16
Genome Structure and Evolution in Drosophila: Applications of the Framework P1 Map
Daniel L. Hartl, Dmitry I. Nurminsky, Robert W. Jones, and Elena R. Lozovskaya
A clone-based physical map consists of a set of ordered, overlapping inserts of cloned genomic DNA. Such a map affords a unique resource for studying the structure and function of the genome. The map facilitates the molecular identification of mutant genes, and the clones in the map provide ready access to substrates for genomic sequencing. A clone-based physical map also opens up new opportunities for studies of genome evolution.
Clone-based physical maps have been assembled for several species chosen as model organisms in the Human Genome Project (Collins and Galas, 1993). These include Escherichia coli (Kohara et al., 1987), Saccharomyces cerevisiae (Olson et al., 1986), Caenorhabditis elegans (Coulson et al., 1986), and Drosophila melanogaster (Hartl, 1992). There is also a first-generation physical map of the human genome (Cohen et al., 1993). In most organisms, sets of overlapping clones covering an uninterrupted stretch of the genome (contigs) are assembled by detecting overlaps by means of shared restriction fragments in fingerprints or shared sequence-tagged sites (STS). Drosophila is unique among model organisms in presenting giant polytene chro-
Daniel L. Hartl is professor of biology and Dmitry I. Nurminsky and Elena R. Lozovskaya are research associates in the Department of Organismic and Evolutionary Biology at Harvard University, Cambridge, Massachusetts. Robert W. Jones is a senior associate in the Department of Biochemistry and Biophysics at Washington University Medical School, St. Louis, Missouri.
mosomes in the larval salivary glands so that clones can be assigned positions in the genome by means of in situ hybridization. There are about 5000 polytene bands, most ranging in DNA content from 5 to 50 kilobase pairs (kb), with an average DNA content of ![]() 20 kb (Heino et al., 1994). The limit of cytological resolution with in situ hybridization is approximately 20 kb (Merriam et al., 1991). One advantage of the hybridization approach is that the approximate locations of clones covering much of the genome can be assembled relatively rapidly (yielding a ''framework map"), although clones that appear to be adjacent in the framework map need not necessarily overlap at the molecular level. The in situ mapping strategy therefore requires that molecular overlaps be determined after the framework map is completed rather than during assembly.
20 kb (Heino et al., 1994). The limit of cytological resolution with in situ hybridization is approximately 20 kb (Merriam et al., 1991). One advantage of the hybridization approach is that the approximate locations of clones covering much of the genome can be assembled relatively rapidly (yielding a ''framework map"), although clones that appear to be adjacent in the framework map need not necessarily overlap at the molecular level. The in situ mapping strategy therefore requires that molecular overlaps be determined after the framework map is completed rather than during assembly.
A physical map of the Drosophila genome based on yeast artificial chromosomes (YACs) ordered by in situ hybridization has been reported previously (Ajioka et al., 1991; Merriam et al., 1991; Hartl, 1992; Hartl and Lozovskaya, 1992; Lozovskaya et al., 1993; Cai et al., 1994). The YAC map includes ![]() 1200 clones with inserts averaging 200 kb that cover 90% of the euchromatic part of the genome. These clones have been grouped into
1200 clones with inserts averaging 200 kb that cover 90% of the euchromatic part of the genome. These clones have been grouped into ![]() 150 "cytological contigs," averaging 650 kb in extent, based on apparent overlaps detected by means of in situ hybridization; the gaps between cytological contigs average
150 "cytological contigs," averaging 650 kb in extent, based on apparent overlaps detected by means of in situ hybridization; the gaps between cytological contigs average ![]() 50 kb (Hartl, 1992). The YAC clones provide molecular access to much of the Drosophila genome, but the vector has a low copy number and it is difficult to separate large quantities of YAC from contaminating yeast genome.
50 kb (Hartl, 1992). The YAC clones provide molecular access to much of the Drosophila genome, but the vector has a low copy number and it is difficult to separate large quantities of YAC from contaminating yeast genome.
A second-level framework map based on bacteriophage P1 clones has now been assembled and is reported here. The map is based on 2461 clones with insert sizes averaging ![]() 80 kb that have been ordered by in situ hybridization. The P1 map includes an estimated 85% of the sites in the euchromatic genome with an average depth of coverage of 1.8. The mapped P1 clones, as well as 6755 additional clones, are being screened with STS markers in order to be arrayed in molecular contigs. The localizations of P1 clones and other information, as well as sources from which P1 clones may be obtained, are available to the scientific community by anonymous file-transfer protocol from ftp.bio.indiana.edu in the directory flybase or by electronic mail requests to flybase@morgan.harvard.edu.
80 kb that have been ordered by in situ hybridization. The P1 map includes an estimated 85% of the sites in the euchromatic genome with an average depth of coverage of 1.8. The mapped P1 clones, as well as 6755 additional clones, are being screened with STS markers in order to be arrayed in molecular contigs. The localizations of P1 clones and other information, as well as sources from which P1 clones may be obtained, are available to the scientific community by anonymous file-transfer protocol from ftp.bio.indiana.edu in the directory flybase or by electronic mail requests to flybase@morgan.harvard.edu.
Materials and Methods
Drosophila Strains and Procedures. Drosophila P1 clones were produced from DNA extracted from nuclei of adult D. melanogaster flies from an
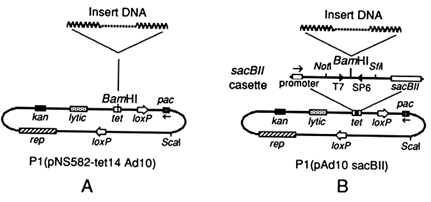
Figure 1 Bacteriophage P1 vectors.
isogenic strain of genotype y; cn bw sp according to the methods described in Smoller et al., (1991) and Lozovskaya et al., (1993). High molecular weight DNA was partially digested with Sau3A1 and fractionated according to molecular weight in sucrose gradients (10–40% sucrose) in order to isolate fragments in the size range 75–100 kb. These fractions were dialyzed, concentrated with 2-butanol, and precipitated in ethanol in preparation for ligation to the P1 vectors.
P1 Cloning Vectors. P1 clones were produced by using the P1 vectors pNS582-tet14 Ad10 (Figure 1A) and pAd10 sacBII (Figure 1B), which differ in the region around the cloning site. In pNS582-tet14 Ad10 (Sternberg, 1990), the BamHI cloning site interrupts the tetracycline-resistance gene; in pAd10 sacBII (Pierce et al., 1992), the BamHI cloning site is flanked on one side by a T7 promoter, a Not I site, and the promoter of the sacBII gene of Bacillus amyloliquefaciens; and it is flanked on the other side by an SP6 promoter, an Sfi I site, and the sacBII structural gene for levansucrase. Large fragments of DNA inserted into the cloning site disrupt expression of the sacBII gene and thereby allow cells of E. coli to survive in medium containing 5% sucrose (Pierce et al., 1992).
Vector arms resulting from digestion of either vector with BamHI and Sca I were treated with calf intestinal alkaline phosphatase, and an equimolar ratio of vector-arm DNA and genomic DNA was ligated in the presence of T4 DNA ligase as described (Smoller et al., 1991). Packaging of the ligated DNA was carried out as described in Sternberg (1990). During packaging, molecules are packaged stepwise in the counterclockwise direction from the pac site (as Figure 1 is drawn) until the phage head has been filled (100–115 kb), after which cleavage occurs.
Following packaging of the vector pNS582-tet14 Ad10, E. coli strain NS3145 (Sternberg, 1990) was infected and plated on LB plates (Miller, 1972) with kanamycin (25 µg/ml). Kanamycin-resistant colonies were tested for tetracycline resistance, and tetracycline-sensitive bacterial colonies were isolated. Following packaging of the vector pAd10 sacBII, E. coli strain NS3529 (Pierce et al., 1992) was infected and plated on LB agar containing kanamycin (25 µg/ml) and 5% sucrose in order to select for insert-containing vectors. Both NS3145 and NS3529 produce a site-specific recombinase that targets the loxP sites (Figure 1) and circularizes the vector by recombination. After isolation, a sample of clones from each vector was isolated and the size of the insert was determined by contour-clamped homogeneous electric field (CHEF) gel electrophoresis (Vollrath and Davis, 1987).
DNA Preparation from P1 Clones. Bacterial isolates containing single P1 clones were inoculated into LB medium containing kanamycin (25 µg/ml) and isopropyl β-D-thiogalactopyranoside (1 mM), which induces the lytic replicon (rep in Figure 1) to amplify the plasmid copy number (Sternberg, 1990), and grown overnight at 37°C. Plasmid DNA was extracted by the alkaline lysis method (Birnboim and Doly, 1979).
Cytological Analysis. Localization of clones was carried out with laboratory strain Oregon RC. Polytene chromosomes were prepared as in Atherton and Gall (1972). Chromosomes were pretreated in 2× SSC at 65°C for 30 min (1× SSC is 0.15 M NaCl/0.015 M sodium citrate, pH 7), dehydrated in 70% and 95% (vol/vol) ethanol, denatured in 0.07 M NaOH for 2.5 min, washed twice in 2× SSC, dehydrated again, and dried in air. DNA from P1 clones was labeled with biotin derivatives of dNTPs (GIBCO/BRL) by means of the random hexamer method (Feinberg and Vogelstein, 1984). Hybridization of labeled DNA to polytene chromosome squashes in situ was carried out overnight at 37°C in 1.4× SSC/7% (wt/vol) dextran sulfate/35% (vol/vol) N,N-dimethylforma-mide containing sonicated denatured salmon sperm DNA (0.6 mg/ml). Hybridization was detected with the Detek I horseradish peroxidase signal-generation system (Enzo Diagnostics) and 3,3'-diaminobenzidine (Sigma). Chromosomes were stained with Giemsa stain and embedded in Permount.
PCR Amplification of Insert–Vector Junctions. STS markers were determined from sequences at the termini of the genomic fragments present in P1 clones after amplification by the polymerase chain reaction (PCR) as described in Nurminsky and Hartl (1993); in this method, a partially double-stranded anchor-adapter oligonucleotide is ligated onto the
genomic fragment near the junction of vector and insert, and a template suitable for PCR amplification is produced by primer extension from a site in the flanking vector sequence. After amplification of the genomic fragment, DNA sequencing was performed with an Applied Biosystems model 373A DNA sequencing system and the Taq DyeDeoxy terminator cycle-sequencing kit.
STS Markers from Known Genes. STS markers were also determined from known Drosophila sequences present in GenBank release 79.0. Oligonucleotides suitable as PCR primers were chosen with an algorithm (Rychlik and Rhoads, 1989) implemented in the program OLIGO (National Biosciences, Plymouth, MN), version 4.0, for the Macintosh. The primer oligonucleotides, ranging from 18-mers to 21-mers, were either synthesized with an Applied Biosystems model 392 DNA Synthesizer or supplied by Research Genetics (Huntsville, AL) or Genset (La Jolla, CA).
PCR Screening of the P1 Library. For PCR screening, the P1 library was organized in 96-well microtiter dishes (arrays of 8 rows × 12 columns). The screening was carried out in two stages. First, a set of 96 reactions was carried out on DNA pools from all the clones present in each 8 × 12 array; each array containing a clone able to support amplification was subjected to a second set of 20 reactions carried out on DNA pools from clones in the 8 rows and 12 columns of the array. The DNA pools were prepared and generously provided by Bill Kimmerly and collaborators at Lawrence Berkeley Laboratories as part of the Drosophila Genome Center described in the acknowledgments. The individual PCRs were carried out in MJ Research (Watertown, MA) PTC-100 thermal cyclers in 96-well, V-bottomed, polycarbonate microtiter plates. PCR was carried out in 20-µl reaction mixtures containing 1–3 mM MgCl2 (optimized for each pair of oligonucleotides) overlaid with mineral oil and subjected to 25 cycles of 3 sec at 96°C, 45 sec at 92°C, 90 sec at the annealing temperature (optimized for each pair of oligonucleotides), and 90 sec at 72°C. The duration of time at the annealing temperature was increased by 1 sec in each cycle, and the duration of time at 72°C was increased by 3 sec in each cycle. The reactions were terminated by holding at 72°C for 5 min and stored at 4°C. PCR products were fractionated by electrophoresis in 1.2% agarose gels.
Results
The Drosophila P1 library presently in use consists of 9216 clones. Approximately 40% of the clones are in pNS582-tet14 Ad10 (Figure 1A);
the remainder, ![]() 60%, are in pAd10 sacBII (Figure 1B). Inserts in pNS582-tet14 Ad10 cluster in the size range 70–100 kb with a mean and standard deviation of 83.0 ± 6.2 kb (n = 25) (Smoller et al., 1991); inserts in pAd10 sacBII cluster in the same size range and have a mean and standard deviation of 82.5 ± 5.8 kb (n = 20) (data not shown).
60%, are in pAd10 sacBII (Figure 1B). Inserts in pNS582-tet14 Ad10 cluster in the size range 70–100 kb with a mean and standard deviation of 83.0 ± 6.2 kb (n = 25) (Smoller et al., 1991); inserts in pAd10 sacBII cluster in the same size range and have a mean and standard deviation of 82.5 ± 5.8 kb (n = 20) (data not shown).
Distribution of Clones by Chromosome. A total of 3104 clones were localized by in situ hybridization with the polytene salivary-gland chromosomes. Among the localized clones, 388 hybridized with the chromocenter, the underreplicated mass consisting largely of the pericentromeric heterochromatin and the Y chromosome, and/or with multiple euchromatic sites (typically, 10–100) without any apparent major euchromatic site of hybridization. A total of 64 clones yielded dual hybridizations (strong signals in two distinct euchromatic sites); these clones have not been investigated further, but some of them may represent chimeric clones containing ligated fragments from two different parts of the genome. An additional 191 clones were deliberate duplicates introduced into the workstream as blind controls in order to verify the accuracy of the procedures and reproducibility of the cytological localizations.
The remaining 2461 localized clones yielded single major sites of euchromatic hybridization. Approximately 10% of these clones also exhibited multiple (typically, 10–100) secondary sites of hybridization in the euchromatin; about half of this class also hybridized with the chromocenter. The multiple sites of hybridization are interpreted as resulting from transposable elements or other types of moderately repetitive, dispersed DNA contained in the cloned insert. With such clones, it is usually not difficult to identify a principal site of hybridization in the euchromatin in which the signal is intense and encompasses several bands, compared to which the multiple sites of hybridization are usually much weaker and present in single bands; the principal site of hybridization is the site to which the clone is assigned. (The distinction between the major site and secondary sites of hybridization can be difficult if the probe is excessive in amount or too heavily labeled.) A final class of clones, ![]() 2% of those localized, hybridized with the chromocenter and also to one principal site of hybridization in the euchromatin, without detectable secondary sites of euchromatic hybridization.
2% of those localized, hybridized with the chromocenter and also to one principal site of hybridization in the euchromatin, without detectable secondary sites of euchromatic hybridization.
Because the average insert size of the clones is ![]() 80 kb, the mapped clones with unique major sites of hybridization include
80 kb, the mapped clones with unique major sites of hybridization include ![]() 200 megabase pairs (Mb) of DNA, or the equivalent of
200 megabase pairs (Mb) of DNA, or the equivalent of ![]() 1.8 copies of the haploid euchromatic genome. If we assume that all euchromatic sites are equally likely to be present in the clones, the proportion of euchromatic sites
1.8 copies of the haploid euchromatic genome. If we assume that all euchromatic sites are equally likely to be present in the clones, the proportion of euchromatic sites
TABLE 1 Distribution of P1 clones in the euchromatic genome
|
Chromosome arm |
No. of P1 clones |
No. of P1 clones per lettered subdivision (mean ± SEM) |
No. of P1 clones per 80 kb |
|
Xa |
317 |
2.7 ± 0.2 |
1.5 |
|
2L |
490 |
4.2 ± 0.3 |
2.1 |
|
2R |
463 |
3.9 ± 0.3 |
1.6 |
|
3L |
531 |
4.6 ± 0.4 |
2.0 |
|
3R |
653 |
5.7 ± 0.4 |
1.9 |
|
a Applying the correction factor of 4/3 for X-linked segments yields an average of 3.6 P1 clones per lettered subdivision on the X chromosome and an average of 2.0 P1 clones per 80 kb. |
|||
expected to be represented at least once among the clones is 85%. The distribution of clones localized in the arms of the large euchromatic chromosomes is given in Table 1. (The X chromosome is acrocentric, chromosomes 2 and 3 are metacentric.) The ![]() 25% deficiency of clones localized to the X chromosome is expected because the libraries were constructed from a mixture of male and female DNA (Ajioka et al., 1991); however, the relative underrepresentation of clones from 2R is statistically significant (P < 0.01). Although a small percentage of the clones are duplicates arising from inadvertent transfer of the same bacterial colony during clone isolation, these are not sufficient in number to account for the nonrandomness. An additional 7 clones were localized to chromosome 4 which, based on its DNA content, would be expected to have
25% deficiency of clones localized to the X chromosome is expected because the libraries were constructed from a mixture of male and female DNA (Ajioka et al., 1991); however, the relative underrepresentation of clones from 2R is statistically significant (P < 0.01). Although a small percentage of the clones are duplicates arising from inadvertent transfer of the same bacterial colony during clone isolation, these are not sufficient in number to account for the nonrandomness. An additional 7 clones were localized to chromosome 4 which, based on its DNA content, would be expected to have ![]() 25; the underrepresentation of chromosome 4 is also statistically significant (P < 0.01).
25; the underrepresentation of chromosome 4 is also statistically significant (P < 0.01).
The large autosome arms are each divided into 20 numbered divisions, and each of these (except for those at the base) is divided into six lettered subdivisions (A–F) with an average DNA content of ![]() 200 kb. The distribution of P1 clones by lettered subdivision is given in Figure 2. Overall, >90% of the lettered subdivisions have at least one P1 clone, and the average number of clones per lettered subdivision is 4.2.
200 kb. The distribution of P1 clones by lettered subdivision is given in Figure 2. Overall, >90% of the lettered subdivisions have at least one P1 clone, and the average number of clones per lettered subdivision is 4.2.
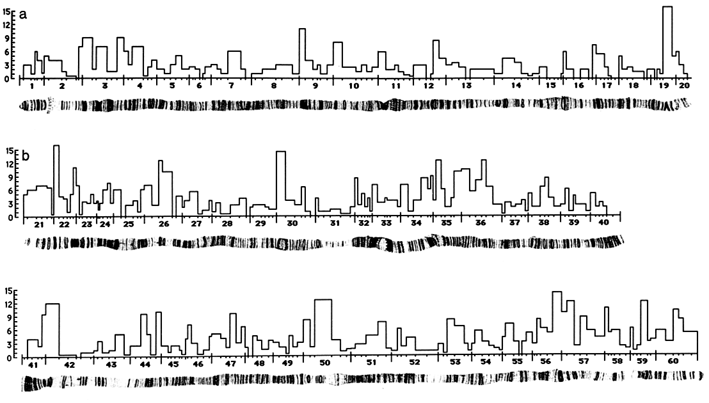
Framework Map. The P1 framework map is summarized in Figure 3. Sections 1–20 comprise the X chromosome, 21–40 and 41–60 the respective left and right arms of chromosome 2, and 61–80 and 81–100 the respective left and right arms of chromosome 3. In each numbered section, the short vertical tick marks, from left to right, set off the lettered subdivisions A–F (unlabeled). The histograms give the number of P1 clones in each lettered subdivision. A clone overlapping two
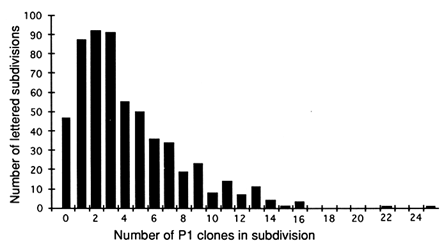
Figure 2 Distribution of P1 clones by lettered subdivision. See text for explanation.
subdivisions is counted as one-half clone in each subdivision. At the level resolution of Figure 3, the alignment between the chromosome bands and the delineators is approximate. Detailed information on the clones is accessible electronically from the flybase database and in printed form in Hartl and Lozovskaya (1994).
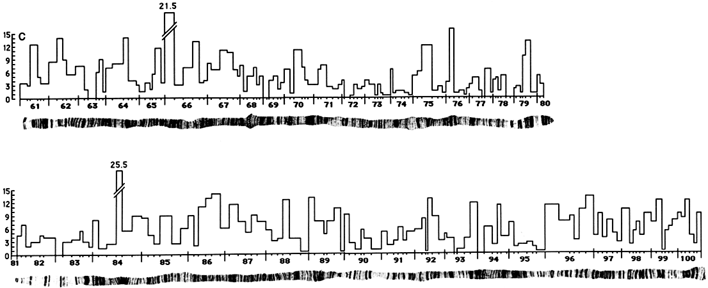
Contig Assembly. While the framework map in Figure 3 was being completed, contig assembly began, using the strategy of STS-content mapping, in which overlapping P1 clones are identified by virtue of their ability to support the PCR amplification of single-copy genomic sequences. The STS markers used for contig assembly are derived from (i) known genes with sequences available in GenBank, (ii) the termini of the genomic fragments present in P1 clones, and (iii) sites of insertion of the transposable element P (Hartl and Palazzolo, 1993). Although genome-wide contig assembly is still in its early stages, progress in one of the targeted regions near the tip of the X chromosome is shown in Figure 4 and illustrates the general strategy.
In Figure 4, the heavy black bars across the top represent the bands in the polytene chromosome. The positions of the STS markers are indicated by vertical dashed lines intersecting with horizontal black bars below that represent P1 clones; the ability of a P1 clone to support amplification of a STS is indicated with a cross at the site of intersection. The P1 contigs are indicated by long horizontal black bars across the bottom. In this region, the STS markers include 7
known genes and 11 termini of P1 inserts (hyphenated names) over ![]() 650 kb. There is a contig of 10 clones extending
650 kb. There is a contig of 10 clones extending ![]() 240 kb from bands 2E2 to 3A1 and another contig of 15 clones extending
240 kb from bands 2E2 to 3A1 and another contig of 15 clones extending ![]() 370 kb from 3A1 to 3B3. Between these contigs there may be a small gap of ≤40 kb, which we have not, as yet, attempted to bridge with additional P1 clones identified by screening the entire library with the ends of the inserts in the flanking P1 clones. The average density of STS markers per P1 clone in this region is 2.5.
370 kb from 3A1 to 3B3. Between these contigs there may be a small gap of ≤40 kb, which we have not, as yet, attempted to bridge with additional P1 clones identified by screening the entire library with the ends of the inserts in the flanking P1 clones. The average density of STS markers per P1 clone in this region is 2.5.
Discussion
The framework map reported here and summarized in Figure 3 affords a unique resource for research in the genetics of Drosophila. The framework map includes ![]() 85% of the euchromatic genome with an average redundancy of coverage of 1.8. Although there is some nonrandomness in the distribution of clones, the coverage of the euchromatic genome is very broad, and DNA from most euchromatic regions of interest should be accessible from the clones in the framework map. Less well represented are clones from the meshlike region of the polytene chromosomes denoted b heterochromatin, constituting the base of each chromosome arm and most of chromosome 4. Unrepresented in the framework map are clones from the chromocenter, comprising the a heterochromatin, which is grossly underreplicated in salivary gland nuclei and which includes the pericentromeric heterochromatin and the entire Y chromosome. Some of this material may be present in the
85% of the euchromatic genome with an average redundancy of coverage of 1.8. Although there is some nonrandomness in the distribution of clones, the coverage of the euchromatic genome is very broad, and DNA from most euchromatic regions of interest should be accessible from the clones in the framework map. Less well represented are clones from the meshlike region of the polytene chromosomes denoted b heterochromatin, constituting the base of each chromosome arm and most of chromosome 4. Unrepresented in the framework map are clones from the chromocenter, comprising the a heterochromatin, which is grossly underreplicated in salivary gland nuclei and which includes the pericentromeric heterochromatin and the entire Y chromosome. Some of this material may be present in the ![]() 15% of P1 clones that hybridize with the chromocenter and that have no major sites of hybridization in the euchromatin.
15% of P1 clones that hybridize with the chromocenter and that have no major sites of hybridization in the euchromatin.
Applications of the Drosophila physical map in studies of genome structure and function have been stressed elsewhere (Kafatos et al. , 1991; Merriam et al., 1991; Hartl et al., 1992; Hartl and Lozovskaya, 1992). Less consideration has been given to the utility of clone-based physical maps in studies of genome evolution. Drosophila has a long history of studies of genome evolution because, within many species groups, the banding patterns of the polytene chromosomes are sufficiently similar that phylogenetic relationships can be inferred. The principal limitation of cytological analysis is that, between species groups, the differences in banding patterns are usually too great for homologous chromosomal regions to be identified reliably, even between species whose morphological similarity implies virtual certainty of close relationship (Stone, 1962). The result is a very incomplete understanding of the patterns, processes, and functional significance of genome evolution in Drosophila. A case in point is the obscura species group, long an object of evolutionary studies, which includes such species as D. pseudoobscura,
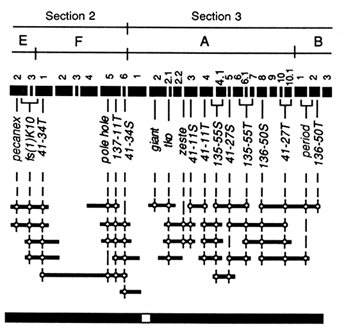
Figure 4 P1 contigs in a 650-kb region near the tip of the X chromosome. Numbered black bars across the top represent the polytene chromosome bands; narrow black bars below represent P1 clones. Positions of STS markers are denoted by dashed vertical lines intersecting P1 clones able to support PCR amplification. STS markers with hyphenated designations are the termini of P1 inserts; others are known genes. The thick horizontal bars across the bottom are two contigs.
D. miranda, D. subobscura, D. guanche, and D. affinis (Lakovarra and Saura, 1982). Although the correspondences between the chromosome arms of D. melanogaster and various species in the obscura group have been ascertained by in situ hybridization with probes from D. melanogaster (Segarra and Aguadé, 1992), no detailed, point-by-point cytological comparison between the genomes is possible because the polytene chromosome band morphologies, after an estimated 30–40 million years of evolutionary divergence (Beverley and Wilson, 1984), are too dissimilar. However, in studies with P1 clones from the framework map of D. melanogaster, >80% yielded strong, single hybridization patterns with the salivary gland chromosomes of D. pseudoobscura (Segarra and Lozovskaya, unpublished observations). No ambiguities resulting from hybridization with transposable elements have been noted, probably because, relative to single-copy genes, the transposable elements are
sufficiently divergent in sequence (de Frutos et al., 1992). Hence, the framework map in Figure 3 provides the material for a detailed alignment of the D. melanogaster and D. pseudoobscura euchromatic genomes with a density of markers of approximately one every 100 kb. Undoubtedly, many of the 80-kb P1 clones will also contain sites of rearrangement breakpoints between the species and thereby provide the material for determining the molecular mechanisms, as well as the patterns, of genome evolution.
In contrast to the situation with D. Pseudoobscura, no reliable hybridization signals with the salivary-gland chromosomes of D. virilis have been possible with P1 clones from D. melanogaster (Lozovskaya et al., 1993). The simplest explanation is that there has been too much DNA sequence divergence in the estimated 60 million years since the existence of a common ancestor of D. virilis and D. melanogaster (Beverley and Wilson, 1984). However, a bacteriophage P1 library of D. virilis has been constructed and initial progress toward a framework P1 map described; the framework map of D. virilis can be put into correspondence with that of D. melanogaster by means of DNA hybridization with suitable single-copy probes (Lozovskaya et al., 1993). Similar to the manner in which the framework map of D. melanogaster affords access to genome organization in D. pseudoobscura and other species in the subgenus Sophophora, it is anticipated that a framework map of D. virilis will afford access to genome organization and evolution in the subgenus Drosophila, including the Hawaiian drosophila. The P1 clones in the D. virilis framework map also provide the materials for large-scale sequencing of selected regions of the genome for comparison with the corresponding regions in D. melanogaster.
Summary
Physical maps showing the relative locations of cloned DNA fragments in the genome are important resources for research in molecular genetics, genome analysis, and evolutionary biology. In addition to affording a common frame of reference for organizing diverse types of genetic data, physical maps also provide ready access to clones containing DNA sequences from any defined region of the genome. In this paper, we present a physical map of the genome of Drosophila melanogaster based on in situ hybridization with 2461 DNA fragments, averaging ![]() 80 kilobase pairs each, cloned in bacteriophage P1. The map is a framework map in the sense that most putative overlaps between clones have not yet been demonstrated at the molecular level. Nevertheless, the framework map includes
80 kilobase pairs each, cloned in bacteriophage P1. The map is a framework map in the sense that most putative overlaps between clones have not yet been demonstrated at the molecular level. Nevertheless, the framework map includes ![]() 85% of all genes in the euchromatic
85% of all genes in the euchromatic
genome. A continuous physical map composed of sets of overlapping P1 clones (contigs), which together span most of the euchromatic genome, is currently being assembled by screening a library of 9216 P1 clones with single-copy genetic markers as well as with the ends of the P1 clones already assigned positions in the framework map. Because most P1 clones from D. melanogaster hybridize in situ with chromosomes from related species, the framework map also makes it possible to determine the genome maps of D. pseudoobscura and other species in the subgenus Sophophora. Likewise, a P1 framework map of D. virilis affords potential access to genome organization and evolution in the subgenus Drosophila.
We thank Lara Brilla for administrative support, Yaping Xu for data base management, and Jie Wei for technical help. The work reported here was supported in part by the National Center for Human Genome Research, Drosophila Genome Center (HG00750), Gerald M. Rubin, Principal Investigator. The Drosophila Genome Center includes investigators at the University of California at Berkeley, Lawrence Berkeley Laboratories, Harvard University, and the Carnegie Institution of Washington in Baltimore. We are grateful to G. M. Rubin, A. C. Spradling, and other participating investigators in the Drosophila Genome Center, especially Bill Kimmerly, Mike Palazzolo, and Chris Martin. David Smoller participated in making the Drosophila P1 libraries, and we are grateful to Nat Sternberg, James Pierce, and Phil Moen for their advice.
References
Ajioka, J. W., Smoller, D. A., Jones, R. W., Carulli, J. P., Vellek, A. E. C., Garza, D., Link, A. J., Duncan, I. W. & Hartl, D. L. (1991) Drosophila genome project: one-hit coverage in yeast artificial chromosomes. Chromosoma 100, 495–509.
Atherton, D. & Gall, J. (1972) Salivary gland squashes for in situ nucleic acid hybridization studies. Drosophila Info. Serv. 49, 131–133.
Beverley, S. M. & Wilson, A. C. (1984) Molecular evolution in Drosophila and the higher diptera II. Time scale for fly evolution. J. Mol. Evol. 21, 1–13.
Birnboim, H. C. & Doly, J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7, 1513–1523.
Cai, H., Kiefel, P., Yee, J. & Duncan, I. (1994) A yeast artificial chromosome clone map of the Drosophila genome. Genetics 136, 1385–1401.
Cohen, D., Chumakov, I. & Weissenbach, J. (1993) A first-generation physical map of the human genome. Nature (London) 366, 698–701.
Collins, F. & Galas, D. (1993) A new five-year plan for the U.S. human genome project. Science 262, 43–49.
Coulson, A., Sulston, J., Brenner, S. & Karn, J. (1986) Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 83, 7821–7825.
de Frutos, R., Peterson, K. R. & Kidwell, M. G. (1992) Distribution of Drosophila melanogaster transpoable element sequences in species of the obscura group. Chromosoma 101, 293–300.
Feinberg, A. P. & Vogelstein, B. (1984) A technique for radiolabeling DNA restriction
endonuclease fragments to high specific activity: Addendum. Anal. Biochem. 137, 266–267.
Hartl, D. L. (1992) Genome map of Drosophila melanogaster based on yeast artificial chromosomes. In Genome Analysis: Strategies for Physical Mapping, eds. Davies, K. E. & Tilghman, S. M. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 4, pp. 39–69.
Hartl, D. L., Ajioka, J. W., Cai, H., Lohe, A. R., Lozovskaya, E. R., Smoller, D. A. & Duncan, I. W. (1992) Towards a Drosophila genome map. Trends Genet. 8, 70–75.
Hartl, D. L. & Lozovskaya, E. R. (1992) The Drosophila genome project: Current status of the physical map. Comp. Biochem. Physiol. B 103, 1–8.
Hartl, D. L., and E. R. Lozovskaya. (1994) The Drosophila Genome Map: A Practical Guide. R. G. Landes, Austin, TX
Hartl, D. L. & Palazzolo, M. J. (1993) Drosophila as a model organism in genome analysis. In Genome Research in Molecular Medicine and Virology, ed. Adolph, K. W. (Academic, Orlando, FL), pp. 115–129.
Heino, T. I., A. O. Sura, and V. Sorsa (1994) Salivary chromosome maps of Drosophila melanogaster. Drosophila Information Service, Vol. 73.
Kafatos, F. C., Louis, C., Savakis, C., Glover, D. M., Ashburner, M., Link, A. J., Siden-Kiamos, I. & Saunders, R. D. C. (1991) Integrated maps of the Drosophila genome: progress and prospects. Trends Genet. 7, 155–161.
Kohara, Y., Akiyama, K. & Isono, K. (1987) The physical map of the whole E. coli chromosome: Application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50, 495–508.
Lakovarra, S. & Saura, A. (1982) Evolution and speciation in the Drosophila obscura group. In The Genetics and Biology of Drosophila , eds. Ashburner, M., Carson, H. L. & Thompson, J. N., Jr. (Academic, New York), pp. 1–59.
Lozovskaya, E. R., Petrov, D. A. & Hartl, D. L. (1993) A combined molecular and cytogenetic approach to genome evolution in Drosophila using large-fragment DNA cloning. Chromosoma 102, 253–266.
Merriam, J., Ashburner, M., Hartl, D. L. & Kafatos, F. C. (1991) Toward cloning and mapping the genome of Drosophila. Science 254, 221–225.
Nurminsky, D. I. & Hartl, D. L. (1993) Amplification of the ends of DNA fragments cloned in bacteriophage P1. BioTechniques 15, 201–208.
Olson, M. V., Dutchik, J. E., Graham, M. Y., Brodeur, G. M., Helms, C., Frank, M., MacCollin, M., Scheinman, R. & Frank, T. (1986) Random-clone strategy for genomic restriction mapping in yeast. Proc. Natl. Acad. Sci. USA 83, 7826–7830.
Pierce, J. C., Sauer, B. & Sternberg, N. (1992) A positive selection vector for cloning high molecular weight DNA by the bacteriophage P1 system: Improved cloning efficacy. Proc. Natl. Acad. Sci. USA 89, 2056–2060.
Rychlik, W. & Rhoads, R. E. (1989) A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 17, 8543–8551.
Segarra, C. & Aguadé, M. (1992) Molecular organization of the X chromosome in different species of the obscura group of Drosophila. Genetics 130, 513–521.
Smoller, D. A., Petrov, D. & Hartl, D. L. (1991) Characterization of bacteriophage P1 library containing inserts of Drosophila DNA of 75–100 kilobase pairs. Chromosoma 100, 487–494.
Sorsa, V. (1988) Chromosome Maps of Drosophila (CRC, Boca Raton, FL), Vols. 1 and 2.
Sternberg, N. (1990) Bacteriophage P1 cloning system for the isolation, amplification,
and recovery of DNA fragments as large as 100 kilobase pairs. Proc. Natl. Acad. Sci. USA 87, 103–107.
Stone, W. S. (1962) The dominance of natural selection and the reality of superspecies (species groups) in the evolution of Drosophila. Univ. Texas Publ. 6205, 507–537.
Vollrath, D. & Davis, R. W. (1987) Resolution of DNA molecules greater than 5 megabases by contour-clamped homogeneous electric fields. Nucleic Acids Res. 15, 7865–7876.