Proc. Natl. Acad. Sci. USA
Vol. 95. pp. 5906–5912. May 1998
Colloquium Paper
This paper was presented at the colloquium “Computational Biomolecular Science,” organized by Russell Doolittle, J.Andrew McCammon, and Peter G.Wolynes, held September 11–13, 1997, sponsored by the National Academy of Sciences at the Arnold and Mabel Beckman Center in Irvine, CA.
Precise sequence complementarity between yeast chromosome ends and two classes of just-subtelomeric sequences
(Saccharomyces cerevisiae/inverted recombination/sequence exchange/telomeres/Y′ and X2 repeats)
ROY J.BRITTEN*
Division of Biology, California Institute of Technology, 101 Dahlia Avenue, Corona del Mar. CA 92625
ABSTRACT The terminal regions (last 20 kb) of Saccha romyces cerevisiae chromosomes universally contain blocks of precise sequence similarity to other chromosome terminal regions. The left and right terminal regions are distinct in the sense that the sequence similarities between them are reverse complements. Direct sequence similarity occurs between the left terminal regions and also between the right terminal regions, but not between any left ends and right ends. With minor exceptions the relationships range from 80% to 100% match within blocks. The regions of similarity are composites of familiar and unfamiliar repeated sequences as well as what could be considered “single-copy” (or better “two-copy”) sequences. All terminal regions were compared with all other chromosomes, forward and reverse complement, and 768 comparisons are diagrammed. It appears there has been an extensive history of sequence exchange or copying between terminal regions. The subtelomeric sequences fall into two classes. Seventeen of the chromosome ends terminate with the Y′ repeat, while 15 end with the 800-nt “X2” repeats just adjacent to the telomerase simple repeats. The just-subterminal repeats are very similar to each other except that chromosome 1 right end is more divergent
Once the complete Saccharomyces cerevisiae DNA sequence became available (1, 2) it appeared worthwhile to see if an insight into the origin of repeated sequences could be obtained, since all repeated sequences of this yeast strain are available for examination in the complete sequence. The initial stage has been to examine the terminal regions, and that is what is reported here. Naturally the results overlap the many previous studies, but they differ from what has been published by the completeness of the examination of the terminal relationships. There have been extensive examinations of yeast telomeres and of pairing and recombination processes revealing extensive regions of subterminal sequence relationships, but they will not be reviewed here and reference is made to previous reviews (3–8).
There is of course a question as to what can be learned by merely examining sequence similarities, so this work is an experiment, but there are new results of significance. Here the telomeric and subtelomeric sequences are referred to together as the terminal regions, which include the last 20 kb of each chromosome. By custom, numbering starts at the left end. Many left terminal region sequences are the reverse complement of some right terminal region sequences, and no cases occur of significant lengths of precise direct sequence similarity between any parts of left and right terminal regions among all of the chromosomes. However this observation does not signify that there is a consistent orientation of the arbitrary historical identification of the ends of the yeast chromosomes. The situation is clarified if, for test purposes, the numbering of a chromosome is reversed. After this test change all left end sequences would still be the reverse complement of the right end sequences. Only a few specific relationships would change, while the reverse complementary pattern as a whole would remain unchanged. The reverse complementary relationships between chromosome ends have been clear to some workers (e.g., ref. 9) but I am not aware of a discussion of their significance.
RESULTS
Initial Tests of Terminal Reverse Complementarity. To test the general occurrence of reverse complementary relationship between chromosome terminal regions, the reverse complement of a 5-kb terminal segment of each of the left ends of the 16 chromosomes was compared with all of the chromosomes, using FASTA (10). Reverse complementary regions were observed at the right end of several chromosomes for each of the 16 searches. Always one or more examples extended all the way to the right end of the chromosome. A similar study was carried out for all chromosomes with probes that were the reverse complement of the terminal 2-kb right ends. Also in every case several chromosomes were found with left terminal regions the reverse complement of the right terminal regions, and in every case the sequence similarity of at least one chromosome extended all the way to the left end. The program that was used, FASTA, selected the best-fitting regions, as it is designed to do, and the regions of similarity were often more extensive than exhibited in this initial search. To avoid missing significant similarities the best-scoring chromosomes found by the left end probes were divided into 1-kb-long fragments to form libraries that were searched with long probes that consisted of the reverse complement of the left ends of each of the 16 chromosomes. In this way all of the significant regions of reverse complementary sequence similarity were determined, often broken by internal nonmatching regions. The results are diagrammed in Fig. 1. Fig. 1 shows the right ends of the chromosomes with the matching region of the left end of the matching chromosome identified and the percent similarity (reverse complement) for 1-kb regions listed.
In 16 of 16 cases there are high quality reverse complement sequence similarities of left terminal regions at the right ends of different chromosomes. Often several chromosomes share in this sequence similarity, since there are extensive direct terminal region similarities among the same ends of the yeast
©
PNAS is available online at http://www.pnas.org.
* | To whom reprint requests should be addressed, e-mail: rbritten@etna.bio.uci.edu. |
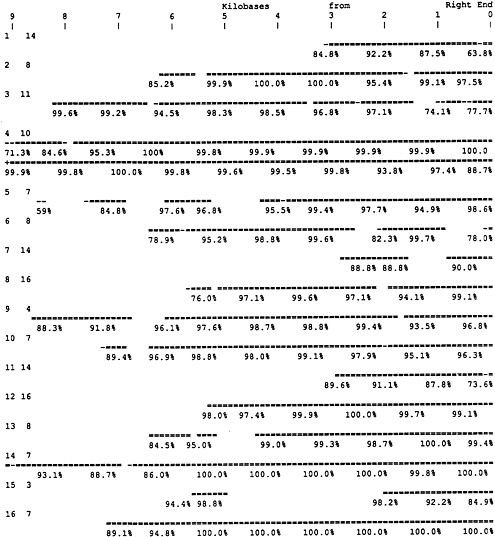
FIG. 1. Selected subset of right end sequences showing extensive similarity (reverse complement) to each of the 16 left ends. Long segments of the left ends of all 16 chromosomes were converted to reverse complement and compared with libraries of 1-kb segments of particular chromosomes, chosen on the basis of previous comparisons. The left column is the number of the chromosome from which the left end probe was extracted. The second column is the number of the chromosome with which it was compared. The top of the figure shows the position in the right end sequence in kilobases. The first example is for chromosome (chr) 1 left end compared with chr 14 and exhibits a just over 3-kb reverse complementary region terminating at the right end of chr 14. The double line shows the length of the matches found by FASTA and the % match is shown below. The next example, left end of chr 2 on right end of chr 8, is a case of Y′ repeat similarity and exhibits minor internal deletions or very poorly matching regions. The next example, chr 3 left end on chr 11, does not involve the Y′ repeat and is mostly made up of single-copy (2 copy) sequences. The next example, chr 4 left end (reverse complement) on chr 10, matches for 18.5 kb and requires two lines for display with the leftward part on the upper line, a very extensive reverse complementary region. This figure displays a subset of reverse complementary regions that does not necessarily include the longest and best-matching regions.
chromosomes. While these terminal regions include repeated sequences, they are not entirely composed of them. They include regions consisting of just the few copies resulting from the terminal relationships reported here. The precision of match often reaches 100% in the best-matching regions as shown in Fig. 1. In many cases the central parts of the overlapping regions have the highest precision, and in almost every case one or another of the end parts of the overlap have
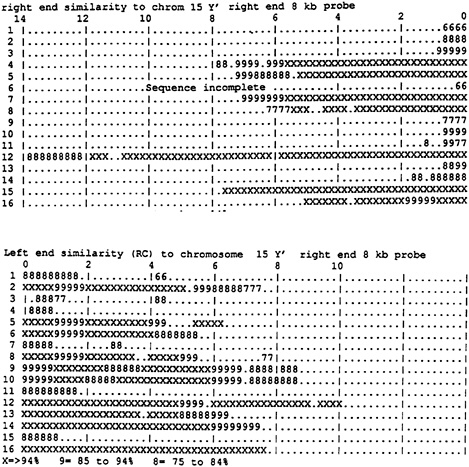
FIG. 2. Terminal region similarity to the Y′ repeat. The probe was 8 kb of the right end of chromosome 15. By using FASTA it was compared with a library of the whole yeast genome divided into 1-kb segments. (Upper) All of the direct sequence matches of the 16 chromosomes. All of the significant matches are at the right terminal regions that are displayed. At the top is a scale exhibiting kilobases from the ends. (Lower) Reverse complementary matches, all of which are in the left terminal regions. The symbols indicate the quality of match over the regions identified by FASTA. XXX represents greater than 94% match, while 999 represents from 85% to 94%. The other symbols follow the same pattern down to 666, which represents between 55% and 65% match. All of the longer matches are with the Y′-containing chromosome ends. The short matches at the ends are the X2 repeats to be described in a later section.
a lower precision. This suggests that multiple events of sequence exchange are responsible. In every case the telomeric end of the sequence similarity ceases only at the end of the known sequence.
A variety of searches indicate that there are no long direct sequence similarities between regions near (within 20 kb) the left and those near the right end of the same or any other chromosome. However, there are a few short and imprecise direct sequence similarities between the opposite ends of different chromosomes (200–400 nt and 70% identity or less). These appear to be members of short repeated sequence families that are near the termini. Their presence is almost certainly the result of a different set of phenomena from the long and precise sequence similarities. Often there are about 5-kb-long precise sequence similarities, both forward and reverse complement, between the TY elements at locations spread through the yeast chromosomes. The terminal regions include isolated long terminal repeats (LTRs) of these elements but none of the complete elements.
Genes in the Duplicated Terminal Regions. The presence or absence of genes in the regions shown in Fig. 1 was explored by using the detailed maps and tabular information of the Stanford Saccharomyces Genome Database (SGD: see acknowledgements). No genes were found that meet the stiff criterion that the gene must be genetically mapped as well as confirmed by the DNA sequence. However many ORFs, ranging up to more than 5 kb in length, are present in the terminal regions. The 18.5-kb inverted sequence similarity between the left end of chromosome 4 and the right end of chromosome 10 (Fig. 1) includes two genes identified on the basis of very good sequence similarity. These are listed in the SGD as related genes in the respective terminal regions. Only one of the regions listed in Fig. 1 does not have an ORF recognized and listed in the SGD, but the reverse complement on the matching chromosome in this case does have a listed ORF. The regions reported below, such as the extensive reverse complementary region between the left end of chromosome 16 and the right end of chromosome 15. share genes that are recognized as closely related in the SGD. In strains that carry the SUC gene it is found in this region between the X and Y′ repeats (3). In addition the Y′ repeats contain conserved ORFs that are expressed in meiosis (3). Recently the duplication of genomic ORFs has been examined (11), and
some of the complementary regions described here are reported in ref. 11 because they include ORFs.
Y′ Repeat Terminal Patterns. It is a matter of interest how much the known repeated sequences contribute to the terminal sequence similarities. Fig. 2 shows the sequence similarities to a particular Y′ repeat that occurs at the right terminal region of chromosome 15. All of the left end copies are the reverse complement of the right end copies, and all of the copies appear to be terminal in occurrence except that in some cases such as chromosome 12 right end there are additional inner copies. The Y′ repeat and other sequences in its region form a part of the similarities observed in this work. In some cases large lengths of Y′ sequence similarity (reverse complement) occur at both ends (chromosomes 5, 8, 12, and 16) but often there is Y′ sequence only at one end. The Y′ repeat occurs at about half of the ends and the expectation, if the chromosome ends that contain Y′ repeats were a random set, is that they would occur at both ends of four chromosomes, as observed. There are also three chromosomes for which both ends lack the Y′ repeats, another observation that suggests randomness. Nine chromosomes include a Y′ repeat at one end but lack it at the other (9).
Terminal Relationships as a Whole. To examine the terminal 20-kb sequence similarities as a whole, a large number of comparisons are required. Each chromosome left end was compared as reverse complement against the full length of all chromosomes. Direct sequence searches were also made with the left and right terminal regions, adding up to 768 comparisons. The results are shown diagramatically in Fig. 3. To make these comparisons the whole yeast genome was divided into about 12,000 1-kb segments in a library. FASTA was used to compare each of the terminal region 20-kb segments with the library. With this method all occurrences of significant sequence similarity are detected. Fig. 3 shows the regions of similarity to the reverse complement of the left end of each of the 16 chromosomes. The upper left block is the set of 16 similarities of the chromosome 1 left terminal 20-kb region (reverse complement). The next block below is the same for chromosome 2 and below that chromosome 3, etc. Fig. 3 Right gives this information for chromosomes 9–16. These similarities all occur in the right terminal regions of the 16 chromosomes. With the aid of a hand lens you will notice that the majority of these similarities are symbolized XXX and thus are better than 95% matches. Many are literally 100% matches. There are also a significant number of 999s, meaning 85–95% matches. The overall view without a hand lens shows the patterns very well. For example, the second diagram in Fig. 3 Left is for chromosome 2 left end (reverse complement), which contains the Y′ repeat, and thus the pattern shows all of the right ends that also contain the Y′ repeat. Thus it is clear that the left ends of chromosomes 2, 5, 6, 8, 9, 10, 12, 13, 14, and 16 contain the Y′ repeat, and similarly it is present on the right ends of chromosomes 4, 5, 7, 8, 12, 15, and 16, which have a consistent relationship to the other Y′ repeats. There is a lot of variation in these patterns and the Y′ repeat is merely a prominent part. Chromosome 1 is exceptional and includes the W′ repeat shown on the first line of Fig. 3 as more than 8 kb of precise reverse complement similarity between the two ends of chromosomes 1. In addition there is an extensive region of direct similarity to chromosome 8 right end (not shown). The long and precise direct sequence relationships of the left ends are restricted to the left ends of the other chromosomes and the extensive direct sequence similarities of the right ends all occur on right ends (not shown). Direct terminal region similarity data will appear on my web page, as there is not space here.
If a left end has few reverse complementary similarities to the right ends of other chromosomes it also has few direct similarities to the left ends of other chromosomes, reflecting the presence or absence of the Y′ repeats. There are many precise matches, including one very extensive match of 18.5 kb between 4 left end and 10 right end, mentioned earlier. At the very end of Fig. 3 is shown a match of nearly 20 kb between left chromosome 15 (reverse complement) and right chromosome 16. These surely resulted from extensive events of recombination or copying between the ends of inverted chromosome pairs. The extent to which the Y′ repeats are involved in exchanges is not obvious, but it seems likely that their precise sequence similarities are the result of both exchanges between opposite ends of different chromosomes and exchanges between same ends. There are several examples of extensive relationship separate from the Y′ sequences. For example, left ends of chromosomes 9 and 10 are direct copies of each other for 20 kb. A separate examination showed that the copying extends for another 2.1 kb toward the centromere.
All of the similarities in the terminal regions are shown in Fig. 3, and the long and precise terminal sequence relationships are restricted to the last 20 kb at each end shown there. In every case there are also examples of short and imperfect sequence similarity occurring in a variety of locations, representing short repetitive sequences that for the most part are members of unknown families. The number of these similarities to a given chromosome terminal region ranges from 3 to 16 except for long terminal repeats (LTRs) of mobile elements, for example on chromosome 2 and chromosome 15 left ends. In these cases delta elements are present that match about 100 other sequences. In addition, at 14 kb on chromosome 15 left end is a 200-bp-long element that matches about 60 other locations and is unknown to me. These short and imprecise and LTR sequence similarities are quite distinct categories of relationship from the major long and precise similarities that are involved in the pattern of exclusively complementary relationships between the left and right terminal regions.
Just-Subtelomeric Sequences and the “X2” Repeat. In attempting to And some indication of the function of the reverse complementary relationship between the ends it seemed that the very terminal sequence patterns might contain clues. Terminal short sequences (2 kb) of all chromosomes were multiply aligned with CLUSTALW, with the left end sequences present as reverse complement. The resulting alignments are quite good almost to the end. The interesting result is that the just-subtelomeric sequences fall into two classes with very good sequence similarity within the classes but none between the classes. The first class is made up of the telomeric end of the Y′ repeat. It occurs as the just-subtelomeric sequence of all of the Y′-containing chromosome ends as listed above. The second class includes the X repeat (3) but is more extensive and thus it has been named the X2 repeat to avoid confusion with previous descriptions of X repeats. It is present on all of the chromosome ends that do not contain the Y′ repeat, and Fig. 4 shows the consensus sequence for the 11 best-matching members. Most members agree with about 90% accuracy with the consensus, but chromosome 1 right end matches only 62%. There are other occurrences of the X2 sequence, which may be important. It occurs centromeric of the Y′ repeat on all of the Y′-containing chromosome ends. Thus the X2 sequence occurs on all chromosome ends, although the example 6 kb in from the left end of chromosome 5 is quite short (125 nt). It is possible that if the X2 repeat has a function it could be carried out from either location. There are small sequences between X and Ys known as STR sequences, some combination of which is found at most ends. These probably form a part of the X2 sequence. Most of the X2 repeats that occur centromeric of the Y′ repeats are well conserved and quite similar to those in just-subtelomeric locations. The conservation of the sequences of 30 of the X2 repeats cannot easily be explained by recombination because of their different locations.
The alignments of the just-subtelomeric sequences are of sufficient precision that they can be used to decide if chromo-
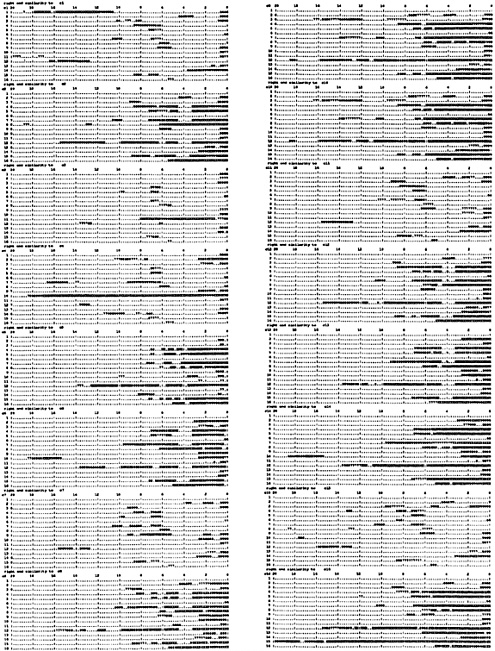
FIG. 3. Partial results of 768 comparisons of 20-kb probes with the complete yeast genome. Twenty-kilobase terminal segments of all 32 chromosome ends are compared as reverse complement with a library of the whole yeast genome divided into 1-kb segments, using FASTA. All of the regions where sequence similarity was recognized are indicated by numbers on the diagrams of the 20-kb terminal regions, each symbol representing 200 bp. The numbers describe the precision of match with the meaning mentioned in the legend of Fig. 2. In each block the 16 chromosomes are arranged in order from top to bottom, numbered at the left. At the top of each block are listed the distance from the end of the chromosomes in kb. The comparisons of left end reverse complements are shown and the other comparisons will be on my web page (www.cco.caltech.edu/~rbritten). This diagram describes the similarities to 20-kb reverse complement probes with a block for each probe, the upper left exhibiting the relationships of a probe from the left end of chromosome 1. The next block below is for a probe from chromosome 2 left end, and this probe contains the Y′ repeat, so that all of the chromosomes with a Y′ at their right ends show large regions of sequence similarity in this block. This relationship establishes the major visible pattern of Fig. 3. but there are many other features. Where a probe does not contain the Y′ repeat as for 1 or 3 the pattern is simpler and includes the just-subterminal similarities where the right end sequence does not contain the Y′ repeat. Where the right end sequence does contain the Y′ repeat, then a small block of similarity appears at about 6 kb in from the end. where the copy of the X2 repeat is present in these chromosomes.

FIG. 4. The X2 repeat consensus. The 11 chromosome ends that include very good copies of the just-subtelomeric X2 repeat were aligned by using CLUSTALW, allowing for the fact that left ends are reverse complements of right ends. A consensus sequence was made showing all positions in uppercase where six or more matched. The X2 consensus begins where there is general agreement between all examples and ends early in the telomerase simple sequence. In this 800-nt region all the 11 sequences match each other with high quality, between 86% and 93%. They are in complete agreement for 63% of the positions. The conserved core of the X repeat (3) begins at position 27 and agrees well to about position 400. The sequence has structure but is not recognizably a tandem repeat of a simpler sequence.
some sequences are complete to the ends. The analysis shows that half a dozen ends are incomplete, missing nearly 200 nt from the just-subtelomeric sequences. Nevertheless, for every end enough of the just-subterminal region is present to permit clear identification of the X2 repeat or the Y′ repeat. Most of the missing sequence ends are from Y′-containing ends apparently due to difficulties of cloning. An alternative could be that some other mechanism besides telomerase stabilizes the ends of the chromosomes and the listed sequences are correct, but for the present it seems safer to assume that the sequences are incomplete.
DISCUSSION
The terminal regions (the last 20 kb) of the yeast chromosomes stand out by sharing blocks of sequence with each other. They share only with each other and not with other regions of the chromosomes, except for short less precise repeats, which apparently have an origin different from the longer high precision sequence similarities. The consistent reverse complementary relationship of the left and right terminal regions suggests that there has been a history of exchange events occurring between inverted pairs of chromosomes. Consider the third block down in Fig. 3 Left, diagramming the similarities to chromosome 3 left end. There is an extensive block of nearly 7 kb of similarity to chromosome 11 right end that is the product of sequence exchange or copying between inverted pairs of chromosomes. However, the similarities to all the other chromosomes are due to the fact that the left end of chromosome 3 contains the X2 repeat. There are the just-subtelomeric similarities to the right ends of chromosomes 1, 2, 3, 6, 9, 10, 11, 13, and 14. Also there are similarities to the X2 repeats at the inner or centromeric ends of the Y′ repeats on the other chromosomes. These X2 repeat similarities must be considered to be due to selection on these sequences rather than exchange. That is because they occur in both the inner and terminal locations, and it is difficult to see how exchange could be limited to these short X2 repeats.
Nevertheless, all of these relationships are reverse complementary between the left and right ends. The pattern observed for chromosome 3 is frequent and is shown in Fig. 3 for all cases where the probe left end sequence does not include a Y′ repeat. What explains the reverse complementary relationship between left and right ends where massive exchange between opposite ends does not seem to have occurred recently? It seems likely that the telomerase simple sequences have a required orientation with respect to the ends and thus must be reverse complements of each other. It is assumed that DNA synthesis proceeds outward toward the telomeres. While this is consistent with the reverse complementary relationships of the whole terminal regions, it does not supply a reason for them except for the telomerase simple sequences. There is probably a functional reason for the orientation of the X2 sequences, having to do with specific protein bindings that are part of the complex control system that stabilizes the chromosome ends.
Candidate models for the exchange process are recombination by breakage and rejoining, sequence conversion, or some unknown mechanism for sequence copying and insertion. The insertions and deletions and mismatches within the similar regions shown in Fig. 1 suggest that in many cases multiple events have occurred and that deletions and base substitutions leading to mismatch have occurred subsequent to the events that created precise inverted sequence matches. It is likely that the process has been regenerative, because once inverted matching sequence regions are present between opposite ends the chromosomes could be aligned by the mechanisms that permit normal recombination to occur between matching pairs of diploid chromosomes. The existence of many near perfect (95–100%) matching regions in Figs. 1, 2, and 3 suggests that the typical event has been quite recent, and only a little base substitution has occurred since. Thus the events are quite frequent on an evolutionary time scale. It is interesting that in many cases the terminal simple sequence created by telomerase is included in the sequence similarity. In every alignment for which both sequences extend into the telomerase simple sequences the alignment extends as far as the sequences go and remains good but not perfect. It may be due to patterns created
by the telomerase, but exchange has to be considered as partially responsible. These exchanges very likely influence the stability of the terminal regions of the yeast chromosomes (4).
It is clear that the known long repeats of the terminal regions—e.g., Y′, X′, and W′—are an intimate part of the process of terminal region sequence exchange that is probably responsible for the patterns shown in Fig. 3. It seems likely that they originate in this process. That leaves open the question as to whether all of these duplications and multiplications are simply the inevitable result of the process of terminal region sequence exchange. Whether they have useful functions is yet uncertain, though the X2 repeat (Fig. 4) is well conserved and present on every chromosome end. There are no examples of precise and long direct sequence similarity between terminal regions on the opposite ends of chromosomes. This finding suggests that the orientation of these sequences or part of them is important to yeast survival. These sequences point outwards (or inwards depending on point of view) from all yeast chromosomes. They are terminated by the simple sequences generated by telomerase. The telomerase sequences are clearly significant to chromosome stability and replication, but there is good evidence that they carry out other functions. Changing their length affects survival (4).
The central issue is, of course, the evolutionary role and potential function of the reverse complementary relationship of the terminal regions, but little can yet be said. Finally, it seems very unlikely that this pattern of asymmetry is restricted to yeast chromosomes. As the human genome project advances so that sufficient lengths of terminal regions are available it will be interesting to see how well the reverse complementary relationship holds in our own genome. The prediction is that it will be very similar to the yeast situation with allowance for different telomerase synthesized sequences and lengths and distinct sets of repeats in the subterminal regions.
The yeast chromosomal sequences were obtained from the Stanford SGD http://genome-www.stanford.edu/Saccharomyces/. Thanks to Ed Louis for preprints. Johnny Williams prepared useful software in Perl language. This work was supported by National Institutes of Health grants.
1. Goffeau, A., Aert, R., Agostini-Carbone, M.L., Ahmed, A., Aigle, M., Alberghina, L., Albermann, K., Albers, M., Aldea, M., Alexandraki, D., et al. (1997) Nature (London) Suppl. 387, 5–105.
2. Pryde, F.E., Gorham, H.C. & Louis, E.J. (1997) Curr. Opin. Genet. Dev. 7, 822–828.
3. Louis, E.J. (1995) Yeast 11, 1553–1573.
4. Zakian, V.A. (1996) Annu. Rev. Genet. 30, 141–172.
5. Kramer, K.M. & Haber, J.E. (1993) Genes Dev. 7, 2345–2356.
6. Wellinger, R.J., Ethier, K., Labrecque, P. & Zakian, V.A. (1996) Cell 85, 423–433.
7. Louis, E.J., Naumova, E.S., Lee, A., Naumov, G. & Haber, J.E. (1994) Genetics 136, 789–802.
8. Flint, J., Bates, G.P., Clark, K., Dorman, A., Willingham, D., Roe, B.A., Micklem, G., Higgs, D.R. & Louis, E.J. (1997) Hum. Mol. Genet. 6, 1305–1314.
9. Louis, E.J. & Borts, R.H. (1995) Genetics 139, 125–136.
10. Pearson, W.R. & Lipman, D.J. (1988) Proc. Natl. Acad. Sci. USA 85, 2444–2448.
11. Coissac, E., Maillier, E. & Netter, P. (1997) Mol. Biol. Evol. 14, 1062–1074.







