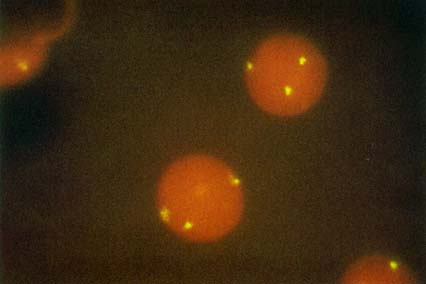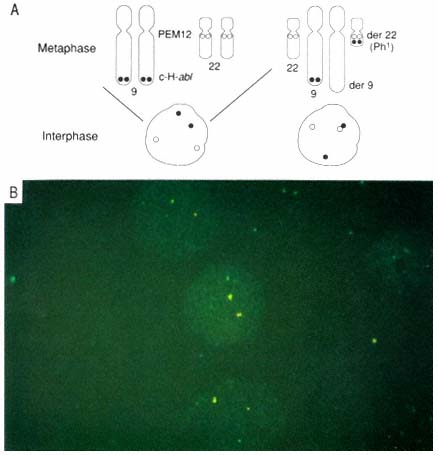
PLATE 1 A. Illustration showing hybridization patterns for normal and CML metaphase and interphase cells. Closed circles represent red signals from c-H-abl (ABL probe) and open circles represent green signals from PEM12 (BCR probe). The left side of the figure shows a normal metaphase with c-H-abl staining near the end of 9q and PEM12 on proximal 22q. The corresponding interphase hybridization pattern shows random placement of all four signals. The right side of the figure shows a classic Ph1 in CML. The BCR-ABL fusion is represented by one set of red and green signals in close proximity in metaphase and interphase. B. Fluorescence in situ hybridization in interphase nuclei with ABL (red) and BCR (green) probes visualized simultaneously through a double band-pass filter. Cells from a CML patient 46,XY,t(9;22)(q34;q11), showing the red-green (yellow) signals resulting from the hybridization to the BCR-ABL fusion gene and the single red and green hybridization signals to the BCR and ABL genes on the normal chromosomes 22 and 9. (Tkachuk, D.C., et al. Detection of bcr-abl fusion in chronic myelogeneous leukemia by in situ hybridization. Science 1990; 250:559-562. Copyright, AAAS. Used with permission.)
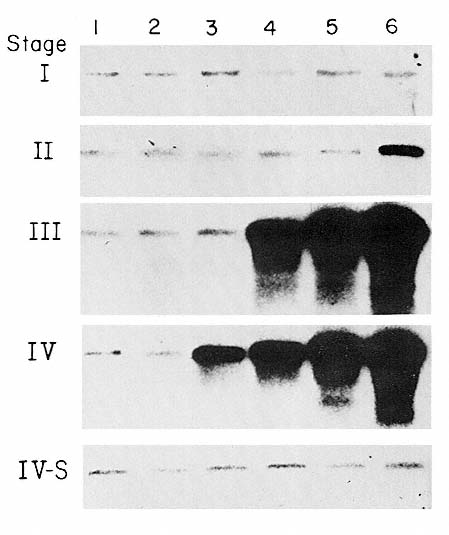
PLATE 3 Autoradiograms of DNA from untreated primary neuroblastomas. After DNA was prepared from the tumors, six tumors representative of each clinical stage were grouped for electrophoresis, blotted, and hybridized with the pNB-1 probe for MYCN. The proportion with MYCN amplification was similar to the proportion in the total 89 tumors tested. Stage I tumors: all have only single-copy level of MYCN. Stage II tumors: only the sixth tumor has MYCN amplification (sixfold). Stage III tumors: three of six have amplification (50-to 200-fold). Stage IV tumors: four of six have amplification (20-to 200-fold). Stage IV-S tumors: none of the first five tumors has MYCN amplification; the last sample in this row is from a ganglioneuroma. [Seeger, S.C., et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. Reprinted, by permission of the New England Journal of Medicine 1985; 313(18):1111-1116.]
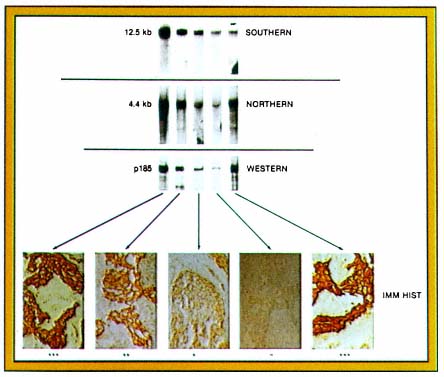
PLATE 4 Correlation between ERBB2 gene amplification and expression. Southern blot analyses show the 12.5-kb ERBB2 band seen with Eco RI cut DNA. Northern blot analyses show the 4.5-kb ERBB2 transcript. Western blot analysess how the 185-kD ERBB2 protein band. Immunohistochemical analysis was performed with anti-ERBB2 specific antibody and frozen sections. (Slamon, D.J., et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989; 244:707-712. Copyright, AAAS. Used with permission.)