4
Building Collaboratories for Molecular Biology
This chapter discusses how to facilitate some aspects of molecular biology research from the viewpoint of a computational biologist or an information scientist interested in constructing biological information systems. Electronic collaboration in molecular biology is facilitated currently by a variety of shared database systems. In addition several prototypes that support collaboration are in use now, and a model collaboratory, the Worm Community System, illustrates a potential future direction for community information systems that support retrieval, analysis, and sharing of essential data and literature.
Molecular biology is the analysis and description of biological organisms at a molecular level. To attempt to relate the observed function to the underlying structure, an immense amount of data must be considered, at many different levels, from studies of many different organisms. Molecular biologists have made many important theoretical advances from such comparisons, notably elucidation of the central role of DNA (deoxyribonucleic acid) as the carrier of genetic information in all free-living organisms.
A DNA molecule provides the "blueprint for life." It is composed of combinations of 3 billion pairs of four chemicals known as nucleotides: adenine and thymine, or guanine and cytosine (Box 4.1). To simplify, the DNA in humans is divided up into 24 different chromosomes. About 10 percent is aggregated into genes, which code for proteins. The other approximately 90 percent has no known function. The complete amount of DNA that defines any living organism is known as its genome.
Molecular biology has traditionally been conducted on a small scale. Most laboratories have been small and have functioned independently, employing a handful of people and producing a moderate amount of data, often on a subject exclusive to each laboratory. Further, many experiments have been easily reproducible, requiring only modest equipment. In contrast to other scientific domains, such as space physics, direct sharing of results from experiments has not been critical, since the experiments could be simply and cheaply rerun. The level of detail in the published literature has been considered sufficient for the sharing of knowledge and for collaboration between laboratories. However, large-scale nucleotide sequencing directed at the human genome (3 billion nucleotides!) may call for centralized, highly automated facilities and new modes of deciding on the division of labor and of disseminating data.
From both a theoretical and practical perspective, molecular biology has been one of the great success stories of modem science. More than 60 percent of all National Institutes of Health grants—covering a wide range of problems in medicine and biology—propose use of the molecular methods of cloning, mapping, or sequencing to investigate problems. More than 70 percent of all articles indexed by the National Library of Medicine contain molecular biology subject headings.
At this time, the field of molecular biology is diverging. One branch addresses a range of integrated biological systems, concentrating on the function of protein molecules in the processes of DNA replication and mutagenesis, transcription, and translation.1 This branch entails a high quota of individual imagination in theory building. The other branch, typified in Walter Gilbert's description of an emerging paradigm shift in biology (Box 4.2), concentrates on the role of DNA in the cell. It is this side of molecular biology that is data intensive and is embodied in the Human Genome Project, a burgeoning scientific endeavor producing vast amounts of new information that researchers must select from to advance their investigations.
|
BOX 4.1 FUNDAMENTALS OF GENOME RESEARCH ''Molecular biology is the discipline that demonstrated the relationship between genes and proteins. Molecular biologists determined that the gene is made of DNA (deoxyribonucleic acid)—that is, DNA is the hereditary material of all species. What is more, in what is now scientific legend, Crick and Watson determined in 1953 that the structure is a double helix and concluded correctly that this specific form is fundamental to DNA's function as the agent of storage and transfer of genetic information. In fact, in biology generally, shape determines properties—that is, structure almost always determines function. ''The DNA double helix is both elegant and simple. Each strand of the DNA double helix is a polymer consisting of four elements called nucleotides: A, T, C, and G (the abbreviations for adenine, thymine, cytosine, and guanine). The two strands of DNA are perfectly complementary: whenever there is a T on one strand, there is an A on the corresponding position on the other strand; whenever there is a G on one strand, there is a C on the corresponding position on the other. That is, T pairs with A, and G pairs with C. This complete redundancy accounts for how a cell can pass on a complete set of genetic information to each of its two daughter cells during cell division: the DNA double helix unravels, and each strand serves as a completely sufficient template upon which a second strand can be synthesized. In addition to providing an easy mechanism for the replication of DNA, the redundancy also provides great resiliency against loss or damage of information during the life of a cell. Such loss or damage of information, when it occurs, is the basis for biological mutations. "From a computer scientist's point of view, the DNA double helix is a clever and robust information storage and transmission system. As Computer scientists accustomed to dealing with a binary alphabet will immediately recognize, the four-letter alphabet of DNA is sufficient for encoding messages of arbitrary complexity. "In brief, particular stretches of the DNA are copied directly into an intermediate molecule called RNA (ribonucleic acid, also composed of A, T, C, and G). RNA is then translated into a protein—which is again a linear chain, but one assembled from 20 different building blocks called amino acids. Each Consecutive triplet of DNA elements specifies one amino acid in the protein chain. In this fashion, biology "reads" DNA (actually, the RNA copy of the DNA) ... [as if it were a computer program]. "Once synthesized, the protein chain folds according to laws of physics into a specialized form, based on the particular properties and order of the amino acids (some of which are hydrophobic, some hydrophilic, some positively charged, and some negatively charged). Although this basic coding scheme is well understood, biologists are not yet able to predict accurately the shape in which the protein will fold. "In total, the human genome (the totality of genetic information in each of us) contains about 3 billion nucleotides. These are distributed among 23 separate strands called chromosomes, each containing about 50 million to 250 million nucleotides. Each chromosome encodes about 10,000 to 50,000 genes. "With the extraordinary advances in molecular biology over the past 20 years, it is now possible to read the specific sequences of individual genes and to predict (by means of the genetic code) the sequence Of the proteins that they encode. A major challenge for molecular biology in the next decade Will be to use this information to predict the actual biological function of these proteins." SOURCE: Reprinted, with permission, from Lander et al. (1991), p. 35. |
|
BOX 4.2 TOWARD A PARADIGM SHIFT IN BIOLOGY "The new paradigm, now emerging, is that all the 'genes' will be known (in the sense of being resident in databases available electronically), and that the starting point of a biological investigation will be theoretical. An individual scientist will begin with a theoretical conjecture, only then turning to experiment to follow or test that hypothesis.... "To use this flood of knowledge [that is resulting from the mapping and sequencing of human genes and the genes of model organisms], which will pour across the computer networks of the world, biologists not only must become computer-literate, but also change their approach to the problem of understanding life. "The next tenfold increase [5 years from now] in the amount of information in the databases will divide the world into haves and have-nots, unless each of us connects to that information and learns how to sift through it for the parts we need.... "We must hook our individual computers into the worldwide network that gives us access to daily changes in the database and also makes immediate our communications with each other. The programs that display and analyze the material for us must be improved—and we must learn how to use them more effectively. Like the purchased kits [of molecular biological reagents], they will make our life easier, but also like the kits, we must understand enough of how they work to use them effectively." SOURCE: Reprinted, with permission, from Gilbert (1991), p. 99. |
GENOME RESEARCH IN MOLECULAR BIOLOGY
Within molecular biology, the importance of sharing data and the scale of research have increased for activities revolving around gene mapping and sequencing. The widespread use of automated DNA sequencing technology2 is now permitting the nucleotide sequence for many genes and chromosome regions to be determined. This enormous amount of information has brought centralized database archives to the fore as major points of collaboration. The main data produced in genome projects are the sequences of DNA base pairs embodying patterns of great evolutionary, physiological, and medical interest. The discovery of meaningful patterns is complicated: first, the data available to support a particular line of research are often sparse, so that many different organisms (e.g., bacteria, yeast, worms, mice, and humans) must be studied, and second, the available theory of the organization of DNA sequences remains scant, so that hands-on experimentation by scientists is required.
As experimental results, DNA sequences themselves are useful to share directly, since similarities in sequence often imply similarities in function. However, the printed literature is no longer adequate for sharing such data, partly because of the economics of charges for the journal pages needed to print the long sequences, but largely because computer examination of the sequences is far more effective than human examination of the data on a printed page. As a result, it has become standard procedure for investigators to use sequence databases such as GenBank and mapping databases such as the Genome Data Base in their research and to submit new results to be incorporated into the databases.
From a computing-oriented perspective, genome analysis is a data-driven science in which researchers must search massive amounts of data for the specific information they need to interpret their experimental results and plan new experiments. The unique product of a gene is a functional protein (such as an enzyme or a structural complex). Many more genes are known than the corresponding proteins; it takes person-years of effort to purify a protein, to understand its function, or to determine its three-dimensional structure. Such research conducted on the current scale would be nearly impossible without computing and information technology.
Such recent major efforts as the Human Genome Project, which seeks to map3 and sequence all human genes, promise to generate data on a scale unprecedented in the history of molecular biology.
Using databases to store, access, compare, and analyze genetic data will thus become even more essential in the future than it is at present. In particular, the rapidly expanding body of sequence data makes it increasingly valuable to analyze the data effectively for functional patterns. There are already striking examples of how sequence analysis has provided insight and stimulated experimentation that would otherwise not have been an obvious line of investigation.
TRENDS IN TECHNOLOGY TO SUPPORT COLLABORATIVE GENOME RESEARCH
The anticipated avalanche of genome information makes it imperative that researchers have tools that will support using the data effectively to conduct research (see Box 4.2). Facilitating the capability to predict new relationships from existing data is an area ripe for the application of computer technology (Lander et al., 1991). Applications so far have concentrated on database management and on simple pattern analysis to enable homology searches. In contrast to space physics and oceanography, modeling and simulation have not been applied extensively in genome analysis.
Given that data from many laboratories are typically required to determine, for example, the part a gene may play in a particular function, communication and some degree of collaboration are necessary among genome researchers. Before computers were used, data were gathered from the literature and by direct contact with colleagues in other laboratories. Today, however, the shared knowledge necessary to achieve progress in genome research is often transferred via communal databases and other electronic modes of dissemination.
The traditional method for sharing new knowledge with other scientists has been to publish articles in the journal literature. In genome-related research, concise presentation of experimental results in fast-publication literature is emphasized. A typical article is 6 pages long and, if accepted, is published 3 to 6 months after submission. In addition to the results published in the traditional journal literature, the results of an experiment may include map or sequence data that are contributed to electronic databases.4 The primary advantages of publishing in electronic format are that it is incrementally less expensive than publication in journals, allows searching and keeping data current to be done more easily, and enables the use of computer-based analysis programs.
In the foreseeable future, advances in support for collaborative genome analysis will likely come from enhancing scientists' interaction with databases. Integrated systems will not only store and retrieve data, but are also expected to facilitate the analysis of data and literature.
CURRENT DATABASE SYSTEMS
The computer and communications support for genome research focuses on networked databases for the submission, editing, retrieval, and analysis of data. Many databases have become essential tools for genome researchers, a status reflected in the creation of national facilities to maintain them. At present, the most important of the standard archival databases each contain a specific type of information. Historically, literature databases (e.g., MedLine) were generated first, followed by data archives for sequences (e.g., GenBank) and for maps (e.g., Genome Data Base).
These archival databases are becoming a new kind of scientific literature, and each has been faced with implementing aspects of the publishing process in electronic form. MedLine developed methods for electronic searching and retrieval of bibliographic information, GenBank developed methods for direct electronic data submission, and the Genome Data Base developed techniques for electronic checking of data by individuals at workstations distributed literally around the world.
Each of these projects costs several million dollars per year to operate. Although each is based at least in part on commercial database technology, all have required development of specific application software running to the hundreds of thousands of lines of code. The costs of operating these databases
reflect what is necessary to develop, maintain, and distribute the application software, to encode and check the data, and to provide appropriate user support and documentation. These databases afford economies of scale for genome researchers.
MedLine
The National Library of Medicine (NLM), a part of the National Institutes of Health, is the world's largest special-collections library. Its primary electronic collection is MedLine, which includes abstracts and bibliographic citations, and permits on-line searching of electronic versions of a wide range of journal articles of interest to practitioners and researchers in medicine and biology. Developed in the 1960s when computing and communications technology first began to enable remote information retrieval, MedLine is now accessed by essentially every molecular biology laboratory in the country as a matter of course, either through a centralized library subscription or an individual laboratory subscription.
MedLine currently includes over 6 million citations and is expanding rapidly; in 1990, for example, some 400,000 citations were added from 3,600 journals selected by an advisory committee. A group of professional indexers examines selected issues and generates citations for each article of biomedical interest (the abstract from the primed article, bibliographic information including author, title, journal, and volume and page numbers, plus classification information such as relevant terms from the standard NLM Medical Subject Heading (MESH) thesaurus). These citations are then entered into the MedLine database.
For genome researchers and other scientists from all over the country who can routinely search the MedLine database, MedLine is a familiar example of an interactive database program with a front end running on their laboratory computers and the back end on a remote machine across the network. The front end is a simple text-based interface, the network is the telephone network accessed with a modem, and the back end provides simple Boolean word searching. Recently, MedLine has been made accessible via Internet. The MedLine database is also made available by other on-line search services such as Dialog and Bibliographic Retrieval Service. With the advent of CD-ROMs, many libraries and even some individual laboratories have purchased readers to run retrievals locally and save on-line charges. Typically, these collections are updated monthly rather than daily.
GenBank
Use of the centralized database GenBank has become routine for genome researchers. Whenever a gene is isolated and sequenced, it is compared with the sequences in the database in the hope of identifying its function by finding another gene of known function and similar sequence. As MedLine's capability for on-line literature searches has become essential to researchers, so also are GenBank's on-line data now essential to scientists who map and sequence genes.
GenBank began in the 1970s as a collection of nucleic acid sequences gathered by physicist Walter Goad, who was interested in sources for testing pattern-analysis algorithms. By the late 1970s, the collection itself was proving useful to biologists for making functional comparisons, and several striking discoveries were made directly from the database (Burks, 1985). In 1982, the Los Alamos Sequence Library became GenBank, the national repository for nucleic acid sequence data. Official responsibility has now been transferred to the National Library of Medicine, where the database will be maintained by the National Center for Biotechnology Information (NCBI) in collaboration with Los Alamos.
Searching of the GenBank database rarely involves manually examining many sequences with a coarse search criterion; instead, an analysis program using some form of similarity matching is utilized. Because such analysis programs tend to be slow and the database is still relatively small, GenBank is
commonly reproduced in its entirety at each local site and the copy is updated weekly or even daily from the central archive. At many large universities, the central biotechnology computing facility maintains a copy of GenBank that researchers can access from a computer in their laboratory over the campus local area network.
Initially, GenBank entries were generated in much the same way as MedLine entries. A group of indexers read the published literature, identified articles containing sequence data, and entered items into the central database. An item was like a citation; it consisted of a sequence plus a variety of annotations, including author, organism, methods, and possible functions. The central annotators typed in the sequence and entered all annotations, an approach that worked well for the first few years. However, the extraordinary growth in the number of sequences generated soon made this strategy less attractive, owing partly to the difficulty of accurately entering a large volume of hard-to-read data and partly to the changing economics of publishing whereby journals began to restrict the amount of sequence data that could be published.
Essentially all submissions to GenBank are now made electronically (Box 4.3). A program available at no cost to scientists provides an interactive form for entering the sequence data and the appropriate annotations and then electronically mailing this information to the central archive. Individual researchers as well as large-scale sequencing projects submit data to GenBank, and essentially every sequence submitted is incorporated into the database. Supporting automated submission has greatly increased the currency and accuracy of the database. In addition, although they exercise no editorial control, GenBank's curators run a series of tests (increasingly automated) on the data to check for accuracy, and they contact the authors when anomalies are found. The combination of electronic submission, which tends to eliminate transcription errors, and electronic checking, which tends to catch further anomalies, has significantly increased the level of review that data receive before being made public.
|
BOX 4.3 ELECTRONIC DATA PUBLISHING AND GENBANK From a review article by the administrators of GenBank, the national repository for DNA sequence data: "Several years ago, in an effort to keep up With an exponentially increasing flow of nucleotide sequence data, we began looking at ways of combining two evolving technologies (namely, database management software and computer networks) into a more effective system for the communication of scientific data. We have designed and implemented a specialized form of electronic publishing, which we term electronic data publishing, where data (in our case DNA sequences and related annotation) are gathered, processed, and distributed electronically, Rather than compete with traditional scientific publications, electronic data publishing is designed both to complement and to support printed publications.... "... At the practical level, electronic data publishing at GenBank has been quite successful. ... At a more conceptual level, however, some larger issues bear discussion. In particular, we must consider questions such as the peer review (and thus the quality) of submitted data and the academic credit that will be associated with database entries.... Our experience indicates that electronic data publishing has actually resulted in a higher level of quality for data in GenBank and therefore also in the journals.... "... [H]aving journal editors encourage or require Submission Of data before publication of a paper has been met with far greater acceptance on the part of authors than might have been anticipated. We are currently receiving approximately 80% of our data as a direct electronic submission before publication of any related paper. The research community will always. decide de facto the relative importance of the generation of data; what we (and Others who have acted as proponents of direct submission) have accomplished is the establishment of a system that researchers are coming to view as the natural mechanism for the communication of large amounts of scientific data." SOURCE: Cinkosky et al. (1991), pp. 1273, 1276. |
The establishment of the paradigm of electronic data publishing for GenBank illustrates some of the broader potential for collaboration technology. The most important aspect of the shift to electronic data publishing is recruiting of the community of researchers to perform the job of sequence entry and annotation and database building. This paradigm supports growth with resources that scale to the size of the community. The scientists who have determined the sequences are also the most appropriate people to annotate their own work, given a controlled semantics and syntax. The responsibility of the database managers is to specify the structure of the data entries and provide the community with the tools to generate entries according to these specifications. These tools also allow automatic checking of data, semantics, and syntax as well as writing a relational transaction for electronically entering data into the database.
Most molecular biology journals will not publish articles discussing sequences without a GenBank accession number, thus guaranteeing that the database is the medium of record. The GenBank staff worked closely with journal editors to develop policies requiring that data be submitted to a database before a related article could appear in print (Cinkosky et al., 1991).
The interval between discussion of a sequence in the journal literature and appearance of the sequence in the GenBank database has shrunk dramatically, from nearly 2 years to a few days. This reduction has paralleled an increase in the fraction of electronically submitted entries from 10 percent to about 90 percent. In 1993, the database will be 20 times the size it was 5 years ago. GenBank currently processes more sequence data in a month than it held in its entire database in 1987. This 10-fold increase in production was made possible by leveraging the efforts of the entire genome research community.
Electronic data publishing has also simplified collaboration with subscribers to other international databanks (e.g., the European Molecular Biology Laboratory Data Library and the Database of Japan, each of which exchanges new entries daily via the Internet. The currency of its database has made the GenBank On-Line Service a very valuable research tool, and daily GenBank updates are the critical reason that many molecular biologists have become part of the Internet community. Hence GenBank, a collaborative resource, has stimulated yet further collaboration.
The GenBank experience has shown that, with appropriate technological support and sufficient sociological incentives (such as the requirement by journals that sequences referenced in papers be deposited in a public database), the biological community itself can become actively involved in submitting information to and retrieving it from an archival database. The Human Genome Project promises to greatly increase the scale not only of data generated but also of scientists involved in direct maintenance of electronic archives. Computer support for a larger portion of the publishing process, in particular, editing and checking functions for quality control, can enhance the currency and accuracy of data made available to the molecular biology community.
Genome Data Base
In its initial phases, the Human Genome Project is concentrating on the sequencing and mapping of model-organism and human genes. The Genome Data Base (GDB) has been designated as the central database for human gene mapping data. Closely associated with the Welch Medical Library and maintained at the Johns Hopkins University Medical School, the GDB is operated in conjunction with the On-line Mendelian Inheritance in Man project, which provides ready access to detailed information about human genetic phenotypes and diseases.
Although the relational database underlying GDB contains more than 250 tables, GDB may be envisioned as containing data on four major classes of biomedical objects (map objects, proposed maps, mapping reagents, and clinical phenotypes) and two classes of supporting objects (references and people). In addition, many dependent object classes are also represented, such as alleles, polymorphisms, and populational allele frequencies. At the moment all of the data are stored in a commercial relational database, as with GenBank, and retrieved via simple text forms. It is expected that more sophisticated
graphical interfaces will become available, both from GDB and from other third-party developers, in the near future.
Data may be entered into the system through a variety of mechanisms, including direct submission by authors, entry by GDB staff, or input by editors or their assistants. The editors then guide the data through an approval process. Editorial control is explicitly supported by the software developed and maintained by the GDB project. A board of editors ensures that the database represents a consensus of checked mapping data. One or more editors act as referees for incoming data on each of the 24 human chromosomes. Other editorial groups address issues involving nomenclature, clinical phenotypes, and comparative mapping. Each editor has electronic access to GDB, which provides special interfaces and underlying functionality to facilitate editorial work. A complex system of approval flags and message files allows editorial groups to communicate with each other regarding aspects of the approval process for a particular piece of data. All editorial changes are logged, and users may call up and view at any time the editorial history of any entry.
Data transmission and interaction within this publishing process are supported electronically, although the actual decisions are made by the (human) editors. Such an environment enables the system to support the many laboratories that are major players in the genome mapping and sequencing project.
FUTURE INFORMATION SYSTEMS-TOWARD A WORKING COLLABORATORY
Steady progress has been made in increasing the functionality of computer support for collaborative research. MedLine, a product of the 1960s, provides remote interaction with a database generated at a central site. GenBank, a product of the 1980s, provides for electronic submission of data generated at laboratories across the international community and then sent to a central site for editing and redistribution. The Genome Data Base, a product of the 1990s, has developed electronic editing, with editorial activities also distributed and supported electronically.
To continue to provide increasing functionality to support collaboration in genome research, future information systems must address a number of needs. First, since an abundance of data is now available on-line, ways are needed to conveniently pass selections in and out of a variety of analysis programs. Second, since many different sources of data are now available, ways of establishing links between related items in different databases are required. Finally, a complete cycle of electronic publishing—from entry to editing to distribution of data—is needed not only for formal archival material but also for the informal community material that is also vital to science. In a sense, such a cycle brings the control of data generation, curatorship, and distribution directly to the scientific community. The next generation of support for collaborative genome research will include comprehensive computer environments for analyzing data. Such systems will additionally begin to support computational biology, in which substantial parts of some experiments can be done via computer programs that access databases and apply analysis packages.
The first step in supporting collaboration is to invoke analysis remotely across a network, as is done in literature searches that retrieve from MedLine abstracts that contain specified keywords.
The next step is to support analysis across multiple sources. Each data source represents a different experimental method, point of view, or level of knowledge. Interrelating these sources is an essential component of genome research, largely because the available knowledge is now so incomplete. To obtain as much information as possible about the biological function of the genes they are studying, researchers consult and interrelate gene lists, genetic maps, DNA sequence data, formal and informal literature, and other sources of information. Often they need to cross-compare among organisms, since different organisms have become models for studying different functions. Interconnecting different sources is accomplished by making associations between similar items from different databases. For example, standardized nomenclature, such as gene names, can be used as keys to associate aspects of the
genetic maps of different organisms. In a more generalized sense, searches that discover semantic similarities can be used to associate related items.
The final step in the next generation of support for collaboration in genome research is to support a complete system for the genome research community that combines the association of information from multiple sources provided by similarity analysis software with the range of knowledge provided by a complete electronic publishing environment. Such a community system, or collaboratory, will capture all of the specialized knowledge needed by a geographically distributed community of researchers, including informal, more detailed sources of information, and the quality-assurance mechanisms needed to check these items. Scientists will then interact transparently with this knowledge across the national networks, both retrieving and analyzing existing information, as well as submitting and publishing new knowledge. The goal is to extend the publishing process into a distributed electronic medium to enable the whole community of genome researchers to contribute to and edit its knowledge. This functionality is available in research prototypes discussed below.
Basic Components and Research Prototypes
Remote Analysis Servers—Blast
To implement a complete analysis environment, it is first necessary to support the transparent application of analysis programs. A remote analysis server enables scientists to use large, remote computers to analyze their data; on-line use of MedLine provides a simple version of such a server. A version with modern technology is commonly used at the National Science Foundation's supercomputer centers to enable scientific visualization: an interactive graphical front end runs on a personal computer to enable the user to issue commands and display results; this is connected via the Internet to a powerful back-end supercomputer running a sophisticated, computationally intensive numerical simulation.
A research version of a remote analysis server for genome research is the "Blast" server maintained by the National Center for Biotechnology Information (NCBI) at the National Library of Medicine. "Blast" is one of the best current sequence analysis programs (Altschul, 1990); given a sequence, it produces a rank-ordered list of the sequences in the database that are most similar under a set of biological heuristics. NCBI has set up a continuously running server for Blast on its fast parallel machine, using the daily updated GenBank database. An Internet electronic mail client enables biologists to submit their sequences, have them mailed electronically to NCBI, and have the results mailed back, usually in a matter of minutes.
A good illustration of how scientific collaboration can be facilitated by computer technology is the expressed sequence tag (EST) project (Box 4.4), which has the Blast server as an essential component (Adams, 1991; Fields, 1992).
Interconnecting Archival Databases—Entrez
A complete analysis environment must enable analysis of the similarity of relationships among items not only within a single database, but also across multiple databases. To ensure that multiple sources can be easily interlinked, it is necessary to define a standard format for passing items between analysis programs.
To facilitate interconnection between the literature database MedLine and the sequence database GenBank, indexers (human librarians) at the National Library of Medicine are placing standard links between related items in the two databases. A field for every MedLine entry specifies referenced sequences, and a field for every GenBank entry specifies referenced literature. The professional indexers
|
BOX 4.4 A NEW COLLABORATION EVERY DAY: IDENTIFYING HUMAN GENES USING AUTOMATED SEQUENCERS AND THE INTERNET Cloning, sequencing, and characterizing the biological function of a gene are long, slow processes requiring many different types of biological expertise. In 1990, J. Craig Venter at the National Institute of Neurological Disorders and Stroke developed a shortcut method for identifying genes: using automated DNA sequencers to sequence a few hundred bases from one end of a complementary DNA (cDNA) clone of the gene's messenger RNA (mRNA) transcript. The resulting "expressed sequence tag" (EST) is a unique identifier for the gene. ESTs generated from randomly chosen cDNA clones effectively provide a Clone and a partial sequence of a gene in a single experiment. In many cases, the gene family to which a newly cloned gene belongs can be identified by comparing the EST sequence to the known sequences contained in databases such as GenBank. The National Center for Biotechnology Information's "Blast" Internet server and Oak Ridge National Laboratory's GRAIL server are key resources in this analysis process. In the last 2 years, the EST project has published sequences of thousands of human genes, developed a database of sequences and analysis results that is available over the Internet, and provided the clones from which these ESTs were derived to the research community via the American Type Culture Collection. Venter's laboratory, now at the Institute for Genomic Research, is sequencing thousands of new ESTs per month. Each EST raises the possibility of a new scientific collaboration to characterize the function and assess the biological and medical significance of a new human gene, and hundreds of requests for sequences, data, clones, or additional information have been answered by the laboratory. In some cases, members of Venter's laboratory participate in a collaborative follow-up analysis, doing additional sequencing or other experiments. Several human genes related to the "discs-large" tumor suppressor gene in drosophilia, for example, are being pursued in Collaboration with colleagues at the University of California, Irvine and San Francisco. In many cases, data and materials are sent for others to pursue independently. Either way, the rapid communication facilities provided by the Internet allow quantities of data that could not feasibly be published in print media to be exchanged quickly and efficiently between colleagues around the United States and in many other countries. SOURCE: Adapted from Fields (1992). |
specify these links when they specify classification and annotation information. Entries in both databases have unique identifiers so that the links can be uniquely and permanently specified. Once the links have been established, software can be written to enable retrieval of related items by following the relationship links. For standard archival sources, such explicit manual specification of interconnections promises great utility.
There are many archival sources in molecular biology, covering the many different experimental aspects of the field. Currently a few such sources, maintained by a single library, have an explicit set of maintained links. For other sources, the links must be maintained automatically or semi-automatically by attempting to match "common" items in different databases. For example, both DNA sequence and protein sequence databases often mention the corresponding gene name for a given organism. These common names could be used automatically as points of interconnection if there were a standard nomenclature for naming genes. These implicit (discovered) links will likely be far more numerous than the explicit (manual) ones.
Standardized formats are also necessary before name matching can be done. The fields representing the annotations of a sequence, for example, must be of the same set before their corresponding values can be matched. A program must be able to tell, for example, that the "gene name" field in a DNA sequence database is the same as the "function name" field in a protein sequence database. To describe data types in molecular biology, the National Center for Biotechnology Information (NCBI) is using the International Organization for Standardization standard language ASN.1 (Abstract Syntax Notation) to describe data fields and their values.
The NCBI ASN. 1 toolkit provides a variety of parsing programs and a set of semantic definitions for the values for the most common types of genome data, such as literature and sequences (Gish et al., 1992). While ASN. 1 provides a standard interface for the programs of the NCBI toolkit, it is not broadly implemented in other information processing environments in molecular biology. However, authors of analysis and display software are gradually adopting ASN. 1 as their external publication format for data exchange, partly as a result of NCBI's stature within the community. Adoption of a standard interface for passing data between analysis servers would greatly facilitate development of a complete analysis environment. For example, any server that manipulates DNA sequence data should be able to pass and return the same representation for a sequence. Ideally, analysis environments should need to provide an invocation procedure only for each data type, rather than for each data server—a feature that is more feasible for genome data given that the number of data types is fairly small.
A good illustration of a software package utilizing this technology is Entrez (Gish et al., 1992), which currently supports interactive cross-comparison of data in MedLine and GenBank. The base data are the data in MedLine and GenBank and the relationship links explicitly established between entries in these two databases. The base software is the Blast server. Users can specify desired keywords and retrieve related documents from MedLine and can then follow related links from the MedLine documents to a set of GenBank sequences, run the sequence searcher to find a set of similar sequences, follow the related links from this new set back to a set of related literature documents, run the text searcher to find a set of related documents, and so on, thus engaging in an iterative process of pattern matching to make connections that could not be discovered using a single database analysis.
Systems like Entrez will soon include a wider variety of databases in molecular biology. For example, NCBI is already expanding Entrez to include protein databases. Increasing the number of databases in integrated environments increases the possibility of discovering important similarity matches or patterns that might not have been apparent otherwise.
Community Information Systems
A complete community information system to support research would encompass an electronic library containing all sources of data, software enabling interactive display and analysis of all data types, and an underlying information infrastructure that transparently supports extensive facilities for browsing, filtering, and sharing knowledge across the international Internet. Building working models of such a complete collaboration system is possible today and is actually being done in an ongoing research project, the Worm Community System.
Basic Elements and Capabilities
The first step in building a community information system is to decide the extent of the knowledge to be captured, which depends in turn on the needs of the particular community whose work is to be facilitated. Not all of the elements in the scientific process described in Chapter 1 are equally important in genome research. For example, investigators rarely wish to examine actual raw data and are satisfied if the maintainers of instruments, e.g., those in the large sequencing projects, keep the raw data streams and release only a final version to the archival databases. Because experiments in genome research typically involve only a few people or only loose cooperation between different laboratories, the need for real-time cooperative work tools is relatively small, and support for collaboration should focus on retrieval and analysis of the archival data and literature, and on extending the same facilities to more informal material that is critical to progress in science.
In genome research, this informal material typically includes results that are not yet publishable in journal articles but may be discussed in newsletters (e.g., giving one- to two-page descriptions of
current experiments) and in conference proceedings (e.g., giving abstracts of new results). It also typically includes data too specialized to be publishable in archival databases—such as strain lists (local sets of mutant organisms) and restriction maps (detailed locations of genes)—yet still extremely useful to other researchers working on similar problems. These informal results are usually stored in individual laboratories inside filing cabinets or taped to the backs of doors and are usually retrieved from the outside only by telephoning the laboratory. Electronic sharing of this knowledge would make it available to a much wider set of potential collaborators.
To collect and check this informal knowledge, the appropriate biological community must be directly involved. The experience with GenBank demonstrates that biologists conducting genome research will electronically submit material that is deemed useful, as does the extensive experience of scientists with electronic bulletin boards such as Netnews. The experience with the Genome Data Base shows that electronic support for distributed editors (human editors to check the quality of materials) is workable with community-chosen curators of the individual data sources. A complete electronic publishing environment will support entry of all the specific types of knowledge and publishing in all the different styles. With such a system, for example, an investigator will be able to run a program that enables interactive specification of a restriction map or preparation of a newsletter article and then automatically submits it to a central archive, which then automatically distributes it to the community. Local maps might be unchecked and newsletter articles might be moderated (checked for topic), as opposed to centrally archived data or literature that must be checked for quality and consistency.
The process of associating and relating items from myriad formal and informal sources involves major technological and sociological complications. Interconnection of informal sources of information depends on implicit associations being made automatically, e.g., by parsing the text of newsletter articles to locate gene names with which to make associations. In a community information system, interconnection is facilitated by community members themselves, who may add most associations. That is, the users of the system can specify their own associations between items, including associations discovered while using the system itself.
A distributed system is required to support sharing of information in a scientific community, since the users and the generators of knowledge are geographically distributed. National networks are expected to be a necessary component of a community information system or analysis environment, even if the total amount of knowledge is small. A community information system should present users with what appears to be a single logical ''database'' for retrieval and storage of data.
A Model Collaboratory—Worm Community System
A model of a complete community information system is under active development in the Community Systems Laboratory at the University of Arizona (Box 4.5). This system represents a substantial operational model of a collaboratory. The community is the collection of molecular biologists who study the nematode worm Caenorhabditis elegans; the system itself is called the Worm Community System. The size of the community and the amount of data are manageable—large enough to be interesting from the perspective of data organization and management yet small enough to be doable. The community's approximately 500 members are spread across the United States, Europe, and Japan. Significant sources of data already exist in electronic format, with a range of types and location. Community members have a long tradition of openness and sharing of data.
The underlying technology of a community information system such as WCS is based on a representation called an information space (Schatz, 1987, 1989) that supports transparent manipulation of objects from multiple, heterogeneous, distributed sources and is thus a form of a federated object-oriented database. The goal is to enable users to manipulate data items from many sources of different types as though the items are uniform units of information. The information space consists of the set of associations among these information units. Thus, a single set of user commands suffices to browse,
filter, and share all the different types of data. This is accomplished internally by packaging data from all external sources as uniform objects with a generic set of operations for publishing, searching, displaying, and associating the data. The system itself also handles retrieval of objects from remote sources across the network and provides the necessary caching policies to make this retrieval speed-transparent. For the types of data common in genome research, the existing Internet has sufficient bandwidth to support interactive retrieval across the country.
|
BOX 4.5 A MODEL COLLABORATORY Genome research is the subject domain for a current example of a functioning national collaboratory. The Worm Community System comprises a digital library containing the data of the community of molecular biologists who study the nematode worm Caenorhabditis elegans, which has become a primary model organism in the Human Genome Project, and a software environment that supports interactive manipulation of this library across the Internet. The current library contains a substantial fraction of the extensive knowledge about the worm, including gene descriptions, genetic maps, physical maps, DNA sequences, formal journal literature, informal newsletter literature, and a wide variety of other informal materials. The current environment enables Users to browse the library by search and navigation, to examine and analyze selected materials, and then to share composed 'hyperdocuments' within the community. The current prototype is running in some 25 worm laboratories nationwide, and there are already instances of users electronically submitting items to the 'central' information space and having these automatically redistributed to other sites. The next release of the system will support electronic publication with editorial levels, as well as invocation of external analysis programs. Subsequent releases will move toward a complete analysis environment, with a large collection of databases and literature accessible transparently across the national network for examination and for analysis. |
The first release of the Worm Community System is now running in worm laboratories across the country and supports a sample range of the necessary knowledge and functionality (Figure 4.1). The second release will be available in 1993 and will support a sample range of publishing mechanisms. As the publishing system and the electronic community evolve, subsequent releases will support deeper knowledge semantics and begin to move toward a generic information infrastructure.
OPPORTUNITIES TO ENHANCE RESEARCH
Genome research in molecular biology has undergone a significant revolution owing to the existence of archival databases such as MedLine, GenBank, and the Genome Data Base. All practicing genome researchers consider it essential to cross-compare their experimental results with existing results by running similarity searches on these databases. Next-generation central archives, now in use in research prototypes, promise even greater utility and opportunities for collaboration. Remote analysis servers (e.g., Blast) will enable rapid, daily comparisons of sequences. Systems for interconnecting multiple archives (e.g., Entrez) will enable rapid comparison across different sources. The fact that such prototypes are being implemented with a standard data exchange format by the National Center for Biotechnology Information at the National Library of Medicine promises that integrated analysis environments for standard archives will become a reality in the foreseeable future.
At the same time, models for a complete collaboratory are also being developed. The Worm Community System illustrates what a complete collaboratory in genome research could become in the foreseeable future. It provides analysis of both formal and informal knowledge and electronic sharing of user-provided knowledge. As a distributed system utilizing the existing network communications infrastructure, it points the way toward a national information infrastructure that will enable scientists to
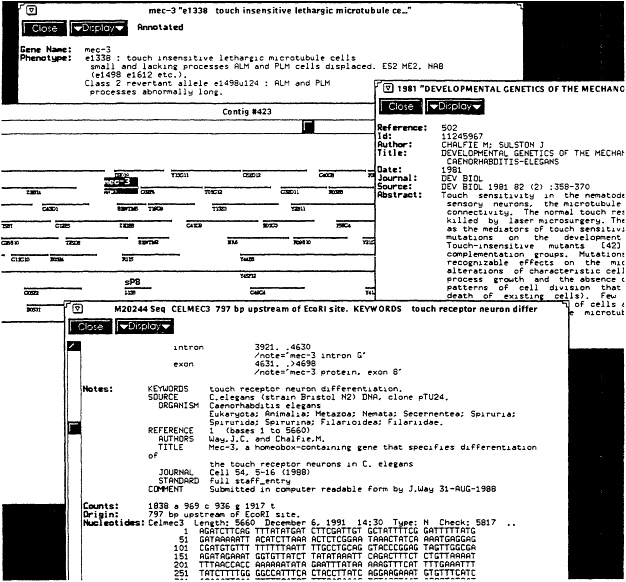
Figure 4.1 Sample session with release 1 of the Worm Community System, illustrating what might occur when a molecular biologist interacts with the community library. Shown are the coverage of both data and literature, and some of the relationship links. The user began with a broad query of the term "sensory," which returned all items from all sources mentioning that term, including the formal literature, informal literature, gene descriptions, sequence annotations, and so on. By browsing through short summaries of these items, the user found a literature item describing a number of mechanosensory genes (shown on the right). The relationship links to this literature article were then followed to retrieve a set of gene descriptions. The gene "mec-3" was of particular interest, as shown at the top left. From this gene description, the physical map was selected and an interactive display of the DNA clones appeared centered around where the gene was located (shown in middle left). This graphical display can be selected and manipulated; in this case a further zoom or link following was done to retrieve further information, which included the DNA sequence shown in the bottom left. Note that each of the items shown (literature, gene, map, sequence) comes originally from its own database, but the community information system enables navigation across all these sources with single, uniform commands. Not shown is a further interaction made possible by using an analysis program on the sequence to display its coding regions. SOURCE: Courtesy of Bruce Schatz, University of Arizona.
manipulate sources of information transparently across the country. However, it is still a preliminary model and will have to be expanded considerably to demonstrate a functional community information infrastructure. For example, the sets of knowledge and analysis must be expanded, a true distributed system across platforms and networks must be developed, and a case-hardened implementation must be evolved before the technology is ready to support standard archives. But with sufficient resources, complete collaboratories for sharing, comparing, and analyzing data will be built for genome research since the need is there and the technology is available. The pattern discovery enabled by "dry-lab" analysis environments promises another significant revolution for the support of research in molecular biology.















