This paper was presented at a colloquium entitled “Genetics and the Origin of Species,” organized by Francisco J.Ayala (Co-chair) and Walter M.Fitch (Co-chair), held January 30–February 1, 1997, at the National Academy of Sciences Beckman Center in Irvine, CA.
Divergent and conserved features in the spatial expression of the Drosophila pseudoobscura esterase-5B gene and the esterase-6 gene of Drosophila melanogaster
NATALIA A.TAMARINA*, MICHAEL Z.LUDWIG†, AND ROLLIN C.RICHMOND‡§
*Division of Vascular Surgery, Department of Surgery, Northwestern University Medical School, 251 East Chicago Avenue, Suite 626, Chicago, IL 60611: †Department of Ecology and Evolution, University of Chicago, 1101 East 57 Street, Chicago, IL 60637; and ‡Department of Ecology and Evolution, State University of New York at Stony Brook, Stony Brook, NY 11794
ABSTRACT The regulatory regions of homologous genes encoding esterase 6 (Est-6) of Drosophila melanogaster and esterase 5B (Est-5B) of Drosophila pseudoobscura show very little similarity. We have undertaken a comparative study of the pattern of expression directed by the Est-5B and Est-6 5'-flanking DNA to attempt to reveal conserved elements regulating tissue-specific expression in adults. Esterase regulatory sequences were linked to a lacZ reporter gene and transformed into D. melanogaster embryos. Est-5B, 5' upstream elements, give rise to a ß-galactosidase expression pattern that coincides with the wild-type expression of Est-5B in D. pseudoobscura. The expression patterns of the Est-5B/ lacZ construct are different from those of a fusion gene containing the upstream region of Est-6. Common sites of expression for both kinds of constructs are the third segment of antenna, the maxillary palps, and salivary glands. In vitro deletion mutagenesis has shown that the two genes have a different organization of regulatory elements controlling expression in both the third segment of antenna and maxillary palps. The results suggest that the conservation of the expression pattern in genes that evolved from a common ancestor may not be accompanied by preservation of the corresponding cis-regulatory elements.
The esterase 5B (Est-5B) gene of Drosophila pseudoobscura and the esterase 6 (Est-6) gene of Drosophila melanogaster derive from a common ancestor and likely are orthologous (1). This hypothesis is based on the observations that Est-5B and Est-6 produce homologous products although with differing tissue specificity (2, 3). Both loci code for two major transcripts in adults. The coding regions of Est-5B and Est-6 are about 80% similar at both the nucleotide and protein levels (3). In contrast, the 5' and 3' flanking regions of these genes have very little similarity. Only about 170–200 bp of the upstream sequences of Est-5B and Est-6 can be aligned to determine similarity, and this alignment requires numerous substitutions and deletions/insertions, indicating that these regions are quite different. Sequences more than 200 bp 5' of the start site of these genes show only occasional similarities. These similarities are typically no longer than 12 bp and occur at different distances from the AUG codon and sometimes in different orientations (1). It is apparent that since the divergence of Est-5B and Est-6, DNA upstream of the coding regions of these two genes has changed to a significantly greater degree than the coding regions of the two genes.
The upstream region of the Est-6 gene is capable of directing the expression of a lacZ reporter gene in mature adult flies (older than 1 day posteclosion) in tissues such as antennae, maxillary palps, salivary glands, trachea and air sacks of head and thorax of males and females, as well as organs of the male reproductive tract (the ejaculatory bulb and anterior ejaculatory duct), lacZ expression in these tissues appears at 12 hr posteclosion. In young flies less than 12 hr posteclosion, we observed lacZ expression only on the prefrons. This expression pattern usually disappeared as the mature pattern described above developed (4). These data and other results derived from analyses of deletion mutants lead to the conclusion that the upstream region of Est-6 is composed of several independently acting, tissue-specific regulatory regions (4).
We report here an analysis of the functional organization of the upstream regulatory region of the Est-5B gene of D. pseudoobscura and add to our previous results on the regulation of the Est-6 gene of D. melanogaster. This analysis allows us to evaluate the degree and kinds of functional differences that arose in the upstream regions of the Est-5B and Est-6 genes during evolution. We placed the upstream DNA of the Est-5B and Est-6 genes 5' of the reporter gene in a P element-based, promotorless vector and monitored the patterns of the expression of the resulting hybrid genes after transformation into D. melanogaster. This approach allowed us to compare the action of the tissue-specific, cis-regulatory elements present in the Est-5B and Est-6 genes in a common genetic background. This experimental design also eliminates the possible influence of species-specific trans-acting factors on the expression patterns of transformed genes.
Our data show that the upstream regions of the Est-6 (1,150 bp) and Est-5B (5,100 bp and 1,224 bp) genes confer different expression patterns on the reporter gene. The common sites of expression for these genes are the third segment of the antenna, maxillary palps, and salivary glands of the adults. The results of in vitro deletion mutagenesis show that the two genes probably have a different organization of regulatory elements that control expression in conserved tissue locations. These results suggest that the conservation of the expression pattern of genes that evolved from a common ancestor is not necessarily accompanied by an evolutionary preservation of the corresponding cis-regulatory elements.
MATERIALS AND METHODS
P Element-Mediated DNA Transformation. P element-mediated germ-line transformation was carried out using
© 1997 by The National Academy of Sciences 0027–8424/97/947735–7$2.00/0 PNAS is available online at http://www.pnas.org.
|
§ |
To whom reprint requests should be addressed. e-mail: Rrichmond@sunysb.edu. |
methods described elsewhere (4). Approximately 200 transformed stocks were checked by Southern blotting to estimate the number of insertions per genome. Southern blotting with digoxigenin-labeled probes was performed as described by Maniatis et al. (5) and according to the instructions provided by the manufacturer of the Genius Kit (Boehringer Mannheim). The frequency of double and triple insertions was less than 10% of the total. All transformed lines analyzed contained only one insertion per stock, and the length of the inserted Est-5B upstream region was not altered as determined by Southern blotting analyses (data not shown).
Histochemical Staining. The procedures used for histochemical staining have been described (4). We made the following modifications to our previously published procedures: the fixing and staining solutions contained 10 mM MgCl2; 1% phosphoric acid was used to adjust the pH of working solutions instead of citric acid; samples were stained for 20 hr; in the majority of experiments, 3- to 8-day-old adults were assayed, although for some transformed stocks, adults younger than 2 days old were used; all histochemical results were treated as qualitative data.
Constructs. A 472-bp HindIII-AflIII fragment from the D. pseudoobscura Est-5B locus (1) containing the Est-5 5' flanking region from 0 to -472 relative to the AUG codon, was subcloned into pGEM3 (Fig. 1). Subsequently the HindIII site of this plasmid was converted into an EcoRI site, and the AflIII site, after treatment by mung bean nuclease, into a BamHI site. This construct is termed 5B-472. Deletion constructs were obtained from 5B-472 by using convenient restriction sites (3). The 10 restriction fragments of the 472-bp Est-5B promotor region were inserted between the EcoRI and BamHI sites of the P element-based vector CaSpeR-AUG-ß-galactosidase (6).
Construct 5B-1224 (Fig. 1) was prepared by inserting a genomic fragment BglII(-1224)–SalI(-58) between the EcoRI and SalI sites of plasmid 5B-472 in pGBM3. This construction required the conversion of the BglII site in the fragment into an EcoRI site. The resulting insert was subcloned into pCaSpeR using EcoRI and BamHI sites.
Construct 5B-5100 was prepared by subcloning a genomic fragment, EcoRI(-5100)–SalI(-58), between the EcoRI and SalI sites of plasmid 5B-472 in pGEM3 in which the BamHI site
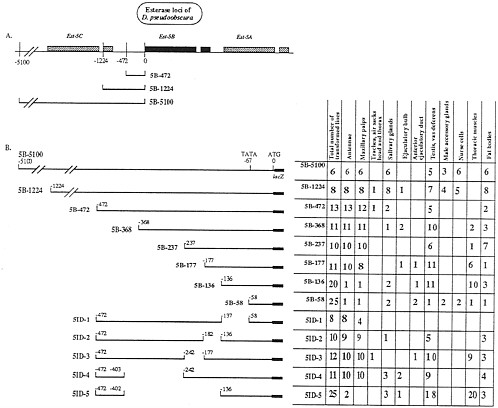
FIG. 1. Sites of expression of Est-5B constructs when transformed into D. melanogaster. (A) Schematic organization of the carboxylesterase loci of D. pseudoobscura showing the extent of three of the constructs used in transformation experiments. Shaded boxes show the extent of the transcribed regions of the gene. All Est-5B genes have a short intron shown as a break in the shaded box in the 3' half of the gene. (B) (Left) Diagram of the constructs used to transform D. melanogaster lines. (Right) Total number of lines transformed with each construct and the number of lines showing expression of each construct in the adult tissues studied.
had been converted into a KpnI site. The resulting insert was subcloned into pCaSpeR using EcoRI and KpnI sites.
All subcloning procedures were performed according to the procedures of Maniatis et al. (5).
RESULTS
We studied the location of elements regulating the expression of the Est-6 and Est-5B genes of D. melanogaster and D. pseudoobscura, respectively, by preparing constructs of the 5' flanking sequences of these genes and inserting them 5' to the lacZ gene in the CaSpeR-AUG-ß-galactosidase transformation vector (6). Figs. 1 and 2 provide diagrams of the genomic regions of the two esterase genes and show the number and extent of the transformation constructs we studied. DNA fragments of 5,100 and 1,224 bp from the Est-5B gene (Fig. 1) or 1,150 bp from the Est-6 gene (see Figs. 2 and 4) that cover the regions 5' of the AUG codon of both genes were used as the bases for constructing transformation vectors. These DNA fragments presumably contain most of the 5' regulatory sequences of both genes (4). The constructs prepared were used to generate stable transformed lines of D. melanogaster by the method of P element-mediated, germ-line transformation. The expression of the lacZ reporter gene in transformed lines was studied by in situ analysis of Escherichia coli ß-galactosidase activity in tissues from adult flies. The number of transformed lines expressing ß-galactosidase in the tissues analyzed is summarized in Figs. 1B and 2B and interpreted in Fig. 4.
ß-Galactosidase Expression Controlled by the Est-5B Constructs. Transformed lines carrying the Est-5B/lacZ gene with 5,100 bp or 1,224 bp of Est-5B, 5' DNA (5B-5100 and 5B-1224, Fig. 1) expressed ß-galactosidase activity in antennae, maxillary palps (Fig. 3 B and E), fat bodies, and salivary glands in adult flies of both sexes and also in nurse cells of females and in testes and vasa deferentia of males (data not shown). In half of the transformed lines, expression in male accessory glands was observed (Fig. 1B).
The major difference between the two larger Est-5B/lacZ constructs was the presence in 5B-1224 transformants of additional sites of expression that differed among independently generated transformed lines with insertions in different chromosomes and genomic locations (as judged by genetic and Southern blot analyses). These additional sites of expression included the tracheae, the ejaculatory bulb (Fig. 1), and many other specific locations in different tissues (data not shown). Such variation also was characteristic of all constructs shorter than 5B-1224. We attribute this variation in the sites of reporter-gene expression in shorter constructs to the influence of different adjacent loci at the different sites of insertion (7). These data suggest that in the region from -1,224 to -5,100 bp there are sequences that obviate the position effect, thus eliminating variation in the tissue specificity of expression.
Deletion analysis (Fig. 1B) suggests that regulatory elements essential for the spatial pattern of expression of Est-5B/lacZ genes in antennae, maxillary palps, salivary glands, fat bodies, testes, vasa deferentia, and male accessory glands are located within 1,224 bp of Est-5B upstream DNA. Positive regulatory elements controlling the expression of Est-5B in nurse cells, male accessory glands, salivary glands, and fat bodies probably are situated in the region between nucleotides -1,224 and -472. The region between nucleotides -403 and -136 contains elements controlling the expression in antennae and maxillary palps. Positive elements controlling ß-galactosidase expression in testes and vasa deferentia presumably are located in the region between nucleotides 136 and -58.
Some of the Est-5B deletion constructs directed the expression of ß-galactosidase in the thoracic muscles of flies of both sexes (Fig. 1B). This phenotype was not seen in flies transformed with larger constructs. Our data suggest that the region between -177 bp and -237 bp contains elements that reduce the expression of ß-galactosidase in thoracic muscles, while the positive control elements are located in the region between nucleotides -136 and -58.
Expression of ß-Galactosidase in Young Flies Carrying Est-5B Constructs. Expression of the Est-5B/lacZ genes in young flies was studied in lines transformed with constructs 5B-472, 5B-237, 5B-177, and 5B-136. In recently emerged flies (<1 day old), the reporter gene is expressed in the hypoderm of head thorax, and abdomen (data not shown). This expression pattern disappears after 1 day of age. A comparison of the
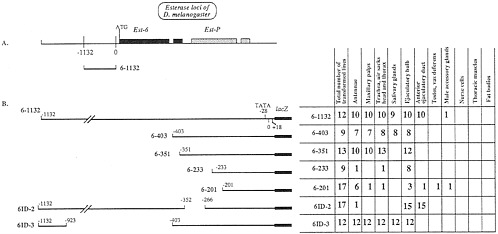
FIG. 2. Sites of expression of Est-6 constructs when transformed into D. melanogaster. (A) Schematic diagram of the genomic organization of carboxylesterase loci of D. melanogaster. Shaded boxes show the extent of the transcribed regions of the gene. Est-6 genes have a short intron shown as a break in the shaded box in the 3' half of the gene. (B) (Left) Diagram, of the constructs used to transform D. melanogaster lines. (Right) Total number of lines transformed with each construct and the number of lines showing expression of each construct in the adult tissues studied.
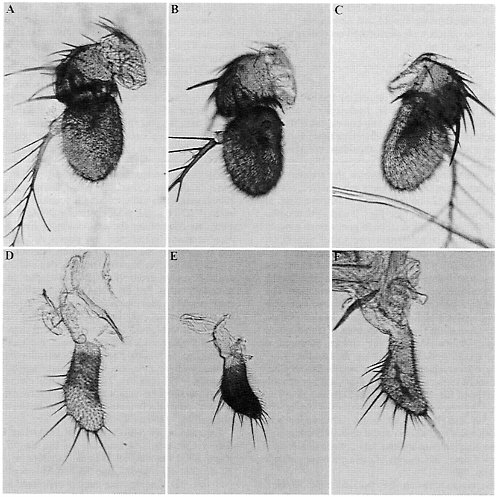
FIG. 3. ß-Galactosidase activity in antennae and in maxillary palps of D. melanogaster transformed with Est-6 and Est-5B deletion constructs. See Figs. 1 and 2 for a description of the nomenclature and extent of each deletion. (A) 6–1132, antenna. (B) 5B-5100, antenna. (C) 5B-136, antenna. (D) 6–1132, maxillary palp. (E) 5B-5100, maxillary palp. (F) 5B-136, maxillary palp.
pattern of expression in young flies in the four constructs studied suggests that construct 5B-136 contains elements that are responsible for the expression in hypoderm of young flies. These results are reminiscent of changes in the expression of loci involved in Drosophila olfaction that were studied by Riesgo-Escovar et al. (8).
ß-Galactosidase Expression Controlled by Est-6 Constructs. Flies transformed with construct 6–1132 containing 1,150 bp of Est-6 upstream sequences (Fig. 2) expressed ß-galactosidase in the antennae (Fig. 3A), maxillary palps (Fig. 3D), tracheae, and air sacks of head and thorax, in salivary glands of flies of both sexes, and in male ejaculatory bulb and anterior ejaculatory duct. These results confirm our previous studies (4).
Fig. 2B shows the deletion constructs used to localize elements regulating Est-6 gene expression. Fig. 4 summarizes the results of experiments reported here and in our previous work (4). We have localized four independently acting Est-6 regulatory regions that direct expression of lacZ in (i) the ejaculatory duct, (ii) the adult salivary glands, (iii) the antennae, the maxillary palps, and respiratory system, and (iv) the ejaculatory bulb (Figs. 2 and 4).
Control of Conservative Modes of Expression of the Est-6/ lacZ and Est-5B/lacZ Genes. The similarity in the patterns of expression of Est-6/lacZ and Est-5B/lacZ in antennae, maxillary palps, and salivary glands provides an opportunity to compare the mechanisms that regulate these conservative modes of expression for each gene. We describe in detail below the localization of the putative regulatory sequences that control the expression of the two esterase genes in similar tissue locations.
Antennae. The pattern of ß-galactosidase expression in the antenna of Est-6/lacZ transformants is shown in Fig. 3A. The positive regulatory element that presumably controls the development of this pattern in antennae was previously localized to the region between nucleotides -328 and -233 upstream of the Est-6 transcription start site (4). Here we localize this element more precisely. Deletion 6ID-2 (Fig. 2B) eliminates reporter-gene expression in the antennae, thus allowing us to locate the positive regulatory elements controlling expression in this organ to the region -328 bp to -266 bp upstream from the transcription start site.
Fig. 3B shows the basic staining pattern in antenna that is characteristic of the Est-5B/lacZ larger constructs, 5B-5100, 5B-1224, and 5B-472. In these transformed lines, ß-galactosidase is expressed in much of the third antennal segment. Particularly intense staining is seen in certain regions such as the sacculus (pit organ, Fig. 3B) of the third segment. This basic staining pattern resembles, although it is more extensive, the pattern observed in lines transformed with the Est-6 gene constructs (Fig. 3A). Reporter-gene expression in the second antennal segment, as was routinely observed for the Est-6 gene
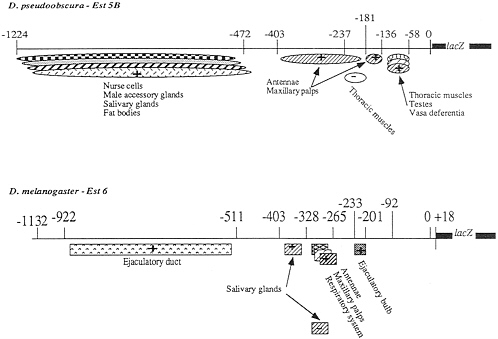
FIG. 4. Diagram summarizing the location of the putative regulatory elements in the 5' regions of the Est-5B and Est-6 genes of D. pseudoobscura and D. melanogaster. The positions of both positive (+) and negative (-) regulatory elements are shown.
constructs (Fig. 3A), was detected only with constructs 5B-237, 5ID-2, and some lines of 5ID-4.
Deletion analysis of the Est-5B promoter region suggests that there are two or more elements that control the basic pattern of expression in antennae. The basic pattern (Fig. 3B) was eliminated completely by deleting the sequences between nucleotides -402 and -136 (5ID-5 and 5B-136, Fig. 1B). Thus, the -402 and -136 region likely contains all the necessary regulatory elements for the control of reporter-gene expression in the antennae in Est-5B. Deletion construct 5B-177 produces flies with the basic pattern of antennal expression. This result suggests that there is a positive regulatory element between nucleotides -136 and -177. However, if nucleotides between -136 and -177 are removed as in deletion 5ID-2, flies transformed with this construct still show the basic pattern of expression in 90% of the transformed lines. This result suggests that the region between -402 and -177 likely contains another element that is capable of producing the basic pattern of antennal expression for the Est-5B gene.
Yet another pattern of expression in antennae (Fig. 3C, not included in Fig. 1B) is apparent when elements controlling the basic expression pattern (Fig. 3B) are removed in deletion constructs 5ID-5 and 5B-136. Transformed lines carrying these constructs exhibit staining of the third antennal segment in a pattern reminiscent of the distribution of major nerve branches in the antennae (9). The resolution of the staining patterns exemplified in Fig. 3C did not permit us to determine which cell types were actually involved. A similar pattern of antennal expression was found in some transformed lines carrying the short Est-6/lacZ constructs, 6–201 and 6–92 (data not shown).
Maxillary Palps. The expression of ß-galactosidase activity in the maxillary palps of flies transformed with Est-6/lacZ or Est-5B/lacZ is shown in Fig. 3 D and E, respectively. The reporter gene appears to be expressed primarily in cells near or on the surface of the palps. Obvious differences in the pattern of expression are apparent between the lines transformed with the Est-6 and Est-5B constructs. Transformation with the Est-6/lacZ constructs results in light staining in the proximal half of each maxilla (Fig. 3D). Transformation with the Est-5B/lacZ constructs results in intense staining of most of the surface of the maxilla (Fig. 3E).
A positive element that presumably controls the expression of the Est-6/lacZ gene in the maxillary palps previously was located in the region -328 to -233 (4). Deletion 6ID-2 (Fig. 2B) of the Est-6 upstream region eliminates ß-galactosidase expression in the maxillary palps. Thus the positive regulatory elements controlling ß-galactosidase expression in this organ most probably lie within the region -328 to -266. This is the same general region controlling antennal expression in D. melanogaster (Fig. 4).
In flies transformed with deletion constructs of the Est-5B/ lacZ gene, the reporter gene was expressed coordinately in both the maxillary palps and antennae (Figs. 1 and 4). This suggests that expression in these two organs may be controlled by the same regulatory elements located at nucleotides -403 to -181 and -181 to 136. However, expression in maxillary palps was observed in only half of the lines transformed with deletion construct 5ID-1 (Fig. 1B), suggesting that region -136 to -58 contains sequences specifically contributing to the maxillar expression.
An alternative pattern of maxillary staining (Fig. 3F, not included in Fig. 1B) similar to that observed in antennae (Fig. 3C) was seen in some lines transformed with Est-5B/lacZ constructs 5B-136 and 5ID-1. ß-Galactosidase expression in
the palps of these lines is observed along the passage of the main nerve branch (9). Lines transformed with Est-6 constructs did not show this pattern of maxillary expression.
Variation in Staining Patterns in Antennae and Maxillary Palps. The appearance of staining patterns in antennae and maxillary palps in flies transformed with Est-6 or Est-5B constructs did vary slightly in individual flies from the same stock. Some flies from the same line produced less intense staining than others. The details of the staining pattern also were variable in flies of the same stock and occasionally in two antennae from the same fly. In our interpretations of these data, we have attempted to use the most common or average pattern.
Salivary Glands. We have shown previously (4) that the positive regulatory element(s) responsible for the expression of esterase 6 enzyme in the salivary glands of D. melanogaster is located in the region between nucleotide positions -402 and -328 of the Est-6 upstream sequence. We identified possible negative elements in the regions -328 to -72 and -1,132 to -511 (4). Here we studied two additional Est-6 constructs 6–351 and ID-2 (Fig. 2B). Flies transformed with these constructs do not express ß-galactosidase in the salivary glands. Thus we conclude that the putative positive regulatory element most likely spans the nucleotide position -351 because the disruption of the promoter sequence at this site apparently leads to inactivation of the element.
The 5B-5100 and 5B-1224 constructs of the Est-5B gene of D. pseudoobscura direct the expression of ß-galactosidase in the salivary glands in 100% of the independently transformed lines (Fig. 1). Constructs containing 472 or fewer bp of the most proximal, 5' Est-5B DNA direct the expression of the reporter gene in only 7% to 18% of the independently transformed lines. This sporadic pattern of expression could be due to a position effect. It follows that the positive element of the Est-5B gene that controls the expression of the reporter gene in the salivary glands probably is located between nucleotides -472 and -1,224 upstream from the initiation codon (Fig. 4).
DISCUSSION
Interspecific gene transfer was used to compare the action of cis-regulatory sequences of homologous esterase genes in a common genetic background. This approach follows a strategy suggested by Cavener and Dickinson (10, 11) and allows us to study variation in the expression of two related genes and to identify conserved, cis-acting regulatory elements.
The Est-5B/lacZ constructs directed expression of ß-galactosidase in accessory glands, parts of testes and vasa deferentia in males, nurse cells in females, third segment of antennae, maxillary palps, salivary glands, and fat bodies in flies of both sexes. This expression pattern reflects features of the normal spatial expression of Est-5B in D. pseudoobscura. Our data are consistent with previous results showing expression of Est-5B of D. pseudoobscura in testes, accessory glands, ovaries, and antenna, and the lack of expression in ejaculatory duct and ejaculatory bulb (1, 3, 4). However, the tissue-specificity of expression of Est-5B/lacZ constructs in transformed flies should be confirmed by studies of Est-5B wild-type expression in D. pseudoobscura by methods such as in situ hybridization of mRNA because we cannot rule out the possibility that some D. melanogaster tissues lacked required, trans-acting factors for Est-5B expression. For example, we did not see any expression of our Est-5B/lacZ constructs in eyes, although expression in this organ was reported by Brady et al. (3). Those authors transformed D. melanogaster with an Est-5B gene that contained 472 bp of upstream sequences, the coding region, its intron, and 1,200 bp the 3' flanking sequences. It is possible that regulatory elements specifying expression in eyes are located in the 3' flanking, noncoding region of the Est-5B gene. Healy et al. (12) report that there are elements 3' of the Est-6 structural gene in D. melanogaster that modify the expression of this locus in transformed lines.
Our results show that many changes have occurred in the information content of the 5' regulatory regions of the Est-6 and Est-5B genes during evolution. Both Est-5B and Est-6 contain numerous cis-regulatory elements within 5'-flanking regions of comparable length (1,150 bp for Est-6 and 1,224 bp for Est-5B) (12). There are pronounced differences in the expression of the two lacZ constructs containing these regions. The sites of ß-galactosidase expression specific for the Est-6/ lacZ construct were the trachea and air sacks of head and thorax, prefrons in young flies, in the ejaculatory bulb and the anterior ejaculatory duct (4). We have not detected Est-6/lacZ expression in accessory glands, testes, vasa deferentia, nurse cells, or fat bodies. Both genes, however, directed expression of the reporter gene in several common tissue locations (salivary glands, maxillary palps, and the third segment of antenna). The data of Healy and her colleagues (12) suggest that another conserved site of expression is the hemolymph of both larvae and adults. Our experimental procedures did not allow us to detect expression of our constructs in hemolymph. All of these observations lead to the conclusion that selection may have acted to retain these common patterns of expression even though the underlying sequence has changed dramatically.
We have localized regions capable of directing the expression of the lacZ reporter gene in two of the three sites common to both D. melanogaster and D. pseudoobscura, the maxillary palps and the third segment of antenna. The putative Est-6 regulatory region is located between -265 and -328 (Fig. 4). The same function is apparently performed by two separate regions located between -136 and -181 and between -181 and -403 for the Est-5B gene (Fig. 4).
We compared the sequences of the three regions that are capable of directing reporter gene expression in the maxillary palps and the third segment of antenna to determine if any sequence similarities could be detected. The larger region in D. pseudoobscura (-403 to -181) has a single 9-bp sequence, CAAATATTT (-373 to -365), that is found in the corresponding Est-6 region at -278 to -270. This element is not found elsewhere in either the Est-5B or the Est-6 5' region. An identical element is located in approximately the same position 5' of the Est-6 loci in D. simulans (-241 to -249) and D. mauritiana (-277 to -269). A second smaller element, AAATCT, is found at position -175 to -170 in D. pseudoobscura and at two positions in D. melanogaster, -508 to -503 and -298 to -293. The latter site overlaps the region we have identified as containing positive regulatory elements for the maxillary palps and the antennae. This element also is found in the same relative positions in both D. simulans and D. mauritiana. The conservation of these elements and their positions relative to the start site of the Est-6 gene in the four species for which data are available suggests that they may be responsible for the coordinated expression of esterase 6 in the antennae and maxillary palps.
The function of these regulatory elements may be a result of the action of several different trans-acting transcription factors that are capable of specifying expression in both antennae and maxillary palps. Indeed, the spatial patterns of the expression produced by the melanogaster and pseudoobscura genes in these organs (see Fig. 3) are similar, but not identical. This suggests that these patterns might be composed of expression in different or overlapping cell populations. Thus evolution may have acted to conserve a spatial pattern of expression using very different regulatory mechanisms. Similar patterns of evolution have been observed in the regulation of the eve locus in Drosophila (13).
It is also possible that these esterase regulatory elements may consist of multiple short consensus sequences (putative transcription factor binding sites) with imperfect similarities.
Enhancers with this kind of structure have been described for many genes (14). Healy et al. (12) have identified putative 5' regulatory elements for Est-6 and related loci in several species that appear to have some of these characteristics. In addition to the antennae and maxillary palp sites identified in this work, Healy et al. (12) have located putative ejaculatory bulb and hemolymph sites in D. pseudoobscura. Our procedures did not allow us to detect expression in the hemolymph, but occasional lines did show expression in the ejaculatory bulb (Fig. 1), suggesting that the element identified by Healy and colleagues may function when other regulatory regions are not present.
The elements essential for the expression of Est-6 in D. melanogaster in both antennae and maxillae lie within 400 bp immediately 5' of the Est-6 coding sequence (Fig. 4). This region of the genome has remarkably low levels of polymorphism and divergence within several species of the melano gaster subgroup (15). Brady et al. (3) found that the 174 bp immediately 5' of the start site in Est-5B was the most highly conserved 5' region between D. melanogaster and D. pseudoobscura. Karotam et al. (15) argue that strong directional selection or functional constraint would be necessary to retain such a low level of sequence variation.
The conservation of esterase expression in the antennae and the maxillary palps suggests that esterase 6 may have a role in olfaction. Vogt and Riddiford (16) identified esterases in the antennae of silk moths, Antheraea polyphemus, that were capable of degrading the female sex pheromone of this species. Carlson and his colleagues (8, 17) have demonstrated that the antennae and maxillary palps of D. melanogaster both are olfactory organs and that a single gene affects the response of these organs to a variety of odorants. The effect of the Est-6 null allele on several mating behaviors (2) and our previous demonstration (18) that esterase 6 is capable of hydrolyzing a Drosophila pheromone suggest that an investigation of the effect of Est-6 alleles on the olfactory responsiveness of antennae and maxillary palps would be fruitful.
Developmental geneticists have proposed that major evolutionary changes in morphology can be the result of a few changes in homeotic genes that can affect the development of body segments (19). Population genetic analysis of the eve gene-controlling segmentation in arthropods and vertebrates (13) supports a neo-Darwinian model that postulates that major changes in morphology are the result of numerous genetic changes both in structural and regulatory genes. Our analyses of the regulation of the Est-6 locus in Drosophila support the neo-Darwinian model for the evolution of gene regulation. While the expression of the Est-6 homologue in D. pseudoobscura is markedly different from that seen in the D. melanogaster group, a number of small changes in elements regulating the expression of these loci can readily account for these differences.
We thank Susan Brandon for technical assistance, T.Kozlova and Bruce Cochrane for many useful discussions, and Marion Healy for sharing unpublished data. This work was supported by a National Science Foundation grant to R.C.R.
1. Brady, J.P. & Richmond, R.C. (1992) J. Mol. Evol. 34, 506–521.
2. Richmond, R.C., Nielsen, K.M., Brady, J.P. & Snella, E.M. (1990) in Ecological and Evolutionary Genetics, eds. Baker, J.S.F., Starmer, T.W. & MacIntyre, R.J. (Plenum, New York), pp. 273–292.
3. Brady, J.P., Richmond, R.C. & Oakeshott, J.G. (1990) Mol. Biol. Evol. 7, 525–537.
4. Ludwig, M.Z., Tamarina, N.A. & Richmond R.C. (1993) Proc. Natl. Acad. Sci. USA 90, 6233–6237.
5. Maniatis, T., Frisch, D.F. & Sambrook, J. (1982) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
6. Thummel, C.S. Boulet, A.M. & Lipshitz, H.D. (1988) Gene 74, 445–456.
7. Wilson, C., Bellen, M.J. & Gehring, W.J. (1990) Annu. Rev. Cell Biol. 6, 679–714.
8. Riesgo-Escovar, J., Woodward, C., Gaines, P. & Carlson, J. (1992) J. Neurobiol. 8, 947–964.
9. Demerc, M., ed. (1965) Biology of Drosophila (Hafner, New York).
10. Cavener, D.R. (1992) BioEssays 14, 237–244.
11. Dickinson, W.J. (1988) BioEssays 8, 204–208.
12. Healy, M.J., Dumancic, M.M., Cao, A. & Oakeshott, J.G. (1996) Mol. Biol. Evol. 13, 784–797.
13. Kreitman, M. & Ludwig, M. (1996) Semin. Cell Dev. Biol. 7, 583–592.
14. Stanojevic, D.T., Hoey, T. & Levine, M. (1989) Nature (London) 341, 331–335.
15. Karotam, J., Delves, A.C. & Oakeshott, J.G. (1993) Genetica 88, 11–28.
16. Vogt, R.G. & Riddiford, L.M. (1981) Nature (London) 293, 161–163.
17. Ayer, R.K. & Carlson, J. (1992) J. Neurobiol. 23, 965–982.
18. Mane, S.D., Tompkins, L. & Richmond, R.C. (1983) Science 222, 419–421.
19. Palopoli, M.F. & Patel, N.H. (1996) Curr. Opin. Genet. Dev. 6, 502–508.







