This paper was presented at the National Academy of Sciences colloquium “Proteolytic Processing and Physiological Regulation,” held February 20–21, 1999, at the Arnold and Mabel Beckman Center in Irvine, CA.
VanX, a bacterial D-alanyl-D-alanine dipeptidase: Resistance, immunity, or survival function?
IVAN A. D. LESSARD AND CHRISTOPHER T. WALSH*
Biological Chemistry and Molecular Pharmacology Department, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115
ABSTRACT The zinc-containing D-alanyl-D-alanine (D-Ala-D-Ala) dipeptidase VanX has been detected in both Gram-positive and Gram-negative bacteria, where it appears to have adapted to at least three distinct physiological roles. In pathogenic vancomycin-resistant enterococci, vanX is part of a five-gene cluster that is switched on to reprogram cell-wall biosynthesis to produce peptidoglycan chain precursors terminating in D-alanylD-lactate (D-AlaD-lactate) rather than D-AlaD-Ala. The modified peptidoglycan exhibits a 1,000-fold decrease in affinity for vancomycin, accounting for the observed phenotypic resistance. In the glycopeptide antibiotic producers Streptomyces toyocaensis and Amylocatopsis orien talis, a vanHAX operon may have coevolved with antibiotic biosynthesis genes to provide immunity by reprogramming cell-wall termini to D-AlaD-lactate as antibiotic biosynthesis is initiated. In the Gram-negative bacterium Escherichia coli, which is never challenged by the glycopeptide antibiotics because they cannot penetrate the outer membrane permeability barrier, the vanX homologue (ddpX) is cotranscribed with a putative dipeptide transport system (ddpABCDF) in stationary phase by the transcription factor RpoS (ss). The combined action of DdpX and the permease would permit hydrolysis of D-AlaD-Ala transported back into the cytoplasm from the periplasm as cell-wall crosslinks are refashioned. The D-Ala product could then be oxidized as an energy source for cell survival under starvation conditions.
Much attention has been focused recently on the alarming increase in antibiotic resistance in bacterial pathogens (1–3). The explosive emergence of vancomycin-resistant enterococci as life-threatening organisms in hospital settings worldwide (4–6) has led to intensive investigation of the molecular determinants of glycopeptide antibiotic resistance (7–11). These investigations have revealed one of the most sophisticated molecular systems of acquired resistance and a paradigm of genetic adaptation (4). Vancomycin resistance uses a strategy of reprogramming the termini of peptidoglycan (PG) intermediates in cell-wall crosslinking steps from D-alanyl-D-alanine (D-Ala-D-Ala) termini to D-alanyl-D-lactate (D-Ala-D-lactate) termini. The modified PG binds vancomycin 1,000-fold less avidly than the D-Ala-D-Ala PG because of the loss of a central hydrogen bond from the NH of the D-Ala-D-Ala moiety to the vancomycin backbone carbonyl, accounting quantitatively for the gain in phenotypic resistance (9) (Fig. 1 A–C). A three-gene operon vanHAX found on a transposable element directs the reprogramming with VanH and VanA proteins acting sequentially to synthesize D-Ala-D-lactate while VanX selectively hydrolyzes D-Ala-D-Ala produced by the host enzyme but not D-Ala-D-lactate, allowing the depsipeptide to accumulate and become incorporated into the growing PG termini (9, 10, 12). The amounts of VanH, -A, and -X in the cells are in turn controlled by a two-component regulatory system involving a transmembrane sensor kinase VanS and a response regulating transcription factor VanR that becomes active when phosphorylated by VanS (11, 13), after the established paradigms for monitoring of environmental cues.
Although all five of the necessary and sufficient proteins, VanR, -S, -H, -A, and -X, have now been characterized, this paper addresses some broader biological questions that have recently arisen around the functions of VanX in diverse bacterial physiology. In particular, VanX homologues have been discovered in the bacteria that produce vancomycin and related glycopeptide antibiotics (14, 15) as well as in Escherichia coli, a Gram-negative bacterium that is intrinsically indifferent to vancomycin because of the failure of the antibiotic to penetrate the outer membrane barrier (15) (Table 1).
Enterococcal VanX (EntVanX): A Zinc-Dependent D-ALA-D-Ala Dipeptidase of Exquisite Specificity. The first indication of function of EntVanX was provided by Reynolds et al. (10) with the observation that overproduction in E. coli led to activity in the crude extract that hydrolyzed D-Ala-D-Ala but not D-Ala-D-lactate in a ß-lactam-insensitive manner. The purification of EntVanX was then undertaken in this laboratory (16, 17) with maltose-binding protein (MBP)-EntVanX fusion under control of the T7 promoter being used to solve problems of protein aggregation, purification, and most notably toxicity to E.coli (17). The substrate specificity was exclusive for D, D-dipeptides with unmodified N and C termini, and catalytic efficiency analysis suggested up to 1010-fold selection for D-Ala-D-Ala hydrolysis compared with D-Ala-D-lactate, a contrathermodynamic selection for amide over ester bond hydrolysis (15, 16) (Table 2). The MBP-EntVanX active site binds one catalytically essential zinc atom (17). Sequence analysis did not detect consensus catalytic zinc-binding motifs, but comparison with the functional homolog zinc-dependent N-acyl-D-Ala-D-Ala carboxypeptidase from Streptomyces albus G and with the zinc-containing N-terminal domain of murine Sonic hedgehog suggested a motif using His-116, Asp-123, and His-184 (EntVanX) as the zinc ligand set with a conserved Glu-181 as a catalytic base. These predictions were validated first by site-directed mutagenesis to correlate zinc content and catalytic activity (17) and most recently by the determination of the x-ray structure of EntVanX by Bussiere et al. at Abbott Laboratories (18) of the free enzyme as well as complexes with D-Ala-D-Ala and a slow binding phosphinate analog (19) of the proposed tetrahedral reaction intermediate (Fig. 2 A and B). The structure indicates that EntVanX is a variant of a metallo aminopeptidase and that the small constricted active site cavity of 150 Å3 may make rational design of inhibitors a significant
|
|
PNAS is available online at www.pnas.org. |
|
|
Abbreviations: A2pm, diaminopimelate; D-Ala-D-Ala, D-alanyl-D-alanine; D-Ala-D-lactate, D-alanyl-D-lactate; EntVanX, enterococcal VanX; DdpX, Escherichia coli VanX homolog; PG, peptidoglycan; StoVanX, Streptomyces toyocaensis VanX homolog. |
|
* |
To whom reprint requests should be addressed. E-mail: walsh@walsh.med.harvard.edu. |
FIG. 1. (A) Vancomycin binds the D-Ala-D-Ala moiety of the growing peptidoglycan and sterically occludes the transglycosylation and transpeptidation steps of cell-wall assembly. The immature cell wall results in cells susceptible to lysis through osmotic shock. (B) The alternative cell-wall biosynthetic pathway of the VanH, -A, -X proteins, producing peptidoglycan intermediates with D-Ala-D-lactate termini in place of the usual D-Ala-D-Ala termini (12). Pyruvate is reduced to D-lactate by the NADP-dependent dehydrogenase VanH, which is then used as substrate for the ATP-dependent D-Ala-D-lactate depsipeptide ligase VanA. The product D-Ala-D-lactate depsipeptide is used by the enzyme MurF to produce the muramyl-peptidyl-D-lactate intermediate and brought forward in subsequent cell-wall biosynthesis. The zinc-dependent D, D-dipeptidase VanX, specifically hydrolyzes the D-Ala-D-Ala dipeptide pool produced by the native D-Ala-D-Ala Ddl ligase without hydrolyzing the D-Ala-D-lactate and in this way effectively shunts the flux of the cell-wall biosynthesis to the ester termini. Substitution of D-Ala by D-lactate does not impair crosslinking of the modified precursors to the growing peptidoglycan chain, resulting in a mechanically strong peptidoglycan layer and cell survival. (C) Structures of the vancomycin complexes with N-acyl-D-Ala-D-Ala and N-acyl-D-Ala-D-lactate (9). Vancomycin binds to the D-Ala-D-Ala termini through a five-hydrogen bond network. The key hydrogen bond between the D-Ala amide NH and the vancomycin backbone carbonyl is lost in the N-acyl-D-Ala-D-lactate complex, resulting in a 1,000-fold reduction in the affinity of the antibiotic.
Table 1. VanX homologs
|
VanX source |
Role |
|
Vancomycin-resistant enterococci Enterococcus faecium Enterococcus faecalis |
Reprogram cell walls for vancomycin resistance in opportunistic pathogens |
|
Glycopeptide producers Streptomyces toyocaensis Amycolatopsis orientalis |
Coevolution of vanHAX operon with antibiotic biosynthesis genes for immunity |
|
Stationary-phase survival mechanism Escherichia coli |
Transport D-Ala-D-Ala from periplasm back to cytoplasm as cell-wall crosslinks are refashioned and use as RpoS-mediated energy source |
|
|
|
medicinal chemistry challenge. The likely mechanism for D, D-dipeptide hydrolysis, is shown in Fig. 2C with Glu-181 acting as catalytic base and Arg-71 as a cationic coordinator both in the ground state and for stabilization of the developing negative charge in the tetrahedral adduct (18, 20). The structure predicted Asp-123, Asp-142, and Tyr-21 residues for the recognition of the D-Ala-D-Ala dipeptide substrate a-NH3+ and Ser-114 for the carboxylate group. Site-directed mutagenesis of active site residues has revealed roles consistent with predictions of recognition and catalysis (20).
VanX Homologs in the Bacteria That Produce Vancomycin and Related Glycopeptide Antibiotics. In many instances, bacteria that produce antibiotics have evolved strategies and mechanisms that provide immunity to the action of the antibiotic, and there is a general supposition that immunity mechanisms will have coevolved with antibiotic biosynthesis genes to protect the producing organisms (21, 22). Streptomyces toyocaensis synthesizes and secretes a vancomycin-type glycopeptide antibiotic (A47934), and the molecular basis of immunity for this organism and most likely for Amylocatopsis orientalis, which produces vancomycin, has been recently deconvolved (14, 15, 23, 24). PCR probes to EntVanX zinc-binding motif revealed an S.toyocaensis VanX homologue (StoVanX) with 63% similarity to EntVanX, and sequencing analysis then indicated a three-gene operon in S.toyocaensis and A.orientalis equivalent and similarly oriented to the vanHAX operon from (Fig. 3A). Expression and purification of the StoVanX confirms it is a high efficiency D,D-dipeptidase with unmodified N and C termini and that it. lacks D-Ala-D-lactate depsipeptide activity (15) (Table 2). These findings suggest a conserved mechanism for the observed intrinsic resistance of the antibiotic producers to the vancomycin class of glycopeptides and that before S.toyocaensis produces the glycopeptide A47934, it has D-Ala-D-Ala peptidoglycan termini and is sensitive to vancomycin-type antibiotics. Furthermore, S.toyocaensis possesses two D-, D-ligases: a D-Ala-D-lactate ligase encoded by the vanHAX operon and a D-Ala-D-Ala ligase encoded by a separate gene on the chromosome (24). The S.toyocaensis vanHAX equivalents may be switched on transcriptionally when the host turns on the cluster of genes to synthesize the glycopeptide A47934 to reprogram PG
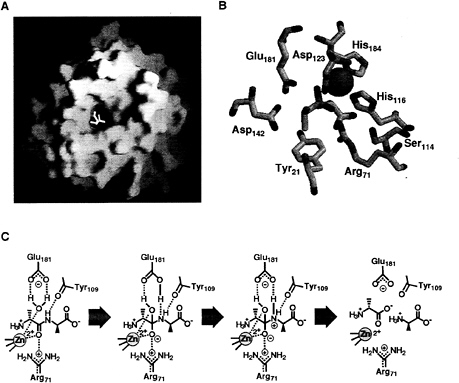
FIG. 2. (A) Structure of EntVanX (18). (B) Active site topology of EntVanX complex with the phosphinate analog (18). The zinc atom is coordinated with His-116, Asp-123, and His-184. The phosphinate analog a-NH3+ hydrogen bonds with Asp-123, Asp-142, and Tyr-21, whereas Ser-114 hydrogen bonds with the carboxylate group. Arg-71 stabilizes the transition state intermediate, represented by the phosphinate analog. Glu-181 is the catalytic base. (C) Proposed mechanism of VanX (20). The water molecule is activated by Glu-181 and attacks the zinc-polarized carbonyl to form a tetrahedral adduct, which is then stabilized by both the zinc atom and the Arg-71. The Glu-181 transfers the proton to the nitrogen, which is hydrogen bonded to the carbonyl group of Tyr-109; peptide bond cleavage follows [C; reprinted from ref. 20 with kind permission from Elsevier Science (Amsterdam)].
termini to end in D-AlaD-lactate, providing in situ resistance to the produced antibiotic (Fig. 3B). This three-gene operon has also been detected in other glycopeptide-producing organisms (14), and one of these operons may have been the origin for the enteroccocal vanHAX genes that are on trans
Table 2. Catalytic efficiencies of VanX homologs on zinc-dependent D-AlaD-Ala dipeptidases (15)
posable elements in most of the VanA clinical phenotypes of vancomycin-resistant enterococci (VRE). Noticeably, the G+ C content of the vanHAX operon in VRE is 5–10% higher than the adjacent vanSR genes and chromosomal genes of enterococci. These findings also exemplify a mechanism for coevolution of glycopeptide antibiotic production and glycopeptide antibiotic resistance, the latter then appropriated by the opportunistic pathogenic enterococci.
The Dilemma for E.coli Strains That Contain and Express the VanX Homolog (ddpX). Analysis of the E.coli genome database turned up a possible VanX homologue [originally referred to as EcoVanX (15) and renamed here DdpX] with 27% similarity to EntVanX. Expression and purification validated the expected activity, although the KM of 14 mM for D-AlaD-Ala was 250- to 3,000-fold elevated compared with the EntVanX and StoVanX enzymes (15), consistent with a purely degradative function for the DdpX (Table 2). All of the active site residues and auxiliary residues that maintain the active-site topology in EntVanX are conserved in DdpX, and kinetic analysis also revealed the same substrate specificity and discrimination between peptide bond cleavage (D-AlaD-Ala)
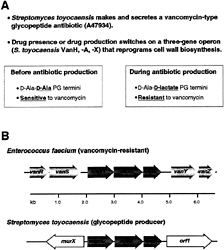
FIG. 3. (A) Comparison of the proposed peptidoglycan termini before and after A47934 antibiotic production by S.toyocaensis. (B) Comparison of the glycopeptide resistance gene operon from vancomycin-resistant enterococci (Enterococcus faecalis) and the glycopeptide producer S.toyocaensis (14).
and the analogous ester-bond cleavage (D-Ala-D-lactate) reported for EntVanX and StoVanX (15). Homology modeling of DdpX with the crystal structure of EntVanX divulges a striking similarity in the overall structure, with the highest identity seen within the key catalytic residues, as expected for the similarity in substrate specificity (15).
It was not immediately apparent why a Gram-negative bacterium such as E.coli would contain a VanX enzyme, because the outer membrane barrier provides effective intrinsic resistance to the glycopeptide class of antibiotics. Further, there was no evidence of VanH or VanA homologs in the genome, so there would be no reprogramming of termini of peptidoglycan intermediates. A further aspect of the dilemma was our prior observation that expression of active EntVanX enzyme in E.coli was toxic and led to cell lysis (17), precisely what would be expected for hydrolytic removal of the key D-Ala-D-Ala building block required for cell-wall synthesis and crosslinking.
The existence of Ddp raised the question whether there was any situation in which E.coli could live or would want to live without D-Ala-D-Ala for cell-wall biosynthesis. Inspection of the ddpX gene suggested two clues. First, immediately downstream was a five-gene cluster (ddpABCDF) withallthe hallmarks of a peptide permease cluster, including periplasmic binding protein, transmembrane proteins, and ABC subunits ATPase ORFs (Fig. 4A). Second, the promoter region of ddpX has two candidates for—10 consensus sequences for the RpoS (ss) alternative sigma factor of RNA polymerase (15, 25). The RpoS subunit is switched on in early stationary phase and is a central regulator of transcription of many genes that contribute to survival of the E.coli cell under starvation conditions (conditions that prevail in nature) (26). Indeed, analysis of the ddpX promoter fused to lacZ verified that ddpX is turned on on entry into stationary phase and furthermore that the mRNA also shown to be produced in stationary phase included the five adjacent candidate permease genes (ddpABCDF) (15). This operon has been named ddpXABCDF (D, D-peptide).
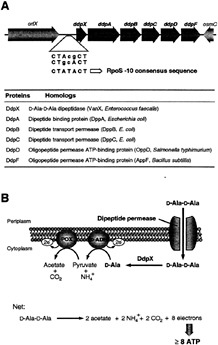
FIG. 4. (A) Gene organization at 33.7 min of the E.coli chromosome. ddpXABCDF, orfX, hypothetical protein gene product; osmC, gene for osmotically inducible protein; dipeptide permease homolog gene products are indicated. The ddpX and dipeptide permease genes (ddpABCDF) form an operon (ddpXABCDF) that is turned on at entrance into stationary phase by the RpoS (ss) transcription factor of RNA polymerase. The consensus (25) and putative RpoS (ss)—10 regions are indicated. (B) Proposed action of the DdpX and the dipeptide permease (DdpABCDF) (15). During stationary phase, periplasmic D-Ala-D-Ala dipeptide is transported by the Ddp permease transport system into the cytoplasm where it could be processed by the DdpX to release two equivalents of D-Ala. The D-Ala monomer is then converted to acetate by the sequential action of D-amino acid dehydrogenase (D-ADH) and pyruvate oxidase (POX) for production of energy during starvation. Periplasmic D-Ala-D-Ala could arise during the A2pm-A2pm crosslink formation, which increases up to 13% of total peptidoglycan crosslinks during stationary phase (26).
When E.coli was assessed for its ability to grow on D-Ala or D-Ala-D-Ala as the sole carbon source, it could use the monomer, oxidized by the membrane enzyme D-amino acid dehydrogenase, but not the dipeptide unless both the ddpX and the five permease genes were specifically up-regulated: the permease can therefore transport D-Ala-D-Ala into the cell (15). At this juncture, the pathway depicted in Fig. 4B can be understood. In stationary phase, the D, D-dipeptide permease, DdpX, and pyruvate oxidase areallproduced under RpoS control to enable the import and net oxidation of the D, D-dipeptide to two molecules of acetate and CO2, while eight electrons are funneled down the respiratory chain to provide energy for survival.
The last question is where free D-Ala-D-Ala in the periplasm comes from under starvation conditions. One possible source is from release during a crosslinking of the peptidoglycan layer.
It has been reported that the frequency of a direct crosslink between two diaminopimelate (A2pm) residues on adjacent PG strands rises from 2% in exponential phase to 13% in stationary phase (27). Although the substrate strands for this unusual crosslink have yet not been identified, if they are the normal pentapeptide strands, this crosslink would release the D-Ala-D-Ala dipeptide. The E.coli A2pm-A2pm transpeptidase has not been identified, but it has been suggested that the L, D-dipeptidylcarboxypeptidase cleaving the muramylpeptidyl-L-A2pm-D-Ala-D-Ala at the L-A2pm-D-Ala peptide bond releasing D-Ala-D-Ala previously described in E.coli (28) could be the long-sought A2pm-A2pm transpeptidase (15). The purpose of the reprogramming of the crosslinks in stationary phase may be driven by the transshipment of the released D, D-dipeptide from periplasm back into cytoplasm to power the cell in starvation mode. Although the cell is catabolizing the cell wall, it cannot decrease the net crosslinking or the mechanical strength will be insufficient to withstand osmotic pressure for lysis, hence the need to switch to A2pm-A2pm linkages.
VanX, a Dipeptidase for All Seasons? The three examples noted in this paper reveal distinct niches for the zinc-dependent D-Ala-D-Ala dipeptidase. It may have arisen in the Gram-positive glycopeptide antibiotic producers at the same time as the ability to biosynthesize these antibiotics, providing selective immunity to the bacteria that could both make the antibiotics and reprogram their cell walls to lower the target affinity. Other Gram-positive bacteria in the soil such as lactobacilli, leuconostoc, and pediococci are intrinsically resistant to vancomycin and were examined (29–32) to have also chosen the D-Ala-D-lactate route. In recent times, the opportunistic enterococci have imported the vanHAX gene operon on transposons and plasmids to gain survival advantage via antibiotic resistance in hospital environments that have seen an order of magnitude increase in the therapeutic use of vancomycin in the past 15 yr (33). The Gram-negative E.coli is not challenged by the impermeable glycopeptide antibiotics and has its own version of VanX, but not VanH or VanA. DdpX is a potentially lethal enzyme, because it removes the necessary metabolite D-Ala-D-Ala during peptidoglycan synthesis and is turned on only in the extreme challenge of stationary phase when starvation threatens and the D-Ala-D-Ala termini of uncrosslinked peptidoglycan strands are retrieved from the periplasm and burned as a metabolic fuel. As additional bacterial genomes are sequenced, more VanX protein homologs are likely to be discovered. Indeed, in the Gram-negative Synechocystis sp PCC6803, a VanX homolog (16% similarity with EntVanX) possessing kinetic parameters similar to DdpX was detected, but notably it hydrolyzes both L, D- and D, D-dipeptides similarly. It is thus proposed to play a role in scavenging both L, D- and D, D-dipeptide products of cell-wall degradation pathways (15). In the Gram-positive pathogen Mycobacterium tuberculosis, the VanX homolog (21% similarity with EntVanX) possesses all the requirements necessary for dipeptide recognition and catalysis but presents an apparent signal sequence and membrane lipoprotein attachment site, suggesting that MtuVanX might reside in the membrane.
We are grateful to Abbott Laboratories for providing the coordinates of the EntVanX crystal structure. We thank members of the Walsh laboratory for helpful and insightful discussions. I.A.D.L. acknowledges the Medical Research Council of Canada for Postdoctoral Fellowship supports. This research was supported in part by National Institutes of Health Grants GM21643 and by funds from Abbott Laboratories.
1. Neu, H.C. (1992) Science 257, 1064–1073.
2. Tomasz, A. (1994) N.Engl. J.Med. 330, 1247–1251.
3. Swartz, M.N. (1994) Proc. Natl. Acad. Sci. USA 91, 2420–2427.
4. Leclercq, R. & Courvalin, P. (1997) Clin. Infect. Dis. 24, 545–554.
5. Murray, E. (1997) Am. J. Med. 102, 284–293.
6. Cunha, B.A. (1995) Med. Clin. N. Am. 19, 817–831.
7. Arthur, M. & Courvalin, P. (1993) Antimicrob. Agents Chemother. 37, 1563–1571.
8. Barna, J.C.J. & Williams, D.H. (1984) Annu. Rev. Microbiol 38, 339–357.
9. Bugg, T.D.H., Wright, G.D., Dutka-Malen, S., Arthur, M., Courvalin, P. & Walsh, C.T. (1991) Biochemistry 30, 10408– 10415.
10. Reynolds, P.E., Depardieu, F., Dutka-Malen, S., Arthur, M. & Courvalin, P. (1994) Mol Microbiol. 13, 1065–1070.
11. Wright, G.D., Holman, T.R. & Walsh, C.T. (1993) Antimicrob. Agents Chemother. 36, 1514–1518.
12. Walsh, C.T., Fisher, S.L., Park, I.-S., Prahalad, M. & Wu, Z. (1996) Chem. Biol. 3, 21–28.
13. Arthur, M., Molinas, C. & Courvalin, P. (1992) J. Bacteriol. 174, 2582–2591.
14. Marshall, C.G., Lessard, I.A.D., Park, I.-S. & Wright, G.D. (1998) Antimicrob. Agents Chemother. 42, 2215–2220.
15. Lessard, I.A.D., Pratt, SD, McCafferty, D.G., Bussiere, D.E., Hutchins, C., Wanner, B.L., Katz, L. & Walsh, C.T. (1998) Chem. Biol 5, 489–504.
16. Wu, Z., Wright, G.D. & Walsh, C.T. (1995) Biochemistry 34, 2455–2463.
17. McCafferty, D.G., Lessard, I.A.D. & Walsh, C.T. (1997) Biochemistry 36, 10498–10505.
18. Bussiere, D.E., Pratt, SD, Katz, L. Severin, J.M., Holzman, T. & Park, C. (1998) Mol. Cell 2, 75–84.
19. Wu, Z. & Walsh, C.T. (1995) Proc. Natl. Acad. Sci. USA 92, 11603–11607.
20. Lessard, I.A.D. & Walsh, C.T. (1999) Chem. Biol. 6, 177–187.
21. Cundliffe, E. (1992) in Secondary Metabolites: Their Function and Evolution, Ciba Foundation Symposium 171 (Wiley, Chichester), pp. 199–214.
22. Cundliffe, E. (1989) Annu. Rev. Microbiol. 43, 207–233.
23. Marshall, C.G., Braodhead, G., Leskiw, B. & Wright, G.D. (1997) Proc. Natl. Acad. Sci. USA 94, 6480–6483.
24. Marshall, C.G. & Wright, G.D. (1997) FEMS Microbiol. Lett. 157, 295–299.
25. Espinosa-Urgel, M., Chamizo, C. & Tormo, A. (1996) Mol. Microbiol. 21, 657–659.
26. Hengge-Aronis, R. (1996) in Escherichia coli and Salmonella, Cellular and Molecular Biology, eds. Neidhardt, F.C., Curtiss, R., Ingraham, J.L., Lin, E.C.C., Low, K.B., Magasanik, B., Reznikoff, W.S., Riley, M., Schaechter, M. & Umbarger, H.E. (Am. Soc. Microbiol., Washington, DC), pp. 1497–1512.
27. Tuomanen, E., Markiewicz, Z. & Tomasz, A. (1988) J. Bacteriol. 170, 1373–1376.
28. Gondré, B., Flouret, B. & van Heijenoort, J. (1973) Biochimie 55, 685–691.
29. Dartois, V., Phalip, V., Schmitt, P. & Divies, C. (1995) Cremoris. Res. Microbiol. 146, 291–302.
30. Elsha, B.G. & Courvalin, P. (1995) Gene 152, 79–83.
31. Park, I.-S. & Walsh, C.T. (1997) J. Biol. Chem. 272, 9210–9214.
32. Billot-Klein, D., Gutmann, L., Sablé, S., Guittet, E. & van Heijenoort, J. (1994) J. Bacteriol. 176, 2398–2405.
33. Kirst, H.A., Thompson, D.G. & Nicas, T.I. (1998) Antimicrob. Agents Chemother. 42, 1303–1304.





