7
Computational Biology and the Cross-Disciplinary Challenges
Deborah A. Joseph
Edward H. Shortliffe
Federal Research and Funding Policies
Deborah A. Joseph
I will begin by talking a bit about the breadth I see in computational biology as a discipline, and then identify some of the challenges I see. Discussions at the Computer Science and Telecommunications Board (CSTB) have made me realize that some of my more computationally oriented colleagues have a narrower view of biology (and computational biology) than I have. Some did not take a biology class after high school, while others graduated from college before DNA sequencing was something that every undergraduate biology student learns about. Computer scientists are well aware that new technology has caused rapid advances in computer science and telecommunications. However, new technology, both biological and computational, is also rapidly influencing work in biology.
Computer scientists often equate computational biology with algorithmic support for genomics and molecular biology. Work in this area is fairly well known by computer scientists, and the computer science conferences often contain papers on genome sequencing and mapping. Computer scientists are fairly familiar with the problem of predicting the secondary and tertiary structure of biological molecules, and applications in drug design. The artificial intelligence community had done some nice work using neural nets and hidden Markov models for finding genetic indicators and genes within genome sequencing data. Problems such as predicting gene linkage are somewhat less well known, but are receiving increasing attention in the algorithms community.
However, beyond the areas of genomics and molecular biology, computational biology is much less understood by computer scientists. Asked what other topics of research are found within computational biology, most computer scientists would probably think back to its origins within mathematical biology. Problems such as population modeling and developmental models (how the zebra get its stripes) would come to mind. Some of the most exciting new research in biology that is benefiting from new technology and computational methods is almost unknown in computer science. I will mention just a few examples.
Currently, an exciting area of research is the quest for a better understanding of life in extreme environments. This work includes the study of deep sea vents, hot springs, extremely acidic environments such as mine tailings, the ice of the Arctic and Antarctic, as well as the possibilities of extraterrestrial life. Studying the organisms of these environments in large part involves studying their DNA. A particular gene, the 16S rRNA gene, has played
a key role in their classification. Computational biology has been central in providing algorithmic techniques for constructing phylogenetic trees based on molecular sequence data for this gene. These phylogenetic trees can then be used to predict the evolutionary relationships between known and newly discovered organisms. Using these techniques scientists have recently discovered an entire new domain of Archaea (bacteria). Some of these organisms appear to be evolutionarily very old and may offer clues to understanding the origins of life.
In other research, computational biologists are building models in areas of biology, ecology, and animal behavior that have typically been very noncomputational. Computational models in some of the areas offer important new possibilities, as well as controversy. For example, new models of animal physiology may in part replace some toxicity and carcinogenicity studies previously done using animals. The acceptable role of computer simulations and models for biological risk assessment is an area of ongoing, and at times heated, discussion.
Computer modeling has been used for many years in agriculture to predict crop yields. More recently an entire area of information intensive agriculture—precision agriculture—has developed. This area brings together many tools: variable-rate applicators, global positioning systems, databases, plant growth models, and models of nitrogen and soil characteristics to improve the way we raise crops. Through total farm management systems we can hope to make farming more efficient and to reduce nonpoint source pollution. Better models of farms as ecological systems will help maintain them in a healthy, efficient, environmentally sensitive way.
The last area that I will mention, tools for research and collaboration, is more familiar to computer scientists. Many of the database and visualization tools developed outside of biology are now being imported into biological research tools. Consider the work I discussed earlier on the construction of phylogenetic trees from molecular sequence data. As one might suspect there are a variety of algorithms that can be used and they sometimes give conflicting results. Today there are visualization tools being developed that will allow one to morph one phylogenetic tree into another. This gives the researcher a way to dynamically, rather than simply statically, visualize the different trees presented by different algorithms.
Systems are also being developed to aid in experiment design and hypothesis testing. Tool kits for building biological models are helping to eliminate the need to go back and start programming from scratch each time. Lastly, large database systems for managing experiment design take the typical laboratory notebook and put it into electronic form. They make it possible to take the data from a large experiment (a genome sequencing project, for example) and make it a flow-through process.
The topics I have mentioned are broad, but are certainly not all-inclusive. Today computational biologists can be found working in all areas of the biological sciences. Just as broad as the problems pursued are the techniques employed. They range from numerical approximation, mathematical programming and numeric solutions of mathematical equations, to neural networks, discrete algorithms and data structures, databases, and visualization.
I have given a very brief overview of some of the exciting work being done today in computational biology. At this point I would like to discuss some of the challenges. Although my talk was to focus on funding challenges, I will broaden what I say to include some more general challenges that I think the field faces. As one might expect, these are frequently interrelated and are challenges that many interdisciplinary fields face.
Federal funding for research in computational biology comes from the National Institutes of Health, National Science Foundation, the Department of Energy, and in more narrowly focused programs from DARPA, the Environmental Protection Agency, the National Institute of Environmental Health Sciences, and the Department of Agriculture. Industrial funding is significant in some areas of biotechnology and agriculture. Within the federal agencies there are two models for funding. One is a model exemplified by the Computational Biology Activity at the National Science Foundation: a specific budget and review process to handle research awards in computational biology. The second model makes funds available for research in computational biology through disciplinary research programs in the biological sciences. Those programs that benefit from particular tools and algorithms are expected to support their development. So, for example, research on modeling of biological molecules might be handled through a program in molecular biology. Each of these models of funding have advantages and disadvantages. I will discuss some of these as I talk a bit about some of the general challenges that computational biology faces.
The first challenge that I will mention is that of disciplinary identity. Computational biology is often described as forming the bridge between biology and computer science. But, is there more to computational biology? Does it form its own discipline, have its own foundation, its own fundamental problems? As scientists, we often judge
fields by their foundation, and by the difficulty and fundamental nature of their research problems. If as computational biologists we describe our research accomplishments solely in the disciplinary terms of computer science and biology, computational biology will fail to develop as an independent discipline. This will have an impact on funding in computational biology and make it difficult to build a cohort of scientists with the interdisciplinary training necessary to push the frontiers of research combining computation and biology.
A second problem is closely related. As scientists, we typically credit depth over breadth, and interpret this as brilliance over diligence. Scientists that work in computational biology must learn the background science and language of both biology and computer science. Further, scientists that work as part of an interdisciplinary research team pay a price in time as communication across disciplinary boundaries (and the physical boundaries of work places) is often slow. Scientists that work in a narrowly focused area simply have more time to delve deeply into their own science. We reward the time spent on science in a way that we tend not to reward the time that a scientist spends learning to speak a different scientific language and interact with other scientists. This affects funding and advancement in computational biology as it comes out in our review process.
A third funding challenge is what I will call the ''glass ceiling" for computational biology. When there are lots of resources or there is a special program in computational biology, funding is not a problem. However, when computational biology lacks its own funding program or when projects grow and outgrow their special program, computational biology projects must compete in disciplinary programs in biology. Many will say that this is as it should be; the disciplinary programs in biology should support the computational research that they benefit from. Nevertheless, when funding becomes short these projects are often the first to go. Even in the best of times, it is difficult to find funding for large computational projects within disciplinary programs. I believe that as computational tools become a more routine part of biological research some of this problem will disappear. Nevertheless, cooperation between funding programs can also ease some of the strain.
The last issue that I will raise is what I will call it the "hacking problem." Whether we are mathematicians proving theorems or computer scientists implementing large software projects, much of our work in science is incremental. We build on our past experiences, our past knowledge and the past knowledge of others. Most new results extend this chain of knowledge; only occasionally are directions radically altered. How we judge this type of incremental research varies. When results in computer science are extended to produce new results in computer science, the review of this work may be quite different from that when a computer scientist applies a novel (but known) date structure in a new way within biology. Too often the cross-disciplinary work is viewed in the negative as "applications" work or "hacking." The ambiguity between implementation and novel extension of an idea often causes problems in the scientific review process.
Although these challenges exist, I am optimistic about the future for computational biology. There are exciting problems to be worked on and an excellent group of students interested in the area.
Finding a Home in Academia
Edward H. Shortliffe
As someone who works at the intersection of two fields, medicine and computer science I have thought a lot about bridging issues that affect interdisciplinary fields, of which computational biology is only one example. The more general discipline that deals with all biomedicine plus computing and communications, known as medical informatics, is the field in which I have worked personally over the years. In recent years, the role of bioinformatics has become increasingly important, and I will tell you a little bit about computational biology as it fits within a medical informatics training and research environment at my own institution.
The challenge of finding a home in academia is my nominal focus today. However, I am going to broaden my charge to discuss issues related to how people who are trained in these intersecting fields find professional outlets in the world beyond, whether in academia, industrial research and development, or other settings where leaders increasingly are recognizing the need for people who have insights and experience that cross disciplines. Much of what I am going to say today harkens back to the CSTB report Academic Careers for Experimental Computer Scientists and Engineers, which was completed in 1994 (Larry Snyder from the University of Washington chaired
that committee); it addressed the issue of how experimental (as opposed to theoretical) computer scientists best fit in this evolving world of computer science within the academic community.
As the field of computer science broadened, we began to hear comments like one that I overheard at a faculty meeting in the computer science department at Stanford in about 1988, "I remember the days when I used to go to departmental seminars and I could understand every talk!" This remark was made by an eminent computer scientist, reminiscing about the good old days when the computer science department was first formed and he could go to any colloquium, understand the content, and ask insightful questions. He realized that it was getting harder to do that. He no longer bothered to go to all of them because some were in areas in which he knew he would have trouble relating to the topic. As computer science both broadened in scope and developed detailed methods within narrowly defined subareas of investigation and application, and as seminars focused on subtopics in areas of specialization, it is hardly surprising that some of these subspecialization areas would involve overlap with other disciplines in which computing was being applied. Biomedicine has been one of these.
One might argue that there is a potential scientific discipline at the intersection of computer science with almost every other field because of the ubiquitous nature of computing and communications as they touch on all aspects of society. Therefore, it is not surprising that we are seeing the evolution of expertise in many of these areas of overlap. The question is, To what extent does interdisciplinary training need to be specifically provided? To what extent can people get good training in computer science and simply choose to apply it in a given area? Or to what extent can people get well trained in the areas of application and then try to pick up enough computing so that they can work effectively at the intersection? Given the complexity of computer science and the tendency either to reinvent the wheel (if you have not trained in computer science) or to apply computing notions naively (if you do not really know the discipline of application), many of us believe that there is an ever-growing need to identify the disciplines that lie at the intersection and to train people explicitly in these areas.
As you might imagine, this was the first question I was asked when, in the early 1980s, I proposed the creation of a new interdisciplinary degree program at Stanford in medical informatics. Understandably, faculty in the computer science department said, "Well, you know we could have a lawyer come in here tomorrow and ask the same question, and then the nutritionists will come in and say that computers and nutrition is a new field, and they need a degree program as well. Where do you draw the line?" Such questions do force you to identify what is unique about an interdisciplinary field that is not being addressed adequately during training in one area or the other in order to justify creating an entirely new degree program. We have, accordingly, tried to characterize the specialized knowledge that lies at the intersection between biomedicine and computer science. Figure 7.1 shows one perspective of the challenge of trying to understand biomedical knowledge and biomedical data, to get them into computer programs, and then to use them for purposes of inference and problem solving. These processes define a set of research areas that explain the focus for much of the current training and research in medical informatics.
I am going to tell you a little about the Stanford informatics degree program to give you a feel for how an effort to train people explicitly at this intersection between biomedicine and computer science has played out over a 15-year period, where people are getting jobs, and some of the lessons that we have learned from this experience.
Our program offers master's and Ph.D. degrees, and it is based in the School of Medicine. The core faculty in the program all have both M.D.s and a Ph.D. in either computer science or medical informatics. We are trying to train people who will be researchers either in industry or in academic positions.1 The program has close ties to
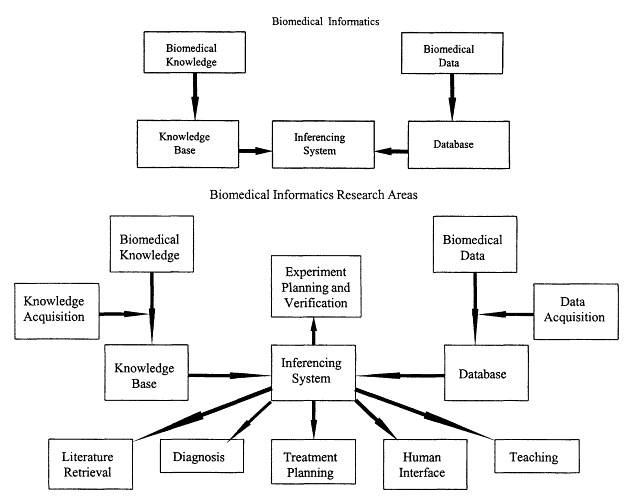
Figure 7.1
Elements in the process of incorporating biomedical date and knowledge into computer programs for use in problem solving. Source: E.H. Shortliffe. 1996. Stanford University.
the computer science department; we happen to be on a campus where the computer science department and the medical school are close to one another. This greatly facilitates the opportunities for interchange between the two environments, allowing students to take a medical school course, hop on a bicycle, and take a computer science course in the next segment of the day's class schedule.
Our emphasis is on rigor coupled with methodological innovation. We decline to give degrees simply because a student builds a clever program, regardless of its size or complexity. Instead, we ask that our graduates be able to describe what the scientific issues are that generalize from the project they worked on to contribute broadly to the work of others. We try to make people think about what they are doing in this context so they can stand up before a computer science colloquium or national meeting and describe their work, playing down the biology issues or the medical issues and focusing instead on the computer science, and be perceived very much as peers when it comes to the nature of the contributions of their work.
Similarly, the same project often needs to be explained in a medical grand rounds, a pediatric seminar, or a molecular biology research seminar. When the same student goes into that setting, he or she needs to be able to play down the computer science, which the typical person in the audience will not understand, and focus instead on what is exciting and challenging about the biology being done in those projects. This is quite a set of demands to
place on people. It is one of the reasons our students tend to be older and actually end up with dual training. About two-thirds of our students are physicians, who then take this kind of degree as their subspecialty training after getting a medical degree and often after completing a residency program.
Since there are many practical issues in biomedical computing, we need to find the right balance between teaching students the theory and giving them the skills that allow them, when they graduate, to have an impact in applied settings. The challenge in building a curriculum for this kind of interdisciplinary field is to think about all the areas students need to know something about (see Figure 7.2). As the bottom of Figure 7.2 indicates, we believe students need to learn something about both clinical medicine and the basic medical sciences. Thus, computational biologists in our program take courses on genetic structure and protein unfolding, and they learn about clinical medicine topics as well. You might ask how much students can learn and retain in any one of these areas if they are trying to learn something in all of them. We believe the answer is that graduate training requires a broad background across a set of fields that one is likely to need to know about, with subsequent focus on an area of specific application or methodology during the period of one's M.S. practicum or Ph.D. dissertation.
In addition to computer science and biomedicine, we add courses in decision sciences, biostatistics, bioengineering, and epidemiology. If people take courses in all of these areas, it is not the same as taking a computer science degree and then applying it in biomedicine. The curriculum provides an unusual mix that did not previously exist in any single degree program. As more schools offer degrees in this area, we are seeing a similar mix of topics being included in their curricula.
The program philosophy rests on the following:
- It is oriented toward research training for academic or industrial careers.
- It has close ties with, and gains inspiration from, the computer science department.
- There is emphasis on rigor, methodological innovation, and an ability to generalize from specific results.
- There is emphasis on verbal and written skills, including an ability to present work to three audiences.
- Training is balanced between theory and practice.
Accordingly, we have five major content areas in our curriculum: biomedical informatics, computer science, decision science, biomedicine, and health policy. Biomedical informatics courses include the incremental offerings that we had to create for our degree candidates. Among these are courses in computational biology, an introduction to medical applications of computing, medical decision support systems, biomedical imaging, and a project course in which students build small systems under the guidance of medical informatics faculty. Our informatics courses are cross-listed in the computer science department, and many computer science and engineer
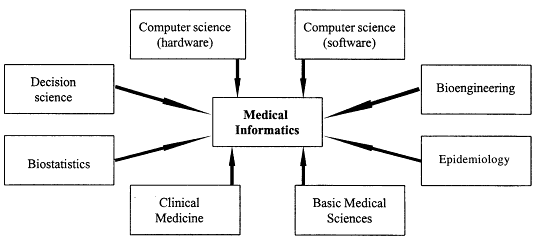
Figure 7.2
Areas of knowledge required in the field of biomedical informatics.
Source: E.H. Shortliffe. 1996. Stanford University.
ing graduate students are coming to the school of medicine to take our offerings. They see biomedicine as an interesting area of application that they often had not realized could be pursued as part of their academic program until they discovered our courses in the computer science portion of the university catalogue.
Let me summarize some of the lessons learned from this training activity. First, the trainees themselves often become tremendous bridge builders. They have brought informatics preceptors into contact with faculty throughout the university that we would not otherwise have known because the students take courses all over the campus. They come back with ideas, oftentimes with strong recommendations about people to whom we should be talking. Sometimes this has played out in very positive ways, resulting in new collaborations and relationships, often outside the medical school. As I mentioned, we have people from other parts of the university taking our courses, learning about the field, and getting excited about it. Some of them end up going into medical informatics or going off to medical school. Although that may not have been their original plan, they realize that there is an emerging career opportunity at the intersection of health care or biology and computer science.
The crucial role of an advanced computing environment is probably obvious to anyone in computer science, but this point is not always obvious to deans of medical schools. It is difficult to provide high-quality training in medical informatics unless you have a computing environment that is just as advanced as one expects to find in a typical engineering school department. Budgets in medical schools seldom take this into account. Thus, we face the challenge of trying to build an advanced computing environment in a school of medicine, which does not have a tradition of supporting such environments from operating funds. We are very dependent on industrial gifts of hardware and software as a result. There is also a tension between the academic and service notions that exist within a school of medicine. Some medical school colleagues assume we should be pulling wires through the walls for them and making sure that their PCs work. Similarly, our affiliated hospitals, realizing that we have talented students who know a lot about computing in the health care setting, often wish we would provide them with technical support for their applications projects.
Some service activities of these types would be healthy for our students; many will be involved with precisely such issues after they graduate. On the other hand, if an academic unit becomes a service organization within a medical environment, it is difficult to maintain the academic standards that are part of the motivation for having the training program in the first place. Thus, we try to find the right balance and to avoid formal service responsibilities to the school or hospitals, just as computer science departments do not run their university computing centers.
With regard to our current status, we have enrolled about 70 students since we started the program in the early 1980s; 33 were physicians when they started, and another 13 were medical students while they studied with us.2 Much more than half of our graduates end up as M.D.-Ph.D.s or M.D.-masters; the majority of these earn Ph.D.s. Essentially all of our current students, 24 out of 26, are Ph.D. candidates.3 It has become clear that there is declining interest in master's degrees in this field, at least among our own trainees. One of the reasons there are currently more master's graduates is that it takes only two years to train a master's graduate and four to six years to train a Ph.D., as in most Ph.D. programs.
People often ask what our students do when they graduate. They do research; they work in academia; they work in industry doing research; they work in operational jobs within medical institutions or, increasingly, in biotech companies that are hiring people who have these kinds of skills. Some of them do clinical, administrative, and educational management. We have one or two that actually run hospital computing groups. Some are working on digital libraries, providing expertise in information retrieval and information science. Seventeen of our graduates are in academia, sixteen in industrial settings, and three in clinical practice, but still doing some informatics as part of their life as practitioners; one is in a hospital working in a clinical computing environment; two work for the government (one as a military chief information officer and the other as a researcher for the National Aeronautics and Space Administration doing biomedical computing); and five are completing their residency programs.
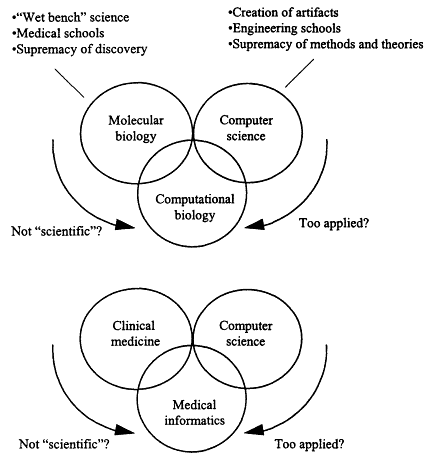
Figure 7.3
Elements of interdisciplinary fields and the challenge of integrating them effectively.
Source: E.H. Shortliffe. 1996. Stanford University.
I would like to close by coming back to a point raised by Professor Joseph. Computational biology can be considered as providing the interface between molecular biology and computer science (Figure 7.3, top), and one can anticipate the emergence of experts who identify this as their professional field; yet such experts must try to maintain credibility in all three of the circled domains in Figure 7.3, moving as fluidly as they can among three rather different environments. The challenge is that oftentimes the molecular biologists ask whether what these people are doing is true science. Can you be doing true science if you are not working with test tubes and gels at the bench? In the culture of a biomedical research environment, it is common for colleagues to define "true scientific research" using such criteria.
Ironically, from a different perspective, computer scientists sometimes see computational biologists as being too applied. They are viewed as being driven too much by applications from the real world. They may not be working at the theoretical level that would more easily validate their role as traditional computer scientists in an academic setting. The same picture applies to those doing medical informatics (Figure 7.3, bottom). Although the relevant area of application is clinical medicine, medical practice and clinical faculty often view "valid" research as focusing on discovering explanations for life processes, with an emphasis on "wet bench" research. Such narrow definitions and high expectations have forced those who work in a field like medical informatics to define their discipline not in terms of their impact on applications but, rather, based on the significant research problems that are associated with the development of new methodologies and solutions. Perhaps all new fields, drawing on
elements from existing disciplines while defining a new one, have had similar challenges in defining and explaining both the scientific content and the practical importance of the problems that they address.
Discussion
DAVID MESSERSCHMITT: Let us assume that in some area such as computational nutrition we decide that it is not appropriate to have a separate program. I can imagine three models. We could have a nutritionist who learns enough computer science but is in the nutrition department, or somebody who is in computer science who gets in that application area, or someone with a joint appointment. Maybe Deborah in particular could comment on the relative merits of these three options.
DEBORAH JOSEPH: I guess there are two issues. One applies to a faculty member or researcher and the other to a student. One of the things that the University of Wisconsin has that I think has worked very well is a National Institutes of Health training grant in biotechnology. We do not offer a degree in biotechnology, but we nevertheless have students who are trained in biotechnology on this grant. These students have advisers in some aspect of the biological sciences and in some aspect of the mathematical computational or engineering sciences. The program has been extremely successful. Students tend to spend a lot of time together, and there tends to be a lot of cross-fertilization between students in the program. This would fulfill some of the need for a program in computational nutrition.
At the faculty and research level, I think that more of it has to do with how we view our colleagues, more than the barriers put in place by universities. The true impediments typically relate to whether you can get through some review process, whether your colleagues respect you, and whether you have colleagues who will write letters of recommendation for you. If we as scientists value cross-disciplinary activities, I do not think that good people who work in computational nutrition will have trouble. If their research is not valued by both their colleagues in
nutrition and their colleagues in computer science, then they have trouble.
MESSERSCHMITT: Can they survive in either department?
JOSEPH: I think at the University of Wisconsin they can, but it depends a lot on the university.
EDWARD SHORTLIFFE: Local culture is very important in this regard. I think there is a great difference among institutions as to how much they are purists with regard to the implied criteria for appointment and promotion, as suggested in my last two diagrams. What we are talking about is credibility at promotion time, especially in the tenured line.
JOHN RIGANATI: One comment is related to the question you just had, but I wonder if either of you knows what the trends are in biology departments around the country about giving credibility to computational biology, as opposed to requiring that the computational aspects be linked directly with wet lab work. I know that a few years ago the percentage was very small, and I do not know if there have been any significant changes as a result of the kinds of things that you describe here. The second question is whether or not biological processes are viewed by either of you as having some basis for practical application in computational devices in the next few years, either literally or in an inspirational sense.
JOSEPH: I guess I could take the last question. Recently DNA and biomolecular computing have received a lot of press. I think we have been overeager in computer science to believe that this is a technology that is right around the corner. It is an interesting idea, but I will reserve judgment on whether we are really going to build supercomputers out of DNA molecules.
SHORTLIFFE: To answer the first question, there has been a growing trend in the recruitment of individuals who really know about computational biology to work in traditional molecular biology, genetics, and biology departments. Frankly, it is impossible now to do modern molecular biology research without local expertise in computing, especially for DNA homology searching and analysis of some of the protein databases and the like.
Molecular biologists can no longer do effective research unless they have somebody in their midst who can help them understand and use the newer technologies. Sometimes professors assign graduate students to go off, learn about computational biology, come back, and basically be the "guru" in the lab. Increasingly there has been a trend toward bringing in people who provide that expertise professionally in-house.
The problem is that they are not necessarily viewed as peers. I think the way we are beginning to see
computational biologists perceived as pure scientists is in the creation of separate academic units within institutions. Here they are recruited as computational biologists, they are evaluated as computational biologists, they are promoted as computational biologists, and then they become collegial collaborators with people in other departments on research projects. The same goes for medical informatics.
MISCHA SCHWARTZ: The issues you raise are common to many interdisciplinary areas. There is one school of thought that says perhaps the way to do this is to learn one discipline well and become a specialist in it and then go on to the other one, rather than spreading yourself too thin. What are your comments on this?
SHORTLIFFE: Well, it was indeed the question I was asked most when we proposed our program. It depends on whether you really believe that there is a body of knowledge at the intersection that warrants full-time focus during training.
There is a difference between learning a lot about medicine and then learning a little about computing, or learning a lot about computing and then a little about medicine, and focusing your entire graduate training at the intersection itself. I believe there is also a significant difference between formal informatics training and having someone first finish a medical degree and then earn a computer science degree in a conventional computer science environment; the connections to medicine are not part and parcel of the way computer science is taught, and understanding the relationships and relevance is thus inherently left as an exercise to the student. We have found that there is a kind of culture in a field that students begin to absorb if they train in an environment where everybody else is also working at the intersection of disciplines that interest them.
I think time has shown that people who actually train explicitly to work at this intersection—individuals who get all their course work and culture in an environment that allows them to interact regularly with others who have the same interdisciplinary interest—develop a special skill set as bridge people and as productive contributors when they get out. Such skills are not easily acquired by earning one degree or the other and then sort of secondarily adding a pure version of what they missed.
WILLIAM WULF: I personally think that the issues raised here about interdisciplinary work between computer science and biology go across the board. Computing intersects absolutely everything, as Ted said. So this is a problem that we computer scientists really have to face up to. I think we can be a model for a lot of other disciplines where the same problems occur, but perhaps there is not quite the same urgency to collaborate at the moment.










