Summary
Characterization, or ''typing," of deoxyribonucleic acid (DNA) for purposes of criminal investigation can be thought of as an extension of the forensic typing of blood that has been common for more than 50 years; it is actually an extension from the typing of proteins that are coded for by DNA to the typing of DNA itself. Genetically determined variation in proteins is the basis of blood groups, tissue types, and serum protein types. Developments in molecular genetics have made it possible to study the person-to-person differences in parts of DNA that are not involved in coding for proteins, and it is primarily these differences that are used in forensic applications of DNA typing to personal identification. DNA typing can be a powerful adjunct to forensic science. The method was first used in casework in 1985 in the United Kingdom and first used in the United States by commercial laboratories in late 1986 and by the Federal Bureau of Investigation (FBI) in 1988.
DNA typing has great potential benefits for criminal and civil justice; however, because of the possibilities for its misuse or abuse, important questions have been raised about reliability, validity, and confidentiality. By the summer of 1989, the scientific, legal, and forensic communities were calling for an examination of the issues by the National Research Council of the National Academy of Sciences. As a response, the Committee on DNA Technology in Forensic Science was formed; its first meeting was held in January 1990. The committee was to address the general applicability and appropriateness of the use of DNA technology in forensic science, the need to develop standards for data collection and analysis, aspects of the
technology, management of DNA typing data, and legal, societal, and ethical issues surrounding DNA typing. The techniques of DNA typing are fruits of the revolution in molecular biology that is yielding an explosion of information about human genetics. The highly personal and sensitive information that can be generated by DNA typing requires strict confidentiality and careful attention to the security of data.
DNA, the active substance of the genes, carries the coded messages of heredity in every living thing: animals, plants, bacteria, and other microorganisms. In humans, the code-carrying DNA occurs in all cells that have a nucleus, including white blood cells, sperm, cells surrounding hair roots, and cells in saliva. These would be the cells of greatest interest in forensic studies.
Human genes are carried in 23 pairs of chromosomes, long threadlike or rodlike structures that are a person's archive of heredity. Those 23 pairs, the total genetic makeup of a person, are referred to as the human "diploid genome." The chemistry of DNA embodies the universal code in which the messages of heredity are transmitted. The genetic code itself is spelled out in strings of nucleotides of four types, commonly represented by the letters A, C, G, and T (standing for the bases adenine, cytosine, guanine, and thymine), which in various combinations of three nucleotides spell out the
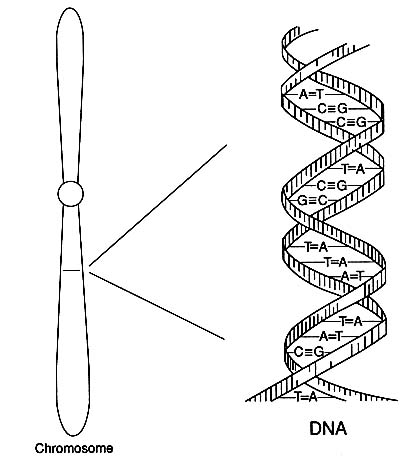
FIGURE 1 Diagram of the double-helical structure of DNA in a chromosome. The line shown in the chromosome is expanded to show the DNA structure.
codes for the amino acids that constitute the building blocks of proteins. A gene, the basic unit of heredity, is a sequence of about 1,000 to over 2 million nucleotides. The human genome, the total genetic makeup of a person, is estimated to contain 50,000-100,000 genes.
The total number of nucleotides in a set of 23 chromosomes—one from each pair, the "haploid genome"—is about 3 billion. Much of the DNA, the part that separates genes from one another, is noncoding. Variation in the genes, the coding parts, are usually reflected in variations in the proteins that they encode, which can be recognized as "normal variation" in blood type or in the presence of such diseases as cystic fibrosis and phenylketonuria; but variations in the noncoding parts of DNA have been most useful for DNA typing.
Except for identical twins, the DNA of a person is for practical purposes unique. That is because one chromosome of each pair comes from the father and one from the mother; which chromosome of a given pair of a parent's chromosomes that parent contributes to the child is independent of which chromosome of another pair that parent gives to that child. Thus, the different combinations of chromosomes that one parent can give to one child is 223, and the number of different combinations of paired chromosomes a child can receive from both parents is 246.
The substitution of even one nucleotide in the sequence of DNA is a variation that can be detected. For example, a variation in DNA consisting of the substitution of one nucleotide for another (such as the substitution of a C for a T) can often be recognized by a change in the points at which certain biological catalysts called "restriction enzymes" cut the DNA. Such an enzyme cuts DNA whenever it encounters a specific sequence of nucleotides that is peculiar to the enzyme. For example, the enzyme HaeI cuts DNA wherever it encounters the sequence AGGCCA. A restriction enzyme will cut a sample of DNA into fragments whose lengths depend on the location of the cutting sites recognized by the enzyme. Assemblies of fragments of different lengths are called "restriction fragment length polymorphisms" (RFLPs), and RFLPs constitute one of the most important tools for analyzing and identifying samples of DNA.
An important technique used in such analyses is the "Southern blot," developed by Edwin Southern in 1975. A sample of DNA is cut with a restriction enzyme, and the fragments are separated from one another by electrophoresis (i.e., they are separated by an electrical field). The fragments of particular interest are then identified with a labeled probe, a short segment of single-stranded DNA containing a radioactive atom, which hybridizes (fuses) to the fragments of interest because its DNA sequence is complementary to those of the fragments (A pairing with T, C pairing with G). Each electrophoretic band represents a separate fragment of DNA, and a given person will have no more than two fragments derived from a partic-
ular place in his or her DNA—one representing each of the genes that are present on the two chromosomes of a given pair. The forms of a given gene are referred to as alleles. A person who received the same allele from the mother and the father is said to be "homozygous" for that allele; a person who received different alleles from the mother and the father is said to be "heterozygous." Many RFLP systems are based on change in a single nucleotide. They are said to be "diallelic," because there are only two common alternative forms. And there are only three genotypes: two kinds of homozygous genotypes and a heterozygous genotype. Another form of RFLP is generated by the presence of variable number tandem repeats (VNTRs). VNTRs are sequences, sometimes as small as two different nucleotides (such as C and A), that are repeated in the DNA. When such a structure is subjected to cutting with restriction enzymes, fragments of varied length are obtained.
It was variation of the VNTR type to which Alex Jeffreys in the United Kingdom first applied the designation "DNA fingerprinting." He used probes that recognized not one locus, but multiple loci, and "DNA fingerprinting" has come to refer particularly to multilocus, multiallele systems. A locus is a specific site of a gene on a chromosome. In the United States, in particular, single-locus probes are preferred, because their results are easier to interpret. "DNA typing" is the preferred term, because "DNA fingerprinting'' is associated with multilocus systems. Discriminating power for personal identification is achieved by using several—usually at least four—single-locus, multiallelic systems.
The entire procedure for analyzing DNA with the RFLP method is diagrammed in Figure 2.
After the bands (alleles) are visualized, those in the evidence sample and the suspect sample are compared. If the bands match in the two samples, for all three or four enzyme-probe combinations, the question is: What is the probability that such a match would have occurred between the suspect and a person drawn at random from the same population as the suspect?
Answering that question requires calculation of the frequency in the population of each of the gene variants (alleles) that have been found, and the calculation requires a databank where one can find the frequency of each allele in the population. On the basis of some assumptions, so-called Hardy-Weinberg ratios can be calculated. For a two-allele system, the ratios are indicated by the expressions p2 and q2 for the frequency of the two homozygotes and 2pq for heterozygotes, p and q being the frequencies of the two alleles and p + q being equal to 1. Suppose that a person is heterozygous at a locus where the frequencies of the two alleles in the population are 0.3 and 0.7. The frequency of that heterozygous genotype in the population would be 2 × 0.3 × 0.7 = 0.42. Suppose, further, that at three
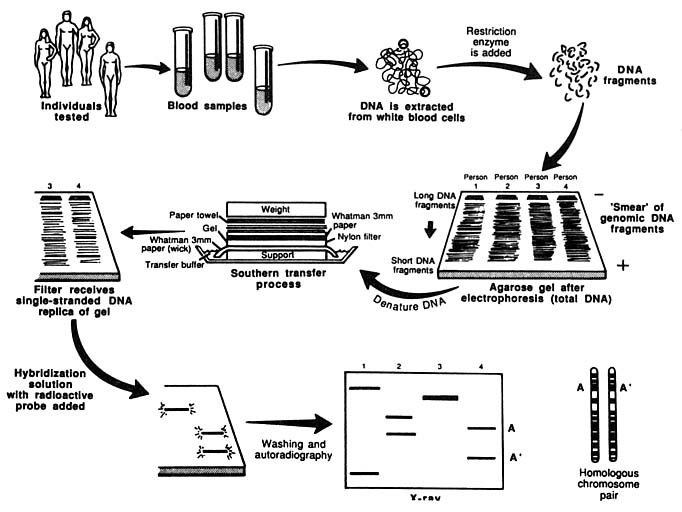
FIGURE 2 Schematic representation of Southern blotting of single-locus, multiallelic VNTR. In example shown here, DNA from four persons is tested. All have different patterns. Three are heterozygous and one homozygous, for a total of seven different alleles. From L. T. Kirby, "DNA Fingerprinting: An Introduction," Stockton Press, New York, 1990. Copyright © 1990 by Stockton Press. Reprinted with permission of W.H. Freeman and Company.
other loci the person being typed has genotypes with population frequencies of 0.01, 0.32, and 0.02. The frequency of the combined genotype in the population is 0.42 × 0.01 × 0.32 × 0.02 or 0.000027, or approximately 1 in 37,000.
That example illustrates what is called the product rule, or multiplication rule. Its use assumes that the alleles at a given locus are inherited independently of each other. It also assumes that there are no subpopulations in which a particular allele at one locus would have a preferential probability of being associated with a particular allele at a second locus.
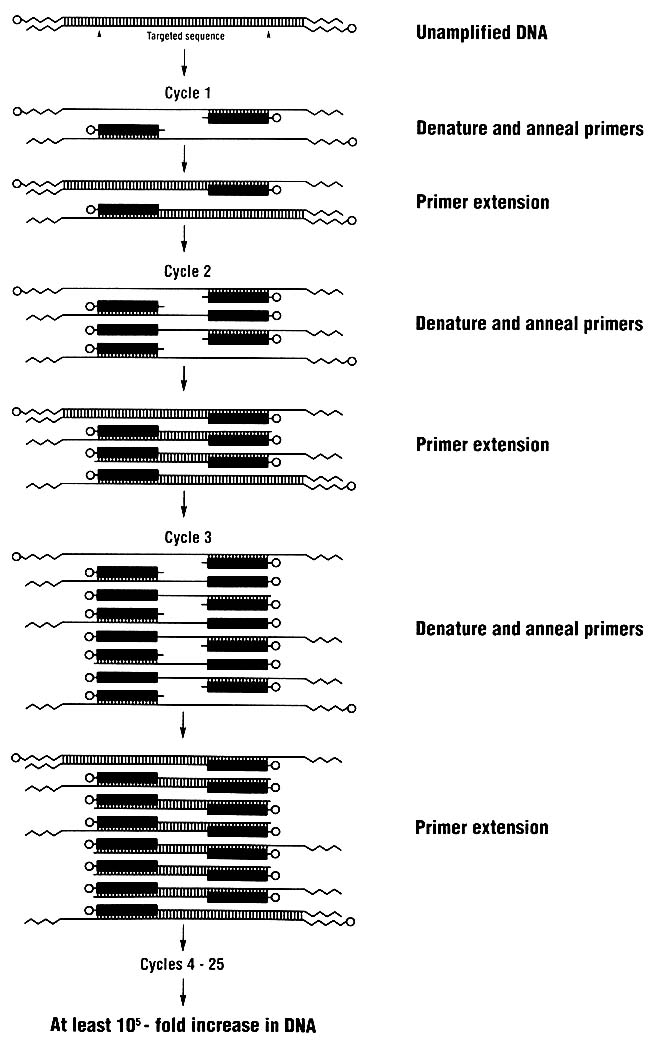
Techniques for analyzing DNA are changing rapidly. One key technique introduced in the last few years is the polymerase chain reaction (PCR), which allows a million or more copies of a short region of DNA to be easily made. For DNA typing, one amplifies (copies) a genetically informative sequence, usually 100-2,000 nucleotides long, and detects the genotype in the amplified product. Because many copies are made, DNA
typing can rely on methods of detection that do not use radioactive substances. Furthermore, the technique of PCR amplification permits the use of very small samples of tissue or body fluids—theoretically even a single nucleated cell.
The PCR process (Figure 3) is simple; indeed, it is analogous to the process by which cells replicate their DNA. It can be used in conjunction with various methods for detecting person-to-person differences in DNA.
It must be emphasized that new methods and technology for demonstrating individuality in each person's DNA are being developed. The present methods explained here will probably be superseded by others that are more efficient, error-free, automatable, and cost-effective. Care should be taken to ensure that DNA typing techniques used for forensic purposes do not become "locked in" prematurely, lest society and the criminal justice system be unable to benefit fully from advances in science and technology.
TECHNICAL CONSIDERATIONS
The forensic use of DNA typing is an outgrowth of its medical diagnostic use—analysis of disease-causing genes based on comparison of a patient's DNA with that of family members to study inheritance patterns of genes or comparison with reference standards to detect mutations. To understand the challenges involved in such technology transfer, it is instructive to compare forensic DNA typing with DNA diagnostics.
DNA diagnostics usually involves clean tissue samples from known sources. Its procedures can usually be repeated to resolve ambiguities. It involves comparison of discrete alternatives (e.g., which of two alleles did a child inherit from a parent?) and thus includes built-in consistency checks against artifacts. It requires no knowledge of the distribution of patterns in the general population.
Forensic DNA typing often involves samples that are degraded, contaminated, or from multiple unknown sources. Its procedures sometimes cannot be repeated, because there is too little sample. It often involves matching of samples from a wide range of alternatives in the population and thus lacks built-in consistency checks. Except in cases where the DNA evidence excludes a suspect, assessing the significance of a result requires statistical analysis of population frequencies.
Each method of DNA typing has its own advantages and limitations, and each is at a different state of technical development. However, the use of each method involves three steps:
-
Laboratory analysis of samples to determine their genetic-marker types at multiple sites of potential variation.
-
Comparison of the genetic-marker types of the samples to determine
-
whether the types match and thus whether the samples could have come from the same source.
-
If the types match, statistical analysis of the population frequencies of the types to determine the probability that a match would have been observed by chance in a comparison of samples from different persons.
Before any particular DNA typing method is used for forensic purposes, precise and scientifically reliable procedures for performing all three steps must be established. It is meaningless to speak of the reliability of DNA typing in general—i.e., without specifying a particular method.
Despite the challenges of forensic DNA typing, it is possible to develop reliable forensic DNA typing systems, provided that adequate scientific care is taken to define and characterize the methods.
Recommendations
-
Any new DNA typing method (or a substantial variation of an existing method) must be rigorously characterized in both research and forensic settings, to determine the circumstances under which it will yield reliable results.
-
DNA analysis in forensic science should be governed by the highest standards of scientific rigor, including the following requirements:
-
Each DNA typing procedure must be completely described in a detailed, written laboratory protocol.
-
Each DNA typing procedure requires objective and quantitative rules for identifying the pattern of a sample.
-
Each DNA typing procedure requires a precise and objective matching rule for declaring whether two samples match.
-
Potential artifacts should be identified by empirical testing, and scientific controls should be designed to serve as internal checks to test for the occurrence of artifacts.
-
The limits of each DNA typing procedure should be understood, especially when the DNA sample is small, is a mixture of DNA from multiple sources, or is contaminated with interfering substances.
-
Empirical characterization of a DNA typing procedure must be published in appropriate scientific journals.
-
Before a new DNA typing procedure can be used, it must have not only a solid scientific foundation, but also a solid base of experience.
-
-
The committee strongly recommends the establishment of a National Committee on Forensic DNA Typing (NCFDT) under the auspices of an
-
appropriate government agency or agencies to provide expert advice primarily on scientific and technical issues concerning forensic DNA typing.
-
Novel forms of variation in the genome that have the potential for increased power of discrimination between persons are being discovered. Furthermore, new ways to demonstrate variations in the genome are being developed. The current techniques are likely to be superseded by others that provide unambiguous individual identification and have such advantages as automatability and economy. Each new method should be evaluated by the NCFDT for use in the forensic setting, applying appropriate criteria to ensure that society derives maximal benefit from DNA typing technology.
STATISTICAL BASIS FOR INTERPRETATION
Because any two human genomes differ at about 3 million sites, no two persons (barring identical twins) have the same DNA sequence. Unique identification with DNA typing is therefore possible, in principle, provided that enough sites of variation are examined. However, the DNA typing systems used today examine only a few sites of variation and have only limited resolution for measuring the variability at each site. There is a chance that two persons have DNA patterns (i.e., genetic types) that match at the small number of sites examined. Nevertheless, even with today's technology, which uses 3-5 loci, a match between two DNA patterns can be considered strong evidence that the two samples came from the same source. Interpreting a DNA typing analysis requires a valid scientific method for estimating the probability that a random person by chance matches the forensic sample at the sites of DNA variation examined. To say that two patterns match, without providing any scientifically valid estimate (or, at least, an upper bound) of the frequency with which such matches might occur by chance, is meaningless. The committee recommends approaches for making sound estimates that are independent of the race or ethnic group of the subject.
A standard way to estimate frequency is to count occurrences in a random sample of the appropriate population and then use classical statistical formulas to place upper and lower confidence limits on the estimate. (Because forensic science should avoid placing undue weight on incriminating evidence, an upper confidence limit of the frequency should be used in court.) If a particular DNA pattern occurred in 1 of 100 samples, the estimated frequency would be 1%, with an upper confidence limit of 4.7%. If the pattern occurred in 0 of 100 samples, the estimated frequency would be 0%, with an upper confidence limit of 3%. (The upper bound cited is the traditional 95% confidence limit, whose use implies that the true value has only a 5% chance of exceeding the upper bound.) Such estimates produced
by straightforward counting have the virtue that they do not depend on theoretical assumptions, but simply on the samples having been randomly drawn from the appropriate population. However, such estimates do not take advantage of the full potential of the genetic approach.
In contrast, population frequencies often quoted for DNA typing analyses are based not on actual counting, but on theoretical models based on the principles of population genetics. Each matching allele is assumed to provide statistically independent evidence, and the frequencies of the individual alleles are multiplied together to calculate a frequency of the complete DNA pattern. Although a databank might contain only 500 people, multiplying the frequencies of enough separate events might result in an estimated frequency of their all occurring in a given person of 1 in a billion. Of course, the scientific validity of the multiplication rule depends on whether the events (i.e., the matches at each allele) are actually statistically independent.
Because it is impossible or impractical to draw a large enough population to test directly calculated frequencies of any particular DNA profile much below 1 in 1,000, there is not a sufficient body of empirical data on which to base a claim that such frequency calculations are reliable or valid. The assumption of independence must be strictly scrutinized and estimation procedures appropriately adjusted if possible. (The rarity of all the genotypes represented in the databank can be demonstrated by pairwise comparisons, however. Thus, in a recently reported analysis of the FBI databank, no exactly matching pairs were found in five-locus DNA profiles, and the closest match was a single three-locus match among 7.6 million pairwise comparisons.)
The multiplication rule has been routinely applied to blood-group frequencies in the forensic setting. However, that situation is substantially different. Because conventional genetic markers are only modestly polymorphic (with the exception of human leukocyte antigen, HLA, which usually cannot be typed in forensic specimens), the multilocus genotype frequencies are often about 1 in 100. Such estimates have been tested by simple empirical counting. Pairwise comparisons of allele frequencies have not revealed any correlation across loci. Hence, the multiplication rule does not appear to lead to the risk of extrapolating beyond the available data for conventional markers. But highly polymorphic DNA markers exceed the informative power of protein markers and so multiplication of their estimated frequencies leads to estimates that are far less than the reciprocal of the size of the databanks, i.e., 1/N, N being the number of entries in the databank.
The multiplication rule is based on the assumption that the population does not contain subpopulations with distinct allele frequencies—that each person's alleles constitute statistically independent random selections from
a common gene pool. Under that assumption, the procedure for calculating the population frequency of a genotype is straightforward:
-
Count the frequency of alleles. For each allele in the genotype, examine a random sample of the population and count the proportion of matching alleles—that is, alleles that would be declared to match according to the rule that is used for declaring matches in a forensic context.
-
Calculate the frequency of the genotype at each locus. The frequency of a homozygous genotype a1/a1 is calculated to be pa12, where pa1 denotes the frequency of allele a1. The frequency of a heterozygous genotype a1/a2 is calculated to be 2pa1pa2, where pa1 and pa2 denote the frequencies of alleles a1 and a2. In both cases, the genotype frequency is calculated by multiplying the two allele frequencies, on the assumption that there is no statistical correlation between the allele inherited from one's father and the allele inherited from one's mother. When there is no correlation between the two parental alleles, the locus is said to be in Hardy-Weinberg equilibrium.
-
Calculate the frequency of the complete multilocus genotype by multiplying the genotype frequencies at all the loci. As in the previous step, this calculation assumes that there is no correlation between the genotypes at the individual loci; the absence of such correlation is called linkage equilibrium. Suppose, for example, that a person has genotype a1/a2, b1/b2, c1/c1. If a random sample of the appropriate population shows that the frequencies of alleles a1, a2, b1, b2, and c1 are approximately 0.1, 0.2, 0.3, 0.1, and 0.2, respectively, then the population frequency of the genotype would be estimated to be [2(0.1)(0.2)][2(0.3)(0.1)][(0.2)(0.2)] = 0.000096, or about 1 in 10,417.
The validity of the multiplication rule depends on the assumption of absence of population substructure. Population substructure violates the assumption of statistical independence of alleles. In a population that contains groups each with different allele frequencies, the presence of one allele in a person's genotype can alter the statistical expectation of the other alleles in the genotype. For example, a person who has one allele that is common among Italians is more likely to be of Italian descent and is thus more likely to carry additional alleles that are common among Italians. The true genotype frequency is thus higher than would be predicted by applying the multiplication rule using the average frequency in the entire population.
To illustrate the problem with a hypothetical example, suppose that a particular allele at a VNTR locus has a 1% frequency in the general population, but a 20% frequency in a specific subgroup. The frequency of homozygotes for the allele would be calculated to be 1 in 10,000 according to the allele frequency determined by sampling the general population, but would actually be 1 in 25 for the subgroup. That is a hypothetical and
extreme example, but illustrates the potential effect of demography on gene frequency estimation.
The key question underlying the use of the multiplication rule—i.e., whether actual populations have significant substructure for the loci used for forensic typing—has provoked considerable debate among population geneticists. Some have expressed serious concern about the possibility of significant substructure. They maintain that census categories—such as North American Caucasians, blacks, Hispanics, Asians, and Native Americans—are not homogeneous groups, but rather that each group is an admixture of subgroups with somewhat different allele frequencies. Allele frequencies have not yet been homogenized, because people tend to mate within their subgroups.
Those population geneticists also point out that, for any particular genetic marker, the actual degree of subpopulation differentiation cannot be predicted in advance, but must be determined empirically. Furthermore, they doubt that the presence of substructure can be detected by the application of statistical tests to data from large mixed populations. Population differentiation must be assessed through direct studies of allele frequencies in ethnic groups.
Other population geneticists, while recognizing the possibility or likelihood of population substructure, conclude that the evidence to date suggests only a minimal effect on estimates of genotype frequencies. Recent empirical studies concerning VNTR loci detected no deviation from independence within or across loci. Moreover, as pointed out earlier, pairwise comparisons of all five-locus DNA profiles in the FBI database showed no exact matches; the closest match was a single three-locus match among 7.6 million pairwise comparisons. Those studies are interpreted as indicating that multiplication of gene frequencies across loci does not lead to major inaccuracies in the calculation of genotype frequency—at least not for the specific polymorphic loci examined.
Although mindful of those opposing views, the committee has chosen to assume for the sake of discussion that population substructure may exist and to provide a method for estimating population frequencies in a manner that would adequately account for it. Our decision is based on four considerations:
-
It is possible to provide conservative estimates of population frequency, without giving up the inherent power of DNA typing.
-
It is appropriate to prefer somewhat conservative numbers for forensic DNA typing, especially because the statistical power lost in this way can often be recovered through typing of additional loci, where required.
-
It is important to have a general approach that is applicable to any loci used for forensic typing. Recent empirical studies pertain only to the population genetics of the VNTR loci in current use. However, we expect
-
forensic DNA typing to undergo much change over the next decade—including the introduction of different types of DNA polymorphisms, some of which might have different properties from the standpoint of population genetics.
-
It is desirable to provide a method for calculating population frequencies that is independent of the ethnic group of the subject.
The committee is aware of the need to account for possible population substructure, and it recommends the use of the ceiling principle. The multiplication rule will yield conservative estimates even for a substructured population, provided that the allele frequencies used in the calculation exceed the allele frequencies in any of the population subgroups. The ceiling principle involves two steps: (1) for each allele at each locus, determine a ceiling frequency that is an upper bound of the allele frequency that is independent of the ethnic background of a subject; and (2) to calculate a genotype frequency, apply the multiplication rule according to the ceiling allele frequencies.
To determine ceiling frequencies, the committee strongly recommends the following approach: (1) Draw random samples of 100 persons from each of 15-20 populations that represent groups relatively homogeneous genetically. (2) Take as the ceiling frequency the largest frequency in any of those populations or 5%, whichever is larger.
Use of the ceiling principle yields the same frequency of a given genotype, regardless of the suspect's ethnic background, because the reported frequency represents a maximum for any possible ethnic heritage. Accordingly, the ethnic background of the individual suspect should be ignored in estimating the likelihood of a random match. The calculation is fair to suspects, because the estimated probabilities are likely to be conservative in their incriminating power.
Some legal commentators have pointed out that frequencies should be based on the population of possible perpetrators, rather than on the population to which a particular suspect belongs. Although that argument is formally correct, practicalities often preclude use of that approach. Furthermore, the ceiling principle eliminates the need for investigating the perpetrator population, because it yields an upper bound to the frequency that would be obtained by that approach.
Although the ceiling principle is a conservative approach, we feel that it is appropriate. DNA typing is unique, in that the forensic analyst has an essentially unlimited ability to adduce additional evidence: whatever power is sacrificed by requiring conservative estimates can be regained by examining additional loci. (Although there might be some cases in which the DNA sample is insufficient to permit typing additional loci with RFLPs, this limitation is likely to disappear with the eventual use of PCR.)
That no evidence of population substructure is demonstrable with the
markers tested so far cannot be taken to mean that such does not exist for other markers. Preservation of population DNA samples in the form of immortalized cell lines will ensure that DNA is available for determining population frequencies of any DNA pattern as new and better techniques become available, without the necessity of collecting fresh samples. It will also provide samples for standardization of methods across laboratories.
Because of the similarity in DNA patterns between relatives, databanks of DNA of convicted criminals have the ability to point not just to individuals but to entire families—including relatives who have committed no crime. Clearly, this raises serious issues of privacy and fairness. It is inappropriate, for reasons of privacy, to search databanks of DNA from convicted criminals in such a fashion. Such uses should be prevented both by limitations of the software for searching and by statutory guarantees of privacy.
The genetic correlation among relatives means that the probability that a forensic sample will match a relative of the person who left it is considerably greater than the probability that it will match a random person.
Especially for a technology with high discriminatory power, such as DNA typing, laboratory error rates must be continually estimated in blind proficiency testing and must be disclosed to juries.
Recommendations
-
As a basis for the interpretation of the statistical significance of DNA typing results, the committee recommends that blood samples be obtained from 100 randomly selected persons in each of 15-20 relatively homogeneous populations; that the DNA in lymphocytes from these blood samples be used to determine the frequencies of alleles currently tested in forensic applications; and that the lymphocytes be ''immortalized" and preserved as a reference standard for determination of allele frequencies in tests applied in different laboratories or developed in the future. The collection of samples and their study should be overseen by a National Committee on Forensic DNA Typing.
-
The ceiling principle should be used in applying the multiplication rule for estimating the frequency of particular DNA profiles. For each allele in a person's DNA pattern, the highest allele frequency found in any of the 15-20 populations or 5% (whichever is larger) should be used.
-
In the interval (which should be short) while the reference blood samples are being collected, the significance of the findings of multilocus DNA typing should be presented in two ways: (1) If no match is found with any sample in a total databank of N persons (as will usually be the case), that should be stated, thus indicating the rarity of a random match. (2) In applying the multiplication rule, the 95% upper confidence limit of the frequency of each allele should be calculated for separate U.S. "racial"
-
groups and the highest of these values or 10% (whichever is the larger) should be used. Data on at least three major "races" (e.g., Caucasians, blacks. Hispanics, Asians, and Native Americans) should be analyzed.
-
Any population databank used to support DNA typing should be openly available for scientific inspection by parties to a legal case and by the scientific community.
-
Laboratory error rates should be measured with appropriate proficiency tests and should play a role in the interpretation of results of forensic DNA typing.
STANDARDS
Critics and supporters of the forensic uses of DNA typing agree that there is a lack of standardization of practices and a lack of uniformly accepted methods for quality assurance. The deficiencies are due largely to the rapid emergence of DNA typing and its introduction in the United States through the private sector.
As the technology developed in the United States, private laboratories using widely differing methods (single-locus RFLP, multilocus RFLP, and PCR) began to offer their services to law-enforcement agencies. During the same period, the FBI was developing its own RFLP method, with a different restriction enzyme and different single-locus probes. The FBI's method has become the one most widely used in public forensic-science laboratories. Each method has its own advantages and disadvantages, databanks, molecular-weight markers, match criteria, and reporting methods.
Regardless of the causes, practices in DNA typing vary, and so do the educational backgrounds, training, and experience of the scientists and technicians who perform the tests, the internal and external proficiency testing conducted, the interpretation of results, and approaches to quality assurance.
It is not uncommon for an emerging technology to go without regulation until its importance and applicability are established. Indeed, the development of DNA typing technology has occurred without regulation of laboratories and their practices, public or private. The committee recognizes that standardization of practices in forensic laboratories in general is more problematic than in other laboratory settings; stated succinctly, forensic scientists have little or no control over the nature, condition, form, or amount of sample with which they must work. But it is now clear that DNA typing methods are a most powerful adjunct to forensic science for personal identification and have immense benefit to the public—so powerful, so complex, and so important that some degree of standardization of laboratory procedures is necessary to assure the courts of high-quality results. DNA typing is capable, in principle, of an extremely low inherent rate of false results, so the risk of error will come from poor laboratory
practice or poor sample handling and labeling; and, because DNA typing is technical, a jury requires the assurance of laboratory competence in test results.
At issue, then, is how to achieve standardization of DNA typing laboratories in such a manner as to assure the courts and the public that results of DNA typing by a given laboratory are reliable, reproducible, and accurate.
Quality assurance can best be described as a documented system of activities or processes for the effective monitoring and verification of the quality of a work product (in this case, laboratory results). A comprehensive quality-assurance program must include elements that address education, training, and certification of personnel; specification and calibration of equipment and reagents; documentation and validation of analytical methods; use of appropriate standards and controls; sample handling procedures; proficiency testing; data interpretation and reporting; internal and external audits of all the above; and corrective actions to address deficiencies and weight their importance for laboratory competence.
Recommendations
Although standardization of forensic practice is difficult because of the nature of the samples, DNA typing is such a powerful and complex technology that some degree of standardization is necessary to ensure high standards.
-
Each forensic-science laboratory engaged in DNA typing must have a formal, detailed quality-assurance and quality-control program to monitor work, on both an individual and a laboratory-wide basis.
-
The Technical Working Group on DNA Analysis and Methods (TWGDAM) guidelines for a quality-assurance program for DNA RFLP analysis are an excellent starting point for a quality-assurance program, which should be supplemented by the additional technical recommendations of this committee.
-
The TWGDAM group should continue to function, playing a role complementary to that of the National Committee on Forensic DNA Typing (NCFDT). To increase its effectiveness, TWGDAM should include additional technical experts from outside the forensic community who are not closely tied to any forensic laboratory.
-
Quality-assurance programs in individual laboratories alone are insufficient to ensure high standards. External mechanisms are needed, to ensure adherence to the practices of quality assurance. Potential mechanisms include individual certification, laboratory accreditation, and state or federal regulation.
-
One of the best guarantees of high quality is the presence of an active
-
professional-organization committee that is able to enforce standards. Although professional societies in forensic science have historically not played an active role, the American Society of Crime Laboratory Directors (ASCLD) and the American Society of Crime Laboratory Directors-Laboratory Accreditation Board (ASCLD-LAB) recently have shown substantial interest in enforcing quality by expanding the ASCLD-LAB accreditation program to include mandatory proficiency testing. ASCLD-LAB must demonstrate that it will actively discharge this role.
-
Because private professional organizations lack the regulatory authority to require accreditation, further means are needed to ensure compliance with appropriate standards.
-
Courts should require that laboratories providing DNA typing evidence have proper accreditation for each DNA typing method used. Any laboratory that is not formally accredited and that provides evidence to the courts—e.g., a nonforensic laboratory repeating the analysis of a forensic laboratory—should be expected to demonstrate that it is operating at the same level of standards as accredited laboratories.
-
Establishing mandatory accreditation should be a responsibility of the Department of Health and Human Services (DHHS), in consultation with the Department of Justice (DOJ). DHHS is the appropriate agency, because it has extensive experience in the regulation of clinical laboratories through programs under the Clinical Laboratory Improvement Act and has extensive expertise in molecular genetics through the National Institutes of Health. DOJ must be involved, because the task is important for law enforcement.
-
The National Institute of Justice (NIJ) does not appear to receive adequate funds to support proper education, training, and research in the field of forensic DNA typing. The level of funding should be re-evaluated and increased appropriately.
DATABANKS AND PRIVACY OF INFORMATION
DNA typing in the criminal-justice system has so far been used primarily for direct comparison of DNA profiles of evidence samples with profiles of samples from suspects. However, that application constitutes only the tip of the iceberg of potential law-enforcement applications. If DNA profiles of samples from a population were stored in computer databanks (databases), DNA typing could be applied in crimes without suspects. Investigators could compare DNA profiles of biological evidence samples with profiles in a databank to search for suspects.
In many respects, the situation is analogous to that of latent finger-prints. Originally, latent fingerprints were used for comparing crime-scene evidence with suspects. With the development of the Automated Fingerprint Identification Systems (AFIS) in the last decade, the investigative use
of fingerprints has dramatically expanded. Forensic scientists can enter an unidentified latent-fingerprint pattern into an automated system and within minutes compare it with millions of person's patterns contained in a computer file. In its short history, automated fingerprint analysis has been credited with solving tens of thousands of crimes.
The computer technology required for an automated fingerprint identification system is sophisticated and complex. Fingerprints are complicated geometric patterns, and the computer must store, recognize, and search for complex and variable patterns of ridges and minutiae in the millions of prints on file. Several commercially available but expensive computer systems are in use around the world. In contrast, the computer technology required for DNA databanks is relatively simple. Because DNA profiles can be reduced to a list of genetic types (hence, a list of numbers), DNA profile repositories can use relatively simple and inexpensive software and hardware. Consequently, computer requirements should not pose a serious problem in the development of DNA profile databanks.
Confidentiality and security of DNA-related information are especially important and difficult issues, because we are in the midst of two extraordinary technological revolutions that show no signs of abating: in molecular biology, which is yielding an explosion of information about human genetics, and in computer technology, which is moving toward national and international networks connecting growing information resources.
Even simple information about identity requires confidentiality. Just as fingerprint files can be misused, DNA profile information could be misused to search and correlate criminal-record databanks or medical-record databanks. Computer storage of information increases the possibilities for misuse. For example, addresses, telephone numbers, social security numbers, credit ratings, range of incomes, demographic categories, and information on hobbies are currently available for many of our citizens in various distributed computerized data sources. Such data can be obtained directly through access to specific sources, such as credit-rating services, or through statistical disclosure, which refers to the ability of a user to derive an estimate of a desired statistic or feature from a databank or a collection of databanks. Disclosure can be achieved through one query or a series of queries to one or more databanks. With DNA information, queries might be directed at obtaining numerical estimates of values or at deducing the state of an attribute of an individual through a series of Boolean (yes-no) queries to multiple distributed databanks.
Several private laboratories already offer a DNA-banking service (sample storage in freezers) to physicians, genetic counselors, and, in some cases, anyone who pays for the service. Typically, such information as name, address, birth date, diagnosis, family history, physician's name and address, and genetic counselor's name and address is stored with samples.
That information is useful for local, independent bookkeeping and record management. But it is also ripe for statistical or correlative disclosure. Just the existence in a databank of a sample from a person, independent of any DNA-related information, may be prejudicial to the person. In some laboratories, the donor cannot legally prevent outsiders' access to the samples, but can request its withdrawal. A request for withdrawal might take a month or more to process. In most cases, only physicians with signed permission of the donor have access to samples, but typically no safeguards are taken to verify individual requests independently. That is not to say that the laboratories intend to violate donors' rights; they are simply offering a service for which there is a recognized market and attempting to provide services as well as they can.
Recommendations
-
In the future, if pilot studies confirm its value, a national DNA profile databank should be created that contains information on felons convicted of particular violent crimes. Among crimes with high rates of recidivism, the case is strongest for rape, because perpetrators typically leave biological evidence (semen) that could allow them to be identified. Rape is the crime for which the databank will be of primary use. The case is somewhat weaker for violent offenders who are most likely to commit homicide as a recidivist offense, because killers leave biological evidence only in a minority of cases.
-
The databank should also contain DNA profiles of unidentified persons made from biological samples found at crime scenes. These would be samples known to be of human origin, but not matched with any known persons.
-
Databanks containing DNA profiles of members of the general population (as exist for ordinary fingerprints for identification purposes) are not appropriate, for reasons of both privacy and economics.
-
DNA profile databanks should be accessible only to legally authorized persons and should be stored in a secure information resource.
-
Legal policy concerning access and use of both DNA samples and DNA databank information should be established before widespread proliferation of samples and information repositories. Interim protection and sanctions against misuse and abuse of information derived from DNA typing should be established immediately. Policies should explicitly define authorized uses and should provide for criminal penalties for abuses.
-
Although the committee endorses the concept of a limited national DNA profile databank, it doubts that existing RFLP-based technology provides an appropriate wise long-term foundation for such a databank. We expect current methods to be replaced soon with techniques that are sim-
-
pler, easier to automate, and less expensive—but incompatible with existing DNA profiles. Accordingly, the committee does not recommend establishing a comprehensive DNA profile databank yet.
-
For the short term, we recommend the establishment of pilot projects that involve prototype databanks based on RFLP technology and consisting primarily of profiles of violent sex offenders. Such pilot projects could be worthwhile for identifying problems and issues in the creation of databanks. However, in the intermediate term, more efficient methods will replace the current one, and the forensic community should not allow itself to become locked into an outdated method.
-
State and federal laboratories, which have a long tradition and much experience in the management of other types of basic evidence, should be given primary responsibility, authority, and additional resources to handle forensic DNA testing and all the associated sample-handling and data-handling requirements.
-
Private-sector firms should not be discouraged from continuing to prepare and analyze DNA samples for specific cases or for databank samples, but they must be held accountable for misuse and abuse to the same extent as government-funded laboratories and government authorities.
DNA INFORMATION IN THE LEGAL SYSTEM
To produce biological evidence that is admissible in court in criminal cases, forensic investigators must be well trained in the collection and handling of biological samples for DNA analysis. They should take care to minimize the risk of contamination and ensure that possible sources of DNA are well preserved and properly identified. As in any forensic work, they must attend to the essentials of preserving specimens, labeling, and the chain of custody and must observe constitutional and statutory requirements that regulate the collection and handling of samples. The Fourth Amendment provides much of the legal framework for the gathering of DNA samples from suspects or private places, and court orders are sometimes needed in this connection.
In civil (noncriminal) cases—such as paternity, custody, and proof-of-death cases—the standards for admissibility must also be high, because DNA evidence might be dispositive. The relevant federal rules (Rules 403 and 702-706) and most state rules of evidence do not distinguish between civil and criminal cases in determining the admissibility of scientific data. In a civil case, however, if the results of a DNA analysis are not conclusive, it will usually be possible to obtain new samples for study.
The advent of DNA typing technology raises two key issues for judges: determining admissibility and explaining to jurors the appropriate standards for weighing evidence. A host of subsidiary questions with respect to how
expert evidence should be handled before and during a trial to ensure prompt and effective adjudication apply to all evidence and all experts.
In the United States, there are two main tests for admissibility of scientific information through experts. One is the Frye test, enunciated in Frye v. United States. The other is a "helpfulness" standard found in the Federal Rules of Evidence and many of its state counterparts. In addition, several states have recently enacted laws that essentially mandate the admission of DNA typing evidence.
The test for the admissibility of novel scientific evidence enunciated in Frye v. United States is still probably the most frequently invoked test in American case law. A majority of states profess adherence to the Frye rule, although a growing minority have adopted variations on the helpfulness standard suggested by the Federal Rules of Evidence.
Frye predicates the admissibility of novel scientific evidence on its general acceptance in a particular scientific field: "While courts will go a long way in admitting expert testimony deduced from a well-recognized scientific principle or discovery, the thing from which the deduction is made must be sufficiently established to have gained general acceptance in the particular field in which it belongs." Thus, admissibility depends on the quality of the science underlying the evidence, as determined by scientists themselves. Theoretically, the court's role in this preliminary determination is narrow: it should conduct a hearing to determine whether the scientific theory underlying the evidence is generally accepted in the relevant scientific community and whether the specific techniques used are reliable for their intended purpose.
In practice, the court is much more involved. The court must determine the scientific fields from which experts should be drawn. Complexities arise with DNA typing, because the full typing process rests on theories and findings that pertain to various scientific fields. For example, the underlying theory of detecting polymorphisms is accepted by human geneticists and molecular biologists, but population geneticists and other statisticians might differ as to the appropriate method for determining the population frequency of a genotype in the general population or in a particular geographic, ethnic, or other group. The courts often let experts on a process, such as DNA typing, testify to the various scientific theories and assumptions on which the process rests, even though the experts' knowledge of some of the underlying theories is likely to be at best that of a generalist, rather than a specialist.
The Frye test sometimes prevents scientific evidence from being presented to a jury unless it has sufficient history to be accepted by some subspecialty of science. Under Frye, potentially helpful evidence may be excluded until consensus has developed. By 1991, DNA evidence had been considered in hundreds of Frye hearings involving felony prosecutions in
more than 40 states. The overwhelming majority of trial courts ruled that such evidence was admissible, but there have been some important exceptions.
In determining admissibility according to the helpfulness standard under the Federal Rules of Evidence, without specifically repudiating the Frye rule, a court can adopt a more flexible approach. Rule 702 states that, "if scientific, technical or other specialized knowledge will assist the trier of fact to understand the evidence or to determine a fact in issue, a witness qualified as an expert by knowledge, skill, experience, training, or education, may testify thereto in the form of an opinion or otherwise."
Rule 702 should be read with Rule 403, which requires the court to determine the admissibility of evidence by balancing its probative force against its potential for misapplication by the jury. In determining admissibility, the court should consider the soundness and reliability of the process or technique used in generating evidence; the possibility that admitting the evidence would overwhelm, confuse, or mislead the jury; and the proffered connection between the scientific research or test result to be presented and particular disputed factual issues in the case.
The federal rule, as interpreted by some courts, encompasses Frye by making general acceptance of scientific principles by experts a factor, and in some cases a decisive factor, in determining probative force. A court can also consider the qualifications of experts testifying about the new scientific principle, the use to which the technique based on the principle has been put, the technique's potential for error, the existence of specialized literature discussing the technique, and its novelty.
With the helpfulness approach, the court should also consider factors that might prejudice the jury. One of the most serious concerns about scientific evidence, novel or not, is that it possesses an aura of infallibility that could overwhelm a jury's critical faculties. The likelihood that the jury would abdicate its role as critical fact-finder is believed by some to be greater if the science underlying an expert's conclusion is beyond its intellectual grasp. The jury might feel compelled to accept or reject a conclusion absolutely or to ignore evidence altogether. However, some experience indicates that jurors tend not to be overwhelmed by scientific proof and that they prefer experiential data based on traditional forms of evidence. Moreover, the presence of opposing experts might prevent a jury from being unduly impressed with one expert or the other. Conversely, the absence of an opposing expert might cause a jury to give too much weight to expert testimony, on the grounds that, if the science were truly controversial, it would have heard the opposing view. Nevertheless, if the scientific evidence is valid, the solution to those possible problems is not to exclude the evidence, but to ensure through instructions and testimony that the jury is equipped to consider rationally whatever evidence is presented.
In determining admissibility with the helpfulness approach, the court should consider a number of factors in addition to reliability. First is the significance of the issue to which the evidence is directed. If the issue is tangential to the case, the court should be more reluctant to allow a time-consuming presentation of scientific evidence that might itself confuse the jury. Second, the availability and sufficiency of other evidence might make expert testimony about DNA superfluous. And third, the court should be mindful of the need to instruct and advise the jury so as to eliminate the risk of prejudice.
Recommendations
-
Courts should take judicial notice of three scientific underpinnings of DNA typing:
-
The study of DNA polymorphisms can, in principle, provide a reliable method for comparing samples.
-
Each person's DNA is unique (except that of identical twins), although the actual discriminatory power of any particular DNA test will depend on the sites of DNA variation examined.
-
The current laboratory procedure for detecting DNA variation (specifically, single-locus probes analyzed on Southern blots without evidence of band shifting) is fundamentally sound, although the validity of any particular implementation of the basic procedure will depend on proper characterization of the reproducibility of the system (e.g., measurement variation) and inclusion of all necessary scientific controls.
-
-
The adequacy of the method used to acquire and analyze samples in a given case bears on the admissibility of the evidence and should, unless stipulated by opposing parties, be adjudicated case by case. In this adjudication, the accreditation and certification status of the laboratory performing the analysis should be taken into account.
-
Because of the potential power of DNA evidence, authorities should make funds available to pay for expert witnesses, and the appropriate parties must be informed of the use of DNA evidence as soon as possible.
-
DNA samples (and evidence likely to contain DNA) should be preserved whenever that is possible.
-
All data and laboratory records generated by analysis of DNA samples should be made freely available to all parties. Such access is essential for evaluating the analysis.
-
Protective orders should be used only to protect the privacy of individuals.
DNA TYPING AND SOCIETY
The introduction of any new technology is likely to raise concerns about its impact on society. Financial costs, potential harm to the interests of individuals, and threats to liberty or privacy are only a few of the worries typically voiced when a new technology is on the horizon. DNA typing technology has the potential for uncovering and revealing a great deal of information that most people consider to be intensely private. Examples might be the presence of genes involved in known genetic disorders or genes that have been linked to a heightened risk of particular major diseases in some populations.
Although DNA technology involves new scientific techniques for identifying or excluding people, the techniques are extensions and analogues of techniques long used in forensic science, such as serological and fingerprint examinations. Ethical questions can be raised about other aspects of this new technology, but the committee does not see it as violating a fundamental ethical principle.
A new practice or technology can be subjected to further ethical analysis by using two leading ethical perspectives. The first examines the action or practice in terms of the rights of people who are affected; the second explores the potential positive and negative consequences (nonmonetary costs and benefits) of the action or practice, in an attempt to determine whether the potential good consequences outweigh the bad.
Two main questions can be asked about moral rights: Does the use of DNA technology give rise to any new rights not already recognized? Does the use of DNA technology enhance, endanger, or diminish the rights of anyone who becomes involved in legal proceedings? In answer to the first question, it is hard to think of any new rights not already recognized that come into play with the introduction of DNA technology into forensic science. The answer to the second question requires a specification of the classes of people whose rights might be affected and what those rights might be.
Concerns about intrusions into privacy and breaches of confidentiality regarding the use of DNA technology in such enterprises as gene mapping are frequently voiced, and they are legitimate ethical worries. The concerns are pertinent to the role of DNA technology in forensic science, as well as to its widespread use for other purposes and in other social contexts. A potential problem related to the confidentiality of any information obtained is the safeguarding of the information and the prevention of its unauthorized release or dissemination; that can also be classified under the heading of abuse and misuse, as well as seen as a violation of individual rights in the forensic context.
Another factor to be weighed in a consequentialist ethical analysis is
whose interests are to count and whether some people's interests should be given greater weight than others'. For example, there are the interests of the accused, the interests of victims of crime or their families in apprehending and convicting perpetrators, and the interests of society. Whether the interests of society in seeing that justice is done should count as much as the interests of the accused or the victim is open to question.
A major issue is the preservation of confidentiality of information obtained with DNA technology in the forensic context. When databanks are established in such a way that state and federal law-enforcement authorities can gain access to DNA profiles, not only of persons convicted of violent crimes but of others as well, there is a serious potential for abuse of confidential information. The victims of many crimes in urban areas are relatives or neighbors of the perpetrators, and these victims might themselves be former or future perpetrators. There is greater likelihood that DNA information on minority-group members, such as blacks and Hispanics, will be stored or accessed. However, it is important to note that use of the ceiling principle removes the necessity to categorize criminals (or defendants in general) by ethnic group for the purposes of DNA testing and storage of information in databanks.
The introduction of a powerful new technology is likely to set up expectations that might be unwarranted or unrealistic in practice. Various expectations regarding DNA typing technology are likely to be raised in the minds of jurors and others in the forensic setting. For example, public perception of the accuracy and efficacy of DNA typing might well put pressure on prosecutors to obtain DNA evidence whenever appropriate samples are available. As the use of the technology becomes widely publicized, juries will come to expect it, just as they now expect fingerprint evidence.
Two aspects of DNA typing technology contribute to the likelihood of its raising inappropriate expectations in the minds of jurors. The first is a jury's perception of an extraordinarily high probability of enabling a definitive identification of a criminal suspect; the second is the scientific complexity of the technology, which results in laypersons' inadequate understanding of its capabilities and failings. Taken together, those two aspects can lead to a jury's ignoring other forensic evidence that it should be considering.
As large felon databanks are created, the forensic community could well place more reliance on DNA evidence, and a possible consequence is the underplaying of other forensic evidence. Unwarranted expectations about the power of DNA technology might result in the neglect of relevant evidence.
The need for international cooperation in law enforcement calls for appropriate scientific and technical exchange among nations. As in other areas of science and technology, dissemination of information about DNA
typing and training programs for personnel likely to use the technology should be encouraged. It is desirable that all nations that will collaborate in law-enforcement activities have similar standards and practices, so efforts should be furthered to exchange scientific knowledge and expertise regarding DNA technology in forensic science.
Recommendations
-
In the forensic context as in the medical setting, DNA information is personal, and a person's privacy and need for confidentiality should be respected. The release of DNA information on a criminal population without the subjects' permission for purposes other than law enforcement should be considered a misuse of the information, and legal sanctions should be established to deter the unauthorized dissemination or procurement of DNA information that was obtained for forensic purposes.
-
Prosecutors and defense counsel should not oversell DNA evidence. Presentations that suggest to a judge or jury that DNA typing is infallible are rarely justified and should be avoided.
-
Mechanisms should be established to ensure accountability of laboratories and personnel involved in DNA typing and to make appropriate public scrutiny possible.
-
Organizations that conduct accreditation or regulation of DNA technology for forensic purposes should not be subject to the influence of private companies, public laboratories, or other organizations actually engaged in laboratory work.
-
Private laboratories used for testing should not be permitted to withhold information from defendants on the grounds that trade secrets are involved.
-
The same standards and peer-review processes used to evaluate advances in biomedical science and technology should be used to evaluate forensic DNA methods and techniques.
-
Efforts at international cooperation should be furthered, in order to ensure uniform international standards and the fullest possible exchange of scientific knowledge and technical expertise.