6
Recent Advances in Developmental Biology
The absence of an incisive understanding of the action of toxicants on development has been in large part attributable to the absence of understanding of development itself. Until a few years ago, there was no understanding of a “developmental mechanism” at the molecular level although there were explanations at the cellular and tissue levels, such as “gastrulation is the mechanism by which the organization of the egg is transformed into the organization of the embryo.” Recent advances in developmental biology have been substantial enough for scientists to be confident for the first time that some aspects of development in some organisms are understood at the molecular level. Protein components are identified, their functions in developmental processes are known, and the time and place in the embryo of expression of the genes encoding them are known. This knowledge greatly benefits elucidating the mechanisms of developmental toxicity.
In this chapter, the committee, in response to its charge, evaluates the state of the science for elucidating mechanisms of developmental toxicity and presents insights of developmental biology. It will show the promise of the subject in the next decade for understanding the action of developmental toxicants.
A BRIEF HISTORY OF DEVELOPMENTAL BIOLOGY
Observations of embryos and embryonic stages were made and recorded in antiquity (e.g., Aristotle, fourth century BC) and with increasing attention in recent centuries (e.g., Malphigi in the 1600s, Wolff in the 1700s, and von Baer in the early 1800s). However, it was only in the late nineteenth century that scientists pursued a detailed description of the embryonic stages of a variety of verte-
brates and invertebrates, aided by the then-recent improvements in light microscopy and in staining methods and stimulated by Darwin’s proposals that the study of ontogeny (i.e., the animal’s embryonic development) holds clues to phylogeny (i.e., its evolutionary origin). Among the highlights during the period of 1880-1940 were the detailed anatomical descriptions of developmental stages of embryos, including the first atlas of human embryos, reconstructed from microscopic sections, published by W. His, Sr., in 1880-1885. In vertebrate embryology, these descriptions revealed the organogenesis of the heart, kidney, limbs, central nervous system (CNS), and eyes. Developmental-fate mapping studies revealed the embryonic sites of the origin of cells of the organs and the rearrangements of groups of cells in morphogenesis. The stages of development were found to include, in reverse order, cytodifferentiation, organogenesis, morphogenesis (gastrulation and neurulation), rapid cleavage, fertilization, and gametogenesis. By the 1940s, anatomical descriptions of the embryos of related animals were integrated into coherent evolutionary schemes, taught in comparative embryology classes, revealing, for example, the modification of the gill slits of jawless fish to the jaw of jawed fish and further modification to the middle ear of mammals. Also, by this time, Haeckel’s oversimplified scheme had been abandoned, namely, that ontogeny merely recapitulates phylogeny.
Experimental embryology also began in the late 1800s. In experimental studies, which mostly involved techniques of cell and tissue transplantation and removal, the central role of cytoplasmic localizations and cell-lineage-restricted developmental fates was recognized in the development of certain invertebrates by the early 1900s. In vertebrate development, the importance of inductions (also called tissue interactions) was recognized in the 1920s, following the stunning organizer transplantation experiments by Spemann and Mangold (1924) on newt embryos. By the 1950s, inductions had been found in every stage and place in the vertebrate embryo, for example, in all the kinds of organogenesis. Vertebrate development, including that of mammals, had become comprehensible as a branching succession of inductive interactions among neighboring members of an increasingly large number of different cell groups of the embryo.
Developmental mechanisms, as understood even in the 1970s, were descriptions of the movements and interactions of cells or groups of cells. They were cellular- or tissue-level mechanisms. The all-important “inducers” were materials of unknown composition released by one cell group and received by another group. Consequently, the recipient cells took a path of development different from the one that would have been taken if they were unexposed. The progression or momentum of development also was recognized: that the individual events of interactions and responses are time-critical, and that certain subsequent aspects of development never occur if one event is prevented.
Molecular mechanisms, however, were not understood at that time. Embryologists encountered the limits of the field in the 1940-1970 period, as they tried to discover the chemical nature of inducers and the responses of cells to them.
The basic information and methods of biochemistry, molecular biology, cell biology, and genetics were not yet available to analyze cell-cell signaling and transcriptional regulation in embryos. In light of discouraging results, some embryologists considered that the organizer concept was faulty and that inducers were an experimental artifact (see later discussion for recent successes in understanding inductions). Although Morgan and other early geneticists had proposed that inducers and cytoplasmic localizations elicit specific gene expression and that development was in large part a problem of ever-changing patterns of gene expression (Morgan 1934), the means were not at hand to pursue those insights. Roux, Spemann, and Harrison had outlined plausible lines of inquiry into determination and morphogenesis in the early part of the twentieth century; however, the means were also not available to pursue those questions at that time.
To many scientists in the 1940-1970 period, the study of development seemed messy and intractable. Researchers turned to more informative subjects such as the new molecular genetics of bacteria and phages (viruses that infect bacteria). From those inquiries came new insights in the 1950-1965 period on the nature of the gene and the code and the processes of replication, transcription, translation, enzyme induction, and enzyme repression. For example, it was only in 1961 that Monod and Jacob described gene regulation in bacteria in terms of promoters, operators, and repressor proteins (Monod and Jacob 1961). Those authors immediately saw the relevance to animal development. All of their insights made possible the invention of techniques for gene isolation and amplification, for in vitro expression of genes, for genome analysis, and, thereafter, for the new developmental biology.
With so little molecular information about developmental processes, there was scarcely any understanding of the action of developmental toxicants. For example, Wilson (1973) in his book Environment and Birth Defects could only raise the following possibilities for connections between inductions and developmental defects:
It has long been accepted that cell interactions (induction) are an important part of normal embryogenesis, despite the fact that specific “inducer substances” have not been identified. [Failures] of normal interactions which may lead to deviations in development include, for example, lack of usual contact or proximity, as of optic vesicle with presumptive lens ectoderm; or the incompetence of target tissue to be activated in spite of its usual relationship with activator tissue, as in certain mutant limb defects; or the inappropriate timing of the interrelation, even though all parts are potentially competent. That the nature of cell-to-cell contacts and the manner of their adhesion are important determinants in both normal and abnormal development has been demonstrated…. Insufficient or inappropriate cellular interactions usually result in arrested or deviant development in the tissue ordinarily induced or activated by the interaction.
This committee will later argue that Wilson’s insight was well directed and is now ready to be pursued.
ADVANCES IN DEVELOPMENTAL BIOLOGY
In the past 15 years, developmental biology has advanced remarkably, perhaps as at no other time in the field’s history. It is now known that the trillions of cells of a large animal, such as a mammal, have the same genotype, which is the same as that of the single-celled zygote (the fertilized egg) from which the animal develops. That is to say, the genetic content of somatic cells does not change during the development of most animals. The recent clonings of Dolly the lamb (Wilmut et al. 1997), the Cumulina mouse family (Wakayama et al. 1998), and a nonhuman primate (Chan et al. 2000) reaffirm the fact that a specialized cell, such as a mammary or cumulus cell, carries the genes for all other kinds of cells of the animal. The scientific advances that led to these clonings were built on earlier nuclear transplantation successes in frogs, first by Briggs and King (1952), but particularly by Gurdon (1960), which had led to similar conclusions for a nonmammalian vertebrate. Despite the same genes, the cells within the individual organism differ greatly in their appearance and functions, meaning that they have the same genotype and different phenotypes. The cell types differ greatly in the ribonucleic acids (RNAs) and proteins contained within them. They differ in which subset of genes they express from their total genomic repertoire. At least 300 cell types are recognized in humans (e.g., red blood cells, Purkinje nerve cells, and smooth or striated muscle cells). The number of cell subtypes is much larger, perhaps numbering tens of thousands, when further differences are taken into account related to the cell’s stage of development and location in the body, as has been discovered in recent years. Development can be viewed as evolution’s crowning example of complex gene regulation. From the single genome, thousands of different gene combinations must be expressed at specific times and places in the developing organism, and from the developing egg the information for the selective use of combinations must be generated.
A major factor in this regulation is the transfer of chemical information (i.e., signals) between cells during development. From recent research, which has built on earlier findings, the following is now realized:
-
Embryonic cells of arthropods and nematodes make many of their developmental decisions based on which chemical signals they receive from other cells just as vertebrate embryonic cells do. Later the embryonic cells of all these organisms will make further decisions based on other signals. The cycles of signaling and responding are repeated over and over as development progresses. With that in mind and the fact that one genotype supports hundreds or thousands of cellular phenotypes, development can be said to rely on “genotype-environment interactions,” where the local environment of each cell is generated by neighboring groups of cells. The genotype and cell’s previous developmental decisions determine its options for responses to the signals currently present (Wolpert 1969).
-
The signaling pathways involved in this information transfer are known to be of 17 types (a few more may remain undiscovered). They are used repeatedly
-
at different times and places in the embryo, from the earliest stages through organogenesis and cytodifferentiation, and even in the various proliferating and renewing tissues of the juvenile and adult (see Appendix C).
-
The signaling pathways are highly conserved across a wide range of phyla of animals (from chordates to arthropods to roundworms), presumably because they were present and already functional in the pre-Cambrian common ancestor of those animals.
-
Many of the kinds of cell responses to signals also are conserved (e.g., responses of selective gene expression, secretion, cell proliferation, or cell migration). The response of developing cells to signals involves activation or repression of the expression of specific genes by transcription factors contained within genetic regulatory circuits. Signaling pathways frequently affect the activity of those factors. Many of the transcription factors and circuits are conserved across a wide range of phyla of animals.
Thus, an effective and general approach to the experimental analysis of developmental processes at all stages has been to inquire about the signaling pathways and transcriptional regulatory circuits that operate in the particular instance of development under study. Different organisms, which differ in aspects of their development, nonetheless use the same conserved signaling pathways and regulatory circuits, but in different combinations, times, and places, and have different genes as the targets of their transcriptional regulatory circuits. Processes of development, which seemed to confront scientists with infinite complexity and variety just a few years ago, now seem interpretable as composites of a small number of conserved elemental processes, namely, those of intercellular signaling, intracellular regulatory circuits, and a limited variety of targeted responses. These conclusions, which were reached by the analysis of development in animals as remote as mice, flies, and nematodes, give great validity to the use of model organisms in studying mammalian development, including that of humans, and in the future analysis of the action of developmental toxicants and in their detection.
Although the signal-response pathways are highly conserved, evolution has produced an increasing complexity of the “community” of pathways in vertebrates. This complexity is evident both in the increased number of closely related pathway components (diversifed protein family members) and in the increased possibilities for cross-talk among pathways. The redundant function of closely related components was made evident by existence of numerous targeted gene-knockout mutations in the mouse that produced little or no identifiable pheno-types—that is, the mice are normal or nearly normal under laboratory conditions (see Table 6-5, later in this chapter). It must be emphasized, however, that functional redundancy provides two advantages. It protects the organism by ensuring that a fundamental process can proceed even in the absence or reduced presence of a critical gene activity. On an evolutionary scale, the multiplicity of overlap-
ping functions provides a basis for generating diversity without losing essential functionality.
The Drosophila Breakthrough
The recent molecular understanding of developmental processes and components was gained from the experimental analysis of a few model organisms such as Drosophila melanogaster (the fruit fly), Caenorhabditis elegans (a free-living nematode), Danio rerio (the zebrafish), Xenopus laevis (a frog), the chick, and the mouse (see Chapter 7 for proposals about their use in the assessment of developmental toxicities). D. melanogaster and C. elegans were chosen by researchers for their amenability to genetic analysis, afforded by their small size (hence, large populations) and short life cycle (hence, many generations). Nüsslein-Volhard and Wieschaus (1980) began a systematic search for developmental mutants of Drosophila in the mid-1970s. They submitted adults to high-frequency chemical mutagenesis and then inspected large populations of offspring for mutant individuals with strong and early developmental defects (before hatching) at discrete locations and discrete stages in the embryo. They discarded mutants with weak or pleiotropic effects as ones too difficult to analyze at their start. They examined mutagenized flies until the same kinds of mutants began to appear repeatedly in their collections. The recurrence was evidence that they had obtained all the different kinds of zygotic mutants (those affected in genes transcribed after fertilization) that mutagenized flies could yield under the conditions of inspection. This procedure is called “saturation mutagenesis,” in which all the susceptible genes whose encoded products are important in development are thought to be revealed. Several laboratories, including those of Nüsslein-Volhard and Wieschaus, were also collecting maternal-effect mutants (those affected in genes transcribed in female germ cells before fertilization) and pursued this search to saturation.
The Drosophila mutants were categorized by phenotype and complementation behavior (putting two mutations together in a heterozygote to see whether they are alike or different) to establish the number of different genes whose mutations give the same phenotypic defect of development. Their categories included those embryos failing to develop the anterior or posterior end, odd or even segments, dorsal or ventral parts, mesoderm, endoderm, or nervous system. Further mutant combinations were made to establish epistasis (the interaction of different gene products, reflected in the dominance of one mutant defect over another) and to deduce plausible developmental pathways in which the actions of the encoded gene products could be related and ordered. By the late 1980s, a solid base of observations of Drosophila mutant phenotypes and gene locations had been built, and ordered pathways of function based on the mutant interactions had been proposed. This information served as the foundation for future molecular genetic analysis. The research was the first systematic and exhaustive approach to under-
standing an organism’s development and to identifying components of developmental processes.
Synergy with Research Advances in Other Areas
Meanwhile, other researchers worldwide made advances in biochemistry, molecular biology, cell biology, and genetics. They learned an enormous amount about the function of proteins in replication, transcription, translation, secretion, uptake, membrane trafficking, cell motility, cell division, the cell cycle, cell adhesion, and apoptosis (programmed cell death), to mention but a few of the cellular processes. Researchers improved the methods to isolate genes, sequence them, manipulate sequences, make transcripts in vitro, detect messenger (m)RNAs in cells by in situ hybridization, translate RNAs to proteins in vitro, and make antibodies to proteins. In situ hybridization, which graphically revealed the time and place of expression of specific genes in the embryo, was to prove particularly important for connecting the new molecular analysis to the older developmental anatomy. Much of the work was initially done with single-celled organisms: bacteria, yeast, or animal cells in culture. Some insights and techniques came from the study of cancer cells in the search for oncogenes.
In the course of that work, many of the processes, protein functions, and protein sequences were found to be strongly conserved among organisms as diverse as yeast and humans or even bacteria and humans. Various proteins of different organisms, and also within the same organism, shared “sequence motifs” by which the protein could be recognized as a member of a protein family with a particular function and descended from a common sequence ancestor. Newly discovered proteins could be assigned a function from just their possession of a particular motif. As more motifs were found, it will be easier to categorize newly discovered proteins. For example, receptor tyrosine kinases were recognizable by their transmembrane hydrophobic motifs and adenosine triphosphate (ATP)-binding domains. G-protein-linked receptors could be distinguished by a seven-pass (serpentine) transmembrane motif. Transcription factors could be recognized by the sequence motifs of their deoxyribnucleic acid (DNA) binding domains (e.g., zinc finger, basic helix-loop-helix, homeodomain, or leucine zipper domains). Of the recently sequenced genomes of yeast and C. elegans, for example, about 40% of the open reading frames (ORFs) are recognizable by known motifs (Chervitz et al. 1998). Function can be assigned, at least preliminarily, to the products of those genes. Plans are afoot to define the function of the missing ORFs of yeast and make the functions of all proteins assignable from sequence. At the same time, there are plans to identify a large number of protein-binding sequences in the regulatory regions of genes to be able to predict better the conditions of expression of genes. These plans are among the aims of “functional genomics,” as described in Chapter 5. All the information on sequences, motifs, and function is stored in databases available
to researchers worldwide (e.g., the Basic Local Alignment Search Tool (BLAST) <http://www.ncbi.nlm.nih.gov/BLAST/>).
Drosophila Development at the Molecular Genetic Level
By the time the Drosophila mutants were characterized in the mid-1980s, techniques were well-suited for molecular genetic analysis of affected genes and gene products. This part of the work moved quickly, thanks to gene-cloning techniques, background information about gene sequence motifs and protein function, and databases available to researchers worldwide. The successful isolation of a gene responsible for a developmental phenotype (when the gene was mutated) could be validated by the rescue of the mutant phenotype by transformation with the wild-type gene (usually as DNA included in a P-element transposon). In situ hybridization, coupled with color stains, readily revealed the normal time and place of expression of the specific genes whose mutations had been isolated. Regarding the function of these developmental genes, many were found to encode proteins with familiar motifs, such as those for receptor tyrosine kinases or various transcription factors. In fact, a surprisingly large number turned out to be transcriptional regulators. Function could be rapidly concluded from sequence data. Other Drosophila genes encoded proteins whose specific functions were unknown, yet they were recognizable generally as secreted proteins by their signal sequences or as new transcription factors by the fact they accumulated in nuclei and could bind to DNA. In the course of this analysis, new intercellular signaling pathways were discovered, such as those involving the Decapentaplegic (DPP), Hedgehog (HH), Wingless (WG), and Notch/Delta ligands. (The whimsical names are those given by researchers to mutants based on the phenotypes.)
Hundreds of laboratories worldwide joined the work on Drosophila mutants, and the picture of early development took on a satisfying coherence and clarity, especially the steps of generation of segmentation and of the overall body organization in the anteroposterior and dorsoventral dimensions. These steps of early development are known collectively as “axis specification.” The following is a brief summary of that picture to illustrate its completeness at the molecular level. The steps are stage-specific mechanisms of development. The mechanisms are now better understood in Drosophila than in any other organism. It is the kind of information scientists would like to have, but do not yet have, for mammalian development.
At the start of Drosophila development, the oocyte is provisioned with hundreds of maternal gene products that are uniformly distributed in the egg during oogenesis. Four gene products are spatially localized in the egg, however, and they provide the initial asymmetries on which the entire anteroposterior and dorsoventral organization of the embryo is built stepwise in development after fertilization. The four gene products include the following:
-
An mRNA located internally at the anterior end (encoding a transcription factor, named Bicoid).
-
An mRNA located internally at the posterior end (encoding an inhibitor of the translation of the mRNA for a transcription factor, named Nanos).
-
An external protein anchored to the egg shell at both ends of the egg (involved in the production of a ligand of a receptor tyrosine kinase in the egg-cell plasma membrane).
-
An external protein also anchored to the egg shell but at the prospective ventral side (involved in the production of a signal ligand of the Toll receptor in the egg-cell plasma membrane).
To exemplify the steps of use of those gene products, only one of the dimensions, the anteroposterior, will be described. The two mRNAs are initially at opposite ends of the egg. They are translated after fertilization, and the encoded proteins diffuse from the ends to form opposing gradients reaching to the middle of the egg. These proteins will act in concert to generate a gradient, high at the anterior end and low at the posterior end, of another transcription factor. The nuclear number increases rapidly in the uncleaved cytoplasm. The graded transcription factors, called members of the “coordinate class” or “egg-polarity class” of gene products, activate at least eight gap genes in nuclei along the egg’s length at different positions, each position unique in terms of the local quantity of transcription factors of the coordinate class. (The terms “coordinate,” “egg polarity,” and “gap” also derive from mutant phenotypes.) The encoded gap proteins, which are all transcription factors themselves, accumulate in a pattern of eight broad and partially overlapping stripes along the egg’s length. The proliferating nuclei are not yet separated by cell membranes—that comes later. These proteins in turn activate at least eight pair-rule genes, all of which also encode transcription factors. Complex cis-regulatory regions of the various pair-rule genes define their expression responses to the spatially distributed gap proteins. The pair-rule proteins then activate at least 12 segment-polarity genes, some of which encode transcription factors and some of which encode secreted protein signals. The pair-rule and gap proteins together also activate eight homeobox (Hox) genes to be expressed in broad stripes, as discussed in the next section. Thus, the early steps of development involve cascades of transcription factors distributed in space according to the initial gradients of a few agents and to the expression rules contained in the complex cis-regulatory regions of genes for yet other transcription factors. These key steps are accomplished in the first 3 hours of development, mostly before cell membranes are formed and gastrulation begins, although the final elaboration of the segment-polarity and Hox genes occurs after cells form.
Once the segment-polarity genes and Hox genes are activated, they maintain their expression in cells by an auto-activating circuitry, in some cases by the encoded transcription factor activating expression of its own gene. The coordi-
nate, gap, and pair-rule proteins are then no longer needed. Their products disappear, and the genes are no longer expressed.
Similar conclusions apply to the development of the termini and the dorsoventral dimension, which also rely on initially asymmetric signals. The developmental mechanisms of the termini and dorsoventral dimension are of additional interest, because the signals bind to transmembrane receptors and activate signal transduction pathways, eventually leading to the activation of transcription factors and new gene expression. These inductions are the first to occur in the developing Drosophila egg. Approximately 100 genes and encoded gene products have been identified as necessary to establish the organization of the early gastrula. Hundreds more participate in the accomplishment of these events, but they are less well described at present. In most cases, these genes probably encode proteins required in numerous developmental processes and, hence, were not recovered under the conditions of the mutant inspections used here.
As shown in Figure 6-1A-D, a coherent scheme of early development was proposed and well supported by 1992, the first of such complexity and completeness at the molecular level for any organism.
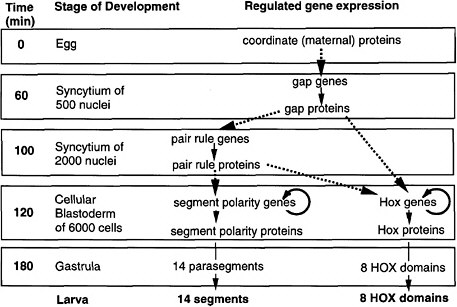
FIGURE 6-1A Outline of anteroposterior development in Drosophila and the steps of regulated gene expression (Ingham 1989). Heavy dashed arrows indicate the activation of specific gene expression by transcription factors. Thin solid arrows indicate transcription and translation. Note that Hox genes are activated by both pair-rule and gap proteins, whereas segment-polarity genes are activated by pair-rule proteins alone. In the anteroposterior dimension, segments and HOX domains are formed. Further explanation is given in Figure 6-1B.

FIGURE 6-1B Anteroposterior development in Drosophila (Nüsslein-Volhard 1991). Figure 6-1B is shown diagrammatically here, for segment formation and HOX compartment formation. The coordinate proteins Bicoid, Nanos, and Cad are translated from mRNAs localized at the two poles of the egg during oogenesis. Translation generates gradients of proteins. Bicoid and Cad are transcription factors, whereas Nanos protein inhibits the translation of another translation factor (Hunchback) in the posterior half of the egg. The graded transcription factors activate eight gap genes, and different factor concentrations activate different gap genes. The gap proteins are also transcription factors. Each diffuses locally and inhibits other gap genes, setting up eight partially overlapping stripes of gap protein along the egg’s length. The gap proteins activate eight pair-rule genes, each of which has a complex cis-regulatory region and is activated by seven combinations of gap proteins, each making seven evenly spaced stripes of protein. Thus, there are 8 × 7 or 56 stripes of pair rules along the egg’s length, arranged in 7-fold repeats. The pair-rule proteins are all transcription factors. These activate eight segment-polarity genes, each of which has a complex cis-regulatory region activated by at least two combinations of pair-rule proteins, to give 14 stripes of expression each. Thus, there are 14 × 8 or 104 stripes of segment-polarity proteins. The 14-fold repeat is the basis for 14 segments of the posterior head, thorax, and abdomen. The pair-rule and gap proteins together activate Hox genes in eight domains in the posterior head, thorax, and abdomen. Cell outlines are not shown, but cells are present in the two lowest panels.
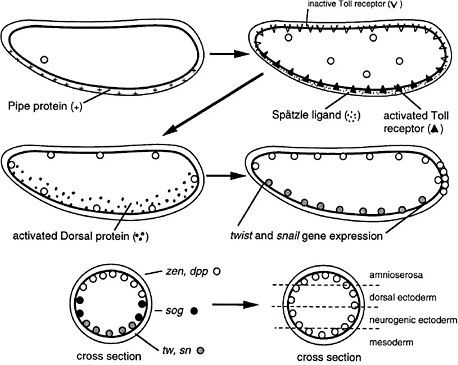
FIGURE 6-1C Dorsoventral development in Drosophila (Nüsslein-Volhard 1991). The egg shell contains Pipe protein on the future ventral side, deposited there during oogenesis. After fertilization, the egg secretes several proteins into the space between the egg shell and plasma membrane. Pipe activates one of the proteins, which then sets off others in a protease cascade, the last member of which cleaves the Spätzle protein, releasing a ligand that binds to the Toll transmembrane receptor, which is uniformly distributed over the egg surface but ligand-activated only on one side. The activated receptor, via several intracellular steps, activates the Dorsal protein, a transcription factor, which enters local nuclei and activates two genes, Twist and Snail, which also encode transcription factors. Those activate other genes for gastrulation and for mesoderm formation on the ventral side. Thus, the Pipe protein is involved in a kind of mesoderm induction. Active Dorsal protein also represses the Zen and Dpp genes on the ventral side. On the dorsal side, Dorsal protein remains inactive and the Zen and Dpp genes are expressed. Laterally, there is enough active Dorsal protein to repress Zen and Dpp but not enough to activate Twist and Snail. Here, the Sog gene is permissively expressed and not repressed, preparatory to neurogenic ectoderm formation. Thus, the dorsoventral dimension of the egg is divided into three domains of gene expression. Later, the Sog protein is secreted and diffuses to the Zen, Dpp region, inhibiting Dpp signaling and allowing the division of that region into two subregions the prospective amnioserosa and prospective dorsal ectoderm.
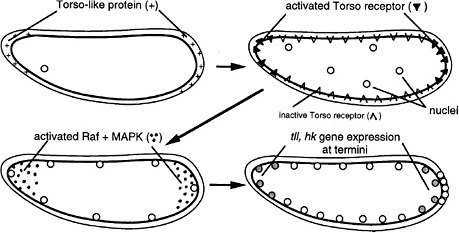
FIGURE 6-1D The development of termini in Drosophila (Nüsslein-Volhard 1991). The Torso-like protein is present in the egg shell at the two ends of the egg, deposited there during oogenesis. After fertilization, the egg secretes several proteins into the space between the plasma membrane and egg shell. The proteins include proteases that are locally activated at the end by the Torso-like protein and release a ligand that binds locally to the transmembrane Torso receptor, a member of the RTK signal transduction family. The activated receptor locally activates Raf and MAPK, which phosphorylate a transcription factor locally, inhibiting its repression of genes and allowing local expression of the Tailless (Tll) and Huckebein (Hkb) genes involved in formation of the endoderm, terminal ectoderm, and gut involution during gastrulation. Thus, the Torso-like protein is involved in endoderm induction.
Hox Genes and the Drosophila Connection to Vertebrate Development
Even though researchers in other areas widely appreciated the breakthroughs in Drosophila development, they questioned the relevance of the information to vertebrate development. Vertebrates, as chordates, were thought to have branched from arthropods long ago and last shared a very simple common ancestor in the pre-Cambrian era (about 540 million years ago). The two groups were thought to have evolved their segmentation and heads independently. One of the first significant similarities between vertebrate and fly development came from work on homeotic genes, now called Hox genes. As mentioned before, the Hox genes are expressed in eight broad bands or spatial compartments in the anteroposterior dimension of the body shortly after gastrulation but prior to organogenesis and cytodifferentiation. Their encoded products make each spatial compartment different from the others.
The study of the eight Hox genes of Drosophila was primarily pioneered by E. Lewis from 1940 to 1970. For his work in that area, he shared the Nobel Prize
with Nüsslein-Volhard and Wieschaus in 1995. Lewis selected Drosophila mutants that exhibited mislocated body parts (e.g., wings in place of halteres (balancing organs) and legs in place of antennae). The term “homeotic” connotes such mislocation without distortion. In the homeotic mutant, the anteroposterior dimension of the animal has fewer anatomical differences along its length. For example, the Ubx mutant has an extra mesothorax located at the normal metathorax position but lacks a metathorax. It has four wings but no halteres, whereas normal Drosophila have two wings and two halteres. When the first two Hox genes (Ubx and Antp) were isolated, their sequences were compared (McGinnis et al. 1984a,b; Weiner et al. 1984), and a shared 60-base sequence was found, the homeobox. The sequence is the same in both genes except for a few bases. That sequence encodes the DNA-binding motif of the encoded proteins, which are members of a large and ancient family of transcription factors. The other six Hox genes were soon isolated from Drosophila, and those too had closely related homeobox sequences. Then the eight genes were shown to exist in a contiguous cluster (actually two subclusters in D. melanogaster but one in another arthropod, Tribolium), probably all tandemly duplicated and diverged from a few founder sequences in an ancestor of arthropods. Furthermore, the members are expressed in stripes in the anteroposterior dimension of the body, in an order identical to their gene order on the chromosome (a correspondence referred to as “colinearity” of gene order and expression).
In the mid-1980s frogs and mice were found to contain similar sequences, also arranged in contiguous gene clusters. Interestingly, their expression in mice showed the same anteroposterior colinearity as that in Drosophila. As an evolutionary explanation, the common ancestor of arthropods and chordates must have had a complex Hox cluster already functioning in its development. Vertebrates, however, differ from arthropods in having at least four multi-member clusters instead of one (Krumlauf 1994). A comparison of gene arrangements and domains of expression in Drosophila and mammalian (mouse) Hox clusters is shown in Figure 6-2.
Such genes are called selector genes because their encoded products, which are transcription factors, select which other genes will be expressed in that spatial compartment of the body. The thousands of target genes of a selector-gene product encode proteins involved in subsequent local development, including the many kinds of organogenesis of different parts of the body. Hox genes have a central role in development. Because of them, the coordinate, gap, and pair-rule proteins of early development do not have to directly activate those thousands of target genes in a region-specific way but activate only the Hox genes, whose encoded proteins then do the job of regulating sets of genes in their respective regions. Methods for the directed knockout of genes in mice were invented by the mid-1980s as a way to test gene function, and the Hox genes of mice were found to control aspects of local development in their compartments, especially in vertebrae, neural tube, and neural crest derivatives. Their selector role was similar to
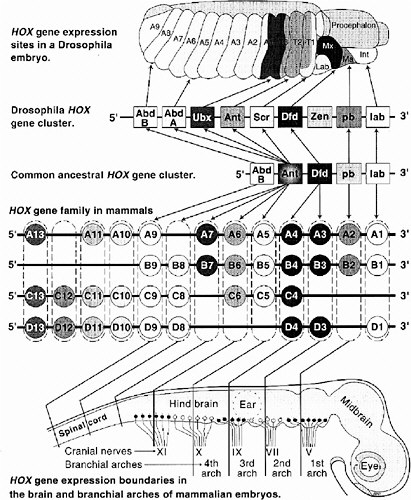
FIGURE 6-2 This figure illustrates the striking similarities of gene organization and expression of Hox clusters in Drosophila and mammalian (mouse) embryos. At the top is a 10-hour Drosophila embryo showing expression zones of individual Hox genes in thoracic (T1-3) and abdominal (A1-9) segments and parts of the head (Lab, labrum; Mx, maxillary; Ma, mandible; Int, intercalary segment). Note the colinearity of Hox gene expression sites along the anterior-posterior body axis to their 3′ to 5′ location along the chromosome. The greatly expanded vertebrate Hox gene family is shown in the middle. These genes are arranged in four clusters (labeled A, B, C, D), each on a separate chromosome. Having arisen by duplications early in chordate evolution, Hox genes in paralogous groups (e.g., A4, B4, C4, D4; shown enclosed in dashed boxes) are more closely related than are adjacent genes (e.g., B3 vs. B4 vs. B5). The four most 5′ paralogous groups have no close equivalent in arthropods; these are expressed in the tail and fins or limbs. Lines extending from each paralogous group to the schematic brain and cranial spinal cord show the rostral limits of expression of members on each group. Note, again, the colinearity between expression sites and relative chromosomal position of most Hox genes. The same is generally true for somites and, in the proximo-distal orientation, for limbs.
that in Drosophila (Behringer et al. 1993). However, many of the target genes of Hox proteins in mice and flies are clearly different.
The Hox clusters of Drosophila and chordates are under intense study. It is now known that genes of four mouse clusters are coordinated in an elaborate circuitry of auto- and cross-activation and repression, in which the genes near the 5′ end of the DNA sequence tend to repress genes near the 3′ end when both are initially expressed in same cell. Equivalent paralogs in different clusters tend to overlap in the target genes they activate and repress, but each has some unique targets, as shown by the phenotypes of single-Hox knockout mutants of the mouse. As a whole, the Hox genes operate as a complex genetic regulatory system rather than as independent members.
More recently, the Hox-like Ems and Otd genes have been discovered in Drosophila as expressed in the head in regions anterior to the expression compartments of the Hox genes. Homologs of these genes (called Emx and Otx) have been found expressed in the head of the frog and mouse anterior to the Hox gene domains of the posterior head, thorax, and trunk. This was a surprise, because evolutionary biologists had thought that the vertebrate head is unique to that group and has little in common with the head of a common ancestor of vertebrates and arthropods. However, even that complexity of body organization, like HOX compartments, must predate the branching of arthropods and chordates.
The Emergence of Caenorhabditis elegans
The free-living nematode Caenorhabditis elegans emerged as an important model system in the 1970s, as the result of pioneering work on its genetics by S. Brenner (1974). Chosen for its short life cycle (3 days) and general amenability for genetic analysis, small size (1-mm length), transparency, and simplicity (only 959 somatic cells), C. elegans quickly attracted a following among developmental biologists and geneticists. In particular, J. Sulston was primarily responsible for first describing the complete cell lineage from fertilization to adulthood (Sulston and Horvitz 1977; Sulston et al. 1983) and then spearheading the physical mapping and DNA sequencing of the genome. C. elegans recently became the first metazoan organism whose genome is completely sequenced (C.elegans Sequencing Consortium 1998). In the meantime, researchers from many laboratories isolated mutants and identified many important genes controlling development, the result being that C. elegans is now the most completely described and one of the best understood models for development (see Chapter 7). In some ways, the development of vertebrates is more similar to that of C. elegans than of Drosophila (e.g., having a cellular rather than a syncyctial early embryo), and in other ways less similar (e.g., having a highly invariant cell lineage and a fixed small number of cells, no Sonic Hedgehog signaling pathway, and few HOX genes). These two model animals complement each other usefully for research into fundamental mechanisms of metazoan development.
Conserved Developmental Processes
Researchers increasingly suspected similarities of development between fruit flies and mice and began to look systematically for homologs of Drosophila developmental genes in mice, frogs, and chicks. In the late 1980s, this was a new research approach. Its success has favored the impression that at a gross level, nematodes, flies, and mice are “all the same organism” and that what is learned about one will have relevance to the others. In a genetically tractable organism, such as Drosophila or C. elegans, a gene is isolated by using a screen for a particular kind of developmental failure, and then the role of its encoded product in development is efficiently deciphered in that organism. Homologs of “developmentally interesting” genes are then sought in vertebrates, such as mice or frogs, in which mutant searches are still daunting due to the comparatively small populations and slow development. The homolog’s function is thereafter studied in the vertebrate, for which the Drosophila or C. elegans information is used as a guide. The mouse is attractive for such studies, because the homologous gene can be knocked out and the phenotype of the null mutant examined to learn about the function of the encoded product.
A surprising array of developmental components and processes is shared between Drosophila and vertebrates (i.e., between arthropods and chordates). In addition to the EMX, OTX, and HOX organization of the body plan, they share the compartments of the dorsoventral dimension (which are thought to be inverted in orientation in one group relative to the other); the presence and mode of organogenesis of limbs (appendages), eyes, heart, visceral mesoderm, and gut; the steps of cytodifferentiation during neurogenesis and myogenesis; and even segmentation. Although the anatomical structures themselves are very different between arthropods and chordates, a number of the underlying steps of development are the same. These are listed in more detail in Table 6-1. The last common ancestor of chordates and arthropods was, it seems, a pre-Cambrian animal of much greater complexity than previously realized. Divergent groups of metazoa (members of the animal kingdom) can be treated as “the same organism” in the experimental analysis of many fundamentals of development. From all of those similarities, the value of model systems for gaining an understanding of difficult basic problems in mammalian development, including that of humans, is undeniable. Humans, flies, and even roundworms are less different than widely thought just 10 years ago.
Signaling Pathways in Development
An important realization to come from the Drosophila research concerns the pervasive use of cell-cell signaling in most aspects of development, starting with the termini and dorsoventral dimension (see Figures 6-1A-D) and extending to organogenesis of many kinds. Inductive signaling was thought to be important in vertebrate development, as mentioned above, but insects and other invertebrates
TABLE 6-1 Similarities of Arthropods and Chordates
had been assumed to develop as composites of independent lineages of cells (“mosaic” development). This is not at all the case. Six signaling pathways are used repeatedly in early Drosophila development: the Hedgehog, Wingless-Int (Wnt), transforming growth factor β (TGFβ), Notch, receptor tyrosine kinase (RTK), and cytokine receptor (cytoplasmic tyrosine kinase) pathways. Comparative studies soon showed that these pathways exist in vertebrates as well, and most also exist in nematodes (except the Hedgehog pathway). Four other conserved pathways in addition to those six are used heavily in later development, mainly in organogenesis, and seven others come into use in the physiological functioning of the organism’s differentiated cell types. The number of known pathways has now reached 17. Each pathway is distinguished by its unique set of transduction protein intermediates. The 17 pathways are listed in Table 6-2. Details of the components and steps of the individual pathways are given in Appendix C.
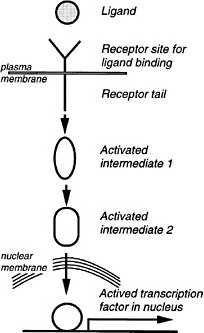
As a generalization, most of the pathways involve transmembrane receptor proteins that bind ligands at the extracellular face, as diagramed in Figure 6-3. Ligands arrive in some cases by free diffusion after secretion from distant neighbor cells. Others diffuse only short distances or remain attached to the surface of the cell of origin, reaching only the contacting cells. Activated receptors of the recipient cell activate the first intracellular component of a signal transduction pathway, and this then activates a subsequent component, and so on. Some pathways are long, with 7-10 intermediates. Others have one or two. The nuclear hormone receptor pathway is the shortest, having only one step. In this case, hydrophobic ligands penetrate the cell membrane on their own and bind to a receptor protein, which also functions as a transcription factor. In the longer pathways, a change of activity is passed along a series of on-off switches, which constitutes an information relay pathway, or signal transduction pathway. Ultimately, in some pathways, a protein kinase is activated at the end of the series, and that enzyme phosphorylates numerous target proteins, which change their activity (activated or inhibited) because of the phosphate addition. The target proteins are components of various basic cell processes, such as transcription, the cell cycle, motility, or secretion. Hence, these processes are turned on or off, and the change of function constitutes the cell’s response to a signal. In many other pathways, a specific transcription factor is activated at the end of the pathway, and this factor is a pathway component. In development, the most frequent target of signaling pathways is indeed transcription. The pathways used in early development tend to have transcription as the only target. That is, particular transcription factors are phosphorylated or proteolyzed as a signal transduction step of the pathway, changing their activity in activating or repressing particular genes.
The pathways are used repeatedly at different times and places of development in Drosophila, nematode, and vertebrates, as listed in Table 6-3. Drosophila null mutants are usually lethal if they lack a step in any of those pathways. Lethality is an indication of the essentiality of those signaling functions. However, in the mouse (and probably all vertebrates), a null mutant for a step of a
TABLE 6-2 The 17 Intercellular Signaling Pathways
pathway is often not lethal. The mutant is born with a limited abnormality of anatomy and sometimes of behavior. The genetic basis for some of these mouse developmental defects is fully known. In some cases, the mutant mice live to adulthood and reproduce.
Are the signaling pathways less important in vertebrate development than in Drosophila? No, the nonlethality in vertebrates reflects the fact that the pathways have a substantial redundancy of signaling components. It is postulated that early in vertebrate evolution (as jawless fish arose), the genome underwent a quadruplication (Holland et al. 1994). In addition, some genes underwent tandem dupli-
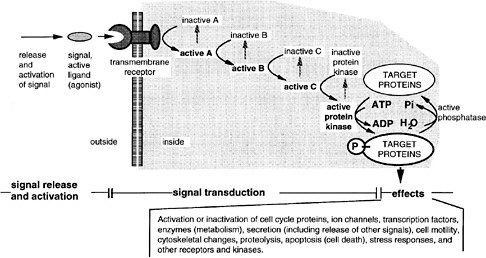
FIGURE 6-3 A generalized signal transduction pathway (information transfer via a series of on-off switches. The active ligand is shown on the left, approaching the transmembrane receptor protein. Inside the cell is a multistep signal transduction pathway composed of switch-like intermediates, A, B, and C, that can exist in active and inactive states. In the absence of signal, the intermediates are inactive. During binding, the receptor becomes active and activates one intermediate that activates the next, and so on in series, until eventually a protein kinase is activated. This pathway transfers information, not energy or materials. The kinase enzyme is specific for transferring phosphate from adesine triphosphate to a serine, threonine, or tyrosine residue of select protein targets within the cell. Phosphorylated target proteins change activity, becoming either active or inactive, and therefore the basic cellular process of which they are a part, changes activity. In this way, the signal has effects (i.e., it triggers cellular responses). Target proteins might be components of the processes of transcription, translation, the cell cycle, cell movement, differentiation, or other signaling pathways. Transcription is a particularly frequent target. Fourteen of the 17 involve transmembrane receptors; the other two involve intracellular receptors and the ligands pass through the plasma membrane readily.
cations. Many of the initially identical genes later diversified their sequences, leading either to diversified functions of encoded products or to diversified cis-regulatory regions setting different conditions of expression. Despite this diversification, extensive redundancy still remains. For example, there are more than 24 TGFβ ligand genes and 11 Wnt ligand genes in mouse, but only 3-5 TGFβ and 1-3 Wnt genes in Drosophila. In mouse development, the various genes for a single step of a single kind of signaling pathway are expressed at different times and places in the embryo. When one gene is knocked out, the defect in development is limited in scope to a few times and places where no related gene is expressed to provide overlapping function.
TABLE 6-3 Various Uses of Signaling Pathways in Vertebrate Development
As shown in Table 6-4 and Table 6-5, many single-null mutants of the mouse are born live and have minimal defects that can be scored as developmental defects (e.g., skeletal abnormalities). Some have motor coordination defects, such as the WNT1 null mutant, which lacks the entire cerebellum. Some live for a few days, and others reach adulthood and are fertile. In Table 6-4, five pathways are surveyed partially (one or two components eliminated per pathway). In Table 6-5, one pathway is surveyed exhaustively (all components eliminated one at a time). Each kind of mutant is defective for one component of one signaling
TABLE 6-4 Survey of Phenotypes of Mouse Mutants Lacking Components of Any of Several Signaling Pathways
|
Signaling Component |
Viability of Null Mutant |
Phenotype of Null Mutant |
References |
|
Wnt pathway |
|
||
|
WNT-1 |
Adulthood |
No midbrain, cerebellum, and rhombomere 1; behavioral deficits |
McMahon et al. 1992 |
|
Axin |
Early lethal (day 8-10) |
Twinning |
Vasicek et al. 1997; Zeng et al. 1997 |
|
Transforming growth factor β pathway |
|
||
|
TGFβ 1 |
Adulthood |
Immune defects, inflammation |
McCartney-Francis et al. 1997 |
|
TGFβ 2 |
Perinatal death |
Defects of heart, lung, spine, limb, and craniofacial and spinal regions |
Sanford et al. 1997 |
|
GDF5 |
Adulthood |
Fused skeletal elements in limbs, one-third of joints missing; like brachypodism mutant. |
Storm and Kingsley 1996 |
|
BMP5 |
Adulthood |
Thin axial bones; abnormal lung, liver, ureter, and bladder; like a short ear mutant |
Mikic et al. 1996 |
|
BMP7 |
Adulthood |
Defects in eye and kidney; skeletal abnormalities; polydactyly of hindlimbs |
Dudley and Robertson 1997 |
|
Noggin |
Juvenile |
Bone hyperplasia, joints not formed; neural tube and somite defects |
Brunet et al. 1998 |
|
Hedgehog pathway |
|
||
|
Sonic Hedgehog (SHH) |
Perinatal death |
Cyclopia, defects of spinal cord, axial skeleton, and limbs |
Chiang et al 1996 |
|
Patched receptor |
Homozygotes, early lethality |
Open neural tube |
Goodrich et al. 1997 |
|
|
Heterozygotes, adulthood |
Rhabdomysarcomas, hindlimb defects, large size. Like Gorlin syndrome in humans. |
Hahn et al. 1998 |
|
Notch pathway |
|
||
|
Notch 1 |
Perinatal death |
Disordered somites. Like Danforth short tail mutation? |
Swiatek et al. 1994; Conlon et al. 1995 |
|
Delta (DII1) |
Perinatal death |
Disordered somites |
Hrabe De Angelis et al. 1997 |
TABLE 6-5 Phenotypes of Mouse Mutants Lacking Components of the Receptor Tyrosine Kinase Pathway
|
Signaling Component |
Time of Death |
Affected Tissues and Organs |
Phenotype |
Human Syndrome or Diseasea |
References |
|
Signals |
|
||||
|
FGF2 |
Viable |
Brain, skin |
Delayed wound healing, lower neuronal density |
|
Ortega et al. 1998 |
|
FGF3 |
Neonatal |
Tail, inner ear |
Developmental defects |
|
Mansour 1994 |
|
FGF4 |
Early post-implantation |
Inner cell mass |
Growth failure after implantation |
|
Feldman et al. 1995 |
|
FGF5 |
Viable |
Hair follicle |
Continuous hair growth |
Hypertrichosis universalisb |
Hebert et al. 1994 |
|
FGF6 |
Viable |
Muscle |
Defective muscle regeneration |
|
Floss et al. 1997 |
|
FGF7 |
Viable |
Keratinocyte and hair follicle |
Rough coat |
|
Guo et al. 1996 |
|
FGF8 |
Gastrulation |
Mesoderm |
No heart and somites |
|
Meyers et al. 1998 |
|
GDNF |
Neonatal |
Kidney, neural crest |
Renal agensis, enteric nervous system defects, neoplasia |
Hirschprung’s disease, multiple endocrine |
Pichel et al. 1996; Sanchez et al. 1996; Moore et al. 1996 |
|
HGF |
Mid-gestation |
Skeletal muscle, liver |
No muscles, small liver |
|
Schmidt et al. 1995 |
|
IGF1 |
Reduced, some neonatal deathc |
Organs, muscle, bone |
Severe growth deficiency |
Dwarfism; neoplasiab |
Powell-Braxton et al. 1993; Liu et al. 1993 |
|
IGFII |
Viable |
Fetus and placenta |
Severe growth deficiency |
Beckwith-Wiedemann syndrome |
DeChiara et al. 1990 |
|
NGF |
Neonatal |
Nervous system |
Lack of sensory and sympathetic neurons |
|
Crowley et al. 1994 |
|
PDGFA |
Mid-gestation or neonatal |
Lung, alveolar, myofibroblasts |
Emphysema |
|
Bostrom et al. 1996 |
|
Receptors |
|
||||
|
EGFR |
Perimplantation, mid-gestation, or postnatalc |
Inner cell mass, placenta, skin, kidney, brain, liver, lungs |
Multiple abnormalities, lung immaturity, wasting |
Cancer |
Threadgill et al. 1995; Miettinen et al. 1997; Sibilia and Wagner 1995; Downward et al. 1984 |
|
EPHA2 |
Viable |
None |
No phenotype |
|
Chen et al. 1996 |
|
EPHA8 |
Viable |
Neurons |
Altered axonal projections |
Park et al. 1997 |
|
|
FIT1 |
Viable |
Vascular endothelium |
Abnormal vascular channels |
Fong et al. 1995 |
|
|
FLK1 |
Mid-gestation |
Vascular endothelium |
No blood or blood vessels |
|
|
|
FLK2 |
Viable |
Hematopoietic stem cells |
Deficient B cells |
||
|
FGFR1 |
Mid-gestation |
Mesoderm |
Abnormal mesoderm patterning |
Pfeiffer syndrome, Acrocephalopoly-syndactyly type ll; stem cell leukemia/lymphoma syndromeb |
Deng et al. 1997; Yamaguchi et al. 1994 |
|
FGFR2 |
Periimplantation |
Inner cell mass |
Growth failure |
Apert, Crouzon, Pfeiffer, Jackson-Weiss, Cutis gyrata syndromeb |
Arman et al. 1998 |
|
FGFR3 |
Viable |
Skeleton, inner ear |
Skeletal overgrowth |
Achondroplasia; hypochandroplasia; thanatophoric dwarfism; deafness; Muenke syndrome |
Colvin et al. 1996 |
|
FGFR4 |
Viable |
None |
Slight growth retardation |
|
Weinstein et al. 1998 |
|
GCSFR |
Viable |
Immune system |
Immune deficiency |
Kostmann disease |
Liu et al. 1996 |
|
HGFR |
Mid-gestation |
Skeletal muscle, liver |
No muscle, small liver |
|
Bladt et al. 1995 |
|
IGF2R |
Perinatal |
Fetus |
Overgrowth, cardiac defects |
Lau et al. 1994; Wang et al. 1994 |
|
|
IR |
Neonatal |
Fetus |
Diabetic ketoacidosis, growth retardation |
Leprechanism |
Joshi et al. 1996 |
|
Signaling Component |
Time of Death |
Affected Tissues and Organs |
Phenotype |
Human Syndrome or Diseasea |
References |
|
LIN12 |
Mid-gestation |
Somites |
Somitogenesis delayed/disorganized |
|
Conlon et al. 1995 |
|
NGFR |
Viable |
Nervous system |
Defective sensory neurons |
|
Lee et al. 1992 |
|
NTRK1 |
Neonatal |
Central nervous system (CNS) |
Neuropathies |
Cancer, congenital neuropathy with anhidrosis |
Smeyne et al. 1994 |
|
PDGFRA |
Mid-gestation |
Skeleton, neural crest |
Spina bifida oculta and other skeletal abnormalities, increased neural crest apoptosis |
|
Soriano 1997; Payne et al. 1997 |
|
RET |
Perinatal |
Kidney, neural crest |
Renal agenesis, enteric nervous system defects |
Hirschprung’s disease, multiple endocrine neoplasia |
Schuchardt et al. 1994, 1996 |
|
Intracellular |
|
||||
|
GAP |
Mid-gestation |
Undefined |
Postimplantation arrest and death |
|
Henkemeyer et al. 1995 |
|
PLCY1 |
Mid-gestation |
Unknown |
Growth retarded |
Ji et al. 1997 |
|
|
PRKM4/MAPKK4 |
Mid-gestation |
Undefined |
Death |
Yang et al. 1997b |
|
|
PTPN11/SHP2 |
Mid-gestation |
Undefined |
Failure to gastrulate |
Saxton et al. 1997 |
|
|
SAPK/ERK1 |
Viable |
T cells |
Altered susceptibility to apoptosis |
Nishina et al. 1997 |
|
|
SERK2/SAPK/ERK/JNK3 |
Viable |
CNS |
Altered response to kainic acid damage |
Yang et al. 1997a |
|
|
H-RAS |
Viable |
None |
None |
Cancer |
Johnson et al. 1997 |
|
K-RAS 2 |
Mid-gestation |
CNS |
Increased cell death |
Cancer |
Koera et al. 1997 |
|
N-RAS |
Viable |
None |
None |
Tumors |
Umanoff et al. 1995 |
|
RAS/GAP |
Mid-gestation |
CNS, heart, allantois |
Disorganized, organogenesis defects |
|
Henkemeyer et al. 1995 |
|
RAS-GRF |
Viable |
CNS |
Impaired long-term memory |
Brambilla et al. 1997 |
|
|
VAV |
Viable |
T cells |
Defective selection of thymocytes |
Turner et al. 1997 |
|
|
Target Proteins |
|
||||
|
C-FOS |
Viable |
Osteoclasts |
Osteopetrosis, toothless |
|
Johnson et al. 1992; Wang et al. 1992 |
|
FOSB |
Viable |
Mammary gland |
Failure to nurture offspring |
Gruda et al. 1996; Brown et al. 1996 |
|
|
C-JUN |
Mid-gestation |
Fibroblasts and others |
Altered response to mitogens |
Hilberg et al. 1993; Johnson et al. 1993 |
|
|
aSources: Online Mendelian Inheritance in Man (<ww3.ncbi.nlm.nih.gov/Omim/>) and reference no. 2. bPossible association between gene and syndrome. cVariable phenotype depending on genetic background. Note: Null phenotypes of mice produced by gene targeting for some genes involved in one signaling pathway, the receptor tyrosine kinase pathway, and human syndromes or diseases associated with mutations or deletions in these genes. In addition to the entries in this table, many of the mouse mutations have been combined to make double or triple mutants. Additional follow-up studies, some using hypomorphic alleles and chimeras, have been reported (see Mouse Knockout Database: <http://tbase.jax.org>). |
|||||
pathway. Of relevance to this report, some mutant phenotypes resemble those of animals treated in embryogenesis with various developmental toxicants. Said otherwise, some toxicant treatments produce phenocopies of mutants. For example, SHH mutants have cyclopia, as do normal embryos treated with cyclopamine (Beachy et al. 1997). Researchers of human developmental defects have benefitted by analysis of these mouse developmental defects for which the genetic defects are accurately known.
Antagonists of Signaling
Although only a small number of signaling pathways operate in early development, these have several regulatory features of relevance to developmental toxicity. One is that many of the pathways engage in “cross-talk” with one another, so that activation of one enhances or suppresses the activation of another. Cross-talk occurs at all levels, from effects on ligand availability to effects on target function. For example, the active RTK pathway can lead to phosphorylation and activation of GSK3 in the Wnt pathway, or phosphorylation and inactivation of a SMAD protein of the TGFβ pathway. Signaling via one pathway, such as the SHH pathway, can lead to repression of gene expression of the components of another pathway, such as the Nodal TGFβ pathway. Signaling via the RTK or Wnt pathways can negatively affect signaling via the Notch pathway.
A second regulatory feature is the production by cells of antagonists of signaling, some of which are listed in Table 6-6. For example, embryonic cells can produce proteins, such as Chordin or FRZB, that bind directly to the BMP4 or WNT8 ligands, respectively, and block their capacity to bind to their receptors. Hence, signaling is prevented even though the signal is present. The Chordin protein is further regulated by a protease that degrades it, an antiantagonist. Thus, the pattern of activity of a signaling pathway is subject to extensive modulation, both positive and negative. Several kinds of adjacent cells can affect the outcome of signaling. It is plausible that some of these antagonist proteins are targets of toxicants. Disrupting the activity balance of agonists and antagonists in a region of the developing embryo would be expected to disrupt development, leading to the over- or under-development of an organ. That could occur without a change in level of the agonist or antagonist.
Finally, in many pathways, the operation of the pathway leads to the generation of self-inhibiting components in the cells receiving signals, a feedback mechanism, which is thought to have importance in the controlled spatial responses of cells to diffusible signals. Toxicants upsetting these feedbacks would upset development.
Molecular-Stress Pathways and Checkpoint Pathways
Molecular-stress pathways and checkpoint pathways are not pathways of intercellular signaling but of intracellular signaling, reflecting an individual cell’s
Table 6-6 Natural Antagonists of Signaling Pathways
sensing of disruptions of normal cell function and development due to either physical or chemical agents of the environment (the molecular-stress pathways) or the cell’s own internal imbalance or errors in its synthetic activities (the checkpoint pathways). These pathways are widely present in single-celled eukaryotes (e.g., yeast) and even prokaryotic cells, as well as in animals. Several molecular-stress and checkpoint pathways are listed in Table 6-7 and illustrated in Appendix C.
Molecular-stress pathways are activated when the cell suffers some chemical alteration, such as damage to DNA (e.g., by X-ray, UV, or alkylating agents) or denaturation of proteins (e.g., by hyperosmotic conditions, oxidation, heat, or alcohol). The cell’s signaled response is one of repair and homeostatic counteraction. In the case of the cytosolic unfolded protein pathway (previously called the “heat shock response”), chaperone proteins, such as Hsp90, help to refold denatured proteins, restoring their activity. These same chaperones play a folding role in the normal synthesis and deployment of intrinsically unstable proteins, such as cell-surface receptors. Recent experiments have suggested that if Hsp90 is partially disabled by mutation or overloaded by stress, variant proteins in some members of a population might be unable to fold correctly, resulting in developmental defects (Rutherford and Lindquist 1998). Another example would be the multidrug transport proteins (P-glycoprotein of mammals) that are induced in the presence of high drug levels and serve to export a wide variety of drugs from the cell.
In checkpoint pathways, the cell’s response is one of delaying certain synthetic processes until other processes are complete. These controls are important in coordinating the timing and extent of cellular processes, such as ensuring the completion of DNA synthesis before mitosis begins or ensuring the attachment of chromosomes to the spindle before anaphase begins. An example of relevance to developmental toxicology is that when colchicine (a Vinca alkaloid) inhibits microtubule formation in a mitotic cell, the cell is prevented from initiating anaphase because of a checkpoint control pathway, which assesses the attachment of kinetochores to spindle microtubules (Rudner and Murray 1996). While chromosomes remain unattached, anaphase is not initiated. When the inhibitor is removed, the cells assemble a spindle and proceed with anaphase. Mutant cells have been isolated that lack components of the control (such as the MAD2 protein), and these cells initiate anaphase in the presence of colchicine, without a spindle. They suffer extensive aneuploidy.
Checkpoint and molecular-stress pathways work together. For example, damaged DNA inside the cell triggers various stress pathways, leading to DNA repair. During the repair, the checkpoint pathways delay DNA synthesis or mitosis until repair is complete. In the context of this committee’s evaluation, there are two relevant points about those widely distributed pathways:
-
They offer possibilties for the detection and analysis of developmental toxicants, because they indicate the cell’s state of stress in the presence of toxi-
Table 6-7 Molecular-Stress Response and Checkpoint Pathways (see Appendix C for illustrations)
|
Checkpoint Pathways |
Function and Cell Response |
|
G1/S checkpoint |
Monitors nutritional state, biosynthetic capacity, and cell adhesion and imposes G1 arrest until cell is prepared for S phase. |
|
G2/M checkpoint |
Monitors completion of DNA synthesis (S phase) and imposes G2 arrest until cell is prepared for M phase. |
|
Metaphase/anaphase checkpoint |
Monitors attachment of chromosomes to the spindle and imposes metaphase arrest until cell is ready for anaphase. |
-
cants. Broadly acting toxicants are likely to show up as triggers of stress responses.
-
The intercellular signaling pathways of metazoa probably arose in evolution as elaborations and reworkings of the more ancient molecular-stress and checkpoint pathways of single-celled eukaryotic ancestors. This is surmised because a number of the intermediates (e.g., protein kinases) of the molecular-stress and checkpoint pathways are also used in the metazoan signaling pathways.
One more kind of molecular-stress and checkpoint pathway should be noted: the apoptosis pathway, which is also a signaling pathway (see Table 6-2). It can be activated by either extracellular or intracellular signals and leads to the “programmed” death and destruction of a cell. It is a tightly controlled process in which a cell is destroyed but neighboring cells are unaffected. Apoptosis is not found in single-celled organisms. It is an invention of metazoa and is used in normal embryonic development as well as in recovery attempts of teratogen-damaged embryos. In normal development, where apoptosis is also known as programmed cell death, it is important in the shaping of tissues and organs (e.g., the elimination of cells from the interdigital spaces of the human hand). Cells undergoing apoptosis are found throughout most embryonic mesenchymal tissues, presumably reflecting the elimination of cells that have not been able to successfully integrate the signals impinging upon them. Some mouse mutants, such as Hammertoe, fail to initiate the normal amount of programmed cell death in normal limb development, and an abnormal limb results (Zakeri et al. 1994).
Apoptosis is also the ultimate molecular-stress and checkpoint pathway, for it eliminates cells too damaged to be restored to a normal state by the various repair and checkpoint pathways. For example, if DNA repair is incomplete and the cell attempts to divide, it is killed and autolyzed. It has been proposed that cell death is less detrimental to the multicellular organism than having live cells with highly modified DNA, perhaps proliferating uncontrollably and interacting aberrantly with other cells. Apoptotic cell death is an early response of embryos to many if not all teratogens (Scott 1977; Knudsen 1997). Often, teratogen-induced cell death occurs in the areas of normal programmed cell death but in an expanded area (Alles and Sulik 1989). If cell death is not too extensive, embryos are thought to recover by compensatory cell proliferation (Sugrue and DeSesso 1982). Excessive teratogen-induced cell death, however, is directly linked to abnormal development. For example, eye defects induced by 2-chloro-2'-deoxyadenosine are associated with excessive teratogen-induced cell death (Wubah et al. 1996).
The intracellular signals of apoptosis are not yet known. Key components in the execution phase of the apoptotic pathway are the intracellular cysteinyl-aspartate proteases known as caspases, particularly caspase-3 (Colussi and Kumar 1999). These enzymes are normally present in all cells as inactive precursors that become activated by cleavage at specific internal motifs, in response to cytochrome c leaked by mitochondria into the cell’s cytoplasm. Once activated, these caspases function to degrade specific target substrates such as poly(ADP-ribose)-polymerase (PARP), DNA-PKs, and lamins. Thereafter, chromosomal DNA is broken down. Treatment of cells with such developmental toxicants as hyperthermia, cyclophosphamide (an alkylating agent), and sodium arsenite (thiol oxidant) leads to the activation of caspase-3, cleavage of PARP, fragmentation of DNA, and cell death (Mirkes and Little 1998). It is not known how cells in the embryo recognize exposure to a developmental toxicant and initiate the apoptotic
response, but perturbation of the redox status of the cell and oxidative stress are often, if not always, involved. As in other pathways, the apoptotic pathway engages in cross-talk, for example, with the nuclear factor-kappaB (NF-kB) and INF and FAS pathways. A recent report demonstrates that heat shock (43°C) can rapidly activate the stress-activated protein kinase pathways mediated by c-JUN terminal kinase (JNK) and p38 (Wilson et al. 1999).
As noted for the drug-metabolizing enzymes discussed in Chapter 5, these molecular-stress and checkpoint pathways deserve attention as elements of the organism’s defense against physical and chemical interventions. It remains to be learned whether polymorphisms of defense components exist in humans, compromising their responses to environmental agents. The extent to which the germ line, gametes, and early embryos operate these molecular-stress and checkpoint pathways is also poorly understood.
Developmental Differences
Although Drosophila and mouse development share more similarities than anyone thought 15 years ago, significant differences do exist. Mice share more aspects of development with other chordates (the chordate phylum includes vertebrates, cephalochordates, and urochordates) than they do with Drosophila, and they share still more aspects with other mammals. There appear to be “nested similarities” of development (i.e., the more recent the common ancestor of two groups, the more shared features of their development). Regarding HOX genes, for example, chordates have four more kinds of genes (HOX 10-13) than do arthropods. These differ slightly in sequence from the others and are located at the 5′ end of each cluster. They are expressed in the postanal tail, which is a chordate structure not shared by arthropods, and also in the developing vertebrate limb. Still, the difference between chordates and arthropods is a modification of a shared feature, namely, the use of HOX genes to divide the anteroposterior dimension of the animal into nonequivalent spatial compartments.
Chordates, but not arthropods, share the development of a dorsal hollow nerve cord, a notochord, and a segmentally arranged pharyngo-branchial apparatus, in addition to a postanal tail. They also share a kind of development involving a centralized “organizer” group of cells, the Spemann organizer, which releases inducers important in the placement, orientation, and scaling of later development by surrounding cells. The inducers secreted by the organizer have now been identified. Several inducers are secreted protein antagonists of the TGFβ and WNT signals and are used by surrounding cells to maintain their ventral posterior paths of development. The inducer antagonists disinhibit and hence release the inherent capacity of the surrounding cells to undertake dorsal anterior kinds of development (e.g., to form the neural tube rather than epidermis) (Harland and Gerhart 1997; Smith and Schoenwolf 1998; Weinstein and Hemmati-Brivanlou 1999). Few researchers would have guessed a few years ago
that this subtle depression of neural development is the organizer’s function in all chordates. Nonetheless, it should be noted that Holtfreter (1947), building on a discovery of Barth (1939), suggested that neural inducers provide little information except to release the inherent capacity of ectoderm cells to develop as neural tissue. This suggestion came from Barth and Holtfreter’s findings that ectoderm would develop neural tissue if merely shocked briefly by ion imbalances or pH extremes.
Even though the organizer mode of development is distinctive to chordates, the components of the process are common to a wide range of other animals. For example, one antagonist, the Chordin protein, exhibits significant homology with the SOG protein of Drosophila. The SOG protein antagonizes a TGFβ inductive signal (called Screw) in Drosophila as part of the development of regions of neural versus epidermal development (Neul and Ferguson 1998). Furthermore, in both Drosophila and frogs, there is a specific metalloproteinase that degrades the signal-antagonist complex, releasing the signal. The chordate and Drosophila inductive processes have deep similarities, though differing in details of time, place, and circumstances of use.
As a final example of differences, the dorsoventral dimension of arthropods looks quite different from that of a mouse, but recent analysis has shown that a number of similar genes are expressed in the nerve cords, hearts, body muscle, visceral mesoderm, and gut of both. It is currently accepted that these organs were present in primordial form in a common ancestor, but the arrangement of the organs in chordates is the inverse of that in arthropods. That is, the nerve cord is dorsal in chordates and ventral in arthropods, and the heart is ventral in chordates and dorsal in arthropods. The inversion of the dorsoventral axis is thought to have occurred in the chordate line after hemichordates split off (Nübler-Jung and Arendt 1996).
Recognizing the fact that Drosophila does not share all details of early development and organogenesis with vertebrates, researchers have begun a systematic collection of developmental mutants of the zebrafish, a small vertebrate with a short life cycle (see Chapter 7), suitable for the production of a large mutant collection. The organs of embryonic zebrafish, more than the organs of Drosophila, resemble those of mammalian embryos in structure and function. In light of the extensive conservation of developmental processes found thus far, it is expected that in most cases what is true for fish development, as learned from those mutants, will be true for mammalian development, down to the level of molecular details of components and processes. That is not meant to deny differences among organisms (e.g., mammals undergo placental development with extensive extra-embryonic tissues not found in a zebrafish), nor to dismiss the possibility that developmental biologists might be misled in some instances by the study of model organisms. The greater part of mammalian development can be understood, however, by the study of other organisms’ development. Ultimately, mammalian development will have to be understood in all the details of its differ-
ences, but even this pursuit will benefit from the context of knowledge of the processes shared with other organisms. For example, unique mammalian processes, such as extra-embryonic tissue formation or more extensive forebrain development, are still expected to entail many of the same signal transduction pathways and genetic regulatory circuits as used elsewhere in development.
The Evolutionary Perspective
In light of the availability of base sequences for a variety of kinds of genes in a variety of organisms, the place of metazoa (the multicellular animals) among the kingdoms of living organisms has been recently re-evaluated. It now appears that animals share a common ancestor more closely with plants (especially fungi) than with protozoa such as ciliates or amoebae. These three multicellular kingdoms arose from a common eukaryotic ancestor (probably single celled) perhaps 1.2 billion years ago, whereas eukaryotic single cells go back 2.2 to 2.7 billion years and prokaryotic life goes back perhaps 3.5 billion years (Feng et al. 1997; Pace 1997). The conservation of basic biochemical, genetic, and cell biological functions has been surprisingly extensive in that long lineage. At least 3 billion years ago, ancient prokaryotes originated the processes of replication, transcription, translation, energy metabolism, and biosynthesis, and those processes have been carried forward to this day with little change in all life forms, including animals. The comparisons of the whole genomes of bacteria, yeast, and now the nematode, C. elegans, show clearly the conservation of the protein-coding sequences of genes. At least 2 billion years ago, single-celled eukaryotes originated the basic cell biological processes of mitosis, meiosis, a cdk-cyclin-based cell cycle, an actin-based cytoskeleton and myosin-based movements, a tubulin-based cytoskeleton and kinesin-dynein-based movements, membrane-trafficking, and membrane-bounded organelles. These processes and structures have been carried forward by the single-celled eukaryotes and animals with little change to this day.
In light of this conservation of ancient processes, what have metazoa added in the past 1.2 billion years? Their innovations include abundant cell-cell signaling, extracellular matrix, cell junctions, and a wide range of responses to intercellular signals based on complex genetic regulatory circuits and protein phosphorylation. The C. elegans genome shows that, compared with yeast, metazoa have greatly expanded the number of genes encoding proteins of signal transduction, the cytoskeleton, and transcriptional regulation and have greatly increased the size and complexity of the cis-regulatory regions of genes. Metazoa seem to have evolved in a regulatory or informational direction, that of determining the time, location, and circumstances within a multicellular population for activating and inhibiting the many conserved biochemical and cell biological processes brought forward from their single-celled ancestors. All these ancient processes have been made contingent on cell-cell signaling.
As mentioned above, the importance for developmental toxicology of the discovery of extensive conservation of components and processes among seemingly disparate animals is the conclusion that the study or testing of toxicants in model animals can provide relevant information about humans, as long as the extrapolation is done within conserved responses, of which there are many.
Organogenesis
Organs are usually defined as containing two or more tissues, each tissue containing differing cell types and cell functions, coordinated in a higher level of organization and function than the independent tissues. Second to the organism’s overall body organization, organs are the most complex level of organization of cells. Organogenesis is the organ-forming phase of embryonic development. It begins once the basic anteroposterior and dorsoventral organization of the embryo is established by gastrulation and neurulation. During organogenesis, cytodifferentiation takes place, and then the organ begins to function.
A fundamental question about organogenesis concerns the means by which the different parts of the organ are brought into complex alignment and integrated function. In the first half of the twentieth century, organ formation was described in detail by light microscopy, and the inductive interactions of different cell groups involved in organ formation were revealed by experimental analysis. In general, the different tissues of the organ were not found to form independently and then come together in perfect apposition. Rather, tissues that are nearby as a result of extensive movement during gastrulation and neurulation interact with each other and also with surrounding tissues. Combinations of signals establish positional identity and initiate the progressive delineation of organ-specific gene activations. Thus, it is not necessary that all participants in early organogenesis have position and cell-type specific information. Cell signaling operates throughout organogenesis. Recently, the local signals and responses have been identified in several kinds of organogenesis, the responses often being experimentally proven by using “marker” or “reporter” genes activated at various stages of the process and visualized by staining specific mRNAs by in situ hybridization. Extensive molecular descriptions and cellular and genetic analyses have defined key regulatory pathways that facilitate the development of many vertebrate systems, including the following:
-
Neural tube: regionalization of forebrain, midbrain, hindbrain, and spinal cord (for reviews, see Wassef and Joyner 1997; Brewster and Dahmane 1999; Dasen and Rosenfeld 1999; Veraksa et al. 2000).
-
Neural tube: dorso-ventral organization of brain and spinal cord (for reviews, see Edlund and Jessell 1999; Lee and Jessell 1999).
-
Sensory systems: optic vesicle and eye, otic vesicle and inner ear, and olfactory epithelium (for reviews, see Fekete 1999; Holme and Steel 1999; Kraus and Lufkin 1999; McAvoy et al. 1999).
-
Neural crest: autonomic and sensory ganglia and glia and melanocytes (for reviews, see Francis and Landis 1999; Gershon 1999a,b; LaBonne and Bronner-Fraser 1999).
-
Neural crest: midfacial and branchial connective tissues and teeth (for reviews, see Francis-West et al. 1998; Peters and Balling 1999; Schneider et al. 1999; Tucker and Sharpe 1999; Vaglia and Hall 1999).
-
Paraxial mesoderm: somites, skeletal muscle, vertebrae, and ribs (for reviews, see Brand-Saberi and Christ 1999; Relaix and Buckingham 1999; Burke 2000; Rawls et al. 2000; Summerbell and Rigby 2000).
-
Intermediate mesoderm: kidneys, gonads, reproductive ducts, and sex determination (for reviews, see Sariola and Sainio 1998; Horster et al. 1999; Parker et al. 1999; Swain and Lovell-Badge 1999).
-
Cardiovascular system: heart, angiogenesis, and hematopoiesis (for reviews, see Baldwin and Artman 1998; Mercola 1999; Morales-Alcelay et al. 1998; Tallquist et al. 1999).
-
Limb: growth and specification of axes (for reviews, see Martin 1998; Ng et al. 1999; Vogt and Duboule 1999).
-
Pharyngeal endoderm: thyroid and thymus (for reviews see Bodey et al. 1999; Missero et al. 1998).
-
Gut tube: lungs, liver, pancreas, stomach, and intestines (for reviews, see Gretchen 1999; St-Onge et al. 1999; Warburton and Lee 1999).
The familiar conserved signaling pathways are used over and over in many different contexts in organogenesis and other steps of development, as listed in Table 6-3. For example, the Sonic Hedgehog (SHH) signaling pathway is involved in establishing asymmetry in the early gastrula, inducing floor plate and motor neurons, separating the single eye field into paired optic primordia, maintaining proliferation in migrating neural crest cells, establishing patterning of the medial and lateral nasal prominences and tooth induction, inducing sclerotome segregation and epaxial muscle formation in somites, development of the prostate gland, determining left-right asymmetry, establishing the anteroposterior (rostrocaudal) axis of the limbs, delineating the tracheo-esophageal diverticulum, and establishing sites of formation and branching patterns in lung and pancreatic epithelia. Comparable matches could be made for members of the fibroblast growth factor (FGF), TGFβ, BMP, and WNT signaling families. Although the signaling pathways involve the same or closely related signaling molecules, the responses made by cells are distinct because of the genes and gene products they express prior to and in response to the many different combinations of these signaling factors.
The Vertebrate Limb: The Best Known Organogenesis Model
The limb is by far the most studied organ rudiment of vertebrates, supported by over 50 years of experimental embryology and intensive recent molecular
genetic analysis. Cell-cell interactions, based on known signaling pathways, are well understood, as are the patterns of cell proliferation within the developing limb. Surprisingly, many homologous genes and signaling pathways are conserved between the vertebrate limb and the developing leg or wing of Drosophila. The researchers who followed the seemingly remote leads from the early Drosophila work on appendages have made rapid progress on vertebrate limb development in the past few years.
Limb development is discussed here to show (1) the importance of precise temporal and spatial signaling for organizing a complex organ rudiment, and (2) the interaction of multiple signaling pathways to establish the organ’s three-dimensional morphology.
The developing limb contains three axes: proximodistal, anteroposterior (rostrocaudal), and dorsoventral. Each axis has its own secreted signals and these are integrated in the limb bud, as shown in Figure 6-4 and as summarized by Johnston and Tabin (1997) and Ferretti and Tickle (1997). The bud originates as a locus of rapidly dividing cells in the somatic mesoderm and overlying epidermal ectoderm of the flank of the trunk. Outgrowth of the paired anterior and posterior limbs involves the maintenance of a high rate of cell division within the bud. Local trunk structures, such as the mesonephros, somites, and notochord, secrete FGF10 onto the flank tissues, and this signal initially keeps the bud proliferating. As outgrowth begins, the ectoderm overlying the bud locally thickens to form a ridgelike structure called the apical ectodermal ridge (AER), which then secretes several FGFs, notably FGF8, and takes over for the flank in maintaining cell proliferation in the adjacent bud mesoderm. The bud is then self-sufficient for this signaling and the flank stops serving as an FGF source. (The bud can be transplanted to a remote site, such as the yolk sac, at this stage and will develop autonomously.) The area of rapid division in the bud is called the progress zone (PZ). It lies just beneath the AER. As cells proliferate in it, some are displaced away from the AER and are no longer exposed to FGF. They slow their division rate and stop changing their specification (i.e., their capacity to develop as one
FIGURE 6-4 Development of the limb bud in tetrapods (the four-legged vertebrates). (Panel A) Cross section of mouse embryo after ventral closure, at the level of the forelimbs. The limb buds emerge as small protrusions from the left and right flank. They consist of a surface layer of epidermal cells (ectoderm) and an internal mass of mesenchymal cells (mesoderm). The latter engage in rapid proliferation. (Panel B) Close-up view of a bud, cross section, dorsal, ventral. The apical ectodermal ridge (AER) secretes FGF onto the underlying mesenchyme. The zone of polarizing activity (ZPA) of the mesenchyme secretes SHH onto the ridge. A reciprocal activation circuit is completed in which the AER and ZPA keep each other active. In the mesenchyme close to the ridge is the progress zone (PZ), a population of rapidly dividing cells. Their division is kept going by FGF and SHH. Dividing cells keep changing their option for a future developmental path, in a progression, proximal (shoulder) to distal (hand). As they divide, some are displaced toward the flank, out of the PZ, too far away to receive FGF signals. Their division slows, and they adopt the developmental path of the step of the progression at which they were when they were last in the PZ. The SHH from the ZPA spreads in a gradient toward the anterior edge of the bud. The anteroposterior differences of the limb are signaled by this gradient, perhaps through a BMP2,4 signaling coupled to SHH signaling. (Panel C) The limb bud in cross section, at right angles to panel B, to show the dorsoventral plane. The dorsal epidermis secretes WNT7A onto the mesenchyme. That signals it to develop dorsally. The ventral epidermis does not secrete WNT7A, because the cells express the En gene encoding a transcription factor inhibiting the Wnt7a gene from expression. See text for further information and references.
part or another of the future limb). As a consequence, cells leave the PZ in a proximal to distal order of specification (i.e., the first to leave have the capacity to form the upper limb, the next to form the lower limb, and the last to form the hand or foot). When cells leave the PZ, they start to differentiate into cartilage, bone, and connective tissue of their specified limb level. Precursor cells of limb muscles migrate into the bud from the adjacent somitic myotomes, and nerves extend in from spinal ganglia and the spinal cord. Interactions between the AER and PZ are reciprocal (i.e., the AER maintains mitosis within the PZ by way of FGF, and the PZ maintains the thickened AER by way of a yet unknown signal).
The second axis of the limb, the anteroposterior axis, is also established through cell-cell interactions and secreted signals. The posterior portion of the limb bud contains a specialized region called the zone of polarizing activity (ZPA). This zone was originally recognized as a signaling center, because when it was transplanted to the anterior side of another limb bud, the bud develops a mirror-image duplicated limb. The ZPA secretes SHH, which diffuses across the anteroposterior dimension of the bud, establishing a gradient and setting off the formation of a second gradient of BMP2 and 4, two kinds of TGFβ signals. SHH is both sufficient and necessary to establish the anteroposterior pattern of the limb. Retinoic acid, acting via a nuclear receptor, might also have a role.
The third axis of the limb, the dorsoventral axis, is established through interactions between the nonridge dorsal ectoderm of the limb and the underlying bud mesoderm. That was initially shown by experiments in which the dorsal and ventral regions of the ectoderm were rotated with respect to the mesoderm. The dorsal ectoderm releases the WNT7A signal, which induces the expression of the Lmx-1 gene in the underlying mesoderm and which suppresses expression of the engrailed-1 gene, thereby restricting its expression to the ventral ectoderm. Mutants defective in WNT7A signaling develop double ventral limbs. Mutants defective in engrailed-1 expression develop double dorsal limbs with double sets of fingernails.
It remains to be learned how the signaling pathways of each axial dimension are coordinated with those of the other dimensions, and how the integration of these pathways leads to the formation of unique skeletal structures in precise locations within the limb. The limb exemplifies the advanced understanding of vertebrate organogenesis at a molecular genetic level (i.e., of the signal pathways and the genetic regulatory circuits involved in changing transcription and regulating cell proliferation). This kind of understanding is prerequisite to understanding the action of toxicants on embryogenesis. On the basis of new information, mechanisms of action of toxicants have been recently proposed, although these have yet to be tested. For example, thalidomide leads to a failure to form proximal parts of the limb (the upper and middle parts of the limb), but the hand or foot is usually formed. That developmental outcome is paradoxical, because other treatments, such as the removal or inactivation of the AER, lead to truncation of the limb in the reverse order; the upper and middle parts are present but the hand
or foot is missing. Tabin (1998) recently proposed that thalidomide reduces cell proliferation in the PZ, by means yet unknown, and the few cells that remain there in prolonged contact with FGF8 secreted by the AER are specified as hand or foot cells, the last normally to emerge from the zone. This is an example of an incisive prediction about a long-known toxicant made on the basis of recent knowledge. Yet, the chemical basis for thalidomide’s specific effect on cell proliferation in the PZ escapes even a proposal at this time, and so the hypothesis is incomplete. Stephens and Fillmore (2000) have suggested that thalidomide interferes with integrin gene expression in limb bud mesenchyme cells and, thereby, with their ability to stimulate angiogenesis at the level needed for continued rapid proliferation.
The capacity of the limb bud to develop normally after injury has been studied. Large numbers of cells can be removed at early stages, and as long as representatives remain of the AER, ZPA, and PZ, development will be normal. Immigrating muscle cells from any myotome will enter and adapt to the limb bud, and nerves from any spinal cord level will enter and make neuromuscular connections, although the CNS circuitry of that level may not be appropriate for normal limb movement. The robustness of limb development, like that of other parts of the body, is substantial, because each of the interactive cell groups is much larger than minimally necessary and is capable of proliferation. Robustness is not unlimited, however, and total removal of a key signaling or responding group is deleterious. Regenerating limbs, such as those of newts, have surmounted even that limit, but they are the exception among vertebrates.
SUMMARY
This committee has been asked to evaluate the state-of the-science for elucidating mechanisms of developmental toxicity. It seems self-evident that the knowledge about the basic processes of development provides developmental biologists with an understanding of normal development not even thought possible a decade ago, and also provides developmental toxicologists with improved tools to understand the mechanisms by which chemicals cause abnormal development.
In the last decade, great advances have been made in the understanding of developmental processes on a molecular level in model organisms, such as Drosophila and C. elegans, and in several vertebrates, including the mouse. For the first time, molecular components and their functional interactions have been identified. Developmental processes can be described for the first time as organized networks of these components and their functions. The examples examined so far primarily concern early development before organogenesis but also organogenesis in a few cases. Cell-cell interactions by way of intercellular signals are pervasively and repeatedly used. In all aspects of development, including organogenesis and cytodifferentiation, signaling is expected to be of central impor-
tance. A small number of signal transduction pathways are used in these interactions. There are approximately 10 kinds in early development and organogenesis. (Seven more are used by differentiated cell types.) The number of allelic variants that exist in these human genes remains to be studied. These pathways are conserved among animal phyla, as are many of the genetic regulatory circuits involved in the responses of cells to signals. In addition, many of the basic cell processes, such as proliferation, secretion, motility, and adhesion, are also highly conserved among animals. This extensive conservation gives strong justification to the use of model animals, including Drosophila, C. elegans, zebrafish, and mouse, to learn about basic aspects of mammalian, even human, development.
The understanding of development is far from complete. Although a number of main components have been identified for early processes, their interactions and use in combinations introduce substantial complexity to an inclusive understanding of development. Few of the many types of mammalian organogenesis have been analyzed, and only a few of the 300 types of cytodifferentiation have been studied. As more components and interactions are revealed, it will become important to establish readily accessible databases containing all the information about development.