments, which now cost about $200,000 and a $2 cost per sample, will be miniaturized to a point-of-care level. âIf you want to move into environmental detection 4 or 5 years from now, you would probably do that much more easily than trying to design a system for this purpose now,â he said. Cindy Bruckner-Lea pointed out that SPAMS could be valuable for monitoring indoor releases because it provides the possibility of real- time detection, which could be used to trigger follow-on analysis. She indicated that a cost-benefit analysis would be needed, however, and a major limitation is the very high cost of these systems. She questioned whether SPAMS would be able to have enough sensitivity for outdoor settings, because the limit-of-detection requirements are more challeng- ing for outdoor compared with indoor releases (unless there is a very high density of detectors outdoors). Thomas Companion raised the point that mass spectrometry is one of the primary technologies used to detect trace explosives, which are of very high molecular weight and with virtually no vapor pressure, so there are some similarities between that and detecting large biomolecules. What DHS found was that how sampling was done was more important than the device that was used to analyze the sample. He suggested that investments should be made in the area of sampling. Snyder said that, given the complexity of environmental samples compared with clinical samples, he supported that idea.
