This paper was presented at the National Academy of Sciences colloquium “Proteolytic Processing and Physiological Regulation,” held February 20–21, 1999, at the Arnold and Mabel Beckman Center in Irvine, CA.
Reverse biochemistry: Use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue
TOSHIHIKO TAKEUCHI*, MARC A. SHUMAN†, AND CHARLES S. CRAIK*‡
*Departments of Pharmaceutical Chemistry and Biochemistry & Biophysics, and †Department of Medicine, University of California, San Francisco, CA 94143
ABSTRACT Serine proteases of the chymotrypsin fold are of great interest because they provide detailed understanding of their enzymatic properties and their proposed role in a number of physiological and pathological processes. We have been developing the macromolecular inhibitor ecotin to be a “fold-specific” inhibitor that is selective for members of the chymotrypsin-fold class of proteases. Inhibition of protease activity through the use of wild-type and engineered ecotins results in inhibition of rat prostate differentiation and retardation of the growth of human PC-3 prostatic cancer tumors. In an effort to identify the proteases that may be involved in these processes, reverse transcription-PCR with PC-3 poly(A)+ mRNA was performed by using degenerate oligonucleotide primers. These primers were designed by using conserved protein sequences unique to chymotrypsinfold serine proteases. Five proteases were identified: urokinase-type plasminogen activator, factor XII, protein C, trypsinogen IV, and a protease that we refer to as membrane-type serine protease 1 (MT-SP1). The cloning and characterization of the MT-SP1 cDNA shows that it encodes a mosaic protein that contains a transmembrane signal anchor, two CUB domains, four LDLR repeats, and a serine protease domain. Northern blotting shows broad expression of MT-SP1 in a variety of epithelial tissues with high levels of expression in the human gastrointestinal tract and the prostate. A His-tagged fusion of the MT-SP1 protease domain was expressed in Escherichia coli, purified, and autoactivated. Ecotin and variant ecotins are subnanomolar inhibitors of the MT-SP1 activated protease domain, suggesting a possible role for MT-SP1 in prostate differentiation and the growth of prostatic carcinomas.
Serine proteases possessing a chymotrypsin fold are of great interest because they provide detailed understanding of their enzymatic properties and their proposed role in a number of physiological and pathological processes. A wealth of information exists on structure-function relationships regarding this large class of enzymes. Moreover, potent and specific inhibitors are readily available for use in dissecting the function of these enzymes. These proteases exist as precursors that are activated by specific and limited proteolysis, allowing regulation of enzyme activity (1). Examples of this type of regulation include blood coagulation (2), fibrinolysis (3), complement activation (4), and trypsinogen activation by enteropeptidase in digestion (5). The precise control of these activation processes is crucial for normal physiological enzymatic function; misregulation of these enzymes can lead to pathological conditions (2–5).
We are interested in studying the role of these chymotrypsin-fold serine proteases in cancer by using a “fold-specific” inhibitor, ecotin (6, 7). Ecotin or engineered versions of ecotin can be introduced into complex biological systems as probes of proteolysis by these chymotrypsin-fold proteases. If effects are observed on treatment with these unique inhibitors, then the large body of knowledge concerning the biochemistry of these proteases can be tapped to understand the structure and function of the target proteases. For example, the molecular cloning, structural modeling, and mechanistic understanding of the enzymes are immediately accessible. We refer to this approach, which is analogous to “reverse genetics,” as “reverse biochemistry,” and we have applied it to identification of specific serine proteases in prostate cancer.
Urokinase-type plasminogen activator (uPA) has been implicated in tumor-cell invasion and metastasis. Cancer-cell invasion into normal tissue can be facilitated by uPA through its activation of plasminogen, which degrades the basement membrane and extracellular matrix (reviewed in refs. 8 and 9). The role of other serine proteases in cancer has been less well characterized.
One useful model system for studying many issues that are pertinent to prostate cancer is the development of the rodent ventral prostate in explant cultures. Macromolecular inhibitors of serine proteases of the chymotrypsin fold, ecotin and ecotin M84R/M85R (6, 7), inhibit ductal branching morphogenesis and differentiation of the explanted rat ventral prostate (F. Elfman, T.T., C.C., G. Cunha, and M.S., unpublished data). Ecotin M84R/M85R is a 2,800-fold more potent inhibitor of uPA than ecotin (1 nM vs. 2.8 µM) (6). However, inhibition of prostate differentiation was seen with both inhibitors, suggesting that uPA and other related serine proteases are involved in the differentiation and continued growth of the rat ventral prostate. Thus, unidentified serine proteases may play a role in growth and prevention of apoptosis in prostate epithelial cells in this system.
Another well characterized model that is derived from human prostate cancer epithelial cells is the PC-3 cell line (10). The PC-3 cell line expresses uPA as assayed by ELISA and by Northern blotting of PC-3 mRNA (11). We found that the primary tumor size in PC-3-implanted nude mice was significantly smaller in both ecotin M84R/M85R and ecotin wild-type treated mice treated for 7 weeks compared with the primary tumor size of PBS-treated mice. Metastasis from the primary tumors were similarly lower in the inhibitor-treated
|
|
PNAS is available online at www.pnas.org. |
|
|
Abbreviations: MT-SP1, membrane-type serine protease 1; CUB, complement factor 1R-urchin embryonic growth factor-bone morphogenetic protein; LDLR, low density lipoprotein receptor; uPA, urokinase-type plasminogen activator; pNA, p-nitroanilide. |
|
|
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. Banklt257050 and AF133086). |
|
‡ |
To whom reprint requests should be addressed. E-mail: craik@cgl.ucsf.edu. |
mice than in PBS-treated mice (O.Melnyk, T.T., C.C., and M.S., unpublished data). Inhibition was not unexpected with ecotin M84R/M85R treatment, because uPA has been implicated in metastasis. However, wild-type ecotin is a poor, micromolar inhibitor of uPA; one interpretation of the data is that the decrease in tumor size and metastasis in the mouse model involves the inhibition of additional serine proteases. Thus, identification of the serine proteases expressed by PC-3 prostate cells may provide insight into the role of these proteases in cancer and prostate growth and development. In this report we have extended the strategy of using PCR with degenerate oligonucleotide primers that were designed by using conserved sequence homology (12–14) to identify additional serine proteases made by cancer cells. Five independent serine protease cDNAs derived from PC-3 mRNA were sequenced, including a novel serine protease, which we refer to as membrane-type serine protease 1 (MT-SP1), and the cloning and characterization of this cDNA that encodes a mosaic, transmembrane protease is reported.
MATERIALS AND METHODS
Materials. All primers used were synthesized on a Applied Biosystems 391 DNA synthesizer. All restriction enzymes were purchased from New England Biolabs. Automated DNA sequencing was carried out on an Applied Biosystems 377 Prism sequencer, and manual DNA sequencing was carried out under standard conditions. N-terminal amino acid sequencing was performed on an ABI 477A by the University of California, San Francisco Biomolecular Resource Center. The synthetic substrates, Suc-AAPX-p-nitroanilide (pNA), [N-succinylalanyl-alanyl-prolyl-Xxx-pNA (Xxx=alanyl, aspartyl, glutamyl, phenylalanyl, leucinyl, methionyl, or arginyl)], and H-Arg-pNA, (arginyl-pNA), were purchased from Bachem. Deglycosylation was performed by using PNGase F (NEB, Beverly, MA). All other reagents were of the highest quality available and purchased from Sigma or Fisher unless otherwise noted.
Isolation of cDNA from PC-3 Cells. mRNA was isolated from PC-3 cells by using the polyATtract System 1000 kit (Promega). Reverse transcription was primed by using the “lock-docking” oligo(dT) primer (15). Superscript II reverse transcriptase (Life Technologies, Grand Island, NY) was used in accordance with the manufacturer’s instructions to synthesize the cDNA from the PC-3 mRNA.
Amplification of MT-SP1 Gene. The degenerate primers used for amplifying the protease domains were designed from the consensus sequences flanking the catalytic histidine (5′ His-primer) and the catalytic serine (3′ Ser-primer), similar to those described (12). The 5′ primer used is as follows: 5′-TGG (AG)TI (CAG)TI (AT)(GC)I GCI (GA)CI CA(CT) TG-3′, where nucleotides in parentheses represent equimolar mixtures and I represents deoxyinosine. This primer encodes at least the following amino acid sequence: W (I/V) (I/V/L/M) (S/T) A (A/T) H C. The 3′ primer used is as follows: 5′-IGG ICC ICC I(GC)(AT) (AG)TC ICC (CT)TI (GA)CA IG(ATC) (GA)TC-3′. The reverse complement of the 3′ primer encodes at least the following amino acid sequence: D (A/S/T) C (K/E/Q/H) G D S G G P.
Direct amplification of serine protease cDNA was not possible by using the above primers. Instead, the first PCR was performed with the 5′ His-primer and the oligo(dT) primer described above, by using the “touchdown” PCR protocol (16), with annealing temperatures decreasing from 52°C to 42°C over 22 rounds and 13 final rounds at 54°C annealing temperature. Cycle times were 1 min (denaturing), 1 min (annealing), and 2 min (extension) and were followed by one final extension time of 15 min after the final round of PCR. The template for the second PCR was 0.5 μl (total reaction volume 50 μL) of a 1:10 dilution of the first PCR mixture that was performed with the 5′ His-primer and the oligo(dT). The second PCR reaction was primed with the 5′ His- and the 3′ Ser-primers and performed by using the touchdown protocol described above. All PCRs used 12.5 pmol of primer for 50-μl reaction volume.
The product of the second reaction was purified on a 2% agarose gel, and all products between 400 and 550 bp were cut from the gel and extracted by using the QIAquick gel extraction kit (Qiagen, Chatsworth, CA). These products were digested with the BamHI restriction enzyme to cut any uPA cDNA, and all 400- to 500-bp fragments were repurified on a 2% agarose gel. These reaction products were subjected to a third PCR by using the 5′ His-primer and the 3′ Ser-primer by using the identical touchdown procedure. These reaction products were gel-purified and directly cloned into the pPCR2.1 vector by using the TOPO TA ligation kit (Invitrogen). DNA sequencing of the inserts determined the cDNA sequence from nucleotides 1,984 to 2,460 (see Fig. 1).
Northern Blot Analysis. 32P-labeled nucleotides were purchased from Amersham Pharmacia. A cDNA fragment containing nucleotides 1,173–2,510 was digested from expressed sequence tag w39209 by using restriction enzymes EcoRI and BsmbI, yielding a 1.3-kilobase nucleotide insert. Labeled cDNA probes were synthesized by using the Rediprime random primer labeling kit (Amersham Pharmacia) and 20 ng of the purified insert. Poly(A)+RNA membranes for Northern blotting were purchased from Origene (Rockville, MD; HB-1002, HB-1018) and CLONTECH (Human II 7759–1, Human Cancer Cell Line 7757). The blots were performed under stringent annealing conditions as described in ref. 17.
Construction of Expression Vectors. The mature protease domain and a small portion of the pro-domain (nucleotides 1,822–2,601) cDNA were amplified by using PCR from expressed sequence tag w39209 and ligated into the pQE30 vector (Qiagen). This construct is designed to overexpress the protease sequence from amino acids (aa) 596–855 with the following fusion: Met-Arg-Gly-Ser-His6-aa596–855. The Histag fusion allows affinity purification by using metal-chelate chromatography. The change from Ser-805, encoded by TCC, to Ala (GCT) was performed by using PCR. The presence of the correct Ser → Ala substitution in the pQE30 vector was verified by DNA sequence analysis.
Expression and Purification of the Protease Domain. The above-mentioned plasmids were separately transformed into Escherichia coli X-90 to afford high-level expression of recombinant protease gene products (18). Expression and purification of the recombinant enzyme from solubilized inclusion bodies was performed as described (19). Protein-containing fractions were pooled and dialyzed overnight at 4°C against 50 mM Tris (pH 8), 10% glycerol, 1 mM 2-mercaptoethanol, and 3 M urea. Autoactivation of the protease was monitored on dialysis against storage buffer (50 mM Tris, pH 8/10% glycerol) at 4°C by using the substrate Spectrozyme tPA (hexahydrotyrosyl-Gly-Arg-pNA, American Diagnostics, Greenwich, CT). Hydrolysis of Spectrozyme tPA was monitored at 405 nM for the formation of p-nitroaniline by using a Uvikon 860 spectrophotometer. Activated protease was bound to an immobilized p-aminobenzamidine resin (Pierce) that had been equilibrated with storage buffer. Bound protease was eluted with 100 mM benzamidine and the protein containing fractions were pooled. Excess benzamidine was removed by using FPLC with a Superdex 70 (Amersham Pharmacia) gel filtration column that was equilibrated with storage buffer. Protein containing fractions were pooled and stored at –80°C. The cleavage of the purified Ser805 Ala protease domain was performed at 37°C by addition of active recombinant protease domain to 10 nM. Cleavage was monitored by using SDS/ PAGE.
Determination of Substrate Kinetics. The purified serine protease domain was titrated with 4-methylumbelliferyl pguanidinobenzoate (MUGB) to obtain an accurate concen-

FIG. 1. Nucleotide sequence of the cDNA encoding human MT-SP1 and predicted protein sequence. Numbering indicates nucleotide or amino acid residue. Amino acids are shown in single-letter code. The termination codon is shown by *. The underlined stop codon at nucleotide 10 is in frame with the initiating methionine. The Kozak consensus sequence (24) at the start codon is underlined at nucleotide 32. The predicted N-glycosylation sites at amino acids 109, 302, 485, and 772 are underlined. A possible polyadenylation sequence (46) at nucleotide 3,120 is also underlined. The catalytic triad in the serine protease domain is highlighted: His-656, Asp-711, and Ser-805.
tration of enzyme active sites (20). Enzyme activity was monitored at 25°C in assay buffer containing 50 mM Tris (pH 8.8), 50 mM NaCl, and 0.01% Tween 20. The final concentration of substrate Spectrozyme tPA ranged from 1 to 400 µM. Enzyme concentrations ranged from 40 to 800 pM. Active-site titrations were performed on a Fluoromax-2 spectrofluorimeter. Measurements were plotted by using the KALEIDAGRAPH program (Synergy Software, Reading, PA), and the Km, Acat, and kcat/Km for Spectrozyme tPA was determined by using the Michaelis-Menten equation.
Inhibition of MT-SP1 Protease Domain with Ecotin and Ecotin M84R/M85R. Ecotin and ecotin M84R/M85R were purified from E.coli as described (6). Various concentrations of ecotin or ecotin M84R/M85R were incubated with the His-tagged serine protease domain in a total volume of 990 µl of buffer containing 50 mM NaCl, 50 mM Tris·HCl (pH 8.8), and 0.01% Tween 20. Ten microliters of Spectrozyme tPA was added, yielding a solution containing 100 µM substrate. The final enzyme concentration was 63 pM, and the ecotin and ecotin M84R/M85R concentration ranged from 0.1 to 50 nM. The data were fit to the equation derived for kinetics of reversible tight-binding inhibitors (21, 22), and the values for apparent Ki were determined.
RESULTS
Cloning of Serine Protease Domain cDNAs from PC-3 Cells and Amplification of MT-SP1 cDNA. PCR amplification of serine protease cDNA was performed by using “consensus cloning”, where the amplification was performed with degenerate primers designed to anneal to cDNA encoding the region about the conserved catalytic histidine (5' His-primer) and the conserved catalytic serine (3' Ser-primer). The consensus primers were designed by using 37 human sequences within a sequence alignment of 242 serine proteases of the chymotrypsin fold that are reported in the SwissProt database. To bias the screen for previously unidentified proteases in the PC-3 cDNA, uPA cDNA was cut and removed by using the known BamHI endonuclease site in the uPA cDNA sequence. The expected size of the cDNA fragments amplified between His-57 and Ser-195 cDNA (standard chymotrypsinogen numbering) is between 400 and 550 bp; statistically, only 1 in 10 cDNAs of that length will be cleaved by BamHI. Thus, cDNAs obtained from the PCR reactions with the 5' His-primer and 3' Ser-primer were size selected for the 400- to 550-bp range, digested with BamHI, and purified from any digested cDNAs. After a subsequent round of PCR, the products were cloned into pPCR2.1 (Fig. 2). Twenty clones were digested with EcoRI to monitor the size of the cDNA insert. Three clones lacked inserts of the correct size. The remaining 17 clones containing inserts between 400 and 550 bp were sequenced. BLAST searches of the resulting sequences revealed that six clones did not match serine protease sequences. The remaining cDNAs yielded clones corresponding to factor XII (two clones), protein C (two clones), trypsinogen type IV (two clones), uPA (one clone), and MT-SP1 (four clones). Additional serine protease sequences may not have been found because they were digested by BamHI, lost in the size selection, or present in lower frequencies.

FIG. 2. Lane 1 shows the PCR products obtained by using degenerate primers designed from the consensus sequences flanking the catalytic histidine (5' His-primer) and the catalytic serine (3' Serprimer). The products remaining between 400 and 550 bp after digestion with BamHI were reamplified by using the same degenerate primers. The products from this second PCR are shown in Lane 2.
Multiple expressed sequence tag sequences were found for the cDNA. Expressed sequence tag accessions aa459076, aa219372, and w39209 were used extensively for sequencing the cDNA starting from nucleotide 746 and 2, 461–3, 142, but no start codon was observed. A sequence was also found in GenBank (accession no. U20428). This sequence also lacks the 5' end of the cDNA but allowed amplification of cDNA from nucleotides 196–745. Rapid amplification of cDNA ends (RACE) (23) was used to obtain further 5' cDNA sequence. Application of RACE did not yield a clone containing the entire 5'-untranslated region, but the sequence obtained contained a stop codon in-frame with the Kozak start sequence (24), giving confidence that the full coding sequence of the cDNA has been obtained. The nucleotide sequence and predicted amino acid sequence are shown in Fig. 1.
The nucleotide sequence surrounding the proposed start codon matches the optimal sequence of ACCATGG for translation initiation sites proposed by Kozak (24). In addition, there is a stop codon in-frame with the putative start codon, which gives further evidence that initiation occurs at that site. The DNA sequence predicts an 855-aa mosaic protein composed of multiple domains (Fig. 3). The coding sequence does not contain a typical signal peptide but does contain a single
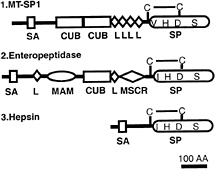
FIG. 3. The domain structure of human MT-SP1 is compared with the domain structure of enteropeptidase (47) and hepsin (25). SA, possible signal anchor; CUB, a repeat first identified in complement components C1r and C1s, the urchin embryonic growth factor and bone morphogenetic protein 1 (27); L, LDLR repeat (29); SP, a chymotrypsin family serine protease domain (40); MAM, a domain homologous to members of a family defined by meprin, protein A5, and the protein tyrosine phosphatase µ (48); MSCR, a macrophage scavenger receptor cysteine-rich motif (29). The predicted disulfide linkages are shown labeled as C–C.
hydrophobic sequence of 26 residues (residues 55–81), which is flanked by a charged residue on each side. This sequence may constitute a signal anchor sequence, similar to that observed in other proteases, including hepsin (25) and enteropeptidase (26). Following the putative signal anchor sequence are two complement factor 1R-urchin embryonic growth factor-bone morphogenetic protein (CUB) domains (27), which are named after the proteins in which the modules were first discovered: complement subcomponents C1s and C1r, urchin embryonic growth factor (Uegf), and bone morphogenetic protein 1 (BMP1). CUB domains have conserved characteristics, which include the presence of four cysteine residues and various conserved hydrophobic and aromatic positions (27). The CUB domain, which has recently been characterized crystallographically (28), consists of 10 ß-strands that are organized into two 5-stranded ß-sheets. Following the CUB domains are four low-density lipoprotein receptor (LDLR) repeats (29), which are named after the receptor ligand-binding repeats that are present in the LDLR. These repeats have a highly conserved pattern and spacing of six cysteine residues that form three intramolecular disulfide bonds. The final domain observed is the serine protease domain. The alignments of these domains with other members of their respective classes are shown in Fig. 4.
Tissue Distribution of MT-SP1 mRNA. Northern blots of human poly(A)+RNA, made by using a 1.3kilobase fragment of MT-SP1 cDNA fragment as a probe, show a ˜3.3-kilobase fragment appearing in epithelial tissues including the prostate, kidney, lung, small intestine, stomach, colon, and placenta, as well as other tissues, including spleen, liver, leukocytes, and thymus. This band was not observed in muscle, brain, ovary, or testis (Fig. 5). Similar experiments performed on a human cancer cell line blot shows that MT-SP1 is expressed in the colorectal adenocarcinoma, SW480, but was not observed in the promyelocytic leukemia HL-60, HeLa cell S3, chronic myelogenous leukemia K-562, lymphoblastic leukemia MOLT-4, Burkitt’s lymphoma Raji, lung carcinoma A549, or melanoma G361 lanes (data not shown). This 3.3-kilobase mRNA fragment is slightly longer than the 3.1-kilobase sequence presented in Fig. 5, suggesting that there may still be sequence in the 5'-untranslated region that has not been identified.
Activation and Purification of His-MT-SP1 Protease Domain. The serine protease domain of MT-SP1 was expressed in E.coli as a His-tagged fusion and was purified from inclusion bodies under denaturing conditions by using metal-chelate affinity chromatography. The yield of enzyme after this step was ˜3 mg of protein per liter of E.coli culture. This denatured protein refolded when the urea was dialyzed from the protein. Surprisingly, the purified renatured protein showed a time-dependent shift on an SDS/PAGE gel (Fig. 6A), with the lower fragment being the size of the mature, processed enzyme lacking the His tag. N-terminal sequencing of the purified, activated protease domain yielded the expected VVGGT activation sequence. When the refolded protein was tested for activity by using the synthetic substrate Spectrozyme tPA, a time-dependent increase in activity was observed (Fig. 6B). In contrast, the protease domain that contains the Ser805 Ala mutation showed neither a change in size on an SDS polyacrylamide gel nor an increase in enzymatic activity under identical conditions (data not shown), suggesting that the catalytic serine is necessary for activation and is not the result of a contaminating protease. To show that the cleavage of the protease domain was a result of His-tagged MT-SP1 protease activity, the inactive Ser805 Ala protease domain was treated with purified recombinant enzyme (Fig. 6C). This treatment results in the formation of a cleavage product that corresponds to the size of the active protease (Fig. 6C, lane 7). Untreated protease domain does not get cleaved (Fig. 6C, lane 8). From these results, it is concluded that the protease autoactivates on
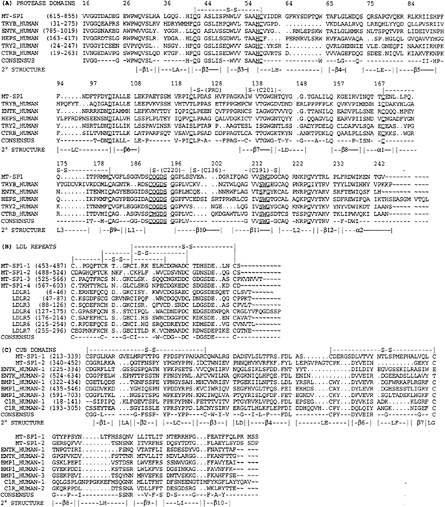
FIG. 4. Multiple sequence alignments of MT-SP1 structural motifs. L, loops; ß, B-sheets; a, a-helices; S-S, disulfides. (A) Multiple sequence alignment of the serine protease domain of MT-SP1 with human trypsinogen B (49), human enterokinase (47), human hepsin (25), human tryptase 2 (50), and human chymotrypsinogen B (51), with standard chymotrypsin numbering. Conserved catalytic and structural residues described in the text are underlined. (B) Alignment of MT-SP1 LDLR with domains of the LDLR (52). (C) Alignment of the CUB domains of MT-SP1 with those found in human enterokinase (48), human bone morphogenetic protein 1 (53), and complement component C1R (54).
refolding. The activated protease was separated from inactive protein and other contaminants by using affinity chromatography with p-aminobenzamidine resin. Purified protein was analyzed by using SDS/PAGE, and no other contaminants were observed. Similarly, immunoblotting with polyclonal antiserum against purified protease domain (raised in rabbits at Berkeley Antibody, Richmond, CA) revealed one band. Under nonreducing conditions, the pro region is disulfide-linked to the protease domain; thus, this purified protein was also immunoreactive with the mAb (Qiagen, Chatsworth, CA) directed against the N-terminal Arg-Gly-Ser-His4 epitope that is contained in the recombinant protease domain, further indicating the purity and identity of the protein (data not shown).
Kinetic Properties of Purified His-MT-SP1 Protease Domain. The enzyme concentration was determined by using an active site titration with MUGB. The catalytic activity of the protease domain was monitored by using pNA substrates.
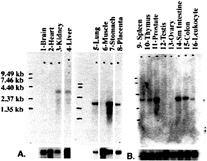
FIG. 5. Tissue distribution of MT-SP1 mRNA levels. Northern blots of human poly(A)+RNA from assorted human tissues was hybridized with radiolabeled cDNA probes as described in Materials and Methods. Upper shows hybridization by using a MT-SP1 1.3-kilobase cDNA fragment derived from expressed sequence tag clone w39209 and exposed overnight. Lower shows the same blot after being stripped and rehybridized with a loading standard ß-actin (A) or human glyceraldehyde phosphate dehydrogenase (GAPDH) (B) cDNA probe exposed for 2 hours. The mobility of RNA size standards is indicated at the left.
Purified protease domain was tested for hydrolytic activity against tetrapeptide substrates of the form Suc-AAPX-pNA, which contained various amino acids at the P1 position (P1-Ala, Asp, Glu, Phe, Leu, Met, Lys, or Arg). The only substrates with detectable activity were those with P1-Lys or P1-Arg. The serine protease domain with the Ser805 Ala mutation had no detectable activity. The activity of the protease domain was further characterized by using the substrate Spectrozyme tPA, yielding: Km=31.4±4.2µM, kcat=2.6×102±6.5 s–1, and kcat/Km=6.9×106±2.3×106 M–1.s–1. Ecotin inhibition of the MT-SP1 His-tagged protease domain fits a tight-binding reversible inhibitory model (21, 22) as observed for ecotin interaction with other serine protease targets (6, 7, 30). Inhibition assays by using ecotin and ecotin M84R/M85R yielded apparent Ki values of 782±92 pM and 9.8±1.5 pM, respectively.
DISCUSSION
Structural Motifs of MT-SP1. In this work, we characterize the expression of chymotrypsin-fold proteases by PC-3 cells and cloned a member of this family we call MT-SP1. The name membrane-type serine protease 1 (MT-SP1) is given to be consistent with the nomenclature of the membrane-type matrix metalloproteases (MT-MMPs; ref. 32). The cDNA likely encodes a membrane-type protein because of the lack of a signal sequence and the presence of a putative SA that is also seen in other membrane-type serine proteases hepsin (25), enteropeptidase (26), and TMPRSS2 (32), and human airway trypsin-like protease (33). We propose that proteins that are localized to the membrane through a SA and that encode a chymotrypsin fold serine protease domain be categorized in the MT-SP family. The membrane localization of MT-SP1 is supported by immunofluorescence experiments that localize the protease domain to the extracellular cell surface (unpublished results).
Following the putative SA are several domains that are thought to be involved in protein-protein interactions or protein-ligand interactions. For example, CUB domains can mediate protein-protein interactions as with the seminal plasma PSP-I/PSP-II heterodimer that is built by CUB-domain interactions (28) and with procollagen C-proteinase enhancer protein and procollagen C-proteinase (BMP-1) (34, 35). Interestingly, most of the proteins that contain CUB domains are involved in developmental processes or are involved in proteolytic cascades (27), which suggests that MTSP1 may play a similar role. The four repeated motifs that follow the CUB domains are known as LDLR ligand-binding repeats, named after the seven copies of repeats found in the LDLR. There are several negatively charged amino acids between the fourth and sixth cysteines that are highly conserved in the LDLR and are also seen in the LDLR repeats of MT-SP1. The conserved motif Ser-Asp-Glu (residues 44–46 in Fig. 4) are known to be important for binding the positively charged residues of the LDLR ligands apolipoprotein B-100 (ApoB-100) and ApoE (29). The ligand-binding repeats of MT-SP1 most likely do not mediate interaction with ApoB-100 or ApoE but may be involved in the interaction with other positively charged ligands. For example, LDLR repeats in the LDLR-related protein have been implicated the binding and recycling of protease-inhibitor complexes such as uPA-plasminogen activator inhibitor-1 (PAI-1) complexes (reviewed in refs. 36 and 37). It also has been shown that the pro domain of enteropeptidase is involved in interactions with its substrate trypsinogen, allowing 520-fold greater catalytic efficiency in the cleavage compared with the protease domain alone (38). By analogy, similar interactions should occur between MT-SP1 and its substrates. Thus, further investigation of MT-SP1 CUB domain or LDLR repeat interactions may yield insight into the function of this protein.
The amino acid sequence of the serine protease domain of MT-SP1 is highly homologous to other proteases found in the family (Fig. 4). The essential features of a functional serine protease are contained in the deduced amino acid sequence of
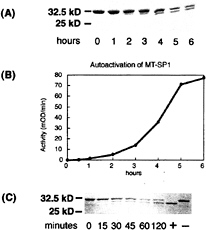
FIG. 6. Activation and purification of His-tagged MT-SP1 protease domain. A representative experiment is shown in A and B. (A) Activation at 4°C was monitored by using SDS/PAGE. The upper band represents inactivated protease domain, and the lower band represents active protease (also verified by N-terminal sequencing). (B) The activation of the protein was monitored by using Spectrozyme tPA as a synthetic substrate for the protease domain. (C) Inactive Ser805 Ala protease domain is cleaved with 10 nM activated His-tagged MT-SP1 protease domain at 37°C. The specific cleavage of active MT-SP1 protease domain is required for proper processing at the activation site. Active protease domain is shown in lane 7 (+), and no cleavage of the untreated inactive protease domain is observed (lane 8, –).
the domain. The residues that comprise the catalytic triad, His-656, Asp-711, and Ser-805, corresponding to His-57, Asp-102, and Ser-195 in chymotrypsin, are observed in MT-SP1 (for reviews, see refs. 39 and 40). The sequence Ser214Trp215Gly216 (Ser825Trp826Gly827), which is thought to interact with the side chains of the substrate for properly orienting the scissile bond is present. Gly-193 (Gly-803) and Gly-196 (Gly-805), which are thought to be necessary for proper orientation of Ser-195 (Ser-805), also are present. Based on homology to chymotrypsin, three disulfide bonds are predicted to form within the protease domain at Cys-44–Cys-58, Cys-168–Cys-182, and Cys-191–Cys-220 (Cys-643–Cys-657, Cys-776–Cys-790, and Cys-801–Cys-830), and a fourth disulfide bond should form between the catalytic and the pro-domain Cys-122–Cys-1 (Cys-731–Cys-604), as observed for chymotrypsin. This predicted disulfide with the pro domain suggests that the active catalytic domain should still be localized to the cell surface via a disulfide linkage. The presence of the catalytic machinery and other conserved structural components described above suggest that all features necessary for proteolytic activity are present in the encoded sequence.
Substrate Specificity of the MT-SP1 Protease Domain. The S1 site specificity (41) of a protease is largely determined by the amino acid residue at position 189. This position is occupied by an aspartate in MT-SP1, suggesting that the protease has specificity for Arg/Lys in the P1 position. In addition, the presence of a polar Gln-192 (Gln-803), as in trypsin, is consistent with basic specificity. Furthermore, the presence of Gly-216 (Gly-827) and Gly-226 (Gly-837) is consistent with the presence of a deep S1 pocket, unlike elastase, which has Val-216 and Thr-226 that block the pocket and thereby contribute to the P1 specificity for small hydrophobic side chains. The specificity at the other subsites is largely dependent on the nature of the seven loops A–E and loops 2 and 3 (Fig. 4). Loop C in enterokinase has a number of positively charged residues that are thought to interact with the negatively charged activation site in trypsinogen, Asp-Asp-Asp-Asp-Lys (26). One known substrate for MT-SP1 (as described below) is the activation site of MT-SP1, which is Arg-Gln-Ala-Arg (residues 611–614). Loop C contains two Asp residues that may participate in the recognition of the activation sequence.
One means of obtaining further data on substrate specificity is by characterization of the activity of the recombinant proteolytic domain. Enterokinase has been characterized from both recombinant (38, 42) and native (43, 44) sources. However, proteolytic activity for the other reported membrane-type serine proteases hepsin (25) and TMPRSS2 (32) are only predicted based on sequence homology. To produce active recombinant MT-SP1, a His-tagged fusion of the protease domain was cloned into an E.coli vector and expressed and purified to homogeneity. Fortuitously, the protease domain refolded and autoactivated after resuspension and purification from inclusion bodies. This activity, coupled with the lack of activity in the Ser195Ala (Ser805Ala) variant, demonstrates that the cDNA encodes a catalytically proficient protease. Autoactivation of the protease domain at the arginine-valine site (Arg614-Val615) shows that the protease has Arg/Lys specificity as predicted by the sequence homology to other proteases of basic specificity. Specificity and selectivity are confirmed by the lack of cleavage of AAPX-pNA substrates that do not have x=R, K. Further characterization with Spectrozyme tPA revealed an active enzyme with kcat=2.6×102 s–1. However, the His-tagged serine protease domain does not cleave H-Arg-pNA, showing that, unlike trypsin, there is a requirement for additional subsite occupation for catalytic activity. This suggests that the enzyme is involved in a regulatory role that requires selective processing of particular substrates rather than nonselective degradation.
MT-SP1 Function. In other studies, we have found that inhibition of serine protease activity by ecotin or ecotin M84R/M85R inhibits testosterone-induced branching ductal morphogenesis and enhances apoptosis in a rat ventral prostate model (F.Elfman, T.T., C.S.C., G.Cunha, and M.A.S., unpublished results). Moreover, the rat homolog of MT-SP1 is expressed in the normal rat ventral prostate (data not shown). Assays of the protease domain with ecotin and ecotin M84R/ M85R showed that the enzymatic activity is strongly inhibited (782±92 pM and 9.8±1.5 pM, respectively), suggesting that rat MT-SP1 is likely to be inhibited at the concentrations of these inhibitors used in our experiments. MT-SP1 inhibition may result in the observed inhibition of differentiation and/or increased apoptosis. Future studies are aimed at definitively resolving the role of MT-SP1 in prostate differentiation. The broad expression of MT-SP1 in epithelial tissues is consistent with the possibility that it is involved in cell maintenance or growth, perhaps by activating growth factors or by processing prohormones.
MT-SP1 may participate in a proteolytic cascade that results in cell growth and/or differentiation. Another structurally similar membrane-type serine protease, enteropeptidase (Fig. 3), is involved in a proteolytic cascade by which activation of trypsinogen leads to activation of downstream intestinal proteases (5). Enteropeptidase is expressed only in the enterocytes of the proximal small intestine, thus precisely restricting activation of trypsinogen. Thus, in contrast to secreted proteases that may diffuse throughout the organism, the membrane association of MT-SP1 should also allow the proteolytic activity to be precisely localized, which may be important for proper physiological function; improper localization of the enzyme, or levels of downstream substrates could lead to disease.
We have found subcutaneous coinjection of PC-3 cells with wild-type ecotin or ecotin M84R/M85R led to a decrease in the primary tumor size compared with animals in whom PC-3 cells and saline were injected (O.Melnyk, T.T., C.S.C. and, M.A.S., unpublished results). Because wild-type ecotin is a poor, micromolar inhibitor of uPA, serine proteases other than uPA likely are involved in this primary tumor proliferation. Both wild-type ecotin and ecotin M84R/M85R are potent, subnanomolar inhibitors of MT-SP1, raising the possibility that MT-SP1 plays an important role in progression of epithelial cancers expressing this protease.
Direct biochemical isolation of the substrates may be possible if MT-SP1 adhesive domains such as the CUB domains or LDLR repeats interact with the substrates. In addition, likely substrates may be predicted and tested for by using knowledge of extended enzyme specificity. For example, the characterization of the substrate specificity of granzyme B allowed the prediction and confirmation of substrates for this serine protease (45). Thus, these complimentary studies should further shed light on the physiological function of this enzyme.
We thank Marion Conn, Robert Maeda, Todd Pray, Ibrahim Adiguzel, and Ralph Reid for technical assistance and helpful discussions. T.T. was supported by a National Institutes of Health postdoctoral fellowship CA71097, and this work was supported by National Institutes of Health Grant CA72006.
1. Neurath, H. & Walsh, K.A. (1976) Proc. Natl. Acad. Sci. USA 73, 3825–3832.
2. Davie, E.W., Fujikawa, K. & Kisiel, W. (1991) Biochemistry 30, 10363–10370.
3. Chandler, W.L. (1996) Crit. Rev. Oncol. Hematol. 24, 27–45.
4. Reid, K.B.M. & Porter, R.R. (1981) Annu. Rev. Biochem. 50, 433–464.
5. Huber, R. & Bode, W. (1978) Acc. Chem. Res. 11, 114–122.
6. Wang, C.-I., Yang, Q. & Craik, C.S. (1995) J. Biol. Chem. 270, 12250–12256.
7. Yang, S.Q., Wang, C.-I., Gillmor, S.A., Fletterick, R.J. & Craik, C.S. (1998) J. Mol. Biol 279, 945–957.
8. Dano, K.Andreasen, P.A., Grondahl-Hansen, J., Kristensen, P., Nielsen, L.S. & Skriver, L. (1985) Adv. Cancer Res. 44, 139–266.
9. Andreasen, P.A., Kjoller, L., Christensen, L. & Duffy, M.J. (1997) Int. J. Cancer 72, 1–22.
10. Kaighn, M.E., Narayan, K.S., Ohnuki, Y., Lechner, J.F. & Jones, L.W. (1979) Invest. Urol. 17, 16–23.
11. Yoshida, E., Verrusio, E.N., Mihara, H., Oh, D. & Kwaan, H.C. (1994) Cancer Res. 54, 3300–3304.
12. Sakanari, J.A., Staunton, C.E., Eakin, A.E., Craik, C.S. & McKerrow, J.H. (1989) Proc. Natl. Acad. Sci. USA 86, 4863– 4867.
13. Wiegand, U., Corbach, S., Minn, A., Kang, J. & Muller-Hill, B. (1993) Gene 136, 167–175.
14. Kang, J., Wiegand, U. & Muller-Hill, B. (1992) Gene 110, 181–187.
15. Borson, N.D., Salo, W.L. & Drewes, L.R. (1992) PCR Methods Appl. 2, 144–148.
16. Don, R.H., Cox, P.T., Wainwright, B.J., Baker, K. & Mattick, J.S. (1991) Nucleic Acids Res. 19, 4008.
17. Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A. & Struhl, K., eds. (1990) Current Protocols in Molecular Biology (Wiley, New York).
18. Evnin, L.B., Vasquez, J.R. & Craik, C.S. (1990) Proc. Natl. Acad. Sci. USA 87, 6659–6663.
19. Unal, A., Pray, T.R., Lagunoff, M., Pennington, M.W., Ganem, D. & Craik, C.S. (1997) J. Virol. 71, 7030–7038.
20. Jameson, G.W., Roberts, D.V., Adams, R.W., Kyle, W.S.A. & Elmore, D.T. (1973) Biochem. J. 131, 107–117.
21. Morrison, J.F. (1969) Biochim. Biophys. Acta 185, 269–286.
22. Williams, J.W. & Morrison, J.F. (1979) Methods Enzymol. 63, 437–467.
23. Frohman, M.A. (1993) Methods Enzymol. 218, 340–356.
24. Kozak, M. (1991) J. Cell Biol. 115, 887–903.
25. Leytus, S.P., Loeb, K.R., Hagen, F.S., Kurachi, K. & Davie, E.W. (1988) Biochemistry 27, 1067–1074.
26. Kitamoto, Y., Yuan, X., Wu, Q., McCourt, D.W. & Sadler, J.E. (1994) Proc. Natl. Acad. Sci. USA 91, 7588–7592.
27. Bork, P. & Beckmann, G. (1993) J. Mol. Biol 231, 539–545.
28. Varela, P.F., Romero, A., Sanz, L., Romao, M.J., Topfer-Petersen, E. & Calvete, J.J. (1997) J. Mol. Biol 274, 635–649.
29. Krieger, M. & Herz, J. (1994) Annu. Rev. Biochem. 63, 601–637.
30. Seymour, J.L., Lindquist, R.N., Dennis, M.S., Moffat, B., Yansura, D., Reilly, D., Wessinger, M.E. & Lazarus, R.A. (1994) Biochemistry 33, 3949–3958.
31. Nagase, H. (1997) Biol Chem. 378, 151–160.
32. Poloni-Giacobino, A., Chen, H., Peitsch, M.C., Rossier, C. & Antonarkis, S.E. (1997) Genomics 44, 309–320.
33. Yamakoka, K., Masuda, K., Ogawa, H., Takagi, K., Umemoto, N. & Yasuoka, S. (1998) J. Biol. Chem. 273, 11895–11901.
34. Kessler, E. & Adar, R. (1989) Eur. J. Biochem. 186, 115–121.
35. Hulmes, D.J.S., Mould, A.P. & Kessler, E. (1997) Matrix Biol. 16, 41–45.
36. Strickl, D.K., Kounnas, M.Z. & Argraves, W.S. (1995) FASEB J. 9, 890–898.
37. Moestrup, S.K. (1994) Biochim. Biopys. Acta 1197, 197–213.
38. Lu, D., Yuan, X., Zheng, X. & Sadler, J.E. (1997) J. Biol Chem. 272, 31293–31300.
39. Perona, J.J. & Craik, C.S. (1995) Protein Sci. 4, 337–360.
40. Perona, J.J. & Craik, C.S. (1997) J. Biol Chem. 272, 29987– 29990.
41. Schecter, I. & Berger, A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162.
42. LaVallie, E.R., Rehmtulla, A., Racie, L.A., DiBlasio, E.A., Ferenz, C., Grant, K.L., Light, A. & McCoy, J.M. (1993) J. Biol. Chem. 268, 23311–23317.
43. Light, A. & Fonseca, P. (1984) J. Biol Chem. 259, 13195–13198.
44. Matsushima, M., Ichinose, M., Yahagi, N., Kakei, N., Tsukada, S., Miki, K., Kurokawa, K., Tashiro, K., Shiokawa, K., Shinomiya, K., et al. (1994) J. Biol Chem. 269, 19976–19982.
45. Harris, J.L., Peterson, E.P., Hudig, D., Thornberry, N.A. & Craik, C.S. (1998) J. Biol Chem. 273, 27364–27373.
46. Nevins, J.R. (1983) Annu. Rev. Biochem. 52, 441–466.
47. Kitamoto, Y., Veile, R.A., Donis-Keller, H. & Sadler, J.E. (1995) Biochemistry 34, 4562–4568.
48. Beckmann, G. & Bork, P. (1993) Trends Biochem. Sci. 18, 40–41.
49. Emi, M., Nakamura, Y., Ogawa, M., Yamamoto, T., Nishide, T., Mori, T. & Matsubara K. (1986) Gene 41, 305–310.
50. Vanderslice, P., Ballinger, S.M., Tam, E.K., Goldstein, S.M., Craik, C.S. & Caughey, G.H. (1990) Proc. Natl. Acad. Sci. USA 87, 3811–3815.
51. Tomita, N., Izumoto, Y., Horii, A., Doi, S., Yokouchi, H., Ogawa, M., Mori, T. & Matsubara, K. (1989) Biochem. Biophys. Res. Commun. 158, 569–575.
52. Sudhof, T.C., Goldstein, J.L., Brown, M.S. & Russell, D.W. (1985) Science 228, 815–822.
53. Wozney, J.M., Rosen, V., Celeste, A.J., Mitsock, L.M., Whitters, M.J., Kriz, R.W., Hewick, R.M. & Wang, E.A. (1988) Science 242, 1528–1534.
54. Leytus, S.P., Kurachi, K., Sakariassen, K.S. & Davie, E.W. (1986) Biochemistry 25, 4855–4863.








